Abstract
Background
Hepatoblastoma (HB) is a highly aggressive paediatric malignancy that exhibits a high presence of cancer stem cells (CSCs), which related to tumour recurrence and chemotherapy resistance. Brain expressed X-linked protein 1 (BEX1) plays a pivotal role in ciliogenesis, axon regeneration and differentiation of neural stem cells. However, the role of BEX1 in metabolic and stemness programs in HB remains unclear.
Methods
BEX1 expression in human and mouse HB was analyzed using gene expression profile data from NCBI GEO and immunohistochemical validation. Seahorse extracellular flux analyzer, ultra-high-performance liquid-chromatography mass spectrometry (LC-MS), flow cytometry, qRT-PCR, Western Blot, sphere formation assay, and diluted xenograft tumour formation assay were used to analyze metabolic and stemness features.
Results
Our results indicated that overexpression of BEX1 significantly enhanced the Warburg effect in HB cells. Furthermore, glycolysis inhibition largely attenuated the effects of BEX1 on HB cell growth and self-renewal, suggesting that BEX1 promotes stemness maintenance of HB cells by regulating the Warburg effect. Mechanistically, BEX1 enhances Warburg effect through the downregulation of peroxisome proliferator-activated receptor-gamma (PPARγ). Furthermore, pyruvate dehydrogenase kinase isozyme 1 (PDK1) is required for PPARγ-induced inhibition of Warburg effect in HB. In addition, BEX1 supports the stemness of HB by enhancing Warburg effect in a PPARγ/PDK1 dependent manner.
Conclusions
HB patients with high BEX1 and PDK1 expression had a poor prognosis. BEX1 promotes the stemness maintenance of HB cells via modulating the Warburg effect, which depends on PPARγ/PDK1 axis. Pioglitazone could be used to target BEX1-mediated stemness properties in HB by upregulating PPARγ.
Subject terms: Cancer stem cells, Mechanisms of disease, Cancer stem cells, Paediatric research
Introduction
Hepatoblastoma (HB) is a rare paediatric liver tumour that mainly affects infants under 3 years of age [1–4]. Although there are differences in the staging systems for HB worldwide, common prognostic factors mainly include the following aspects: intrahepatic tumour extension, hepatic vascular invasion, multifocality, and distant metastasis [5]. In addition, histopathological criteria are still controversial [6]. To date, complete surgical resection or liver transplantation combined with chemotherapy can significantly improve the prognosis of HB patients, with a 3-year event-free survival (EFS) higher than 80% and a 5-year survival rates averaging 75% [7, 8]. However, for those patients with clinically advanced HB, treatment options are very limited, with a 3-year EFS of only 34% [4]. Moreover, survivors of HB patients may suffer from the side effects of chemotherapy and immunosuppression [9]. Therefore, new and effective therapeutic approaches are urgently needed for patients with advanced HB.
Liver cancer stem cells (LCSCs) have high tumour-initiating potential and are currently considered to be one of the main reasons for tumour aggressiveness and chemoresistance [10]. HB is a highly aggressive malignancy that exhibits a high presence of LCSCs, however, the mechanism underlying their self-renewal maintenance remains unclear [11–13]. Recently, Marayati et al. found that PIM3 promotes tumourigenesis and stemness maintenance in human HB cells. PIM3 knockout resulted in decreased tumour sphere formation and decreased expression of LCSCs markers such as Oct4, Nanog, Sox2, nestin and CD133 [14]. In addition, the dysregulation of self-renewal/developmental pathways in normal hepatic progenitor/stem cell also play a key role in hepatocarcinogenesis [15]. As one of the major developmental pathways associated with progenitor/stem cells, Wnt/β-catenin has a mutation rate as high as 90% in HB, suggesting Wnt/β-catenin pathway is involved in the stemness maintenance of HB [15–17]. Of note, the oncoprotein MYC, one of the downstream targets of the Wnt/β-catenin pathway, can induce human HB-like tumours in mice [18, 19]. Moreover, the imprinted genes such as brain expressed X-linked protein 1 (BEX1) [20], insulin-like growth factor 2 (IGF2), Delta-like homolog 1 (DLK1), Paternally-expressed gene 3 (PEG3) and PEG10 were strongly overexpressed in mouse Myc-induced HB tumours, indicating that these genes may be involved in the progression and stemness maintenance of HB [6].
Previous studies have shown that BEX1 plays a key role in the activation and expansion of liver progenitor cells during liver regeneration [21]. BEX1 is also involved in axon regeneration, differentiation of neural stem cells, and repair of skeletal muscles after injury [22, 23]. In addition, BEX1 is also closely related to the development and progression of various human cancers [24–28]. Our previous study demonstrated that BEX1 promoted the self-renewal of HB cells by activating the Wnt/β-catenin pathway [26]. However, the role of BEX1 in the metabolic-related stemness programs in HB is still not clear. In this study, we found that HB cells mainly relied on the enhanced glycolysis to maintain stemness properties. Furthermore, BEX1 supports the stemness of HB cells by facilitating Warburg effect in a PPARγ/PDK1 dependent manner. This suggests that BEX1 promotes the self-renewal of HB cells through multiple mechanisms. Given that CSCs are associated with chemotherapy resistance and tumour recurrence, BEX1 may be a potential drug target for HB.
Materials and methods
Reagents
The PT3-EF1α-c-Myc plasmid was a gift from Xin Chen and was obtained from Addgene (Cambridge, MA, USA, Addgene plasmid #92046). pCMV/SB10 was kindly gifted by Perry Hackett and was obtained from Addgene (Addgene plasmid #24551) (Supplementary Table S1). The antibodies used and their concentrations are detailed in Supplementary Table S2 and Supplementary Table S3. See Supplementary Materials for information on cell lines and other reagents.
Microarray data
Three HB-related datasets with the accession number GSE131329 [29], GSE132037 [11] and GSE133039 [11] were downloaded from the NCBI Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) [30]. One mice HB dataset with the accession number PRJNA721822 [26] were collected from NCBI (https://www.ncbi.nlm.nih.gov/bioproject/).
Western blot analysis
For western blotting, total protein (30 µg) in lysed cells was separated by SDS-PAGE and transferred to PVDF membranes (Invitrogen, Grand Island, NY). Membranes were blocked with nonfat dry milk (5%) for 1 h and then incubated with specific antibodies overnight at 4 °C. Membranes were then incubated with horseradish peroxidase-conjugated secondary antibody for 1 h at room temperature. Protein bands were detected using an enhanced chemiluminescence system (Pierce, USA).
Quantitative real-time PCR (qRT-PCR) analyses
Total RNA was extracted with Trizol reagent (Invitrogen, USA), and cDNA synthesis was performed with TaKaRa PrimeScript RT reagent kit (TaKaRa Biotechnology, Dalian, China). The real-time PCR analysis was performed using SYBR Premix Ex Taq II (TaKaRa) following the manufacturer’s instructions. GAPDH was used as an internal control and the primer sequences are listed in Supplementary Table S4.
Nude mice xenograft model
The mice xenograft model was established as described previously [31]. The male Balb/c nude mice were randomly divided into experimental groups and control groups. BEX1-knockout HuH-6 cells and BEX1-overexpressing HepG2 cells were injected subcutaneously into the right back of 6-week-old male Balb/c nude mice respectively. Mice were sacrificed 40 days later and the wet weight of excised tumours was measured. All animal experiments were approved by the ethics committee of The First Affiliated Hospital of Zhengzhou University.
Statistical analysis
All analyses in this study were performed using SPSS software version 17.0 (SPSS). The differences between groups were compared using student’s t-test. The Spearman correlation analysis was used to analyze the measured variables. Survival analysis were performed using the Kaplan–Meier method and compared using the log-rank test. The p-values less than 0.05 were considered statistically significant. Further details on the materials and methods can be found in the Supplementary Materials.
Results
BEX1 promotes the self-renewal of HB cells
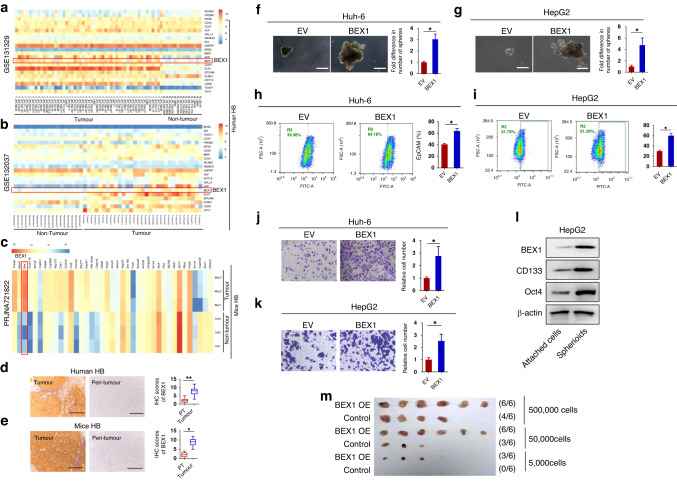
To understand the role of BEX1 in HB, we first compared the mRNA level of BEX1 in two human HB datasets (GSE131329 and GSE132037) and one mice HB dataset (PRJNA721822). The results show that BEX1 is elevated in both human and mice HB tissues compared to non-tumour tissues (Fig. 1a–c). Also, we performed IHC staining analysis to determine the protein expression levels of BEX1 in our own cohort (n = 58). Our results further confirmed that BEX1 was upregulated in HB compared with peri-tumour samples (Fig. 1d). Previous studies have shown that the type of c-Myc-driven mouse liver cancer is HB [6]. Moreover, Myc-induced HB-like tumours in mice strikingly resembled the human immature HB subtype [6]. The c-Myc-driven HB-like mouse model has been widely used in liver cancer research [32, 33]. tumours detected in the c-Myc-driven mouse model were confirmed to be HB by pathologists in this study. Likewise, BEX1 was also significantly elevated in c-Myc-driven HB-like liver tumours in mice (Fig. 1e).
