Abstract
We present a phylogenetic analysis of nine strains of symbiotic nitrogen-fixing bacteria isolated from nodules of tagasaste (Chamaecytisus proliferus) and other endemic woody legumes of the Canary Islands, Spain. These and several reference strains were characterized genotypically at different levels of taxonomic resolution by computer-assisted analysis of 16S ribosomal DNA (rDNA) PCR-restriction fragment length polymorphisms (PCR-RFLPs), 16S-23S rDNA intergenic spacer (IGS) RFLPs, and repetitive extragenic palindromic PCR (rep-PCR) genomic fingerprints with BOX, ERIC, and REP primers. Cluster analysis of 16S rDNA restriction patterns with four tetrameric endonucleases grouped the Canarian isolates with the two reference strains, Bradyrhizobium japonicum USDA 110spc4 and Bradyrhizobium sp. strain (Centrosema) CIAT 3101, resolving three genotypes within these bradyrhizobia. In the analysis of IGS RFLPs with three enzymes, six groups were found, whereas rep-PCR fingerprinting revealed an even greater genotypic diversity, with only two of the Canarian strains having similar fingerprints. Furthermore, we show that IGS RFLPs and even very dissimilar rep-PCR fingerprints can be clustered into phylogenetically sound groupings by combining them with 16S rDNA RFLPs in computer-assisted cluster analysis of electrophoretic patterns. The DNA sequence analysis of a highly variable 264-bp segment of the 16S rRNA genes of these strains was found to be consistent with the fingerprint-based classification. Three different DNA sequences were obtained, one of which was not previously described, and all belonged to the B. japonicum/Rhodopseudomonas rDNA cluster. Nodulation assays revealed that none of the Canarian isolates nodulated Glycine max or Leucaena leucocephala, but all nodulated Acacia pendula, C. proliferus, Macroptilium atropurpureum, and Vigna unguiculata.
Rhizobia, currently comprising the genera Azorhizobium, Bradyrhizobium, Mesorhizobium, Rhizobium, and Sinorhizobium, are widely distributed and abundant soil bacteria belonging to the alpha subclass of the Proteobacteria. Infective rhizobia are phenotypically well characterized by their ability to induce nitrogen-fixing nodules on leguminous plants (39). Rhizobia can be easily isolated from nodules, which are often occupied by single strains. Phylogenetically, these bacteria constitute a heterogeneous group, as revealed by 16S rDNA sequence analysis. Some of their members are more closely related to nonsymbiotic Proteobacteria, including denitrifying, pathogenic, and phototrophic strains of several genera (e.g., Afipia, Agrobacterium, Bartonella, Blastobacter, Brucella, Rochalimaea, and Rhodopseudomonas), than they are to other rhizobia (46). DNA sequence analysis of 16 rDNA regions has revealed a much greater diversity than previously recognized (23, 27, 48), leading to important revisions in the taxonomy and systematics of this group of bacteria and in the description of new genera and species (for recent reviews see references 22, 23, and 49).
Assessment of rhizobial genotypic diversity relevant to ecologically oriented studies requires a higher level of taxonomic resolution than can be achieved by 16S rDNA sequencing (7). Isolates of the same species, even of the same serotype, can significantly differ in their N2-fixing efficiencies and in their abilities to occupy nodules in competition with other closely related strains (35, 37). PCR-based genomic fingerprinting methods provide a much finer taxonomic resolution than 16S ribosomal DNA (rDNA) sequencing. Genomic fingerprints generated with short arbitrary primers (randomly amplified polymorphic DNA-PCR) (44) or primers binding to interspersed repetitive sequences (repetitive extragenic palindromic PCR [rep-PCR]) (41) give the highest level of taxonomic resolution currently achievable by PCR methods (5, 20, 40). The high degree of reproducibility of the rep-PCR approach has recently been discussed (38).
Here we present an analysis of the genotypic diversity and phylogeny of a set of nine rhizobial strains isolated from nodules of plants of the Chamaecytisus proliferus taxonomic complex (10) and from other endemic woody Fabaceae of the Canary Islands (3). Tagasaste, the best known taxon of the C. proliferus complex, is the only nonornamental endemic species of the Canarian flora that has substantial agronomic relevance. This fodder-legume is cultivated not only in the Canaries but also in New Zealand and Australia due to its outstanding forage value and rapid growth (8, 36). The combination of high N2 fixation efficiency (26) and responsiveness to mycorrhizal inoculation (42), renders this plant a very promising candidate for soil conservation programs and low-input agroforestry in regions with a Mediterranean climate.
