Abstract
The past decade has witnessed a revolution in cancer treatment by the shift from conventional drugs (chemotherapies) towards targeted molecular therapies and immune-based therapies, in particular the immune-checkpoint inhibitors (ICIs). These immunotherapies selectively release the host immune system against the tumour and have shown unprecedented durable remission for patients with cancers that were thought incurable such as advanced non-small cell lung cancer (aNSCLC). The prediction of therapy response is based since the first anti-PD-1/PD-L1 molecules FDA and EMA approvals on the level of PD-L1 tumour cells expression evaluated by immunohistochemistry, and recently more or less on tumour mutation burden in the USA. However, not all aNSCLC patients benefit from immunotherapy equally, since only around 30% of them received ICIs and among them 30% have an initial response to these treatments. Conversely, a few aNSCLC patients could have an efficacy ICIs response despite low PD-L1 tumour cells expression. In this context, there is an urgent need to look for additional robust predictive markers for ICIs efficacy in thoracic oncology. Understanding of the mechanisms that enable cancer cells to adapt to and eventually overcome therapy and identifying such mechanisms can help circumvent resistance and improve treatment. However, more than a unique universal marker, the evaluation of several molecules in the tumour at the same time, particularly by using multiplex immunostaining is a promising open room to optimise the selection of patients who benefit from ICIs. Therefore, urgent further efforts are needed to optimise to individualise immunotherapy based on both patient-specific and tumour-specific characteristics. This review aims to rethink the role of multiplex immunostaining in immuno-thoracic oncology, with the current advantages and limitations in the near-daily practice use.
Subject terms: Molecular medicine, Medical research
Introduction
The immuno-oncology (IO) field has witnessed a remarkable booming in the past decade, after years of controversial dogmas and inconsistent findings. The upgraded comprehension of the cancer-immune system interactions and the tremendous technological progress has revived the hope of curing cancer with immune-based therapies. The target of these treatments has shifted from the tumour itself to the host’s immune system, to mobilise immune cells to recognise and eventually eliminate the cancer cells [1]. Hallmarks of immunotherapy are the long-lasting responses, most likely linked to the memory of the adaptive immune system contributing to long-term survival for a subset of patients, as well as the specificity of the trained immune system to recognise and target cancer cells [2]. Immune-checkpoint inhibitors (ICIs) have proven remarkable clinical effects in a wide range of metastatic tumour types. In particular, the programmed cell death-1/programmed cell death-ligand 1 (PD-1/PD-L1) blocking antibodies act through the reactivation of effector pre-existing tumour-infiltrating lymphocytes (TILs) [3]. Likewise, the landscape of treatments of patients diagnosed with an advanced non-small cell lung cancer (aNSCLC) has dramatically evolved in the last ten years, in particular with the approval of ICIs in either first- or second-line settings, with or without chemotherapy, and irrespective of the histological subtypes of aNSCLC.
Since the first IO clinical trials, the only keypredictive marker used to date as a companion diagnostic in thoracic oncology approved in routine clinical practice, is the expression of PD-L1 on tumour/immune cells assessed by conventional immunohistochemistry (IHC) on paraffin-embedded formalin-fixed (FFPE) tissue sections. In addition, many efforts are conducted in order to identify potential predictive markers of ICIs in aNSCLC. For instance, the tumour mutational burden (TMB) gave promises to be a predictive marker for ICIs response, but has multiple technical issues (e.g., variability of TMB calculation between assays, variable reproducibility of bioinformatics algorithms, a trend for race-dependent increases in TMB scores) and still needs to be better evaluated as a predictive marker. Of note, only the FoundationOne CDx assay has been approved by the Food and Drug Administration (FDA) as a companion diagnostic for second-line pembrolizumab monotherapy for solid tumours including aNSCLC with high TMB [4]. However, despite these recent advances in immunotherapy strategies, allowing durable clinical responses and increased overall survival in a subset of patients, most patients with aNSCLC do not benefit or do not respond to ICIs, mainly because of imperfect associated companion diagnostic testing. Moreover, one major challenge is also to determine as well as possible the best treatment decision-making in order to avoid exposing non-responders to unnecessary side effects, but also unnecessary costs.
In this context, there is an urgent need in the IO era in thoracic oncology for the development and clinical validation of predictive markers. In daily practice, a predictive marker can be detected in situ, mainly by immunohistochemistry (IHC) or fluorescent in situ hybridisation (FISH) or by using molecular genetic testing. Since many years, IHC has been used in pathology laboratories worldwide, is easy to use and cost-effective. Conventional IHC is currently the “gold standard” method for diagnosis in pathology. Therefore, this approach is used for the assessment of PD-L1 expression in tumour tissue sections as the major predictive assay in IO until now. However, conventional IHC suffers from several limitations, including the reduced labelling capacity of a single marker per tissue section. In the context of the complexity and heterogeneity of the immune system in patients treated with ICIs, this major drawback results in missed opportunities to harvest essential predictive information from patient samples [5].
Even if single predictive markers have been or are currently evaluated in IO, the possibility to combine different in situ analyses in tumour tissues, including looking at the same time different protein or RNA expressions both on tumour and immune cells, has opened many rooms for the near establishment of sensitive and specific predictive molecular signatures for ICIs. Therefore, the emergence of multiplex immunohistochemistry/immunofluorescence (mIHC/IF) techniques has provided a great opportunity to overcome many of the current conventional IHC challenges [6]. These approaches facilitate the investigation of multiple markers on a single-tissue section as well as the exploration of tissue-level biology, classification of cell-cell interactions and spatial analysis, enhanced interrogation of immune cells phenotyping as well as the identification of rare cellular phenotypes, and not the least the preservation from tissue exhaustion. mIHC/IF is also a powerful supplement to technologies such as next-generation sequencing (NGS), suitable for digital analysis and accurate scoring. As such, mIHC/IF holds the great potential to revolutionise the assessment of predictive markers in thoracic oncology in order to better select patients for immunotherapy.
This review aims to provide recent information on promising in situ predictive markers for ICIs, notably by using mIHC/IF, including the description of the current advantages and limitations of these latter methods.
Pd-L1 and Tmb: two current and imperfect predictive markers in thoracic immuno-oncology
PD-L1 immunohistochemistry: from hope to weakness
PD-L1 IHC had initially opened many hopes for using a simple approach, easy to use in all pathology laboratories worldwide, allowing rapid results for a better assessment of predictive response to ICIs and thus to give the go/no go to the physician for an optimal treatment decision-making. PD-L1 IHC presents advantages such as low cost and application on formalin-fixed, paraffin-embedded (FFPE) tissue or cytological samples. Therefore, this test is now worldwide used routinely for most of the ICIs indication. However, despite the clinical benefit, one of the caveats is that even when patient selection is driven by PD-L1 expression, treatment response rates ranged from 27 to 45% in the first-line setting and 19% in the second-line refractory setting [7, 8]. Thus, despite some advantages, both the pathologists and the oncologists calling into question the usefulness of PD-L1 IHC as a predictive marker have progressively highlighted many limitations. The PD-L1 IHC assay, the only recognised and validated marker used worldwide for predicting anti-PD-1/PD-L1 response, has also demonstrated several essential limitations [9, 10].
Among them, the use of different detection IHC assays, antibody clones, intra-observer variability, subjective interpretation, particularly with the evaluation of malignant cells and various immune cell staining expression leading to high inter-observer variability, semi-quantitative assessment, temporal and intra-tumour heterogeneity expression, difficulty to set up an automatic staining assessment using different software, restricted labelling potential and insufficient availability of samples for testing, notably when having small tissue biopsies [9]. Above all, the discrepancies between the percentage of PD-L1-positive tumour cells and the anti-PD-L1/PD-1 therapeutic response or non-response in many patients, lead to an urgent need to find predictive markers in this setting [11]. Thus, PD-L1 expression is likely not an effective standalone marker for treatment decisions in routine clinical practice in aNSCLC [12].
Finally, “hot” and “cold” tissue microenvironment (TME) cannot be distinguished based on PD-L1 status alone instead requires measurement of tumour-infiltrating lymphocytes (TILs) abundance and IFN-γ signalling which are not routinely evaluated in daily practice [12].
Tumour mutational burden: a tricky predictive marker to be used in daily practice
Addition to to PD-L1 IHC, the TMB, for the number of non-synonymous single nucleotide variants, has been proposed to be a complementary predictive marker for ICIs, but except issues due to the difficulty to its large implementation in pathology laboratories, the use of TMB in this setting is not worldwide recognised and available nowadays [13, 14]. Recent studies suggested that high TMB does not always predict response to ICIs across several types of cancer [15–17]. Moreover, original studies linked TMB level with response to ICIs using whole-exome sequencing, which is costly and a labour-intensive technique that would not be able to set up in routine clinical practice [18, 19]. Limitation of using targeted gene sequencing is the need to extrapolate the TMB from a small number of genes [11]. Finally, a limitation is the availability of many different panels, which vary in the genes targeted, number of genes tested and genomic space sequenced. Therefore, this leads to a barrier to establishing a single standardised technique [20]. In addition, one limitation can be the presence of intra-tumoral heterogeneity. A high intra-tumoral heterogeneity usually indicates that the neoantigen may only be present on a subset of cells and so the immune response generated against the neoantigen may not be effective against the whole tumour. In addition, this heterogeneity may result in a lower dosage of each neoantigen, reducing CD8 + T-cell activation. Moreover, the combination of reduced neoantigen dosage and targeting only a subset of tumour cells means that is can be difficult to establish a TMB cut-off for response to PD-1 pathway blockage across all patients [12]. In addition, recent data showed that the integration of TMB with immune cell infiltration and inflammatory T-cell expression signatures might be able to identify patients with the greatest likelihood of response to ICIs across PD-L1 expression subgroups [21]. Overall, these findings emphasise the importance to develop, combine and validate additional predictive markers that capture the complexity of the TME.
Potential predictive in situ markers in thoracic immuno-oncology
Deciphering the resistance of certain tumour clones and metastases under immune pressure is a crucial step toward tailoring ICIs for long-term disease-free and overall survival. It is challenging to average metastatic illness among patients since each metastasis is a distinct immunological disease. Between parent and child metastases, inside a single metastasis, and over time, the tumour-immune ecosystems fluctuate. For any clinical modality, the fact that neither a single biopsy nor a single metastasis is adequate to generalise the tumour setting and the patient’s response to ICIs presents significant hurdles [22].
“Immune contexture” refers to the combination of immune factors associating the nature, density, immune functional orientation and topography if immune cells within the TME. These immune contexture parameters are associated with long-term survival and prediction of response to ICIs [23, 24]. Thus, better understanding and capture of the immune architecture of a tumour is becoming essential for evaluating the therapeutic responses, particularly for ICIs [23].