Fig. 1. BEX1 overexpression promotes the self-renewal of HB cells.
Analyses of BEX1 expression in (a, b) two human HB datasets and (c) one mice HB dataset. These heatmap figures were generated using three public datasets (GSE131329, GSE132037, and PRJNA721822). Specifically, stemness related genes were extracted, and the signal values were converted by log2(exp+1) for pheatmap clustering analysis. The false discovery rate (FDR) was used to determine the threshold of the p-value in multiple tests. A threshold of the FDR ≤ 0.05 was used to judge the significance of gene expression differences. In these heatmap data, BEX1 was elevated in both human and mouse HB tissues compared to non-tumour tissues, which has a statistically significant difference (FDR ≤ 0.05). d IHC analysis of BEX1 expression in clinical HB samples (n = 58). e IHC analysis of BEX1 expression in c-Myc-driven HB-like liver tumours in mice (n = 6). Scale bars: 50 μm. f, g Spheroid formation assays were performed in Huh-6 and HepG2 cells with treatments as indicated. Scale bars: 20 μm. h, i Flow cytometric analysis of the EpCAM+ cell population in Huh-6 and HepG2 cells with treatments as indicated. j, k The invasion of HB cells was examined by performing an invasion chamber assay. l Levels of BEX1, CD133, Oct4 in spheroids and attached cells were detected by western blotting. β-actin was used as a loading control. m Limiting dilution xenograft formation of HepG2 cells infected with Lv-BEX1 or Lv-Control (n = 6 per group). IHC immunohistochemistry. *P < 0.05, **P < 0.01.
It is known that shRNA knockout could cause off-target effect. Our results are unlikely to be the off-target effects of the shRNA oligos, since the introduction of BEX1 cDNA into the knockdown cells attenuated the effects of the shRNA oligos (Supplementary Fig. S1). Moreover, both gain of function and loss of function approaches indicate that BEX1 can promote the proliferation of HB cells (Supplementary Fig. S2 and Supplementary Fig. S3). HB is an embryonal tumour that has previously been shown to have strong CSC properties [13]. The relationship between BEX1 and CSC markers was further evaluated in three HB dataset. Studies over the past decades have demonstrated several markers for identifying CSC populations in liver cancer. In this study, we used several established CSC markers in HB, including EpCAM [34–36], DLK1 [37, 38], AFP [39], SALL4 [40, 41], CD24 [42, 43], and LGR5 [13, 44], to analyze the relationship between BEX1 and these CSC markers. A positive correlation between BEX1 and CSC markers was confirmed in these three datasets (Supplementary Fig. S4). Our previous results showed that BEX1 co-localized with the CSC marker SALL4 in c-Myc-driven HB tumour tissues [26]. Spheroid formation analysis revealed that BEX1 overexpression significantly increased the number of spheroids in both Huh-6 and HepG2 cells. In contrast, BEX1 knockdown exhibited an opposite effect (Fig. 1f, g and Supplementary Fig. S5a, b). EpCAM has been used as a marker for detecting cancer stem cells (CSCs) in the context of HB [34–36]. Flow cytometry analysis showed that BEX1 overexpression increased the proportion of EpCAM+ cells in both Huh-6 and HepG2 cells. In contrast, BEX1 knockdown exhibited an opposite effect (Fig. 1h, i and Supplementary Fig. S5c, d). In addition, BEX1 overexpression promoted HB cell invasion, whereas BEX1 knockdown inhibited HB cell invasion (Fig. 1j, k and Supplementary Fig. S5e, f). Moreover, the expression levels of BEX1 and other stemness markers such as CD133 and Oct4 were significantly increased in tumour spheroids compared with attached cells (Fig. 1l and Supplementary Fig. S5g). Importantly, in vivo limiting dilution assays demonstrated that BEX1-overexpressing HepG2 cells showed increased tumour-initiating capacity in BALB/c nude mice when compared to the capacity exhibited by control cells (Fig. 1m). These results suggest that BEX1 promotes the self-renewal of HB cells.
BEX1 promotes the stemness maintenance of HB cells via modulating the Warburg effect
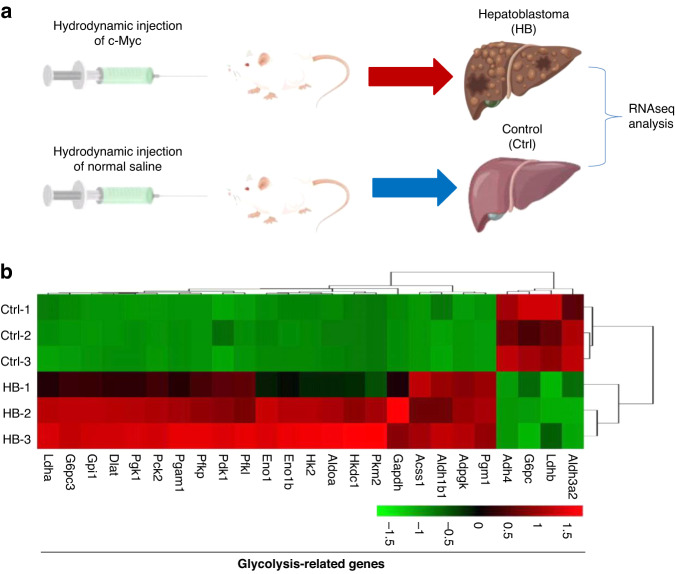
Metabolic reprogramming endows tumour cells with the ability to modulate metabolic pathways to sustain diverse biological processes [32, 45]. We analyzed the expression of glycolysis-related genes in a c-Myc-driven HB-like mouse model using RNAseq (Fig. 2a). The results showed that the expression levels of most glycolysis-related genes were significantly increased in HB tissues compared with normal liver tissues (Fig. 2b), suggesting that glycolysis is a metabolic programme preferred by HB cells.
Fig. 2. Glycolysis is a metabolic programme preferred by mouse HB cells.
a Study design. b A heatmap showing glycolysis-related genes with at least a 2-fold change in transcript levels between mouse HB tissues compared with normal liver tissues. The differentially expressed genes (DEGs) were conducted using edger. The false discovery rate (FDR) was used to determine the threshold of the p-value in multiple tests. A threshold of the FDR ≤ 0.05 was used to judge the significance of gene expression differences. Each gene in the heatmap data had a statistically significant difference (FDR ≤ 0.05).
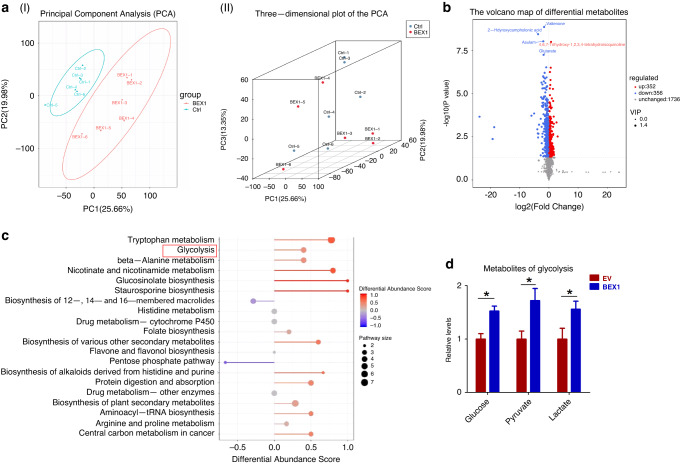
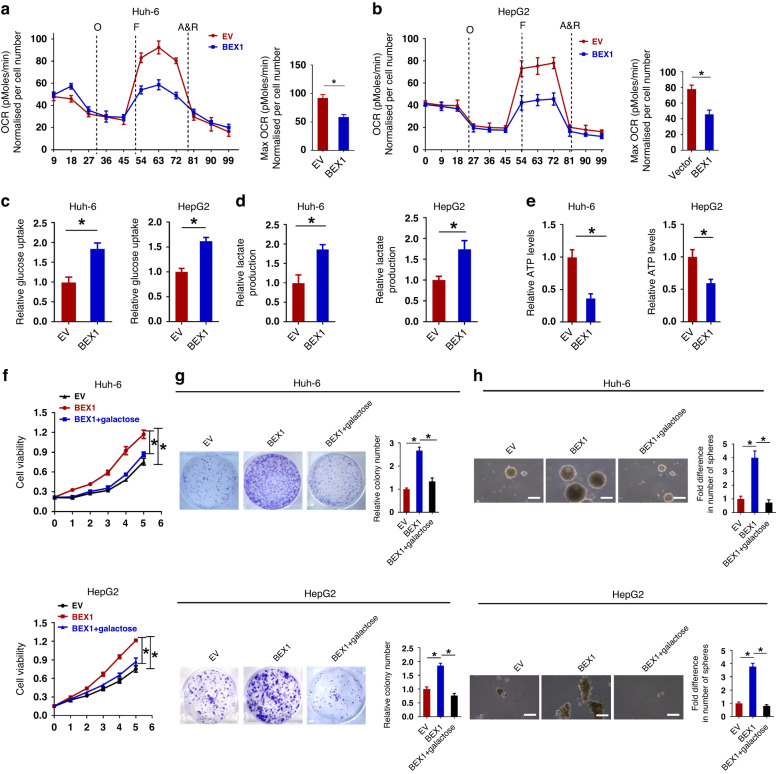
We next explain about genes downregulated in the HB group. Alcohol dehydrogenase 4 (ADH4), as an important member of the ADH family, can metabolize many substrates including ethanol and retinol. Our heatmap shows that ADH4 is significantly reduced in HB tissues. Similarly, previous studies have shown that ADH4 is significantly reduced in hepatocellular carcinoma (HCC) tissues and correlates with the survival of HCC patients [46]. The low expression of ADH4 was positively correlated with the signaling pathways promoting tumourigenesis, such as ATR pathway, NOTCH pathway, and mTOR pathway [47]. Our heatmap shows that glucose-6-phosphatase catalytic subunit (G6PC) is significantly reduced in HB tissues. G6PC, which catalyzes the final step of glycogenolysis, is frequently downregulated to increase glucose storage in premalignant HCC cells [48]. Studies have shown that increased glycogen storage accelerates liver carcinogenesis. Furthermore, G6PC deficiency in humans and mice leads to glycogen storage disease, as well as liver enlargement and tumourigenesis in a Yap-dependent manner [48]. Our heatmap shows that lactate dehydrogenase subunit B (LDHB) is significantly reduced in HB tissues. Consistently, Hong et al. found that LDHB knockdown increased inhibitory phosphorylation of pyruvate dehydrogenase (PDH) through lactate-mediated activation of PDH kinase (PDK), thereby attenuating oxidative phosphorylation activity in HCC. They further showed that LDHB inhibition is a key mechanism to enhance glycolysis and maintain mitochondrial dysfunction in HCC progression through the lactate release [49]. At last our heatmap shows that aldehyde dehydrogenase 3a2 (Aldh3a2) is significantly decreased in HB