León-Barrios and collaborators (21, 31) have phenotypically characterized several nodule isolates from tagasaste and other endemic Canarian legumes, including eight of the strains analyzed here. Each of these isolates was classified as a Bradyrhizobium sp. based exclusively on phenotypic traits, mainly the long generation time (above 6 h), alkalization of growth media, NAD-dependent phosphogluconate dehydrogenase activity, flagellation type, and symbiotic host range (21). Lipopolysaccharide electrophoretic patterns and serotyping revealed a high degree of phenotypic diversity within the strain collection (31).
The aim of this work was to determine the phylogenetic positions of the Canarian isolates and selected reference strains by means of amplified rDNA restriction analysis (ARDRA), a technique that is suitable to group strains at the genus or species level of taxonomic resolution (14). The results provided by ARDRA were confirmed by partial sequencing of the 16S rDNA region. To analyze the diversity of these strains at a finer taxonomic resolution intergenic spacer (IGS) PCR-restriction fragment length polymorphisms (PCR-RFLPs) and rep-PCR genomic fingerprints were generated and used to show that rDNA IGS restriction patterns and even very dissimilar rep-PCR patterns can be efficiently clustered into phylogenetic groupings when combined with 16S rDNA RFLPs by computer-assisted pattern analysis.
MATERIALS AND METHODS
Isolation and cultivation of bacterial strains.
Bacterial strains marked with an asterisk in Table 1 were isolated by one of the authors from mature, fan-shaped, indeterminate nodules present on the roots of plants belonging to different taxa of the C. proliferus taxonomic complex growing in natural or cultivated stands on several islands of the Canarian archipelago. Bacteroids were isolated from surface-sterilized nodules following a standard protocol (34). Isolates inducing nodules on tagasaste were stored as 25% (vol/vol) glycerol stock cultures at −70°C in yeast mannitol broth (YMB) (34). The origin of the isolates and the sources of the reference strains used in this study are listed in Table 1.
TABLE 1.
Sources of the Canarian isolates and reference strains used in this study
| Straina (synonym) | Original hostb | Geographic origin | Sourcec |
|---|---|---|---|
| BC-C1* (BTA-5) | C. proliferus ssp. proliferus var. canariae | Gran Canaria | This study |
| BC-C2* (BTA-6) | C. proliferus ssp. proliferus var. palmensis | Gran Canaria | This study |
| BC-P5* (BTA-8) | C. proliferus ssp. proliferus var. palmensis | La Palma | This study |
| BC-P6* (BTA-9) | C. proliferus ssp. proliferus var. palmensis | La Palma | This study |
| BC-P7* | C. proliferus ssp. proliferus var. calderae | La Palma | This study |
| BES-1 | C. proliferus ssp. angustifolius | Tenerife | M. León-Barrios, ULL |
| BGA-1 | Teline stenopetala | La Palma | M. León-Barrios, ULL |
| BRE-1 | Teline canariensis | Tenerife | M. León-Barrios, ULL |
| BTA-1 | C. proliferus ssp. proliferus var. palmensis | Tenerife | M. León-Barrios, ULL |
| B. japonicum USDA 110spc4 | Glycine max | United States | H. Hennecke, ETH Zurich |
| Bradyrhizobium sp. strain (Centrosema) CIAT 3101 | Centrosema plumieri | Colombia | M. C. Bolaños, CIAT |
| Rhizobium tropici CIAT 899T | Phaseolus vulgaris | Colombia | E. Martínez, UNAM |
| R. etli CFN 42T | Phaseolus vulgaris | Mexico | E. Martínez, UNAM |
| Agrobacterium tumefaciens C58 | M. M. López, IVIA | ||
| Pseudomonas fluorescens ATCC 12983 | ATCC |
Strains without generic or specific epithets correspond to the Canarian isolates. Strains labeled with an asterisk were isolated by one of the authors (P.V.). Synonyms are taken from the work of Santamaría et al. (31). T, type strain.