The TME can be classified into T-cell-inflamed “hot” versus non-T-cell-inflamed “cold” environments with the former being further classified based on the activation status of the T lymphocytes [25]. Notably, “hot” tumours are most often accompanied by an IFN-γ–driven adaptive immune resistance phenotype characterised by up-regulation of immune-regulatory pathways including immune checkpoints, such as PD-1, LAG-3, TIM-3, VISTA and TIGIT, other inhibitory molecules, such as IDO-1, TGFβ1, and INOS, or expansion of immune suppressive cells, such as regulatory T cells (Tregs) and myeloid-derived suppressor cells (MDSCs). The potential for LAG-3, VISTA and TIM-3 or TIGIT-driven regulation of immune responses is now being explored in early-phase clinical trials for aNSCLC [26–30]. Among them, it has been demonstrated in translational research programmes and in clinical trials, that some molecules can be used as potential predictive markers, according to the corresponding ICIs [31, 32]. The presence and the level of expression of these different molecules can be evaluated individually by IHC with specific antibodies in NSCLC tissue sections. In addition to these different specific targets, looking for the presence of some intra and/or peripheral tumour-immune cells can be assessed by conventional single IHC. One promising predicting marker during ICI treatment could be the high levels of TILs, in particular CD8-positive TILs, that can be assessed an evaluated by IHC on tumour tissue sections [33–36]. Moreover, different subpopulations of immune cells, which can be all assessed by IHC using different antibodies can be associated more or less with ICIs efficacy [37, 38]. However, looking for a single marker is certainly not sufficient for a robust signature of ICIs responsiveness. For example, most studies examining TILs in the TME have focused on CD8 + T cells without knowledge of their functional activity, but is has been shown that the abundance of B cells or plasma cells within the tumour may predict outcomes to PD-L1 blockade in aNSCLC [39, 40]. The evaluation of T-cell dysregulation could also include the assessment of a variety of markers including those for antigen-based activation (CD28, CD39, CD103, CD137, PD-1), regulation (checkpoint: PD-1, LAG-3, TIM-3), differentiation (TCF1, T-bet, EOMES) and apoptosis (FAS, TRAIL) [41–43].
Thus, it is impossible to cover all the various targets important for ICIs response prediction by using conventional IHC. The complexity of studying multiple markers simultaneously and on a single-tissue section, especially those associated to the TME, requires the development of new techniques that empower more in-depth analysis of multiple cell phenotypes as well as their spatial and temporal patterns and interactions. Multiplex IHC is the only imaging method that yields quantitative information on tumour-infiltrating immune cells with spatial resolution. Automated morphometric analysis may then be used to further examine the images acquired using this approach [44]. The simultaneous identification of multiple distinct immune subtypes using this method is now being refined, which will facilitate the ability to analyse their spatial extent and proximity [44]. As a result, multiplexed image analysis technologies labelling could help to stratify patients for appropriate ICIs and identify patients who are most likely to derive benefit or not from immunotherapy. Interestingly, a recent meta-analysis that examined the use of PD-L1 IHC, TMB, gene expression profiling, and mIHC/IF assays to determine objective response to ICIs demonstrated that mIHC/IF and multimodality marker strategies were associated with improved performance over the other approaches alone [6].
Multiplexed stained methodologies and platforms for in situ predictive markers identification
Multiplexed bright-field approaches
Three main staining methods were developed in this setting, the multiplexed immunohistochemical consecutive staining on a single slide (MICSSS) [45], the sequential immunoperoxidase labelling and erasing (SIMPLE) method [46], and a fully automated mIHC technology using tyramide signal amplification (TSA) along with heat deactivation process [47, 48].
Briefly, the MICSSS method is a series of sequential cycles of staining, image scanning, and destaining of chromogenic substrate than can be performed on FFPE tissue samples [45]. This multiplex staining approach uses conventional chromogenic IHC staining, followed by a scanning process of the destained chromogenic substrate in organic solvent that can completely remove the staining [49]. The MICSS method can allow up to ten different antibodies on one single-tissue section and it needs to be supported by third-party scanning and image analysis system. The slow process of the technique is a limitation of the MICSSS, but as the authors mentioned, this limitation could be easily resolved with the automation of the process. Until then, the long processing time is still a limitation that requires improvement. However, although this methodology was tested on limited data, it showed the versatility and potential of the process to study and analyse the complexity of the tumour microenvironment [50].
The SIMPLE method is other staining approach using a sequential labelling bleaching technique that enables simultaneous marker visualisation, combining five to twelve markers using the alcohol-soluble peroxidase substrate 3-amino-9-ethylcarbazole with a fast, non-destructive method for antibody–antigen separation [46]. This method requires the creation of a virtual slide after each labelling using third-party scanner. After digitisation, the staining is erased (or washed) and a new cycle of staining–digitisation–erasing can be performed. Staining erasing is a difficult technique that uses an antibody elution technique intended to preserve tissue and antigen epitopes for the next staining. By correcting certain parameters and including a new elution process, this methodology was recently improved and successfully used with antigens expressed in the same cellular compartment [51]. This method has shown the ability to erase the results of a single stain while preserving tissue antigenicity for repeated rounds of labelling, including image analysis software; this staining process looks very promising for future markers discoveries.
The Discovery Ultra (Roche Diagnostics, Basel, Switzerland) is an alternative IHC research instrument that enables manual and fully automated staining experiments for research purposes. This instrument overcomes some of the problems associated with IHC by performing multiplexed analysis of multiple markers, using tyramine chemistry to combine a vast spectrum of new chromogenic dyes [47, 48]. This is particularly useful for in situ analysis with conventional bright-field microscopes and third-party analysis software. This approach employs covalently deposited chromogens (CDCs) relying on enzymatic activation of dyes conjugated with tyramide and quinone methide precursors to produce stains covalently bound to cellular and tissue components surrounding the sites of targeted proteins [52, 53]. CDCs have the significant advantage of rapid and easy development of new chromogens with desired spectral characteristics, thereby allowing the development of chromogen stains with narrow and well-separated absorbance bands, similar to fluorophores [52]. The dyes, useful both individually and blended to generate novel colours; provide signals like the conventional 3,3′-diaminobenzidine (DAB) chromogen. They may also enable the analysis of co-localised markers [47]. These chromogens have broad absorbance spectra, which produce dark staining patterns that are supposedly easy to distinguish during light microscopy. In addition, this method employs a monochrome camera and sequential illumination with narrow bands of illumination matched to the absorbance bands of the discrete chromogens. Illumination is provided by a filter wheel fitted with narrow bandpass filters in combination with a continuous light source, or by a collection of light-emitting diodes (LEDs) paired with narrow bandpass filters, for further spectral definition. Coordinated rapid LED pulsing and image acquisition provides the basis for high-speed multiplex imaging. While this approach requires optical filtering, like fluorescence, or LEDs, the equipment is simplified relative to fluorescence by not requiring epifluorescence optics, dichroic mirrors and emission filters [48]. More importantly, conventional scanners can acquire images of such stained slides, facilitating marker research and the possibility of in vitro diagnostics product development. In the Discovery Ultra bright-field setting, pathologists can assess the mIHC/IF without any particular software or visualisation tool, which is clearly an advantage. However, recognising more than 2–3 colours for co-localisation of markers in the same cellular compartment might be beyond what the human eye can achieve [54]. Finally, with this approach, clinically relevant four- and five-chromogen multiplex IHC approach was demonstrated in NSCLC [55, 56].
Table 1 lists the main benefits and drawbacks of the most widely used commercialised platforms.
Table 1.
Advantages and disadvantages of the most popular staining methodologies and mIHC/mIF platforms for single-cell analysis.
| Multiplex bright-field approaches | Advantages | Disadvantages |
|---|---|---|
| MICSSS |
- Simple technique using conventional chromogenic IHC staining - Up to 10 different antibodies per tissue section - Whole slide images for each marker - Fairly priced - No autofluorescence |
- Long processing time - No automation - Unable to assess marker intensity - Tissue damage by coverslip removal - Single-cell spatial resolution not yet achieved - No proprietary image analysis software - No availability of a commercial clinical use - No validation studies for routine use |
| SIMPLE |
- Simple technique using conventional chromogenic IHC staining - Whole slide images for each marker - Fairly priced - No autofluorescence |
- No automation - Limited multiplexed capacity up to 5 - Tissue damage - Single-cell spatial resolution not yet achieved - No proprietary image analysis software - No availability of a commercial clinical use - Low publication record |
| Discovery Ultra |
- Readily automated - Fluorescent and chromogenic based - Established protocols - Easy use and interpretation - Clinical relevant, end-to-end solution |
- Co-expression studies are limited and require carefully selected chromogen pairs - Dynamic range of marker intensity is also - Limited - Expensive - No CE-IVD labelling |
| Multiplex immunofluorescence approaches | ||
| MELC |
- Bleaching cycles of up to 50 epitopes in the same tissue site - Highly flexible multiplex detection system - Robotic whole-cell imaging technology - Multiplexing up to dozens of proteins - Generation of a protein collocation map, which can be summarised as a toponome |
- Multiprobe image is limited to a single microscopic medium-to-high power field - PFA-fixed tissue samples - Relatively expensive - Tissue autofluorescence - No proprietary image analysis software - Low publication record |
| MultiOmyx & t-CyCIF |
- High level of multiplexing up to 60 markers per section - Eliminates the need for expensive multispectral instruments - Whole tissue sections using simple three-colour whole slide scanning microscopes |
- Very labour-intensive - Relatively slow process - Low publication record |
| Antibody labelled oligo barcoding platforms (PhenoCycler and PhenoCycler-Fusion) |
- High level of multiplexing up to 40 markers - No spectral overlap - Eliminates autofluorescence - PhenoCycler-Fusion use conventional slides - Single-cell data - High resolution - Bundled image acquisition and analytical software |
- Whole-slide imaging possible but very costly and time-consuming - BSA-free antibodies used (Expensive) - The PhenoCycler use coverslips to place the tissue for staining. - Low publication record - Whole-image analysis software is not included in the PhenoCycler-Fusion - No proprietary image analysis software for whole section analysis |
| Tyramide Signal Amplification platform (PhenoImager HT) |
- Up to 6 different antibodies per tissue section - Signal amplification of biomarkers with low expression - Regular commercial antibodies - Potential assay to be applied for clinical use - Automated staining with partnership to third-party autostainer - Until 6 markers whole slides imaging is allowed - Proprietary image analysis software for region of interest analysis |
- Up to 6 antibody stain is limited to region of interest analysis - Needs autofluorescence extraction - Cross talking and umbrella effect need to be verified during the panel optimisation |
| Imaging Mass spectrometry-based methods | ||
| MALDI MSI |
- In situ visualisation of multiple cell phenotypes, while preserving tissue architecture - Tissue is place on regular slides - Data can be used for spatial and nearest-neighbour analyses - Allows data collection across the entire tissue section |
- Limited amount of markers - Spectral overlap can hinder co-localisation analysis and biomarker quantification - Complex sample preparation protocol - Tissue composition may affect extraction efficiency |
| MIBI platform |
- >40 markers simultaneously - No autofluorescence and spectral overlap - Single-cell data - ~0.5 μm resolution - Bundled image acquisition software |
- Tissue need to be place on gold slides - Sampling time and small area sampling - BSA-free antibodies used (Expensive) - Availability of antibodies - Long imaging times - Extensive training - Need third-party of image analysis |
Main advantages and disadvantages of the leading commercialized mIHC/IF platforms.
Multiplexed immunofluorescence approaches
Multiplexed staining bleaching techniques were created with different platforms to study tumour tissue specimens. Briefly, multi-epitope-ligand cartography (MELC) is a bleaching or erasure staining technique that is capable of co-localising the locations of up to 50 epitopes in the same tissue section using consecutive rounds of conjugate markers with fluorescent detection [57]. In each cycle the sample is incubated with several tags and the image is acquired using a high-resolution digital camera, before bleaching by soft multi-wavelength excitation (e.g., 485 nm for FITC and 546 nm), followed by the construction of co-localisation maps and toponome maps. One limitation of the MELC technique is that the photobleaching step can only be applied to the microscope’s field of view, meaning that the multiprobe image is limited to a single microscopic medium-to-high power field. The MELC needs third-party image analysis software.