tissues. Consistently, endogenous aldehyde accumulation caused by reduced ALDH3A2 expression produces genotoxicity and promotes cancer progression [50]. Moreover, we have analyzed another human HB dataset (GSE133039). The results support that most glycolysis related genes are also highly expressed in human HB (Supplementary Fig. S6). We next explored whether BEX1 promotes HB progression by regulating glycolysis. To this end, we carried out metabolomics analysis in HepG2 cells using untargeted ultra-high-performance liquid-chromatography mass spectrometry (LC-MS). Principal component analysis (PCA) and three-dimensional PCA map of BEX1 overexpression group and control group were performed. The results showed that the variation within groups was small, which met the requirements of subsequent analysis (Fig. 3a). Volcano plots showed the overall differences in metabolites between BEX1- overexpressing HepG2 cells and control cells (Fig. 3b). The following KEGG analyses and biochemical assays showed that glycolysis were significantly changed in response to BEX1 overexpression (Fig. 3c, d). Furthermore, we compared important cellular metabolic parameters between HB cell with BEX1 overexpression or knockdown and control cells. Our results showed that overexpression of BEX1 significantly decreased basal and maximal oxygen consumption rates (OCR) in HB cells, whereas knockdown of BEX1 obviously increased OCR in HB cells (Fig. 4a, b and Supplementary Fig. S7a, b). Moreover, overexpression of BEX1 resulted in the increased glucose uptake and extracellular lactate levels, along with a decreased cellular ATP levels. In contrast, BEX1 knockdown led to the decreased glucose uptake and extracellular lactate levels, whilst cellular ATP levels were significantly increased (Fig. 4c–e and Supplementary Fig. S7c–e). As a poor substrate for glycolysis, galactose cannot directly enter the glycolysis pathway [51]. To investigate whether the Warburg effect was responsible for progression and stemness maintenance in HB, glucose in the culture medium for BEX1-overexpressing Huh-6 and HepG2 cells were replaced by galactose to inhibit glycolysis. As a result, galactose largely inhibited the effects of BEX1 on HB cell growth and self-renewal, indicating that BEX1 promotes stemness maintenance of HB cells by regulating the Warburg effect (Fig. 4f–h).
Fig. 3. BEX1 regulates glycolysis in HB cells.
a–d Untargeted LC-MS analysis of differential abundance in metabolites in HepG2 cells with BEX1 overexpression. aI Principal component analysis (PCA) and (aII) three-dimensional PCA map of BEX1- overexpressing group and control group. The results showed that the variation within groups was small, which met the requirements of subsequent analysis. b Volcano plots showing the overall differences in metabolites between BEX1- overexpressing HepG2 cells and control cells. c KEGG pathway analysis for different abundant metabolites. X axis represents differential abundance score, and y axis represents the pathway enrichment. Darker colors and larger sizes represent higher pathway impact values and increased pathway enrichment, respectively. d Effect of BEX1 expression on the abundance of glycolysis intermediates measured by LC-MS in HepG2 cells (n = 6). *P < 0.05.
Fig. 4. BEX1 promotes the stemness maintenance of HB cells via modulating the Warburg effect.
a, b Effects of BEX1 on oxygen consumption ratio (OCR) in Huh-6 and HepG2 cells with treatments as indicated. O oligomycin, F FCCP (carbonyl cyanide 4-[trifluoromethoxy] phenylhydrazone), A&R antimycin A and rotenone. c Relative glucose uptake was determined in Huh-6 and HepG2 cells with treatment as indicated. d Relative lactate production was determined in Huh-6 and HepG2 cells with treatment as indicated. e Relative ATP production was determined in Huh-6 and HepG2 cells with treatment as indicated. f MTS assays in Huh-6 and HepG2 cells with treatment as indicated (EV empty vector, BEX1 expression vector encoding BEX1, galactose, glucose in the culture medium was replaced by galactose). g Colony formation assays in Huh-6 and HepG2 cells with treatment as indicated. h Spheroid formation assays were performed in Huh-6 and HepG2 cells with treatments as indicated. LC-MS: ultra-high-performance liquid-chromatography mass spectrometry. Scale bars: 20 μm.*P < 0.05.
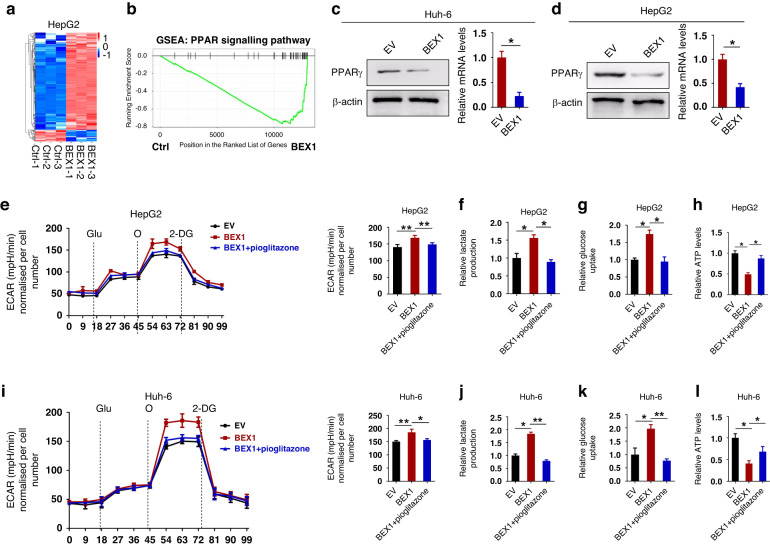
BEX1 enhances Warburg effect via downregulating PPARγ in HB
We then investigated the mechanism by which BEX1 enhances the Warburg effect. The effect of BEX1 on the global gene expression pattern of HepG2 cells at the transcriptomic level was examined by performing RNA sequencing (RNA-seq) analysis (Fig. 5a). Gene set enrichment analysis (GSEA) showed that BEX1 was negatively correlated with the PPAR signalling but positively correlated with the glycolysis pathway in HepG2 cells (Fig. 5b and Supplementary Fig. S8a). Likewise, the negative correlation between BEX1 and PPARγ was confirmed in another study, which is consistent with our results [21]. As a tumour-suppressor gene in the liver [52], PPARγ plays a key role in glucose homeostasis [53]. Western blot and real-time PCR showed that BEX1 overexpression significantly downregulated the expression of PPARγ in both Huh-6 and HepG2 cells, whereas BEX1 knockdown exhibited an opposite effect (Fig. 5c, d and Supplementary Fig. S8b, c). Previous research has shown that PPARγ can inhibit glycolysis in some cancers. For example, PPARγ inhibits glycolysis by suppressing the glycolytic enzymes PGK1 and PKM2 in breast cancer [54]. PPARγ also mediates the inhibitory effect of Cloxiquine on the glycolysis of melanoma cells by downregulating glycolytic genes such as PKM2, LDHA and HK2 [55]. Next, we explored whether PPARγ is involved in the Warburg effect regulated by BEX1 in HB cells. As expected, the extracellular acidification rate (ECAR) of BEX1-overexpressing HepG2 or Huh-6 cells was significantly decreased after treatment with the PPARγ agonist pioglitazone. Moreover, decreased lactate production and glucose consumption as well as an increased ATP production were also observed in BEX1-overexpressing HepG2 or Huh-6 cells upon treatment with pioglitazone (Fig. 5e–l). In contrast, the ECAR of BEX1-knockout Huh-6 or HepG2 cells was significantly increased after treatment with the PPARγ inhibitor GW9662. Meanwhile, the lactate production and glucose consumption were increased and ATP production were decreased in BEX1-knockout Huh-6 or HepG2 cells upon treatment with GW9662 (Supplementary Fig. S8d–k). Therefore, the possible mechanism of action of PPARγ agonist pioglitazone lies in the inhibition of glycolysis [56–58], as shown in decreased glycolytic bioenergetics parameters (Fig. 5e–l). On the contrary, the possible mechanism of action of PPARγ inhibitor GW9662 lies in the activation of glycolysis (Supplementary Fig. S8d–k), which is consistent with previous reports [55]. Collectively, these results indicate that BEX1 enhances glycolysis in HB cells by downregulating PPARγ.