The nomenclature of the C. proliferus complex is taken from the study of Acebes-Ginovés et al. (1). ssp., subspecies.
ATCC, American Type Culture Collection; CIAT, Centro Internacional de Agricultura Tropical, Cali, Colombia; ETH, Eidgenössische Technische Hochschule, Zurich, Switzerland; IVIA, Instituto Valenciano de Investigaciones Agrarias, Valencia, Spain; UNAM, Universidad Nacional Autónoma de México, Cuernavaca, México; ULL, Universidad de La Laguna, Santa Cruz de Tenerife, Spain.
Isolation of DNA.
Genomic DNA of the bacteria was prepared from liquid cultures harvested at late exponential phase by a standard, cetyltrimethylammonium bromide protocol (2). Purified DNA was dissolved in 10 mM Tris-HCl buffer containing 1 mM EDTA (pH 8.0), and its concentration was adjusted spectrophotometrically to 50 μg/ml.
PCR protocols.
All primers used for the PCR experiments were synthesized by MWG-BIOTECH GmbH, Ebersberg, Germany. All PCRs were carried out with Taq polymerase and buffer (U.S. Biochemicals [USB]-Amersham International, Little Chalfont, England) in a Perkin-Elmer 2400 thermal cycler.
ARDRA.
Primers fD1 and rD1 were used to amplify nearly full-length 16S rRNA genes (43). PCR was performed in 50-μl reaction mixtures containing 1× PCR buffer, 1.5 mM MgCl2, 5% dimethylsulfoxide, 200 μM each nucleotide (Boehringer GmbH, Mannheim, Germany), 15 pmol of each primer, 1 U of Taq polymerase, and 50 ng of purified template DNA. The temperature profile was as follows: initial denaturation at 95°C for 3 min 30 s; 35 cycles of denaturation at 94°C for 1 min 10 s, annealing at 56°C for 40 s, and extension at 72°C for 2 min 10 s; and final extension at 72°C for 6 min 10 s. PCR products were purified with Promega’s Wizard PCR-Prep columns (Promega, Mannheim, Germany) and digested with the tetrameric restriction endonucleases CfoI, DdeI, and MspI (Boehringer) and MboI (USB-Amersham International), as recommended by the manufacturers. The digests were resolved by electrophoresis with 7-cm-long 2% Metaphor agarose gels (Biozym, Hess. Oldendorf, Germany) in Tris-borate-EDTA (30) at 55 V for 3 h. A 100-bp ladder (GIBCO BRL, Eggenstein, Germany) was run at both sides and in the central lane of each gel.
16S-23S rDNA IGS RFLP analysis.
The 16S-23S rDNA IGS region was amplified with primer pair FGPS1490/FGPL132′ (20) by using the same PCR mixture described above for ARDRA and the following program: 95°C for 3 min 30 s, followed by 30 cycles at 93.5°C for 1 min, 55°C for 40 s, and 72°C for 1 min and a final extension at 72°C for 5 min.
The PCR products were digested with restriction enzymes DdeI, HaeIII, and MspI (Promega) and resolved on 2% Metaphor agarose gels as described for ARDRA.
rep-PCR genomic fingerprinting.
The cycling programs and reaction mixture composition (25 μl) were as previously described (41), except that 50 ng of template DNA and 2 U of Taq polymerase were used. Six microliters of the reaction mixtures was loaded onto 18-cm-long 1.5% agarose gels and run at room temperature in Tris-acetate-EDTA (TAE) buffer for 4.2 h at 4 V/cm. A 1-kb ladder (GIBCO BRL) was included as a size reference, as described above.
Computer-assisted analysis of rDNA restriction patterns and rep-PCR genomic fingerprints.