MultiOmyx is a multiplex direct immunofluorescence approach where up to 50 antibodies can be interrogated from a single FFPE section [58]. It uses primary conjugated antibodies with fluorochromes to stain different markers of interest in batches of two or four at one time. Methodology development by General Electric Global Research was licensed to NeoGenomics, and it is commercially available by NeoGenomics Laboratories, Inc. (Fort Myers, FL, USA)Another similar method, called t-CyCIF for tissue-based cyclic immunofluorescence (t-CyCIF) can create highly multiplexed images using an iterative process in which conventional low-plex fluorescence images are repeatedly collected from the same sample and then assembled into a high-dimensional representation [59]. The t-CyCIF cycles involve antibody staining against protein antigens, nuclear staining (same fluorophore per cycle), image scanning (low and high magnification),fluorophore bleaching steps and need to be supported by third-party image analysis software
The PhenoCycler [formerly Co-Detection by Indexing (CODEX)] and PhenoCycler-Fusion (Akoya Biosciences, Marlborough, MA) use antibody labelled with specific oligonucleotide tags (barcode) cocktail methodology and dye oligonucleotides (reporters) that are sequentially hybridised and dehybridized across multiple cycles fluorescent-based imaging approach [60, 61]. The oligonucleotide duplexes encode uniquely designed sequences with 5’ overhangs. Fresh frozen tissue and isolated cells were used to validate this methodology, but its application on FFPE tissue. Cells, fresh frozen and FFPE tissue are stained with a cocktail containing all conjugated antibodies (up to 40 antibodies) at the same time. The PhenoCycler platform can be performed on any three-colour fluorescence microscope enabling the conversion of a regular fluorescence microscope into a tool for multidimensional tissue rendering and cell. The Fusion scanner use a multispectral imaging technology which allows for easy detection of multiple overlapping markers on a single tissue and whole-image tissue without any interference of autofluorescence or fluorophore crosstalk. Regions of Interest (ROIs) from the QPTIFF images can be analysed using the InForm software (Akoya Bioscience) or the whole image using a third-party image analysis software. Another barcoding is the DNA exchange imaging (DEI) technique that overcomes speed restrictions by allowing for single-round immunostaining with DNA-barcoded antibodies. It is an easy multiplexed technique that can be adapted to diverse imaging scanners, including standard resolution exchange-confocal and various super-resolution scanners [60, 61]. The DEI need to be supported by third-party image analysis software. Modified-hapten-based technology is another staining technique that allows simultaneous detection of multiplex markers using a standard two-step procedure. The technique is antibody species independent and the signals of the markers can be stronger than those usually observed with direct flour-labelled secondary antibody detection of multiplex. TSA is an enzyme-linked signal amplification method that is using to detect and localise the low copy number of proteins present in tissue by the conventional IHC protocol, using, most commonly, the alkaline phosphatase or horseradish peroxidase enzymatic reaction to catalyse the deposition of tyramide labelled molecules at the site of the probe or epitope detection. Tyramides are conjugated to biotin or fluorescent labels and revealed by the streptavidin–HRP system. Akoya Bioscience developed the Opal™ workflow, which allows the simultaneous staining of multiple markers within a single FFPE tissue using the TSA system. In partnership with Leica Biosystems (Wetzlar, Germany), the staining can be automated using the Leica Bond RX. Using the multispectral image scanner (PhenoImager HT, Akoya Biosciences) and the InForm image analysis software is one of the most complete multiplexed image platforms. The detection of the proteins is more than 10-times greater than standard biotin-based staining methods. Some methods used specially coated nanocrystals (around 1–10 nm in diameter) called quantum dots instead of chromogen. Nanocrystal quantum dots have the property of being excited by any type or wavelength of light to emit light in a very thin fluorescence spectrum. The use of these fluorescent markers in combination with a third-party multispectral imaging technology has a particular utility for multiplexed detection when used as a fluorescent probe bound to different antibody markers. Despite the favourable optical properties of nanocrystal quantum dots, as a fluorescence-based method, they can avoid the endogenous autofluorescence associated with tissue sections, have high photostability, and have a symmetric emission spectrum. An important reported limitation of using nanocrystal quantum dots is the limited number of nanocrystals that possess the proper chemistry to attach themselves to their targeted molecules [60, 61] and the need to use a third-party image analysis software.
The MACSima Imaging Platform that use an automated high multiplex image cycling staining (MICS) enables the simultaneous analysis of hundreds of markers on a single sample based on fluorescence microscopy. It uses the principle of iterative staining with different fluorochrome-conjugated antibodies to acquire microscopy data for many parameters without harming the sample. This iterative process comprises three main steps by the instrument: fluorescent staining, image acquisition, and erasure of the fluorescence signal in a fully automated manner [62]. The instrument is commercialised by Miltenyi Biotec (Bergisch Gladbach, North Rhine-Westphalia, Germany) can be used with FFPE tissues, fresh frozen tissues, or cells fixed with PFA or acetone. The MACSima platform also provides an image analysis system named MACS® Qi Tissue View Software.
Imaging mass spectrometry-based approaches
Mass Spectrometry Imaging (MSI) is defined as the visual representation of the elemental or molecular component of fixed cells or tissues by mass spectrometry [63]. MSI is a technique that uses a mass spectrometer to visualise the spatial distribution of compounds, markers, metabolites, proteins, peptides, or small molecules by their molecular masses. The workflow of MSI includes pre-processing using routine IHC protocols, laser beam, ablated sample aerosol directly transported to the CyTOF by argon and helium gas flow, single isotope signals are plotted using coordinates of each single laser shot, and finally cell features are computationally segmented using the watershed algorithm [64]. Spectrometry started to be applied to biological cells as a chemometric methodology to study the cellular surface composition and the discriminations between normal and neoplastic cells, an issue that can be challenging in cases where neoplastic morphological features may not be evident. Matrix-Assisted Laser Desorption/Ionisation (MALDI), a type of molecular imaging technology, was evolving in the same way as SIMS and LDI, with improved resolution. One of the most exciting applications of MALDI MSI is the analysis of the proteomic pattern composition of tumour cells and the determination of unique profiles that can actually differentiate normal cells from neoplastic cells, even between different subtypes of tumour cells and between primary and metastatic tumours, an approach that has already been explored in non-small cell lung cancer [65].
Multiplexed Ion Beam Imaging (MIBI) platform is another image mass spectrometry that use an antibody-cocktail tagged with monoisotopic metal reporters to be imaging using secondary ion mass spectrometry [66]. MIBI methodology allows the simultaneous detection of more than 40 markers at subcellular resolution in FFPE or frozen tissues, enabling single-cell segmentation, cell-type classification and spatial analysis of the cells present in the TME. In this platform, non-specific binding between antibodies and epitopes can make the validation and standardisation of markers a challenge and proper controls are needed for staining and imaging [67]. The platform is supported by MIBIScope a mass spectrometry imaging commercialised by IONPath (Menlo Park, CA, USA).
Current limitations for daily practice use
Bright-field microscopy is the current clinical standard for cancer diagnostics, prognostics and predictive outcome and has long been the choice of pathologists worldwide for interpreting suspicious tissues, with haematoxylin and eosin stain and IHC being the cornerstones for clinical diagnostics in solid tumours [48]. While IHC remains highly practical and cost-effective, this single-marker method cannot tell the whole story of a complex immune microenvironment. Despite strong promises to improve the use of mIHC for better improvement of predictive IO markers, several constraints and limitations exist currently in order to use mIHC in routine clinical practice (Table 2).
Table 2.
Challenges and current limitations of multiplex IHC/IF approaches.
| Challenge | Current limitations |
|---|---|
| Preanalytical variables | Fixation, cold or hot ischaemic time, sectioning, storage. |
| The choice of the methodology approach and platform |
- Because of the number of methodologies of staining and multiplexed image platforms is challenging to choose which approach would be best for a lab. - Batch controls which are used to normalise quantitative expression signal levels for staining variability. - Pathologists need to consider the advantages and disadvantages of these techniques and the most appropriate use of each technology to be applied for translational research or clinical use. |
| Choice between chromogenic and immunofluorescence multiplex approach |
- The vast majority of pathologists routinely use bright-field microscopes. This approach is certainly more accessible to most pathology laboratories but also has limitations, in particular for the number of possible antibodies to be used per tissue section (on average not more than 5). - Chromogenic approach is not optimal for the analysis of co-expression or precise cellular localisations (nucleus, cytoplasm, membrane). - mIF is better for 3+ plexing, when expression level quantitation and dynamic range is required, and when marker pixel-level colocalization is expected, such as is often the case of cell surface markers. mIHC is better for 1–3 markers that do not colocalize and is primarily for visual assessments, since umbrella affects are significant and hard to mitigate through staining methods. |
| The choice or identify valuable biomarkers within the tumour | - Given the large amount of recent data, in several solid tumours, the use of several dozen antibodies could be required, but it is impossible to ensure exhaustiveness according to the different molecules of interest. |
| Real standards for clinical validation |
- Optimisation and validation of markers for clinical use can be established only across multiple clinical trials. - Quantification criteria need to be established for accuracy and reproducibility. - Validation of artificial intelligence algorithms is required to have a robust platform. |
| Turnaround time | - In the context of the treatment of patients, the time required to obtain the results of these complex analyses must be evaluated. |
| External quality assurance | - Within the framework of external quality assurance controls, it will be necessary to find organisations able to propose external validation tests or to have the possibility of carrying out ring trial studies. |
| Accreditation/certification | - The accreditation/certification of these different approaches will certainly be a long step to achieve and the issue of the In Vitro Diagnostic Regulation (IVDR) of the European Union will have to be integrated. |
| Costs | - The fees and reimbursements of these tests will have to be considered promptly in order to allow their diffusion. |
To begin, the preanalytical variables that can affect conventional IHC assays, such as inadequate fixation, cold or hot ischaemic time, additional factors that can occur through processing from sectioning to storage are just as relevant in multiplex assays [68, 69].