Fig. 5. BEX1 enhances Warburg effect via downregulating PPARγ in HB.
a A heatmap showing genes with at least a 2-fold change in transcript levels between BEX1-overexpressing and control HepG2 cells. Each gene in the heatmap data had a statistically significant difference (FDR ≤ 0.05). b Gene set enrichment analysis (GSEA) showed that the PPAR signaling pathway was significantly negatively correlated with BEX1 in HepG2 cells. c, d Western blotting and real-time PCR analysis the protein and mRNA expression of PPARγ in BEX1-overexpressing Huh-6 and HepG2 cells. e The extracellular acidification rate (ECAR), (f) lactate production, (g) glucose consumption and (h) ATP production were measured in the presence of PPARγ agonist pioglitazone in HepG2 cells. Glu glucose, O oligomycin, 2-DG 2-deoxyglucose. i The ECAR, (j) lactate production, (k) glucose consumption and (l) ATP production were measured in the presence of PPARγ agonist pioglitazone in Huh-6 cells. *P < 0.05; **P < 0.01.
PDK1 is required for PPARγ -induced inhibition of Warburg effect in HB
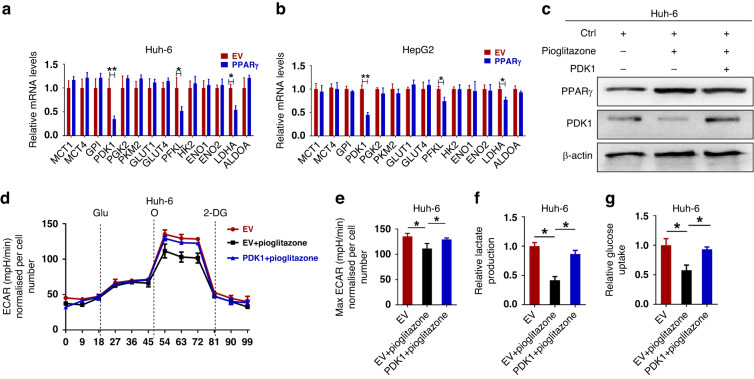
To further study the mechanism of PPARγ in the regulation of glycolysis, we evaluated the role of PPARγ on the expression of key glycolytic enzymes. The real-time PCR results showed that the expression levels of most enzymes involved in glycolysis did not change significantly, except for pyruvate dehydrogenase kinase (PDK1), PFKL and LDHA, and the expression level of PDK1 changed most significantly (Fig. 6a, b and Supplementary Fig. S9a, b). So, we tested the levels of PPARγ and PDK1 in the presence of PPARγ agonist pioglitazone or inhibitors GW9662. As shown in Fig. 6c, pioglitazone significantly increased the protein levels of PPARγ and decreased the PDK1 levels in Huh-6 cells. On the contrary, GW9662 decreased the protein levels of PPARγ and increased the PDK1 levels in HepG2 cells (Supplementary Fig. S9c). In addition, another study also reported that pioglitazone can increase protein levels of PPARγ [59], which is consistent with our results. As expected, the significant reduction in pioglitazone-mediated ECAR, lactate production and glucose consumption were reversed after PDK1 overexpression in Huh-6 cells (Fig. 6d–g). In contrast, GW9662-mediated increases in ECAR, lactate production, and glucose consumption were reversed following PDK1 knockdown in HepG2 cells (Supplementary Fig. S9d–g). These data suggest that PDK1 is required for PPARγ-induced inhibition of Warburg effect in HB cells.
Fig. 6. PDK1 is required for PPARγ -induced inhibition of Warburg effect in HB cells.
a, b The mRNA levels of glycolysis-related genes in PPARγ-overexpressing Huh-6 and HepG2 cells compared to relative control cells. c Protein levels of PPARγ and PDK1 were analyzed using Western blot analysis in the indicated cell lysates after treatment with PPARγ agonist pioglitazone. d, e The ECAR, (f) lactate production, and (g) glucose consumption were measured in the presence of PPARγ agonist pioglitazone in Huh-6 cells. Glu glucose, O oligomycin, 2-DG 2-deoxyglucose. *P < 0.05; **P < 0.01.
BEX1 enhances the stemness maintenance of HB cells in a PPARγ/PDK1 dependent manner
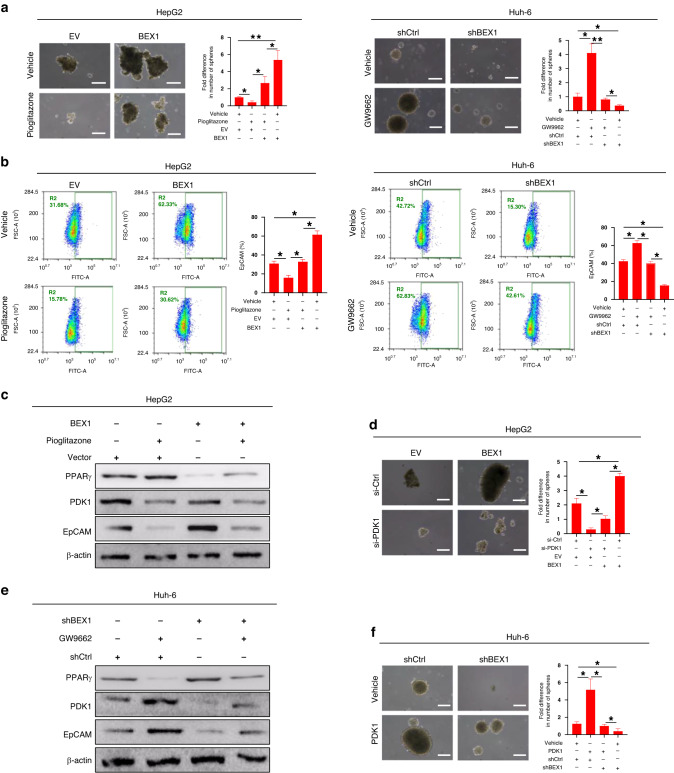
We next investigated the role of the PPARγ/PDK1 axis in BEX1-mediated stemness maintenance in HB cells. Spheroid assays confirmed that pioglitazone treatment reduced the enhancement of BEX1-mediated spheroid formation in HepG2 cells. In contrast, the reduction of spheroid formation in BEX1-knockout Huh-6 cells was reversed after GW9662 treatment (Fig. 7a). Flow cytometry analysis showed that pioglitazone decreased the proportion of EpCAM+ cells in BEX1-overexpression HepG2 cells. In contrast, GW9662 increased the proportion of EpCAM+ cells in BEX1-knockout Huh-6 cells, indicating that BEX1 promotes HB stemness maintenance by inhibiting PPARγ (Fig. 7b). Next, we further verified the effect of PPARγ/PDK1 axis on the stemness maintenance of HB cells. Western blot results showed that pioglitazone increased the expression of PPARγ, and decreased the expression of PDK1 and EpCAM in HepG2- empty vector (EV) and BEX1-overexpressing HepG2 cells, respectively (Fig. 7c). Spheroid assays demonstrated that PDK1 knockout reduced the enhancement of BEX1- mediated spheroid formation in HepG2 cells (Fig. 7d). In contrast, GW9662 decreased PPARγ expression and increased PDK1 and EpCAM expression in Huh-6- control (Ctrl) and BEX1-knockout Huh-6 cells, respectively (Fig. 7e). We have performed quantitative analysis of the protein levels and provided detail on statistical differences in Supplementary Fig. S10. Moreover, the reduction of spheroid formation in BEX1-knockout Huh-6 cells was reversed after PDK1 overexpression (Fig. 7f). Taken together, these results indicate that BEX1 enhances the stemness maintenance of HB cells in a PPARγ/PDK1 dependent manner. However, the effects of BEX1 modulation were only partially reverted by the PPARγ agonist/inhibitor. We hypothesized that BEX1 might promote the stemness of HB through multiple mechanisms. On the one hand, BEX1 promotes the stemness maintenance of HB cells via modulating the Warburg effect, which depends on PPARγ/PDK1 axis. On the other hand, our previous study demonstrated that BEX1 promoted the self-renewal of HB cells by activating the Wnt/β-catenin pathway[26]. BEX1 may also promote the stemness of HB through other unknown mechanisms, which still need to be explored.
Fig. 7. BEX1 enhances the stemness maintenance of HB cells in a PPARγ/PDK1 dependent manner.
a Spheroid formation assays were performed in BEX1-overexpressing HepG2 and BEX1- knockout Huh-6 cells with treatments as indicated. Scale bars: 20 μm. b Flow cytometric analysis of the EpCAM+ cell population in BEX1-overexpressing HepG2 and BEX1-knockout Huh-6 cells with treatments as indicated. c Protein levels of PPARγ, PDK1 and EpCAM were analyzed using Western blot analysis in HepG2-empty vector (EV) and BEX1-overexpressing HepG2 cells after treatment with PPARγ agonist pioglitazone. d Spheroid formation assays were performed in BEX1-overexpressing HepG2 and control cells with treatments as indicated. Scale bars: 20 μm. e Protein levels of PPARγ, PDK1 and EpCAM were analyzed using Western blot analysis in Huh6 -empty vector and Huh6-shBEX1 cells after treatment with PPARγ inhibitors GW9662. f Spheroid formation assays were performed in Huh6-shBEX1 cells and control cells with treatments as indicated. Scale bars: 20 μm. *P < 0.05; **P < 0.01.