Gel images were digitized with a charge-coupled device video camera (INTAS, Göttingen, Germany) and stored to disk as TIFF files. These were converted, normalized with the above-mentioned molecular size markers, and analyzed with GelCompar software (version 4.0; Applied Maths, Kortrijk, Belgium). The “rolling disk” background subtraction method was applied. For ARDRA and IGS RFLP analysis, a band-matching algorithm was selected to calculate pair-wise similarity matrices with the Dice coefficient (14). A band-matching tolerance of 0.75% was chosen. To analyze rep-PCR patterns, as well as combined 16S rDNA restriction patterns with rep fingerprints, similarity matrices of whole densitometric curves of the gel tracks were calculated by using the pair-wise Pearson’s product-moment correlation coefficient (r value), an approach that compares the whole densitometric curves of the fingerprints (12, 28). For this type of analysis zones of gels containing primer bands or not fully restricted DNA bands were excluded by defining appropriate “active zones” on the digitized images to be included in the analysis. The rep-PCR genomic fingerprints from 200 bp to 12 kb were compared. 16S rDNA RFLP patterns were compared by using ranges from 70 bp to 1.4 kb for CfoI restrictions, 70 to 750 bp for DdeI restrictions, 70 bp to 1.3 kb for MboI restrictions, and 70 to 800 bp for MspI restrictions. Cluster analysis of similarity matrices was performed by the unweighted pair group method using arithmetic averages (UPGMA) (33).
Cloning of partial 16S rDNA sequences.
Partial 16S rDNA sequences, corresponding to positions 44 to 337 in the Escherichia coli numbering system (4), were amplified with primers Y1ES (5′-ccgaattcgtcgacaacTGGCTCAGAACGAACGCTGGCGGC-3′; this study) and Y2HBX (5′-cccgggatccaagcttCCCACTGCTGCCTCCCGTAGGAGT-3′; this study). These primers are derivatives of Y1/Y2 (48) and have linker sequences at their 5′ ends containing several restriction sites, permitting directional cloning of the PCR products. The compositions of the PCR mixtures were as described for the amplification of 16S rDNAs. PCR consisted of an initial denaturation at 94°C for 3 min, followed by 30 cycles of 94°C for 45 s, 62°C for 40 s, and 72°C for 2 min and a final extension at 72°C for 3 min 30 s. PCR products were restricted with EcoRI and BamHI. The restriction fragments were purified from low-gelling-point agarose (Biozym) gels with Promega’s Wizard PCR-Prep columns (Promega) and ligated into pBluescript KS(+) (Stratagene, La Jolla, Calif.) with T4 ligase (USB-Amersham). The ligation mixture was used to transform CaCl2-competent E. coli DH5α cells (30).
DNA sequencing of partial 16S rDNA fragments.
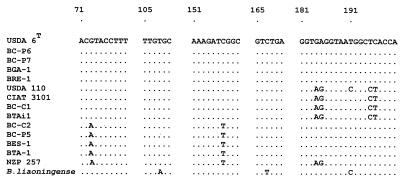
DNA sequencing of partial 16S rDNA fragments was performed with a Li-COR automatic DNA sequencer, model 4000, by using the M13 universal infrared (IR) detectable primers M13fIR and M13rIR (24) and the Thermosequenase fluorescence-labeled primer cycle sequencing kit with 7-deaza-dGTP (Amersham International, Little Chalfont, England), as recommended by the manufacturers. Partial 16S rDNA sequences of both strands were obtained by direct sequencing of PCR products by a nested-PCR strategy previously described (40). Primers Y1 and Y2 (48) were used for the first PCR, the products being diluted 1/100 for the second nested PCR with primer combinations Y1M13f/Y2 and Y1/Y2M13f to obtain the templates for cycle sequencing, which were gel purified as described above. IR-detectable primer M13fIR was used for the cycle sequencing reaction. Both strands of the cloned fragments (two clones per strain) were sequenced with IR-detectable primers M13fIR and M13rIR. The following partial 16S rDNA sequences were retrieved from sequence databases to prepare Fig. 6: Bradyrhizobium liaoningensis (X86065), Bradyrhizobium sp. strain (Aeschynomene) BTAi1 (M55492), and Bradyrhizobium sp. strain (Lotus) NZP 2257 (M55486). For Bradyrhizobium japonicum USDA 6T and USDA 110, the corresponding 264-bp fragment was derived from the sequences with accession no. U69638 and Z35330, respectively.
FIG. 6.
Alignment of sequence stretches containing variable positions within the 264-bp 16S rDNA segment amplified by primer pair Y1/Y2 from Canarian and reference bradyrhizobia (see Table 1). GenBank accession numbers of the complete sequences are listed in Materials and Methods.
Plant material and nodulation assays.