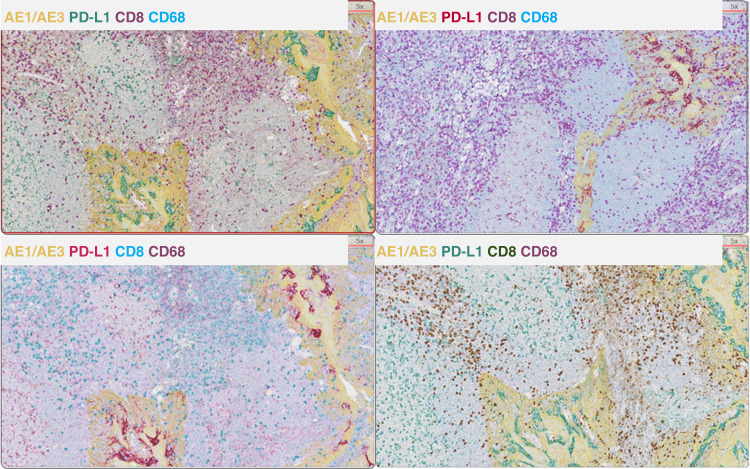
In comparison to standard IHC, multiplex techniques are generally slower and more labour-intensive [69]. They also require expensive, specialised equipment, platforms for image analysis, and data storage that are not always available in all laboratories. Despite the fact that clinical IHC is primarily bright-field-based, multiplexing is often carried out using IF because bright-field multiplexing has proven to be challenging [48, 70]. There are comparatively fewer chromogens than there are fluorophores available for IF, and their absorbance spectra are typically wider than those of fluorophores. In contrast to fluorophores, which exhibit absorbance peaks with full width at half maximum (FWHM) typically between 30 and 60 nm in solution, with some broadening when deposited, common chromogens have FWHM of 200 nm or more. Examples include 3,3’-diaminobenzidine and Fast Red [48]. Accordingly, a combination of two or three chromogens may completely occupy the visible spectrum, with significant spectral overlap between chromogens [48]. Investigation is needed to assess the extent to which a multi-colour background with colour overlap can alter the visual perception and evaluation of key markers, particularly for the heterogeneous expressions. Only a few chromogenic stains successfully detect marker co-expression when applied simultaneously to a single slide. Because the dynamic range of the marker intensity is limited, chromogenic stains are frequently employed to merely assess expression as positive or negative, or to calculate a semi-quantitative H-score [55, 56]. This could lead to additional costs for time-consuming panel development (Fig. 1).
Fig. 1. Representative examples of multiplexed images using bright-field mIHC methodologies.
Example of 4 plex bright-field mIHC panel using different chromogens and cell co-localisations. Images generated using Discovery Ultra platform (Roche Diagnostics, Basel, Switzerland).
The second issue with bright-field microscopy is that commercial colour cameras can only discern the three broadly specified colours—red, green, and blue—that are fixed spectrally to mimic human vision [48]. This restricts the number of dyes that may be properly resolved to three and necessitates that these three dyes have spectral properties that are consistent with the three-colour channels of an RGB camera. This includes dyes that may overlap because of the co-expression of a marker. A monochrome camera and many filter sets that match the absorbance and emission properties of each fluorophore are commonly used for fluorescence imaging. While this enables imaging and resolution of as many fluorophores as spectral separation allowed, it has the drawback that switching between filter sets, collecting, and processing each filtered sample independently takes time [48, 69].
Bright-field measurements are considerably faster than fluorescence since fluorescence light intensities are much weaker than transmitted light. Still, pathologists will need to accept the additional hardware, electronics, and software needed to achieve the higher levels of multiplexing in bright-field [71]. The study of immune contexture is increasingly using fluorescence-based multiplexed IHC staining, which introduces new variables [24]. These variables include the type of staining techniques (sequential versus simultaneous), the localisation of the marker under evaluation, antibody cross-reactivity, antigen retrieval method, spectral overlap between multiple labels, photobleaching, tissue autofluorescence, and signal quenching. Due to the possibility of false positive results, this is extremely critical when the markers are being quantitatively tested for staining intensity [69].
While it is faster and involves less data capture to evaluate a few regions of interest rather than the entire slide, this approach might not be as representative of the entire tumour slide [72].
Finding thresholds for patient stratification based on the expression of markers is the major challenge facing translational scientists by using mIHC/IF to help clinical decisions. [73]. Since all parameter staining results are presented as continuous variables, the appropriate cut-off values should be chosen based on the staining intensity distribution, absolute number, or both. The threshold value of the continuous covariate distribution that best distinguishes between low- and high-risk individuals based on response to outcome is referred to as the “optimal” cut-off point. An actionable image analysis scoring system to determine the optimum calculation is substantially complicated and still needs to be explored in translational studies. In addition, parameters such as percentage positivity within a cell type and the spatial geographic distribution of the immune cells, including their proximity, relation with malignant, and densities, need to be explored to find an immune scoring system to help immune therapy prediction.
Image acquisition and digital pathology
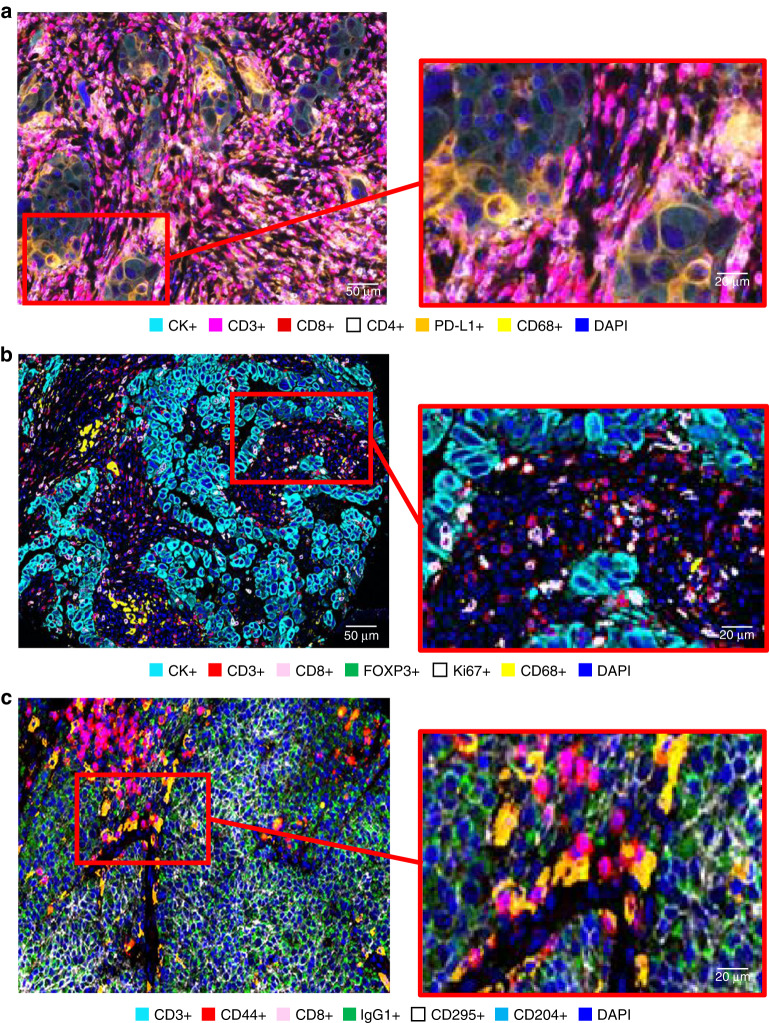
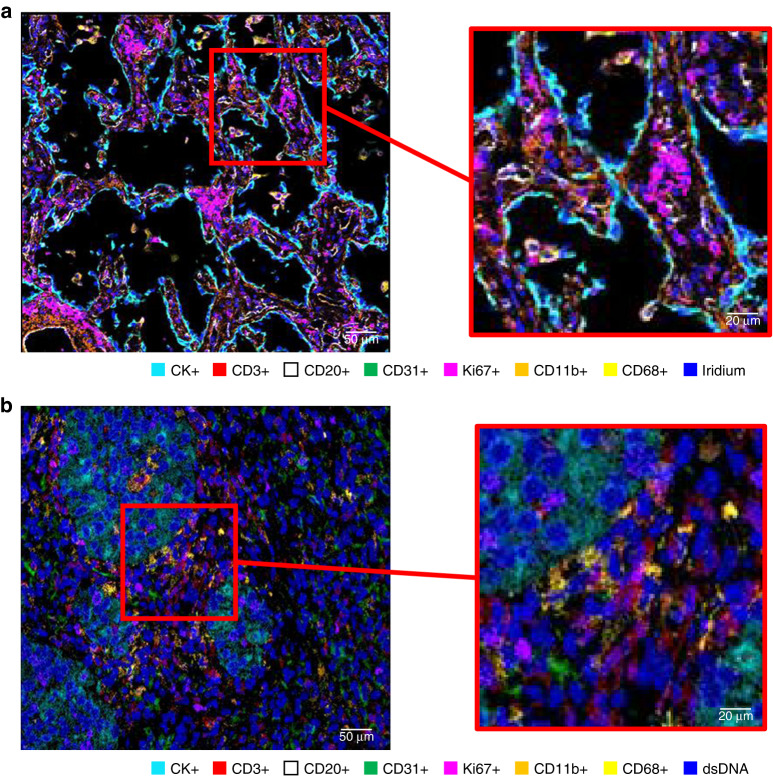
The main reason for performing a multiplexed assay is to obtain a high volume of tumour biological information through multidimensional data related to tissue architecture, the spatial distribution of multiple cell phenotypes, co-expression of markers, and rare cell-type detection [74] (Figs. 2 and 3). The main scanner solutions and image analysis softwares, commercially available and open sources, were previously reviewed recently [73, 74] (Tables 3 and 4).
Fig. 2. Representative examples of multiplexed images from lung cancer tissue.
Composite spectral mixing image and detail from six marker multiplex immunofluorescence panel using tyramide signal amplification (a), an image showing six important markers extracted from a 26 marker hi-plex methodology using barcoding base-oligonucleotides methodology and detail (b), and seven representative markers from a panel of 95 markers using Imaging Cyclic Staining platform and detail (c). a The image was generated using Vectra-Polaris 1.0.13 scanner system and InForm 2.4.8 image analysis software (Akoya Biosciences), b the image was generated using the PhenoCycler system and the QuPath (open sources), c the image was generated using the MACSima Imaging System and Qi Tissue™ Image Analysis Software (Miltenyi Biotec).
Fig. 3. Representative examples of multiplexed images from lung cancer tissue using image mass spectrometry methodology.
Image generated from a 36-plex cocktail panel showing seven important markers for immunoprofiling cancer tissues and detail (a). Similarly, an image generated from a 24-plex cocktail panel showing seven important markers for immunoprofiling cancer tissues and detail (b). a Image generated by Hyperion Imaging System (FLUIDIGM) and HALLO image analysis software (Indica Lab). b Image generated by MIBI scope I (IONpath) and HALLO image analysis software.
Table 3.