BEX1 positively correlates with PDK1 in HB patients
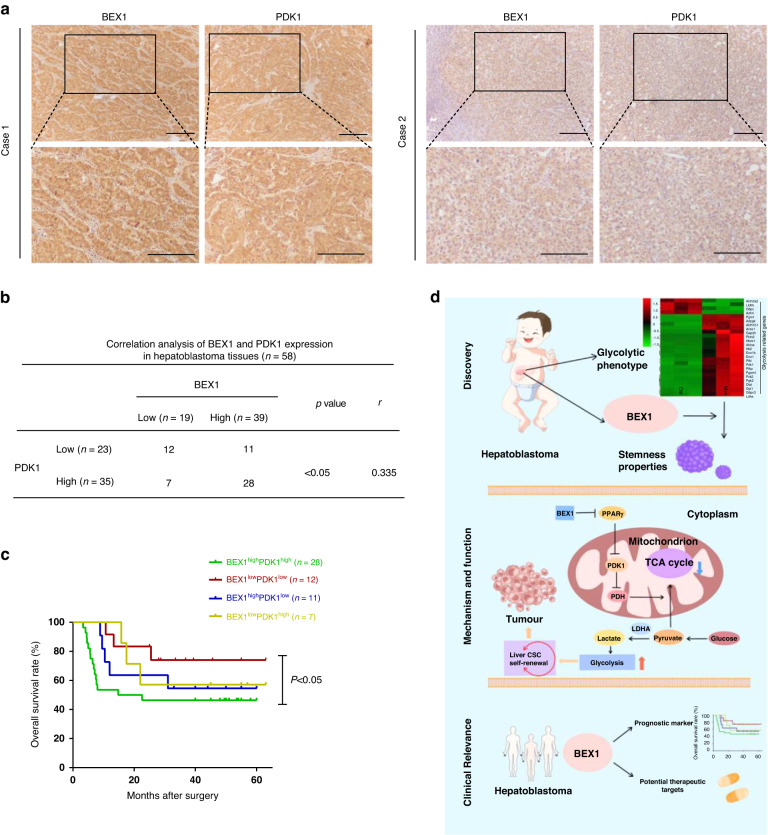
To explore the prognostic values of PDK1 and BEX1 in HB, we analyzed their expression by immunohistochemistry in a cohort of 58 HB patients. The representative images in Fig. 8a show that tissue samples with strong IHC staining for BEX1 also have strong PDK1 staining, and vice versa. Spearman’s rank correlation analysis showed that the expression of these two proteins was significantly positively correlated (r = 0.335, p < 0.05) (Fig. 8b). Kaplan–Meier analysis showed that HB patients with high BEX1 expression and high PDK1 expression had the lowest overall survival (Fig. 8c). These results suggest that PDK1 plays an important role in BEX1-mediated HB progression.
Fig. 8. BEX1 positively correlates with PDK1 in HB patients.
a IHC analysis of BEX1 and PDK1 expression in clinical HB samples (n = 58). Upper scale bars: 50 μm; lower scale bars: 20 μm. b Spearman’s rank correlation analysis of BEX1 and PDK1 expression in clinical HB samples. c Kaplan–Meier analysis of the prognostic value of combining BEX1 and PDK1 in HB patients. p < 0.05 refers to BEX1highPDK1high group vs BEX1lowPDK1low group. d Schematic diagram showing that BEX1 supports the stemness of hepatoblastoma by facilitating Warburg effect in a PPARγ/PDK1 dependent manner.
Discussion
BEX1 is a key molecule that regulates liver development during embryonic period [6, 21]. As a new oncofetal gene, its abnormal expression is closely related to the initiation and progression of liver cancer [26]. For example, Sagawa et al. showed that BEX1 was upregulated in preneoplastic lesions of Cx32ΔTg rats with nonalcoholic steatohepatitis and promoted the growth of HCC cells through NF-κB and SAPK/JNK signaling pathways [20]. Similarly, Uehara et al. demonstrated that embryonic genes such as AFP, H19, and BEX1 were significantly increased in DEN + CCl4-induced mouse HCC tissues [60]. Our previous study demonstrated that BEX1 promoted the self-renewal of HB cells by activating the Wnt/β-catenin pathway [26]. The findings of the present study suggested that BEX1 supports the stemness of HB by enhancing Warburg effect in a PPARγ/PDK1 dependent manner. Considering that cancer stemness is closely related to tumour recurrence and chemoresistance, BEX1 may be an ideal candidate target for intervening HB progression and chemoresistance.
Typically, differentiated cells rely on mitochondrial oxidative phosphorylation for their energy needs. Instead, most cancer cells rely heavily on aerobic glycolysis to generate the energy needed for cellular processes. This phenomenon is known as the “Warburg effect”, which can be caused by the overexpression of glucose transporters or enzymes during glycolysis mediated by oncogene activation [61, 62]. The activation of oncogenes can cause dysregulation of intracellular signalling pathways that affect tumour cell metabolism and promote growth [63]. For example, in intrahepatic cholangiocarcinoma, BEX2 has been reported to inhibit mitochondria-related oxygen consumption and is required for the dormant cancer stem cell maintenance [64]. In this study, we found that overexpression of BEX1 significantly enhanced the Warburg effect in HB cells. Furthermore, glycolysis inhibition largely attenuated the effects of BEX1 on HB cell growth and self-renewal, suggesting that BEX1 promotes stemness maintenance of HB cells by regulating the Warburg effect. Indeed, the glycolytic phenotype actually favours the stemness properties. For example, metabolic reprogramming switch from OXPHOS to glycolysis in induced pluripotent stem cells (iPSCs) is crucial for their efficient acquisition of a pluripotent stem state [65]. In addition, glycolysis was also found to be a metabolic programme preferred by CD133 + CD49f+ tumour-initiating cells (TICS) in HCC [66], radioresistant spheroid-forming cells in nasopharyngeal carcinoma [67], and CD44+CD24lowEPCAM+ CSCs in breast cancer [68]. Interestingly, high expression of oncogenic MYC in the above three cancers was identified as a major factor driving CSC properties [69, 70], suggesting there is a close link between MYC-driven glycolytic phenotype and stemness [71]. As a key transcription factor, MYC is also overexpressed in HB [6]. Our previous study demonstrated that MYC can directly promote the expression of BEX1 [26], and we therefore postulated that MYC/BEX1 axis promotes stemness maintenance of HB through glycolysis. However, OXPHOS has been demonstrated as the preferred energy production mode for CSCs in some other cancer types [72], such as leukaemia [69], pancreatic ductal adenocarcinoma [73], glioblastoma [74] and lung cancer [75]. Differences in tumour types may be one of the reasons why CSCs rely primarily on glycolysis or preferentially on OXPHOS.
PPARγ plays a key role in glucose homeostasis, lipid metabolism, insulin sensitivity and cell fate [72]. Pioglitazone and GW9662 are selective PPARγ agonist or antagonist that directly bind to PPARγ with high affinity [76]. In addition to increasing PPARγ protein levels [59], pioglitazone can also affect the post-translational modification of PPARγ and thus affect PPARγ activity [77–79]. It has been demonstrated that PPARγ is composed of different functional domains, including an N-terminal trans-activation domain (AF1), a C-terminal ligand-binding domain (LBD) and a highly conserved DNA binding domain (DBD) [80]. Recent studies have shown that PPARγ is regulated by post-translational modification such as phosphorylation [81]. Notably, some anti-diabetic PPARγ ligands such as pioglitazone and rosiglitazone have been reported to have a unique biochemical function: blocking the phosphorylation of PPARγ by cyclin dependent kinase 5 (Cdk5) at serine 273 [77–79]. Specifically, pioglitazone directly binds to the LBD of PPARγ and cause a conformational change that interferes with Cdk5’s ability to phosphorylate serine 273, which in turn increase the activity of PPARγ. In contrast, the PPARγ antagonist GW9662 promotes CDK5-mediated phosphorylation of PPARγ at serine 273, thereby reducing PPARγ activity [79]. Together, these studies suggest that CDK5-mediated phosphorylation of PPARγ is a direct target of pioglitazone and GW9662. PPARγ has been shown to be a tumour suppressor gene in the liver, which inhibits HCC cell growth by reducing cell proliferation, migration, and angiogenesis [52, 82, 83]. Moreover, PPARγ inhibits glycolysis by suppressing the glycolytic enzymes PGK1 and PKM2 in breast cancer [54]. PPARγ mediates the inhibitory effect of Cloxiquine on the glycolysis of melanoma cells by down-regulating glycolytic genes such as PKM2, LDHA and HK2 [55]. Our results suggest that BEX1 enhances the Warburg effect by downregulating PPARγ in HB. We further analyzed the effect of PPARγ on the expression of key glycolytic enzymes in HB cells. PPARγ did not alter the expression of most glycolytic enzymes except for PDK1 and LDHA, and the expression level of PDK1 changed most significantly. PDK1 promotes glycolysis by phosphorylating the PDH complex, which prevents the conversion of pyruvate to acetyl-CoA into mitochondria [84, 85]. Overexpression of PDK1 promotes the progression of many human cancers, such as liver [86], breast [87], kidney [88] and colon cancers [89]. Our results showed that the reduction in PPARγ-mediated ECAR, lactate production and glucose consumption was reversed after PDK1 overexpression in HB cells, indicating that PDK1 is required for PPARγ-induced inhibition of Warburg effect in HB.
Our results indicate that BEX1 enhances the stemness maintenance of HB cells in a PPARγ/PDK1 dependent manner. BEX1 inhibits the expression of PPARγ, resulting in the overexpression of its downstream molecule PDK1 in HB. We next investigated the role of the PPARγ/PDK1 axis in BEX1-mediated stemness maintenance in HB. Our results revealed the reduction of spheroid formation in BEX1-knockout HB cells was reversed after GW9662 treatment, indicating that BEX1 promotes HB stemness maintenance by inhibiting PPARγ. Moreover, the reduction of spheroid formation in BEX1-knockout HB cells was reversed after PDK1 overexpression, suggesting PDK1 promotes stemness maintenance of HB cells. Consistent with our results, PDK1 also promotes stemness maintenance in iPSCs [90], embryonic stem cell [91] and many other tumours such as glioma [92], HCC [86], and breast cancer [93].
In conclusion, BEX1 supports the stemness of HB by enhancing the Warburg effect in a PPARγ/PDK1-dependent manner (Fig. 8d). Given that CSCs are associated with tumour recurrence and chemotherapy resistance, BEX1 may be a promising target for new therapeutic strategies for HB.
Supplementary information
Acknowledgements
We thank Professor Xin Chen and Professor Perry Hackett for providing the plasmids. The schematic diagram in this article was drawn by Figdraw. We thank Ms. Jie Chen for her help in designing the schematic diagram.