Tagasaste [C. proliferus subsp. proliferus var. palmensis (L.fil.) Link] seeds were collected from cultivated stands on La Palma, the island of its origin (9). Seeds of Acacia pendula, Glycine max cv. McCall and Peking, Lotus corniculatus, Macroptilium atropurpureum, and Vigna unguiculata cv. Red Caloona, kindly provided by S. G. Pueppke, were surface sterilized in a 3% sodium hypochlorite solution for 3 min, washed thoroughly with sterile water, imbibed for 1 h, and plated for germination on 1% (wt/vol) LN agar plates (45). Tagasaste seeds were similarly treated, but to overcome seed coat dormancy, embryos were excised from sterilized, scarified, and imbibed seeds (29) and incubated in darkness at 20°C on 1% (wt/vol) LN agar. Seedlings were transplanted for nodulation assays after 6 days. Leucaena leucocephala cv. Cunningham seeds (Commonwealth Scientific and Industrial Research Organisation, Canberra, Australia) were scarified for 30 min in concentrated H2SO4, thoroughly rinsed in sterile tap water, and plated on LN plates for germination. The nodulation assays were performed in plastic growth pouches or in Leonard jars with vermiculite-perlite (1:1 [vol/vol]) as the substrate and N-free LN nutrient solution buffered at pH 6.8 with 10 mM MES (morpholineethanesulfonic acid). The inoculum was derived from YMB cultures of the relevant rhizobial strains. Seedlings were briefly dipped into the inoculum suspension and transferred to Leonard jars (woody legumes) or growth pouches (herbaceous legumes) at a density of two seedlings per cultivation unit. Plants were grown in controlled-environment chambers (a cycle of 15 h of light and 9 h of darkness at 25 and 18°C, respectively, and 70% relative humidity) for 21 days, except for A. pendula, C. proliferus, L. leucocephala, and L. corniculatus plants, which were grown for 42, 35, 35, and 42 days, respectively.
After harvest, plant roots were checked for nodulation. For each plant-strain combination, nodules were cut in half and observed under the binocular microscope for visual determination of leghemoglobin content and other characteristics.
Nucleotide sequence accession numbers.
The sequences for strains BC-C1, BC-C2, BC-P5, BC-P6, BC-P7, BES-1, BGA-1, BRE-1, BTA-1, and CIAT 3101 were deposited in the GenBank sequence database under accession no. AF000550 to AF000559, respectively.
RESULTS
Analysis of combined CfoI, DdeI, MboI, and MspI 16S rDNA restriction patterns.
Nearly full-length 16S rDNAs of nine nodule isolates from the Canaries and of six reference strains (Table 1) were amplified via PCR with universal primers rD1 and fD1 (43). The PCR products were restricted with enzymes CfoI, DdeI, MboI, and MspI. The individual RFLP patterns are shown in Fig. 1. The observed sizes of the restriction fragments for each type of pattern in the normalized gels are listed in Table 2, which also shows the expected sizes of the restriction fragments for strains USDA 6T and USDA 76T (the type strains for B. japonicum and Bradyrhizobium elkanii, respectively) and USDA 110spc4 based on the sequences with accession no. U69638, U35000, and Z35330, respectively. The average deviation between observed and expected sizes of the 19 restriction fragments detected for USDA 110spc4 with the four enzymes was 1.8 bp (Table 2).
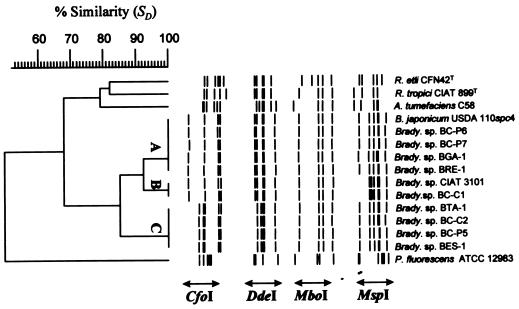
FIG. 1.
Dice/UPGMA cluster analysis of combined CfoI, DdeI, MboI, and MspI restriction patterns of amplified 16S rDNA of nodule isolates of endemic woody legumes of the Canary Islands and reference strains (see Table 1). The individual RFLPs are shown as bands defined on the actual restriction fragments. The sizes of the restriction fragments for each type of pattern are presented in Table 2. Clusters A, B, and C contain all the Canarian isolates and the two reference Bradyrhizobium strains. R. etli, Rhizobium etli; R. tropici, Rhizobium tropici; A. tumefaciens, Agrobacterium tumefaciens; P. fluorescens, Pseudomonas fluorescens.