Different scanners used for multiplexed image technologies.
| Instrument (company) | Slide loading capacity | Scanning speed | Resolution | Scanning capacity (whole slide WS vs. ROIs) | Multispectral Range |
|---|---|---|---|---|---|
|
Pannoramic 250 FLASH III (3DHistech, Budapest, Hungary) |
250 |
5 min at 31× 15 min at 62× |
0.172 and 0.087/ 0.325 and 0.162 pixels |
WS | Undisclosed |
| Aperio GT 450 (Leica Biosystems, Illinois, USA) | 450 | 32 sec (15 mm × 15 mm area at 40x) | 0.26 μm/pixel at 40× | WS | Undisclosed |
| PhenoImager TH (Akoya Biosciences, Marlborough, MA, USA) | 80 |
Fluorescence WS (1.5 cm × 1.5 cm sample at 20×): 11 slides/hour (7 Colour Opal, unmixed) Bright-field WS (1.5 cm × 1.5 cm sample at 20X): 10 slides/hour |
10 × (1.0 m/pixel), 20 × (0.5 μm/pixel) and 40 × (0.25 µm/pixel) | WS for until 6 markers/ROI for more than 6 markers staining | 440–780 nm |
| NanoZoomer S360 (Hamamatsu, Shizuoka, Japan) | 360 (82 slides/h) | Approx. 30 s |
20 × (0.46 μm/pixel) 40 × (0.23 μm/pixel) |
WS | No fluorescence |
| Axioscan 7 (Zeiss, Jena, Germany) | 100 |
Brightfield 10 × 10 mm, 20 ×, 1:13 min Fluorescence 10 × 10 mm, 20 × , 4 FL channels (10 ms exposure each) 5:23 min |
10 × 0.345 μm/pixel 20 × 0.173 μm/pixel 40 × 0.086 μm/pixel |
WS | 380 nm – 770 nm |
| VENTANA DP 600 (Roche/Ventana, Arizona, USA) | 240 |
<49 s at 20× <85 s at 40× (15 mm × 15 mm AOIs) |
40 × 0.23 μm/pixel | WS | No fluorescence |
| MACSima™ Imaging Platform (Miltenyi Biotec, Gaithersburg, USA) | 4 | Epifluorescence microscope, 30 to 35 min per cycle |
2 × 0.06 μm/pixel 20 × 0.75 μm/pixel |
WS | 386–725 |
| PhenoCicler-Fusion (Akoya Biosciences, Marlborough, MA, USA) | 4 | Fluorescence, Brightfield. 1 cycle in 10 min | 10 × (1.0 m/pixel), 20 × (0.5 μm/pixel) and 40 × (0.25 µm/pixel) | WS | 440–780 nm |
| Orion™ Instrument (RareCyte, INC., Seattle, WA, USA | 2 | Brightfield, Fluorescence, 75 min per cm2 at 20× | sCMOS 2 K x 2 K pixels 6.5 μm pixel size 95% QE 1.0 electron read noise | WS | 400–890 nm |
| COMETTM (Lunaphore technologies, Tolochenaz, Switzerland) | 4 | Fluorescence, Average time of 40 min every 2 markers | 20 × (0.5 μm/pixel) | WS | Undisclosed |
Table 4.
Image analysis softwares, commercially available and open sources, for multiplexed staining.
| Company | Software package | Modules | Data visualisation | Availability |
|---|---|---|---|---|
| Akoya Biosciences | InForm/Phenopther Packet | Colour-Based Co-localisation, Tissue Segmentation, Cell/Object Segmentation, Cell Phenotyping, Scoring and Automated Quantitation using Batch Analysis |
Density Raw Data Spatial plots, Bar graphics, Head maps |
Licensed |
| CompuCyte | iCyte/iBroser/iNovator | Nucleus Segmentation or Phantom Contouring, Measures Associated Signals | Density Raw Data | Licensed |
| HistoRx | AQUAnalysis | Signal Intensity Quantification Per Unit Area and Per Layer | Density Raw Data | Licensed |
| Indica Labs | HALO | Area Quantification, Co-localisation, Immune Cell Proximity, Spatial Analysis, Tissue Classifier | Density Raw Data, Percentage, H-score, Spatial Plot, Histogram | Licensed |
| Leica Biosystems | Aperio ImageScope | Pixel-Based Analysis, Cellular identification, Area Quantification and Positive Pixel Count IF Algorithm | Density Raw Data | Licensed |
| Media Cybernetics | Image-Pro | Colour-Based, Nuclear segmentation, Cell quantification, Macro-enabled Advanced Image Processing Solution | Density Raw Data | Licensed |
| NeoGenomics | MultiOmyx Quantification Programme | Epithelial tissue reconstruction, Cellular and Subcellular Segmentation, Cell Phenotyping, Quantification Algorithms | Density Raw Data | Licensed |
| NIH | Image J | Colour-Based, User Interactive Segmentation | Histograms and Profile Plots | Open |
| SlidePath | SlidePath’s Tissue Image Analysis | Membrane, Nuclear and Positive Pixel Quantification | Density Raw Data | Licensed |
| TissueGnostics | HistoQuest/TissueQuest/StrataQuest | Nuclei-Based Segmentation of Tissues, Cell Phenotyping | Density Raw Data | Licensed |
| Visiopharm | Visimoph Tissuemorph | Signal Intensity, Area, Counting Objects, Spatial Analysis, Clustering Statistical Analysis, Batch Analysis and Algorithm Creator. | Density Raw Data, Phenotypic Matrix, t-SNE Plots | Licensed |
| Miltenyi Biotec | Qi Tissue™ Image Analysis Software | Cell segmentation algorithms, Flexible gating and clustering cell phenotypes | Density Raw Data, Ploting tools as t-SNE and UMAP plots, distance mapping clustering and other visualisation tools such as heatmaps, scatter and histogram plots | Licensed |
| https://qupath.github.io | QuPath/CytoMap | View Measurements in Context by Colour Coding Objects According to Their Features, Flexible Object Classification, Trainable Cell Classification and Quantification | Density Raw Data, t-SNE plots, distance mapping clustering and other visualisation tools such as heatmaps, scatter and histogram plots | Open |
| https://cellprofiler.org/ | Cell Profiler/Cell Analyst | Based and Colour Object Identification, Size, Shape, Colour Intensity, Texture, and Number Neighbour Quantification. | Density Plot, Histogram | Open |
Recent advances in technology have allowed pathologists to digitise whole slide images of mIHC slides that allows for interpretation and management of the tumour sample in an image-based environment [75, 76]. The scan time of FFPE slides can increase with higher magnification, larger tissue size, number of field of views or tiles, number of channels used in multiplexed stains, tissue section density, low signal strength and low signal-to-noise ratio [77].
However, mIHC/IF requires specialised expensive equipment, image analysis platforms, and data storage that are not widely available in laboratories worldwide [49, 68].
Technical limitations in scanning can lead to imaging artefacts, such as lower image resolution, heterogeneous staining intensities and patterns, poorly focussed scans, improper stitching of lines or tiles, or overlapping signals from multiplexed spectra, all of which directly affect data accuracy and reproducibility [78]. While efforts have been made toward the harmonisation of a few multiplexed image methodologies across institutions [79, 80], the reproducibility of multiplexed techniques is still testing, and several parameters will need to be established, careful, time-consuming optimisation and validation are required to ensure that marker in the multiplex assays performs the same as a single-marker assay [73, 81].
Establishment of a routine digital pathology workflow therefore requires additional considerations above the traditional histology workflow [82]. These include the need for adequate staffing, proper training of personnel and pathologists, setting up pathologists’ workstations, additional quality control steps, availability and timely maintenance of equipment (e.g., scanner), adequate information technology infrastructure (e.g., server and computer), integration with LIS, standard operating procedures and guidelines for managed workflow [73, 81, 82]. Moreover, there are already a growing number of image acquisition platforms in the market that combine image analysis with automated slide scanning to support bright-field/fluorescence multiplexed staining [83].
The integration and interoperability of digital pathology is one of the significant obstacles to clinical routine adoption of image analysis or digital pathology. Digital pathology or image analysis adoption in clinical settings becomes increasingly expensive, labour- and time-intensive to establish, and requires adjustment to standards and practices [77]. Closed hardware and software systems offer less flexibility for hospital integration. Opening up a hardware and software technology with universal standard for data movement and management may facilitate more rapid marker identification, although more closed systems may be required for integration of these workflows in digital pathology with respect to recent regulations, such as the General Data Protection Regulation (GDPR) legislation and In-vitro Diagnostic Medical Device Regulation (IVDR) [84]. Finally, developed and manufactured quality systems, demonstration of inter- and extra-site reproducibility, staining variability normalisation through control tissues, highly accurate image analysis that tolerates sample heterogeneity and staining variability, demonstrating accuracy, sensitivity equivalent to the clinical standard DAB IHC, are basic essential items to support pathologist annotation, review, and clinical practice.
Perspectives
How to boost multiplex in situ analysis practice? The digital pathology and artificial intelligence (A.I.) development
In light of growing health economic concerns in the field of immuno-oncology and the need for tests that precisely predict responses to costly immunotherapies/cell therapies, the translation of mIHC/IF into clinical practice is of paramount importance. For the integration of immunostaining results from multiplexed analyses, combining digital pathology and artificial intelligence are undoubtedly important for the future approach to handle the abundant number of information obtained by the different methods described above [85]. Thus, future pathology will involve a much greater amount of digitisation, and the next generation of pathologists will have many more computational tools available to them to evaluate the predictive markers, notably in thoracic oncology [86]. The ability to acquire large data from multiple complementary label-free methods should provide powerful new insight in improving predictive markers assessment [87]. In addition, with the help of AI tools, prognostic models maybe created using multidimensional information. However, standardisation across different platforms is strongly needed to enable access to a wider pool of data that, with the appropriate data tools, may offer more precise predictive results [88]. The fusion of multiplex immunostainings, images, and AI data modalities may be a promising strategy to improve performance a single immunostaining modality model [89]. However, the potential improvements have to be confirmed by different external validation and adequate statistical analysis.
From research and development to routine clinical practice: how to fill the gap?
Developing the new methods described above requires standardisation, and optimisation. For further implementation in routine clinical practice to become possible, multicentre studies, need to be performed for validation [90, 91]. Moreover, multidisciplinary teams, including pathologists, oncologists, immunologists, bioinformaticians, and engineers, should be established to most effectively and fully develop this quantitative approach to cancer immunotherapy [73, 81].
The mIHC/IF methodologies enable the simultaneous mapping of around ten markers in one FFPE section. Although these techniques remain limited in the number of markers compared with high-multiplexed technologies but seem more adapted to predictive marker validation in clinics, provided that they meet the recent guidelines on best practices for mIHC/IF [73, 81, 92].
In contrast, hi-multiplexed, spatially resolved technologies are dedicated to marker research discovery and can accommodate around 100 markers using a wide array of chemistries and signal detection systems [93]. Furthermore, complexity, analytical, and clinical validation effort increases non-linearly with number of markers, and reliability probably goes down as number of markers increases. Thus, lower plex assays may be better suited to clinical applications, whereas hi-plex assays are better suited to research and discovery [55]. In addition, an even greater chance for clinical applicability could be achieved by using mIF/mIHC approaches than hi-multiplexed that require specialised and costlier platforms [11, 56].
The future of the integrative biopathology
Currently, there is an urgent need to focus on essential questions and define roadblocks to the fundamental understanding and clinical progress in the immune-oncology field. Recently, ten key challenges still facing cancer immunotherapy were defined, which range from the development of pre-clinical models that translate to human immunity, by understanding the molecular and cellular drivers of primary versus secondary resistance to identifying optimal combinations of immune-based therapies for any given patient [94]. Addressing these challenges will require the combined efforts of researchers and clinicians, and the focusing of resources to accelerate understanding of the complex interactions between cancer and the immune system and the development of improved treatment options for patients with cancer. Together the resistance mechanisms instigated the search for combinatory therapeutic strategies that would act at various molecular and cellular levels to maximise anti-tumour effects and to allow a better tumour control.
The drivers of primary immune resistance have probably evolved for a long time and are the product of the endogenous host immune response. It allows the study of primary resistance mechanisms from pre-treatment tumour specimens and may make these factors more dependent on the affected organs and the cell-type biology from which the cancer originated [94]. This may differ from secondary immune resistance, which is likely to develop after active immunotherapy over shorter intervals. Studying the secondary escape drivers was more complicated, mostly due to a lack of systematic selection of tumour tissue both before start of treatment and at progression time points [94]. Thus, the strategy in identifying the mechanisms of response and resistance to ICIs involves the assessment of serial tumour specimens throughout the course of treatment, together with the development of minimally invasive markers (e.g., liquid biopsy, PBMCs, microbiome) [2, 95]. This approach is important because it encompasses traditional static time points research and aims to recognise superior markers of diagnosis by analysing dynamic responses to ICIs. Recent advances in computational power and the advent of whole slide digitisation have led to several recent studies investigating quantitative approaches to interrogate the TME on high-resolution histology images [96–98]. Research has focused on leveraging artificial intelligence and digital pathology approaches toward answering clinically relevant questions related to detection, diagnosis, prognosis, and treatment response [99, 100]. Therefore, it makes sense that these approaches will be even more crucial in assisting clinicians in selecting appropriate treatments and determining the effectiveness of ICIs. For instance, an artificial intelligence–powered assessment of whole slide images for multiple TME markers and immune phenotypes that correlate with ICIs response may help to optimise treatment selection in clinical practice [101].