Author contributions
Conception and design: GW, XH, TY, QW. Development of methodology: JL, Y, SL. Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): QW, QH, LS, LT. Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): QW, CL, NL. Writing, review, and/or revision of the manuscript: GW, XH, TY. Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): CZ, YT. Study supervision: GW, TY, QW, XH.
Funding
This work was supported by grants from the National Natural Science Foundation of China (No. 82103664), Joint Construction Project of Henan Medical Science and Technology (LHGJ20200281) and Guangzhou Basic and Applied Basic Research Project (No. 202102021274).
Data availability
All of the relevant data are included in supplemental information. RNA-seq data have been submitted to the SRA database (SRA: PRJNA721822). All data are available upon request to the corresponding authors.
Ethics approval and consent to participate
All animal experiments were approved by the Institutional Animal Care and Use Committee of Zhengzhou University in accordance with NIH guidelines.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Qian Wang, Ning Liang, Chaoxu Liu, Jing Li.
Contributor Information
Qian Wang, Email: 1211072068@qq.com.
Xianli He, Email: hexianli999@126.com.
Tao Yang, Email: ttjrkyt@163.com.
Gang Wang, Email: wanggangfmmu@163.com.
Supplementary information
The online version contains supplementary material available at 10.1038/s41416-023-02418-4.
References
- 1.Sumazin P, Peters TL, Sarabia SF, Kim HR, Urbicain M, Hollingsworth EF, et al. Hepatoblastomas with carcinoma features represent a biological spectrum of aggressive neoplasms in children and young adults. J Hepatol. 2022;77:1026–37. doi: 10.1016/j.jhep.2022.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Trobaugh-Lotrario A, Katzenstein HM, Ranganathan S, Lopez-Terrada D, Krailo MD, Piao J, et al. Small cell undifferentiated histology does not adversely affect outcome in hepatoblastoma: a report from the children’s oncology group (COG) AHEP0731 study committee. J Clin Oncol. 2022;40:459–67. doi: 10.1200/JCO.21.00803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Loesch R, Caruso S, Paradis V, Godard C, Gougelet A, Renault G, et al. Deleting the beta-catenin degradation domain in mouse hepatocytes drives hepatocellular carcinoma or hepatoblastoma-like tumour growth. J Hepatol. 2022;77:424–35. doi: 10.1016/j.jhep.2022.02.023. [DOI] [PubMed] [Google Scholar]
- 4.Semeraro M, Branchereau S, Maibach R, Zsiros J, Casanova M, Brock P, et al. Relapses in hepatoblastoma patients: clinical characteristics and outcome–experience of the International Childhood Liver Tumour Strategy Group (SIOPEL) Eur J Cancer. 2013;49:915–22. doi: 10.1016/j.ejca.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 5.Perilongo G, Shafford E, Plaschkes J. SIOPEL trials using preoperative chemotherapy in hepatoblastoma. Lancet Oncol. 2000;1:94–100. doi: 10.1016/S1470-2045(00)00018-8. [DOI] [PubMed] [Google Scholar]
- 6.Cairo S, Armengol C, De Reynies A, Wei Y, Thomas E, Renard CA, et al. Hepatic stem-like phenotype and interplay of Wnt/beta-catenin and Myc signaling in aggressive childhood liver cancer. Cancer Cell. 2008;14:471–84. doi: 10.1016/j.ccr.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 7.Aronson DC, Czauderna P, Maibach R, Perilongo G, Morland B. The treatment of hepatoblastoma: its evolution and the current status as per the SIOPEL trials. J Indian Assoc Pediatr Surg. 2014;19:201–7. doi: 10.4103/0971-9261.142001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ortega JA, Douglass EC, Feusner JH, Reynolds M, Quinn JJ, Finegold MJ, et al. Randomized comparison of cisplatin/vincristine/fluorouracil and cisplatin/continuous infusion doxorubicin for treatment of pediatric hepatoblastoma: a report from the children’s cancer group and the pediatric oncology group. J Clin Oncol. 2000;18:2665–75. doi: 10.1200/JCO.2000.18.14.2665. [DOI] [PubMed] [Google Scholar]
- 9.Rougemont AL, McLin VA, Toso C, Wildhaber BE. Adult hepatoblastoma: learning from children. J Hepatol. 2012;56:1392–403. doi: 10.1016/j.jhep.2011.10.028. [DOI] [PubMed] [Google Scholar]
- 10.Yang T, Liang N, Li J, Hu P, Huang Q, Zhao ZF, et al. MDSCs might be “Achilles heel” for eradicating CSCs. Cytokine Growth Factor Rev. 2022;65:39–50. doi: 10.1016/j.cytogfr.2022.04.006. [DOI] [PubMed] [Google Scholar]
- 11.Carrillo-Reixach J, Torrens L, Simon-Coma M, Royo L, Domingo-Sabat M, Abril-Fornaguera J, et al. Epigenetic footprint enables molecular risk stratification of hepatoblastoma with clinical implications. J Hepatol. 2020;73:328–41. doi: 10.1016/j.jhep.2020.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jagadisan B, Dhawan A. Emergencies in paediatric hepatology. J Hepatol. 2022;76:1199–214. doi: 10.1016/j.jhep.2021.12.027. [DOI] [PubMed] [Google Scholar]
- 13.Liang N, Yang T, Huang Q, Yu P, Liu C, Chen L, et al. Mechanism of cancer stemness maintenance in human liver cancer. Cell Death Dis. 2022;13:394. doi: 10.1038/s41419-022-04848-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marayati R, Stafman LL, Williams AP, Bownes LV, Quinn CH, Markert HR, et al. CRISPR/Cas9-mediated knockout of PIM3 suppresses tumourigenesis and cancer cell stemness in human hepatoblastoma cells. Cancer Gene Ther. 2022;29:558–72. doi: 10.1038/s41417-021-00334-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mavila N, Thundimadathil J. The Emerging Roles of Cancer Stem Cells and Wnt/Beta-Catenin Signaling in Hepatoblastoma. Cancers (Basel) 2019;11:1406. doi: 10.3390/cancers11101406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Monga SP. Beta-catenin signaling and roles in liver homeostasis, injury, and tumourigenesis. Gastroenterology. 2015;148:1294–310. doi: 10.1053/j.gastro.2015.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Russell JO, Monga SP. Wnt/beta-catenin signaling in liver development, homeostasis, and pathobiology. Annu Rev Pathol. 2018;13:351–78. doi: 10.1146/annurev-pathol-020117-044010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang H, Zhang S, Zhang Y, Jia J, Wang J, Liu X, et al. TAZ is indispensable for c-MYC-induced hepatocarcinogenesis. J Hepatol. 2022;76:123–34. doi: 10.1016/j.jhep.2021.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang G, Wang Q, Liang N, Xue H, Yang T, Chen X, et al. Oncogenic driver genes and tumour microenvironment determine the type of liver cancer. Cell Death Dis. 2020;11:313. doi: 10.1038/s41419-020-2509-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sagawa H, Naiki-Ito A, Kato H, Naiki T, Yamashita Y, Suzuki S, et al. Connexin 32 and luteolin play protective roles in non-alcoholic steatohepatitis development and its related hepatocarcinogenesis in rats. Carcinogenesis. 2015;36:1539–49. doi: 10.1093/carcin/bgv143. [DOI] [PubMed] [Google Scholar]
- 21.Gu Y, Wei W, Cheng Y, Wan B, Ding X, Wang H, et al. A pivotal role of BEX1 in liver progenitor cell expansion in mice. Stem Cell Res Ther. 2018;9:164. doi: 10.1186/s13287-018-0905-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khazaei MR, Halfter H, Karimzadeh F, Koo JH, Margolis FL, Young P. Bex1 is involved in the regeneration of axons after injury. J Neurochem. 2010;115:910–20. doi: 10.1111/j.1471-4159.2010.06960.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koo JH, Smiley MA, Lovering RM, Margolis FL. Bex1 knock out mice show altered skeletal muscle regeneration. Biochem Biophys Res Commun. 2007;363:405–10. doi: 10.1016/j.bbrc.2007.08.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Ronde JJ, Lips EH, Mulder L, Vincent AD, Wesseling J, Nieuwland M, et al. SERPINA6, BEX1, AGTR1, SLC26A3, and LAPTM4B are markers of resistance to neoadjuvant chemotherapy in HER2-negative breast cancer. Breast Cancer Res Treat. 2013;137:213–23. doi: 10.1007/s10549-012-2340-x. [DOI] [PubMed] [Google Scholar]
- 25.Foltz G, Ryu GY, Yoon JG, Nelson T, Fahey J, Frakes A, et al. Genome-wide analysis of epigenetic silencing identifies BEX1 and BEX2 as candidate tumour suppressor genes in malignant glioma. Cancer Res. 2006;66:6665–74. doi: 10.1158/0008-5472.CAN-05-4453. [DOI] [PubMed] [Google Scholar]
- 26.Wang Q, Liang N, Yang T, Li Y, Li J, Huang Q, et al. DNMT1-mediated methylation of BEX1 regulates stemness and tumourigenicity in liver cancer. J Hepatol. 2021;75:1142–53. doi: 10.1016/j.jhep.2021.06.025. [DOI] [PubMed] [Google Scholar]
- 27.Ding K, Su Y, Pang L, Lu Q, Wang Z, Zhang S, et al. Inhibition of apoptosis by downregulation of hBex1, a novel mechanism, contributes to the chemoresistance of Bcr/Abl+ leukemic cells. Carcinogenesis. 2009;30:35–42. doi: 10.1093/carcin/bgn251. [DOI] [PubMed] [Google Scholar]
- 28.Doi T, Ogawa H, Tanaka Y, Hayashi Y, Maniwa Y. Bex1 significantly contributes to the proliferation and invasiveness of malignant tumour cells. Oncol Lett. 2020;20:362. doi: 10.3892/ol.2020.12226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hiyama E, Ueda Y, Kurihara S, Kawashima K, Ikeda K, Morihara N, et al. Gene expression profiling in hepatoblastoma cases of the Japanese study group for pediatric liver tumours-2 (JPLT-2) trial, Eur. J. Mol. Cancer. 2019;1:2–8. [Google Scholar]
- 30.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–10. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang G, Wang Q, Huang Q, Chen Y, Sun X, He L, et al. Upregulation of mtSSB by interleukin-6 promotes cell growth through mitochondrial biogenesis-mediated telomerase activation in colorectal cancer. Int J Cancer. 2019;144:2516–28. doi: 10.1002/ijc.31978. [DOI] [PubMed] [Google Scholar]
- 32.Wang D, Tian J, Yan Z, Yuan Q, Wu D, Liu X, et al. Mitochondrial fragmentation is crucial for c-Myc-driven hepatoblastoma-like liver tumours. Mol Ther. 2022;30:1645–60. doi: 10.1016/j.ymthe.2022.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu P, Ge M, Hu J, Li X, Che L, Sun K, et al. A functional mammalian target of rapamycin complex 1 signaling is indispensable for c-Myc-driven hepatocarcinogenesis. Hepatology. 2017;66:167–81. doi: 10.1002/hep.29183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yun WJ, Shin E, Lee K, Jung HY, Kim SH, Park YN, et al. Clinicopathologic implication of hepatic progenitor cell marker expression in hepatoblastoma. Pathol Res Pract. 2013;209:568–73. doi: 10.1016/j.prp.2013.06.015. [DOI] [PubMed] [Google Scholar]
- 35.Wu JF, Ho MC, Ni YH, Hsu HY, Lee PH, Chang MH, et al. Dysregulation of liver developmental microRNA contribute to hepatic carcinogenesis. J Formos Med Assoc. 2020;119:1041–51. doi: 10.1016/j.jfma.2019.09.018. [DOI] [PubMed] [Google Scholar]
- 36.Ward SC, Thung SN, Lim KH, Tran TT, Hong TK, Hoang PL, et al. Hepatic progenitor cells in liver cancers from Asian children. Liver Int. 2010;30:102–11. doi: 10.1111/j.1478-3231.2009.02126.x. [DOI] [PubMed] [Google Scholar]
- 37.Xu X, Liu RF, Zhang X, Huang LY, Chen F, Fei QL, et al. DLK1 as a potential target against cancer stem/progenitor cells of hepatocellular carcinoma. Mol Cancer Ther. 2012;11:629–38. doi: 10.1158/1535-7163.MCT-11-0531. [DOI] [PubMed] [Google Scholar]
- 38.Falix FA, Aronson DC, Lamers WH, Hiralall JK, Seppen J. DLK1, a serum marker for hepatoblastoma in young infants. Pediatr Blood Cancer. 2012;59:743–5. doi: 10.1002/pbc.24024. [DOI] [PubMed] [Google Scholar]
- 39.Ilmer M, Garnier A, Vykoukal J, Alt E, von Schweinitz D, Kappler R, et al. Targeting the Neurokinin-1 receptor compromises canonical Wnt signaling in hepatoblastoma. Mol Cancer Ther. 2015;14:2712–21. doi: 10.1158/1535-7163.MCT-15-0206. [DOI] [PubMed] [Google Scholar]
- 40.Oikawa T, Kamiya A, Zeniya M, Chikada H, Hyuck AD, Yamazaki Y, et al. Sal-like protein 4 (SALL4), a stem cell biomarker in liver cancers. Hepatology. 2013;57:1469–83. doi: 10.1002/hep.26159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zeng SS, Yamashita T, Kondo M, Nio K, Hayashi T, Hara Y, et al. The transcription factor SALL4 regulates stemness of EpCAM-positive hepatocellular carcinoma. J Hepatol. 2014;60:127–34. doi: 10.1016/j.jhep.2013.08.024. [DOI] [PubMed] [Google Scholar]
- 42.Lee TK, Castilho A, Cheung VC, Tang KH, Ma S, Ng IO. CD24(+) liver tumour-initiating cells drive self-renewal and tumour initiation through STAT3-mediated NANOG regulation. Cell Stem Cell. 2011;9:50–63. doi: 10.1016/j.stem.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 43.Wang R, Li Y, Tsung A, Huang H, Du Q, Yang M, et al. iNOS promotes CD24(+)CD133(+) liver cancer stem cell phenotype through a TACE/ADAM17-dependent Notch signaling pathway. Proc Natl Acad Sci USA. 2018;115:E10127–36. doi: 10.1073/pnas.1722100115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cao W, Li M, Liu J, Zhang S, Noordam L, Verstegen MMA, et al. LGR5 marks targetable tumour-initiating cells in mouse liver cancer. Nat Commun. 2020;11:1961. doi: 10.1038/s41467-020-15846-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wei Z, Jia J, Heng G, Xu H, Shan J, Wang G, et al. Sirtuin-1/mitochondrial ribosomal protein S5 Axis enhances the metabolic flexibility of liver cancer stem cells. Hepatology. 2019;70:1197–213. doi: 10.1002/hep.30622. [DOI] [PubMed] [Google Scholar]
- 46.Wei RR, Zhang MY, Rao HL, Pu HY, Zhang HZ, Wang HY. Identification of ADH4 as a novel and potential prognostic marker in hepatocellular carcinoma. Med Oncol. 2012;29:2737–43. doi: 10.1007/s12032-011-0126-3. [DOI] [PubMed] [Google Scholar]
- 47.Liu X, Li T, Kong D, You H, Kong F, Tang R. Prognostic implications of alcohol dehydrogenases in hepatocellular carcinoma. BMC Cancer. 2020;20:1204. doi: 10.1186/s12885-020-07689-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu Q, Li J, Zhang W, Xiao C, Zhang S, Nian C, et al. Glycogen accumulation and phase separation drives liver tumour initiation. Cell. 2021;184:5559–76.e5519. doi: 10.1016/j.cell.2021.10.001. [DOI] [PubMed] [Google Scholar]
- 49.Hong SM, Lee YK, Park I, Kwon SM, Min S, Yoon G. Lactic acidosis caused by repressed lactate dehydrogenase subunit B expression down-regulates mitochondrial oxidative phosphorylation via the pyruvate dehydrogenase (PDH)-PDH kinase axis. J Biol Chem. 2019;294:7810–20. doi: 10.1074/jbc.RA118.006095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Antonowicz S, Bodai Z, Wiggins T, Markar SR, Boshier PR, Goh YM, et al. Endogenous aldehyde accumulation generates genotoxicity and exhaled biomarkers in esophageal adenocarcinoma. Nat Commun. 2021;12:1454. doi: 10.1038/s41467-021-21800-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gao T, Zhang X, Zhao J, Zhou F, Wang Y, Zhao Z, et al. SIK2 promotes reprogramming of glucose metabolism through PI3K/AKT/HIF-1alpha pathway and Drp1-mediated mitochondrial fission in ovarian cancer. Cancer Lett. 2020;469:89–101. doi: 10.1016/j.canlet.2019.10.029. [DOI] [PubMed] [Google Scholar]
- 52.Yu J, Shen B, Chu ES, Teoh N, Cheung KF, Wu CW, et al. Inhibitory role of peroxisome proliferator-activated receptor gamma in hepatocarcinogenesis in mice and in vitro. Hepatology. 2010;51:2008–19. doi: 10.1002/hep.23550. [DOI] [PubMed] [Google Scholar]
- 53.Picard F, Auwerx J. PPAR(gamma) and glucose homeostasis. Annu Rev Nutr. 2002;22:167–97. doi: 10.1146/annurev.nutr.22.010402.102808. [DOI] [PubMed] [Google Scholar]
- 54.Shashni B, Sakharkar KR, Nagasaki Y, Sakharkar MK. Glycolytic enzymes PGK1 and PKM2 as novel transcriptional targets of PPARgamma in breast cancer pathophysiology. J Drug Target. 2013;21:161–74. doi: 10.3109/1061186X.2012.736998. [DOI] [PubMed] [Google Scholar]
- 55.Zhang W, Shao W, Dong Z, Zhang S, Liu C, Chen S. Cloxiquine, a traditional antituberculosis agent, suppresses the growth and metastasis of melanoma cells through activation of PPARgamma. Cell Death Dis. 2019;10:404. doi: 10.1038/s41419-019-1644-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Benit P, Pelhaitre A, Saunier E, Bortoli S, Coulibaly A, Rak M, et al. Paradoxical inhibition of glycolysis by pioglitazone opposes the mitochondriopathy caused by AIF deficiency. EBioMedicine. 2017;17:75–87. doi: 10.1016/j.ebiom.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Morris NL, Michael DN, Crotty KM, Chang SS, Yeligar SM. Alcohol-induced glycolytic shift in alveolar macrophages is mediated by hypoxia-inducible Factor-1 alpha. Front Immunol. 2022;13:865492. doi: 10.3389/fimmu.2022.865492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu Y, Ji JJ, Wang XD, Sun XJ, Li M, Wei Q, et al. Periostin promotes arterial calcification through PPARgamma-related glucose metabolism reprogramming. Am J Physiol Heart Circ Physiol. 2021;320:H2222–39. doi: 10.1152/ajpheart.01009.2020. [DOI] [PubMed] [Google Scholar]
- 59.Yang Y, Zhao LH, Huang B, Wang RY, Yuan SX, Tao QF, et al. Pioglitazone, a PPARgamma agonist, inhibits growth and invasion of human hepatocellular carcinoma via blockade of the rage signaling. Mol Carcinog. 2015;54:1584–95. doi: 10.1002/mc.22231. [DOI] [PubMed] [Google Scholar]