Integrative biopathology can be defined as a multi-parametric holistic approach to integrate qualitative, quantitative, dynamic and static data in order to generate a coherent model for a reliable understanding of a disease and a reproducible prediction of its expected course and its response to different therapeutic options [102]. The integration of biomedical imaging, histopathology and genomic assays to predict immunotherapy response is still in a preliminary phase, but recent proof of principle studies tried to demonstrate the value of multimodal integration [103, 104]. Although recent evidence suggests that a combination of markers can give us a better prediction of response and outcomes to ICIs, the varieties of markers need to be tested and compared in prospective studies, ideally in clinical trials [11].
Conclusions
Analysis of the spatial distribution of cells within the tumour microenvironment by mIHC/IF provides important, clinically relevant information; however, with the availability of multiple spatial analysis tools, choosing a methodology or platform remains a challenge. As such, the development of reproducible, standardisation of analysis pipelines and consensus on their application is necessary to fully exploit the benefits of these approachs. In-depth spatial analysis of FFPE patient samples using mIHC/IF may can facilitate patient stratification for immunotherapy, as well as identification of prognostic and predictive immune markers. Thus, a thorough understanding of different techniques and cellular biology are necessary to achieve reproducible and optimum high-quality data with mIHC/IF. There is still a strong need for standardised, reproducible image analysis tools for understanding disease pathology and combating the propensity for bias associated with manual analysis. Implementation of such tools may be informative for the improvement and design of novel therapeutic strategies. The importance of standardisation and quality control include the importance of refining, standardising and validating the mIHC/IF workflow at the preanalytical, analytical and post-analytical stages. It is necessary to emphasise the importance of antibody selection, optimisation and validation as well as the need for an extensive review of panel design and multiplex staining. It is pivotal to assess tumour content, sample size, and percentages of necrosis and fibrosis for pathology quality control.
Overall, while hi-multiplexed assays are better suited to research and discovery purposes reduced plex assays might be more appropriate for clinical applications. Multiplex IHC/mIF, which are more generally accessible, may have an even higher possibility of clinical applicability than hi-multiplexed methods, which call for expensive lab cost and highly, specialised platforms. Finally, the predictive performance of multiplexed assays has to be investigated and compared in prospective studies, ideally in clinical trials, despite some evidence suggesting that a combination of markers provides a superior prediction of response and outcomes.
Acknowledgements
The authors received no additional support for the research or authorship of this article.
Author contributions
EP, MI, IW and PH and wrote the manuscript.
Funding
There was no financial support for the research or authorship of this article.
Data availability
Not applicable.
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
This article does not contain studies involving human participants or animals that were performed by the authors.
Consent for publication
This article does not contain any individual person’s data in any form.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12:252–64. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sharma P, Hu-Lieskovan S, Wargo JA, Ribas A. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell. 2017;168:707–23.. doi: 10.1016/j.cell.2017.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sharpe AH, Pauken KE. The diverse functions of the PD1 inhibitory pathway. Nat Rev Immunol. 2018;18:153–67.. doi: 10.1038/nri.2017.108. [DOI] [PubMed] [Google Scholar]
- 4.Marcus L, Fashoyin-Aje LA, Donoghue M, Yuan M, Rodriguez L, Gallagher PS, et al. FDA approval summary: pembrolizumab for the treatment of tumor mutational burden-high solid tumors. Clin Cancer Res. 2021;27:4685–9. doi: 10.1158/1078-0432.CCR-21-0327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fincham REA, Bashiri H, Lau MC, Yeong J. Editorial: multiplex immunohistochemistry/immunofluorescence technique: the potential and promise for clinical application. Front Mol Biosci. 2022;9:831383. doi: 10.3389/fmolb.2022.831383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu S, Stein JE, Rimm DL, Wang DW, Bell JM, Johnson DB, et al. Comparison of biomarker modalities for predicting response to PD-1/PD-L1 checkpoint blockade: a systematic review and meta-analysis. JAMA Oncol. 2019;5:1195–204. doi: 10.1001/jamaoncol.2019.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reck M, Rodriguez-Abreu D, Robinson AG, Hui R, Csoszi T, Fulop A, et al. Updated analysis of KEYNOTE-024: pembrolizumab versus platinum-based chemotherapy for advanced non-small-cell lung cancer with PD-L1 tumor proportion score of 50% or greater. J Clin Oncol. 2019;37:537–46.. doi: 10.1200/JCO.18.00149. [DOI] [PubMed] [Google Scholar]
- 8.Paz-Ares L, Luft A, Vicente D, Tafreshi A, Gumus M, Mazieres J, et al. Pembrolizumab plus chemotherapy for squamous non-small-cell lung cancer. N Engl J Med. 2018;379:2040–51.. doi: 10.1056/NEJMoa1810865. [DOI] [PubMed] [Google Scholar]
- 9.Ilie M, Hofman V, Dietel M, Soria JC, Hofman P. Assessment of the PD-L1 status by immunohistochemistry: challenges and perspectives for therapeutic strategies in lung cancer patients. Virchows Arch. 2016;468:511–25. doi: 10.1007/s00428-016-1910-4. [DOI] [PubMed] [Google Scholar]
- 10.Doroshow DB, Bhalla S, Beasley MB, Sholl LM, Kerr KM, Gnjatic S, et al. PD-L1 as a biomarker of response to immune-checkpoint inhibitors. Nat Rev Clin Oncol. 2021;18:345–62.. doi: 10.1038/s41571-021-00473-5. [DOI] [PubMed] [Google Scholar]
- 11.Mino-Kenudson M, Schalper K, Cooper W, Dacic S, Hirsch FR, Jain D, et al. Predictive biomarkers for immunotherapy in lung cancer: perspective from the International Association for the Study of Lung Cancer Pathology Committee. J Thorac Oncol. 2022;17:1335–54.. doi: 10.1016/j.jtho.2022.09.109. [DOI] [PubMed] [Google Scholar]
- 12.Havel JJ, Chowell D, Chan TA. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat Rev Cancer. 2019;19:133–50.. doi: 10.1038/s41568-019-0116-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berland L, Heeke S, Humbert O, Macocco A, Long-Mira E, Lassalle S, et al. Current views on tumor mutational burden in patients with non-small cell lung cancer treated by immune checkpoint inhibitors. J Thorac Dis. 2019;11:S71–S80. doi: 10.21037/jtd.2018.11.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anagnostou V, Bardelli A, Chan TA, Turajlic S. The status of tumor mutational burden and immunotherapy. Nat Cancer. 2022;3:652–6. doi: 10.1038/s43018-022-00382-1. [DOI] [PubMed] [Google Scholar]
- 15.Chan TA, Yarchoan M, Jaffee E, Swanton C, Quezada SA, Stenzinger A, et al. Development of tumor mutation burden as an immunotherapy biomarker: utility for the oncology clinic. Ann Oncol. 2019;30:44–56. doi: 10.1093/annonc/mdy495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marabelle A, Fakih M, Lopez J, Shah M, Shapira-Frommer R, Nakagawa K, et al. Association of tumour mutational burden with outcomes in patients with advanced solid tumours treated with pembrolizumab: prospective biomarker analysis of the multicohort, open-label, phase 2 KEYNOTE-158 study. Lancet Oncol. 2020;21:1353–65.. doi: 10.1016/S1470-2045(20)30445-9. [DOI] [PubMed] [Google Scholar]
- 17.McGrail DJ, Pilie PG, Rashid NU, Voorwerk L, Slagter M, Kok M, et al. High tumor mutation burden fails to predict immune checkpoint blockade response across all cancer types. Ann Oncol. 2021;32:661–72.. doi: 10.1016/j.annonc.2021.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Samstein RM, Lee CH, Shoushtari AN, Hellmann MD, Shen R, Janjigian YY, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet. 2019;51:202–6. doi: 10.1038/s41588-018-0312-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–8. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heeke S, Benzaquen J, Hofman V, Long-Mira E, Lespinet V, Bordone O, et al. Comparison of three sequencing panels used for the assessment of tumor mutational burden in NSCLC reveals low comparability. J Thorac Oncol. 2020;15:1535–40.. doi: 10.1016/j.jtho.2020.05.013. [DOI] [PubMed] [Google Scholar]
- 21.Ricciuti B, Wang X, Alessi JV, Rizvi H, Mahadevan NR, Li YY, et al. Association of high tumor mutation burden in non-small cell lung cancers with increased immune infiltration and improved clinical outcomes of PD-L1 blockade across PD-L1 expression levels. JAMA Oncol. 2022;8:1160–8. doi: 10.1001/jamaoncol.2022.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Angelova M, Mlecnik B, Vasaturo A, Bindea G, Fredriksen T, Lafontaine L, et al. Evolution of metastases in space and time under immune selection. Cell. 2018;175:751–65 e16. doi: 10.1016/j.cell.2018.09.018. [DOI] [PubMed] [Google Scholar]
- 23.Galon J, Bruni D. Tumor immunology and tumor evolution: intertwined histories. Immunity. 2020;52:55–81. doi: 10.1016/j.immuni.2019.12.018. [DOI] [PubMed] [Google Scholar]
- 24.Banik G, Betts CB, Liudahl SM, Sivagnanam S, Kawashima R, Cotechini T, et al. High-dimensional multiplexed immunohistochemical characterization of immune contexture in human cancers. Methods Enzymol. 2020;635:1–20. doi: 10.1016/bs.mie.2019.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Guillebon E, Dardenne A, Saldmann A, Seguier S, Tran T, Paolini L, et al. Beyond the concept of cold and hot tumors for the development of novel predictive biomarkers and the rational design of immunotherapy combination. Int J Cancer. 2020;147:1509–18.. doi: 10.1002/ijc.32889. [DOI] [PubMed] [Google Scholar]
- 26.Schoffski P, Tan DSW, Martin M, Ochoa-de-Olza M, Sarantopoulos J, Carvajal RD, et al. Phase I/II study of the LAG-3 inhibitor ieramilimab (LAG525) +/- anti-PD-1 spartalizumab (PDR001) in patients with advanced malignancies. J Immunother Cancer. 2022;10:e003776. doi: 10.1136/jitc-2021-003776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thakkar D, Paliwal S, Dharmadhikari B, Guan S, Liu L, Kar S, et al. Rationally targeted anti-VISTA antibody that blockades the C-C’ loop region can reverse VISTA immune suppression and remodel the immune microenvironment to potently inhibit tumor growth in an Fc independent manner. J Immunother Cancer. 2022;10:e003382. doi: 10.1136/jitc-2021-003382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Curigliano G, Gelderblom H, Mach N, Doi T, Tai D, Forde PM, et al. Phase I/Ib clinical trial of sabatolimab, an anti-TIM-3 antibody, alone and in combination with spartalizumab, an anti-PD-1 antibody, in advanced solid tumors. Clin Cancer Res. 2021;27:3620–9. doi: 10.1158/1078-0432.CCR-20-4746. [DOI] [PubMed] [Google Scholar]