- 60.Uehara T, Ainslie GR, Kutanzi K, Pogribny IP, Muskhelishvili L, Izawa T, et al. Molecular mechanisms of fibrosis-associated promotion of liver carcinogenesis. Toxicol Sci. 2013;132:53–63. doi: 10.1093/toxsci/kfs342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–33. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen JQ, Russo J. Dysregulation of glucose transport, glycolysis, TCA cycle and glutaminolysis by oncogenes and tumour suppressors in cancer cells. Biochim Biophys Acta. 2012;1826:370–84. doi: 10.1016/j.bbcan.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 63.Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 64.Tamai K, Nakamura-Shima M, Shibuya-Takahashi R, Kanno SI, Yasui A, Mochizuki M, et al. BEX2 suppresses mitochondrial activity and is required for dormant cancer stem cell maintenance in intrahepatic cholangiocarcinoma. Sci Rep. 2020;10:21592. doi: 10.1038/s41598-020-78539-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sancho P, Barneda D, Heeschen C. Hallmarks of cancer stem cell metabolism. Br J Cancer. 2016;114:1305–12. doi: 10.1038/bjc.2016.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen CL, Uthaya Kumar DB, Punj V, Xu J, Sher L, Tahara SM, et al. NANOG metabolically reprograms tumour-initiating stem-like cells through tumourigenic changes in oxidative phosphorylation and fatty acid metabolism. Cell Metab. 2016;23:206–19. doi: 10.1016/j.cmet.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shen YA, Wang CY, Hsieh YT, Chen YJ, Wei YH. Metabolic reprogramming orchestrates cancer stem cell properties in nasopharyngeal carcinoma. Cell Cycle. 2015;14:86–98. doi: 10.4161/15384101.2014.974419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dong C, Yuan T, Wu Y, Wang Y, Fan TW, Miriyala S, et al. Loss of FBP1 by Snail-mediated repression provides metabolic advantages in basal-like breast cancer. Cancer Cell. 2013;23:316–31. doi: 10.1016/j.ccr.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lagadinou ED, Sach A, Callahan K, Rossi RM, Neering SJ, Minhajuddin M, et al. BCL-2 inhibition targets oxidative phosphorylation and selectively eradicates quiescent human leukemia stem cells. Cell Stem Cell. 2013;12:329–41. doi: 10.1016/j.stem.2012.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gabay M, Li Y, Felsher DW. MYC activation is a hallmark of cancer initiation and maintenance. Cold Spring Harb Perspect Med. 2014;4:a014241. doi: 10.1101/cshperspect.a014241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Folmes CD, Martinez-Fernandez A, Faustino RS, Yamada S, Perez-Terzic C, Nelson TJ, et al. Nuclear reprogramming with c-Myc potentiates glycolytic capacity of derived induced pluripotent stem cells. J Cardiovasc Transl Res. 2013;6:10–21. doi: 10.1007/s12265-012-9431-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Desvergne B, Wahli W. Peroxisome proliferator-activated receptors: nuclear control of metabolism. Endocr Rev. 1999;20:649–88. doi: 10.1210/edrv.20.5.0380. [DOI] [PubMed] [Google Scholar]
- 73.Sancho P, Burgos-Ramos E, Tavera A, Bou Kheir T, Jagust P, Schoenhals M, et al. MYC/PGC-1alpha balance determines the metabolic phenotype and plasticity of pancreatic cancer stem cells. Cell Metab. 2015;22:590–605. doi: 10.1016/j.cmet.2015.08.015. [DOI] [PubMed] [Google Scholar]
- 74.Janiszewska M, Suva ML, Riggi N, Houtkooper RH, Auwerx J, Clement-Schatlo V, et al. Imp2 controls oxidative phosphorylation and is crucial for preserving glioblastoma cancer stem cells. Genes Dev. 2012;26:1926–44. doi: 10.1101/gad.188292.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ye XQ, Li Q, Wang GH, Sun FF, Huang GJ, Bian XW, et al. Mitochondrial and energy metabolism-related properties as novel indicators of lung cancer stem cells. Int J Cancer. 2011;129:820–31. doi: 10.1002/ijc.25944. [DOI] [PubMed] [Google Scholar]
- 76.Ahmadian M, Suh JM, Hah N, Liddle C, Atkins AR, Downes M, et al. PPARgamma signaling and metabolism: the good, the bad and the future. Nat Med. 2013;19:557–66. doi: 10.1038/nm.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Choi JH, Banks AS, Kamenecka TM, Busby SA, Chalmers MJ, Kumar N, et al. Antidiabetic actions of a non-agonist PPARgamma ligand blocking Cdk5-mediated phosphorylation. Nature. 2011;477:477–81. doi: 10.1038/nature10383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Choi JH, Banks AS, Estall JL, Kajimura S, Bostrom P, Laznik D, et al. Anti-diabetic drugs inhibit obesity-linked phosphorylation of PPARgamma by Cdk5. Nature. 2010;466:451–6. doi: 10.1038/nature09291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Quan Q, Qian Y, Li X, Li M. Pioglitazone reduces beta amyloid levels via inhibition of PPARgamma phosphorylation in a neuronal model of Alzheimer’s disease. Front Aging Neurosci. 2019;11:178. doi: 10.3389/fnagi.2019.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Poulsen L, Siersbaek M, Mandrup S. PPARs: fatty acid sensors controlling metabolism. Semin Cell Dev Biol. 2012;23:631–9. doi: 10.1016/j.semcdb.2012.01.003. [DOI] [PubMed] [Google Scholar]
- 81.van Beekum O, Fleskens V, Kalkhoven E. Posttranslational modifications of PPAR-gamma: fine-tuning the metabolic master regulator. Obesity (Silver Spring) 2009;17:213–9. doi: 10.1038/oby.2008.473. [DOI] [PubMed] [Google Scholar]
- 82.Hsu HT, Sung MT, Lee CC, Kuo YJ, Chi CW, Lee HC, et al. Peroxisome proliferator-activated receptor gamma expression is inversely associated with macroscopic vascular invasion in human hepatocellular carcinoma. Int J Mol Sci. 2016;17:1226. doi: 10.3390/ijms17081226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cao LQ, Shao ZL, Liang HH, Zhang DW, Yang XW, Jiang XF, et al. Activation of peroxisome proliferator-activated receptor-gamma (PPARgamma) inhibits hepatoma cell growth via downregulation of SEPT2 expression. Cancer Lett. 2015;359:127–35. doi: 10.1016/j.canlet.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 84.Roche TE, Baker JC, Yan X, Hiromasa Y, Gong X, Peng T, et al. Distinct regulatory properties of pyruvate dehydrogenase kinase and phosphatase isoforms. Prog Nucleic Acid Res Mol Biol. 2001;70:33–75. doi: 10.1016/S0079-6603(01)70013-X. [DOI] [PubMed] [Google Scholar]
- 85.Zuo Q, He J, Zhang S, Wang H, Jin G, Jin H, et al. PPARgamma coactivator-1alpha suppresses metastasis of hepatocellular carcinoma by inhibiting warburg effect by PPARgamma-dependent WNT/beta-Catenin/Pyruvate dehydrogenase kinase isozyme 1 Axis. Hepatology. 2021;73:644–60. doi: 10.1002/hep.31280. [DOI] [PubMed] [Google Scholar]
- 86.Bamodu OA, Chang HL, Ong JR, Lee WH, Yeh CT, Tsai JT. Elevated PDK1 expression drives PI3K/AKT/MTOR signaling promotes radiation-resistant and dedifferentiated phenotype of hepatocellular carcinoma. Cells. 2020;9:746. doi: 10.3390/cells9030746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dupuy F, Tabaries S, Andrzejewski S, Dong Z, Blagih J, Annis MG, et al. PDK1-dependent metabolic reprogramming dictates metastatic potential in breast cancer. Cell Metab. 2015;22:577–89. doi: 10.1016/j.cmet.2015.08.007. [DOI] [PubMed] [Google Scholar]
- 88.Baumunk D, Reichelt U, Hildebrandt J, Krause H, Ebbing J, Cash H, et al. Expression parameters of the metabolic pathway genes pyruvate dehydrogenase kinase-1 (PDK-1) and DJ-1/PARK7 in renal cell carcinoma (RCC) World J Urol. 2013;31:1191–6. doi: 10.1007/s00345-012-0874-5. [DOI] [PubMed] [Google Scholar]
- 89.Koukourakis MI, Giatromanolaki A, Harris AL, Sivridis E. Comparison of metabolic pathways between cancer cells and stromal cells in colorectal carcinomas: a metabolic survival role for tumour-associated stroma. Cancer Res. 2006;66:632–7. doi: 10.1158/0008-5472.CAN-05-3260. [DOI] [PubMed] [Google Scholar]
- 90.Wang XQ, Lo CM, Chen L, Ngan ES, Xu A, Poon RY. CDK1-PDK1-PI3K/Akt signaling pathway regulates embryonic and induced pluripotency. Cell Death Differ. 2017;24:38–48. doi: 10.1038/cdd.2016.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ling LS, Voskas D, Woodgett JR. Activation of PDK-1 maintains mouse embryonic stem cell self-renewal in a PKB-dependent manner. Oncogene. 2013;32:5397–408. doi: 10.1038/onc.2013.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang Z, Xu X, Liu N, Cheng Y, Jin W, Zhang P, et al. SOX9-PDK1 axis is essential for glioma stem cell self-renewal and temozolomide resistance. Oncotarget. 2018;9:192–204. doi: 10.18632/oncotarget.22773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Peng F, Wang JH, Fan WJ, Meng YT, Li MM, Li TT, et al. Glycolysis gatekeeper PDK1 reprograms breast cancer stem cells under hypoxia. Oncogene. 2018;37:1062–74. doi: 10.1038/onc.2017.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All of the relevant data are included in supplemental information. RNA-seq data have been submitted to the SRA database (SRA: PRJNA721822). All data are available upon request to the corresponding authors.