- 29.Aroldi F, Saleh R, Jafferji I, Barreto C, Saberian C, Middleton MR. Lag3: from bench to bedside. Cancer Treat Res. 2022;183:185–99.. doi: 10.1007/978-3-030-96376-7_6. [DOI] [PubMed] [Google Scholar]
- 30.Niu J, Maurice-Dror C, Lee DH, Kim DW, Nagrial A, Voskoboynik M, et al. First-in-human phase 1 study of the anti-TIGIT antibody vibostolimab as monotherapy or with pembrolizumab for advanced solid tumors, including non-small-cell lung cancer(☆) Ann Oncol. 2022;33:169–80.. doi: 10.1016/j.annonc.2021.11.002. [DOI] [PubMed] [Google Scholar]
- 31.De Mello RA, Zhu JH, Iavelberg J, Potim AH, Simonetti D, Silva JA, Jr., et al. Current and future aspects of TIM-3 as biomarker or as potential targeted in non-small cell lung cancer scope: is there a role in clinical practice? Transl Lung Cancer Res. 2020;9:2311–4. doi: 10.21037/tlcr-20-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bai R, Lv Z, Xu D, Cui J. Predictive biomarkers for cancer immunotherapy with immune checkpoint inhibitors. Biomark Res. 2020;8:34. doi: 10.1186/s40364-020-00209-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sholl LM. Biomarkers of response to checkpoint inhibitors beyond PD-L1 in lung cancer. Mod Pathol. 2022;35:66–74. doi: 10.1038/s41379-021-00932-5. [DOI] [PubMed] [Google Scholar]
- 34.Shirasawa M, Yoshida T, Imabayashi T, Okuma K, Matsumoto Y, Masuda K, et al. Baseline PD-L1 expression and tumour-infiltrated lymphocyte status predict the efficacy of durvalumab consolidation therapy after chemoradiotherapy in unresectable locally advanced patients with non-small-cell lung cancer. Eur J Cancer. 2022;162:1–10. doi: 10.1016/j.ejca.2021.11.013. [DOI] [PubMed] [Google Scholar]
- 35.Gettinger SN, Choi J, Mani N, Sanmamed MF, Datar I, Sowell R, et al. A dormant TIL phenotype defines non-small cell lung carcinomas sensitive to immune checkpoint blockers. Nat Commun. 2018;9:3196. doi: 10.1038/s41467-018-05032-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anagnostou V, Luke JJ. Quantitative spatial profiling of TILs as the next step beyond PD-L1 testing for immune checkpoint blockade. Clin Cancer Res. 2022;28:4835–7. doi: 10.1158/1078-0432.CCR-22-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim HR, Park SM, Seo SU, Jung I, Yoon HI, Gabrilovich DI, et al. The ratio of peripheral regulatory T cells to Lox-1(+) polymorphonuclear myeloid-derived suppressor cells predicts the early response to anti-PD-1 therapy in patients with non-small cell lung cancer. Am J Respir Crit Care Med. 2019;199:243–6. doi: 10.1164/rccm.201808-1502LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khatir W, Humbert O, Benzaquen J, Bontoux C, Neels J, Berland L, et al. Identification of a circulating immunological signature predictive of response to immune checkpoint inhibitors in patients with advanced non-small cell lung cancer. Clin Transl Med. 2022;12:e1018. doi: 10.1002/ctm2.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Helmink BA, Reddy SM, Gao J, Zhang S, Basar R, Thakur R, et al. B cells and tertiary lymphoid structures promote immunotherapy response. Nature. 2020;577:549–55.. doi: 10.1038/s41586-019-1922-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patil NS, Nabet BY, Muller S, Koeppen H, Zou W, Giltnane J, et al. Intratumoral plasma cells predict outcomes to PD-L1 blockade in non-small cell lung cancer. Cancer Cell. 2022;40:289–300 e4. doi: 10.1016/j.ccell.2022.02.002. [DOI] [PubMed] [Google Scholar]
- 41.Philip M, Schietinger A. CD8(+) T cell differentiation and dysfunction in cancer. Nat Rev Immunol. 2022;22:209–23.. doi: 10.1038/s41577-021-00574-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Datar I, Sanmamed MF, Wang J, Henick BS, Choi J, Badri T, et al. Expression analysis and significance of PD-1, LAG-3, and TIM-3 in human non-small cell lung cancer using spatially resolved and multiparametric single-cell analysis. Clin Cancer Res. 2019;25:4663–73.. doi: 10.1158/1078-0432.CCR-18-4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eiva MA, Omran DK, Chacon JA, Powell DJ., Jr Systematic analysis of CD39, CD103, CD137, and PD-1 as biomarkers for naturally occurring tumor antigen-specific TILs. Eur J Immunol. 2022;52:96–108. doi: 10.1002/eji.202149329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fridman WH, Zitvogel L, Sautes-Fridman C, Kroemer G. The immune contexture in cancer prognosis and treatment. Nat Rev Clin Oncol. 2017;14:717–34.. doi: 10.1038/nrclinonc.2017.101. [DOI] [PubMed] [Google Scholar]
- 45.Remark R, Merghoub T, Grabe N, Litjens G, Damotte D, Wolchok JD, et al. In-depth tissue profiling using multiplexed immunohistochemical consecutive staining on single slide. Sci Immunol. 2016;1:aaf6925. doi: 10.1126/sciimmunol.aaf6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Glass G, Papin JA, Mandell JW. SIMPLE: a sequential immunoperoxidase labeling and erasing method. J Histochem Cytochem. 2009;57:899–905. doi: 10.1369/jhc.2009.953612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang W, Hubbard A, Jones T, Racolta A, Bhaumik S, Cummins N, et al. Fully automated 5-plex fluorescent immunohistochemistry with tyramide signal amplification and same species antibodies. Lab Invest. 2017;97:873–85.. doi: 10.1038/labinvest.2017.37. [DOI] [PubMed] [Google Scholar]
- 48.Morrison LE, Lefever MR, Behman LJ, Leibold T, Roberts EA, Horchner UB, et al. Brightfield multiplex immunohistochemistry with multispectral imaging. Lab Invest. 2020;100:1124–36.. doi: 10.1038/s41374-020-0429-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Akturk G, Sweeney R, Remark R, Merad M, Gnjatic S. Multiplexed immunohistochemical consecutive staining on single slide (MICSSS): multiplexed chromogenic IHC assay for high-dimensional tissue analysis. Methods Mol Biol. 2020;2055:497–519. doi: 10.1007/978-1-4939-9773-2_23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsujikawa T, Kumar S, Borkar RN, Azimi V, Thibault G, Chang YH, et al. Quantitative multiplex immunohistochemistry reveals myeloid-inflamed tumor-immune complexity associated with poor prognosis. Cell Rep. 2017;19:203–17.. doi: 10.1016/j.celrep.2017.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moles Lopez X, Barbot P, Van Eycke YR, Verset L, Trepant AL, Larbanoix L, et al. Registration of whole immunohistochemical slide images: an efficient way to characterize biomarker colocalization. J Am Med Inf Assoc. 2015;22:86–99. doi: 10.1136/amiajnl-2014-002710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Day WA, Lefever MR, Ochs RL, Pedata A, Behman LJ, Ashworth-Sharpe J, et al. Covalently deposited dyes: a new chromogen paradigm that facilitates analysis of multiple biomarkers in situ. Lab Invest. 2017;97:104–13.. doi: 10.1038/labinvest.2016.115. [DOI] [PubMed] [Google Scholar]
- 53.Polaske NW, Kelly BD, Ashworth-Sharpe J, Bieniarz C. Quinone methide signal amplification: covalent reporter labeling of cancer epitopes using alkaline phosphatase substrates. Bioconjug Chem. 2016;27:660–6. doi: 10.1021/acs.bioconjchem.5b00652. [DOI] [PubMed] [Google Scholar]
- 54.Ilie M, Beaulande M, Ben Hadj S, Chamorey E, Schiappa R, Long-Mira E, et al. Chromogenic multiplex immunohistochemistry reveals modulation of the immune microenvironment associated with survival in elderly patients with lung adenocarcinoma. Cancers. 2018;10:326. doi: 10.3390/cancers10090326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ilie M, Beaulande M, Hamila M, Erb G, Hofman V, Hofman P. Automated chromogenic multiplexed immunohistochemistry assay for diagnosis and predictive biomarker testing in non-small cell lung cancer. Lung Cancer. 2018;124:90–4. doi: 10.1016/j.lungcan.2018.07.037. [DOI] [PubMed] [Google Scholar]
- 56.Ilie M, Beaulande M, Long-Mira E, Bontoux C, Zahaf K, Lalvee S, et al. Analytical validation of automated multiplex chromogenic immunohistochemistry for diagnostic and predictive purpose in non-small cell lung cancer. Lung Cancer. 2022;166:1–8. doi: 10.1016/j.lungcan.2022.01.022. [DOI] [PubMed] [Google Scholar]
- 57.Schubert W, Bonnekoh B, Pommer AJ, Philipsen L, Bockelmann R, Malykh Y, et al. Analyzing proteome topology and function by automated multidimensional fluorescence microscopy. Nat Biotechnol. 2006;24:1270–8. doi: 10.1038/nbt1250. [DOI] [PubMed] [Google Scholar]
- 58.Wang N, Li X, Wang R, Ding Z. Spatial transcriptomics and proteomics technologies for deconvoluting the tumor microenvironment. Biotechnol J. 2021;16:e2100041. doi: 10.1002/biot.202100041. [DOI] [PubMed] [Google Scholar]
- 59.Lin JR, Izar B, Wang S, Yapp C, Mei S, Shah PM, et al. Highly multiplexed immunofluorescence imaging of human tissues and tumors using t-CyCIF and conventional optical microscopes. eLife. 2018;7:e31657. doi: 10.7554/eLife.31657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cheng Y, Burrack RK, Li Q. Spatially resolved and highly multiplexed protein and rna in situ detection by combining CODEX with RNAscope in situ hybridization. J Histochem Cytochem. 2022;70:571–81.. doi: 10.1369/00221554221114174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Black S, Phillips D, Hickey JW, Kennedy-Darling J, Venkataraaman VG, Samusik N, et al. CODEX multiplexed tissue imaging with DNA-conjugated antibodies. Nat Protoc. 2021;16:3802–35.. doi: 10.1038/s41596-021-00556-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Herbel C, Reiß S, Jungblut M, Eckardt D, Bosio A. MACSima™ Imaging Platform provides new insights into cancer biology and target discovery by cyclic immunofluorescence-based imaging. MACS More. 2019;18:16–20. [Google Scholar]
- 63.Berghmans E, Van Raemdonck G, Schildermans K, Willems H, Boonen K, Maes E, et al. MALDI mass spectrometry imaging linked with top-down proteomics as a tool to study the non-small-cell lung cancer tumor microenvironment. Methods Protoc. 2019;2:44. doi: 10.3390/mps2020044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Francisco-Cruz A, Parra ER, Tetzlaff MT, Wistuba II. Multiplex immunofluorescence assays. Methods Mol Biol. 2020;2055:467–95.. doi: 10.1007/978-1-4939-9773-2_22. [DOI] [PubMed] [Google Scholar]
- 65.Kriegsmann M, Zgorzelski C, Casadonte R, Schwamborn K, Muley T, Winter H, et al. Mass spectrometry imaging for reliable and fast classification of non-small cell lung cancer subtypes. Cancers. 2020;12:2704. doi: 10.3390/cancers12092704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ptacek J, Locke D, Finck R, Cvijic ME, Li Z, Tarolli JG, et al. Multiplexed ion beam imaging (MIBI) for characterization of the tumor microenvironment across tumor types. Lab Invest. 2020;100:1111–23.. doi: 10.1038/s41374-020-0417-4. [DOI] [PubMed] [Google Scholar]
- 67.Tan WCC, Nerurkar SN, Cai HY, Ng HHM, Wu D, Wee YTF, et al. Overview of multiplex immunohistochemistry/immunofluorescence techniques in the era of cancer immunotherapy. Cancer Commun. 2020;40:135–53.. doi: 10.1002/cac2.12023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cooper WA, Lantuejoul S, Mino-Kenudson M. Predicting response to programmed cell death protein-1 or programmed death-ligand 1 blockade in NSCLC—is multiplex immunohistochemistry or immunofluorescence the answer? J Thorac Oncol. 2021;16:1247–9. doi: 10.1016/j.jtho.2021.06.007. [DOI] [PubMed] [Google Scholar]
- 69.Shakya R, Nguyen TH, Waterhouse N, Khanna R. Immune contexture analysis in immuno-oncology: applications and challenges of multiplex fluorescent immunohistochemistry. Clin Transl Immunol. 2020;9:e1183. doi: 10.1002/cti2.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stack EC, Wang C, Roman KA, Hoyt CC. Multiplexed immunohistochemistry, imaging, and quantitation: a review, with an assessment of Tyramide signal amplification, multispectral imaging and multiplex analysis. Methods. 2014;70:46–58. doi: 10.1016/j.ymeth.2014.08.016. [DOI] [PubMed] [Google Scholar]
- 71.Choi YJ, Nakano K, Ide T, Sakae T, Ichikawa R, Hizawa T, et al. Demonstrating a Filter-free wavelength sensor with double-well structure and its application. Biosens. 2022;12:1033. doi: 10.3390/bios12111033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hofman P, Badoual C, Henderson F, Berland L, Hamila M, Long-Mira E, et al. Multiplexed immunohistochemistry for molecular and immune profiling in lung cancer-just about ready for prime-time? Cancers. 2019;11:283. doi: 10.3390/cancers11030283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Laberiano-Fernandez C, Hernandez-Ruiz S, Rojas F, Parra ER. Best practices for technical reproducibility assessment of multiplex immunofluorescence. Front Mol Biosci. 2021;8:660202. doi: 10.3389/fmolb.2021.660202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Parra ER, Francisco-Cruz A, Wistuba II. State-of-the-art of profiling immune contexture in the era of multiplexed staining and digital analysis to study paraffin tumor tissues. Cancers. 2019;11:247. doi: 10.3390/cancers11020247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mungenast F, Fernando A, Nica R, Boghiu B, Lungu B, Batra J, et al. Next-generation digital histopathology of the tumor microenvironment. Genes. 2021;12:538. doi: 10.3390/genes12040538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Surace M, Rognoni L, Rodriguez-Canales J, Steele KE. Characterization of the immune microenvironment of NSCLC by multispectral analysis of multiplex immunofluorescence images. Methods Enzymol. 2020;635:33–50. doi: 10.1016/bs.mie.2019.07.039. [DOI] [PubMed] [Google Scholar]
- 77.Bankhead P. Developing image analysis methods for digital pathology. J Pathol. 2022;257:391–402. doi: 10.1002/path.5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Evans AJ, Salama ME, Henricks WH, Pantanowitz L. Implementation of whole slide imaging for clinical purposes: issues to consider from the perspective of early adopters. Arch Pathol Lab Med. 2017;141:944–59.. doi: 10.5858/arpa.2016-0074-OA. [DOI] [PubMed] [Google Scholar]
- 79.Akturk G, Parra ER, Gjini E, Lako A, Lee JJ, Neuberg D, et al. Multiplex tissue imaging harmonization: a multicenter experience from CIMAC-CIDC immuno-oncology biomarkers network. Clin Cancer Res. 2021;27:5072–83.. doi: 10.1158/1078-0432.CCR-21-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Taube JM, Roman K, Engle EL, Wang C, Ballesteros-Merino C, Jensen SM, et al. Multi-institutional TSA-amplified multiplexed immunofluorescence reproducibility evaluation (MITRE) study. J Immunother Cancer. 2021;9:e002197. doi: 10.1136/jitc-2020-002197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Taube JM, Akturk G, Angelo M, Engle EL, Gnjatic S, Greenbaum S, et al. The Society for Immunotherapy of Cancer statement on best practices for multiplex immunohistochemistry (IHC) and immunofluorescence (IF) staining and validation. J Immunother Cancer. 2020;8:e000155. doi: 10.1136/jitc-2019-000155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fraggetta F, L’Imperio V, Ameisen D, Carvalho R, Leh S, Kiehl TR, et al. Best practice recommendations for the implementation of a digital pathology workflow in the anatomic pathology laboratory by the European Society of Digital and Integrative Pathology (ESDIP) Diagnostics. 2021;11:2167. doi: 10.3390/diagnostics11112167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rojas F, Hernandez S, Lazcano R, Laberiano-Fernandez C, Parra ER. Multiplex immunofluorescence and the digital image analysis workflow for evaluation of the tumor immune environment in translational research. Front Oncol. 2022;12:889886. doi: 10.3389/fonc.2022.889886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Coulter C, McKay F, Hallowell N, Browning L, Colling R, Macklin P, et al. Understanding the ethical and legal considerations of digital pathology. J Pathol Clin Res. 2022;8:101–15.. doi: 10.1002/cjp2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bera K, Schalper KA, Rimm DL, Velcheti V, Madabhushi A. Artificial intelligence in digital pathology—new tools for diagnosis and precision oncology. Nat Rev Clin Oncol. 2019;16:703–15.. doi: 10.1038/s41571-019-0252-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Berry S, Giraldo NA, Green BF, Cottrell TR, Stein JE, Engle EL, et al. Analysis of multispectral imaging with the AstroPath platform informs efficacy of PD-1 blockade. Science. 2021;372:eaba2609. doi: 10.1126/science.aba2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bosisio FM, Van Herck Y, Messiaen J, Bolognesi MM, Marcelis L, Van Haele M, et al. Next-generation pathology using multiplexed immunohistochemistry: mapping tissue architecture at single-cell level. Front Oncol. 2022;12:918900. doi: 10.3389/fonc.2022.918900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ghaffari Laleh N, Ligero M, Perez-Lopez R, Kather JN. Facts and hopes on the use of artificial intelligence for predictive immunotherapy biomarkers in cancer. Clin Cancer Res. 2022;29:316–23. [DOI] [PubMed]
- 89.Cheng JY, Abel JT, Balis UGJ, McClintock DS, Pantanowitz L. Challenges in the development, deployment, and regulation of artificial intelligence in anatomic pathology. Am J Pathol. 2021;191:1684–92.. doi: 10.1016/j.ajpath.2020.10.018. [DOI] [PubMed] [Google Scholar]
- 90.Hoyt CC. Multiplex immunofluorescence and multispectral imaging: forming the basis of a clinical test platform for immuno-oncology. Front Mol Biosci. 2021;8:674747. doi: 10.3389/fmolb.2021.674747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Locke D, Hoyt CC. Companion diagnostic requirements for spatial biology using multiplex immunofluorescence and multispectral imaging. Front Mol Biosci. 2023;10:1051491. doi: 10.3389/fmolb.2023.1051491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Parra ER, Jiang M, Solis L, Mino B, Laberiano C, Hernandez S, et al. Procedural requirements and recommendations for multiplex immunofluorescence tyramide signal amplification assays to support translational oncology studies. Cancers. 2020;12:255. doi: 10.3390/cancers12020255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lewis SM, Asselin-Labat ML, Nguyen Q, Berthelet J, Tan X, Wimmer VC, et al. Spatial omics and multiplexed imaging to explore cancer biology. Nat Methods. 2021;18:997–1012. doi: 10.1038/s41592-021-01203-6. [DOI] [PubMed] [Google Scholar]
- 94.Hegde PS, Chen DS. Top 10 challenges in cancer immunotherapy. Immunity. 2020;52:17–35. doi: 10.1016/j.immuni.2019.12.011. [DOI] [PubMed] [Google Scholar]
- 95.Hofman P, Heeke S, Alix-Panabieres C, Pantel K. Liquid biopsy in the era of immuno-oncology: is it ready for prime-time use for cancer patients? Ann Oncol. 2019;30:1448–59.. doi: 10.1093/annonc/mdz196. [DOI] [PubMed] [Google Scholar]
- 96.Coudray N, Ocampo PS, Sakellaropoulos T, Narula N, Snuderl M, Fenyo D, et al. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat Med. 2018;24:1559–67.. doi: 10.1038/s41591-018-0177-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Courtiol P, Maussion C, Moarii M, Pronier E, Pilcer S, Sefta M, et al. Deep learning-based classification of mesothelioma improves prediction of patient outcome. Nat Med. 2019;25:1519–25.. doi: 10.1038/s41591-019-0583-3. [DOI] [PubMed] [Google Scholar]
- 98.Wang X, Barrera C, Bera K, Viswanathan VS, Azarianpour-Esfahani S, Koyuncu C, et al. Spatial interplay patterns of cancer nuclei and tumor-infiltrating lymphocytes (TILs) predict clinical benefit for immune checkpoint inhibitors. Sci Adv. 2022;8:eabn3966. doi: 10.1126/sciadv.abn3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jiang Y, Yang M, Wang S, Li X, Sun Y. Emerging role of deep learning-based artificial intelligence in tumor pathology. Cancer Commun. 2020;40:154–66.. doi: 10.1002/cac2.12012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Baxi V, Edwards R, Montalto M, Saha S. Digital pathology and artificial intelligence in translational medicine and clinical practice. Mod Pathol. 2022;35:23–32. doi: 10.1038/s41379-021-00919-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Park S, Ock CY, Kim H, Pereira S, Ma M, Choi S, et al. Artificial intelligence-powered spatial analysis of tumor-infiltrating lymphocytes as complementary biomarker for immune checkpoint inhibition in non-small-cell lung cancer. J Clin Oncol. 2022;40:1916–28.. doi: 10.1200/JCO.21.02010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hanna MG, Ardon O, Reuter VE, Sirintrapun SJ, England C, Klimstra DS, et al. Integrating digital pathology into clinical practice. Mod Pathol. 2022;35:152–64.. doi: 10.1038/s41379-021-00929-0. [DOI] [PubMed] [Google Scholar]
- 103.Vanguri RS, Luo J, Aukerman AT, Egger JV, Fong CJ, Horvat N, et al. Multimodal integration of radiology, pathology and genomics for prediction of response to PD-(L)1 blockade in patients with non-small cell lung cancer. Nat Cancer. 2022;3:1151–64.. doi: 10.1038/s43018-022-00416-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chen RJ, Lu MY, Williamson DFK, Chen TY, Lipkova J, Noor Z, et al. Pan-cancer integrative histology-genomic analysis via multimodal deep learning. Cancer Cell. 2022;40:865–78 e6. doi: 10.1016/j.ccell.2022.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.