TABLE 2.
Observed and expected fragment sizes of 16S rDNA PCR-RFLPs of bradyrhizobia
| Enzyme | Observed sizes (bp) of fragments of pattern:
|
Expected sizesc (bp) of fragments of sequence from:
|
|||
|---|---|---|---|---|---|
| Aa | Bb | USDA 110spc4 | B. japonicum USDA 6T | B. elkanii USDA 76T | |
| CfoI | 452, 355, 331, 135, 115 | 810, 334, 137, 115 | 813, 334, 137, 116, 115 | 813, 334, 137, 116, 115 | 452, 361, 334, 139, 115, 114 |
| DdeI | 370, 295, 257, 243, 113 | 413, 367, 262, 248, 111 | 409, 365, 263, 242, 111 | 365, 296, 263, 242, 113, 111 | 409, 367, 261, 128, 114, 111 |
| MboI | 708, 237, 183, 88 | 708, 237, 183, 88 | 708, 235, 182, 176, 83, 54 | 708, 235, 182, 176, 83, 54 | 710, 235, 174, 133, 83, 54 |
| MspI | 289, 269, 260, 224, 172, 157, 90 | 508, 289, 222, 170, 158, 90 | 502, 289, 222, 170, 156, 91, 81 | 502, 289, 222, 170, 156, 91, 81 | 495, 289, 222, 170, 122, 91, 79 |
For CfoI, DdeI, and MboI, the fragment sizes are those of strain BC-C2; for MspI, the fragment sizes are those of strain BC-C1.
Observed sizes of restriction fragments for strain USDA 110spc4 for all four enzymes.
Only fragment sizes greater than 50 bp, on the basis of the 16S rRNA gene sequence, are shown. These sequences were aligned and then modified at their ends to include the sequences of primers fD1 and rD1, which were used for ARDRA, to make precise size comparisons between those from RFLP and those from sequence data possible. T, type strain. Accession numbers for strains USDA 110spc4, B. japonicum USDA 6T, and B. elkanii USDA 76T are Z35330, U69638, and U35000, respectively.
The CfoI digestion revealed two genotypes with five and four well-resolved bands (patterns A and B in Table 2, respectively). The DdeI restrictions yielded five fragments, with strains from the Canaries falling into the same two Bradyrhizobium groups, A and B, identified from the CfoI restriction pattern. All Canarian and reference bradyrhizobia analyzed displayed a monomorphic MboI restriction pattern with four detectable bands, indicating that no B. elkanii strains were present in our collection and that they are phylogenetically related to strains in the B. japonicum 16S rDNA cluster (see Table 2). The MspI digestion resulted again in two types of patterns for the bradyrhizobia, with six (pattern B) and seven (pattern A) bands.
The combined CfoI, DdeI, MboI, and MspI restriction patterns of the amplified 16S rDNAs were used for cluster analysis by UPGMA. This analysis revealed three groups, A, B, and C (Fig. 1), which were consistent with those obtained by the analysis of individual RFLP patterns. All Canarian isolates clustered together with the two Bradyrhizobium reference strains USDA 110spc4 and CIAT 3101, making a coherent “Bradyrhizobium spp.” cluster at a linkage level (SD) of 84.9%. The CfoI and DdeI restriction patterns divided these strains into two well-defined groups. Group A comprised strains BC-P6, BC-P7, BGA-1, and BRE-1 together with B. japonicum USDA 110. Group C comprised only Canarian isolates, namely, BC-C2, BC-P5, BES-1, and BTA-1. The MspI restriction resolved a third genotype comprising strains BC-C1 and CIAT 3101 (group B in Fig. 1). The Agrobacterium, Pseudomonas, and Rhizobium reference species formed well-defined separate branches.
Analysis of combined rDNA IGS RFLPs.
All the Canarian isolates and Bradyrhizobium sp. strain CIAT 3101 yielded single IGS PCR products of ∼930 bp, while strain USDA 110spc4 yielded a slightly larger fragment of about 970 bp. The IGS PCR fragments of the other reference strains were substantially larger (data not shown) and were not included in the RFLP analysis.
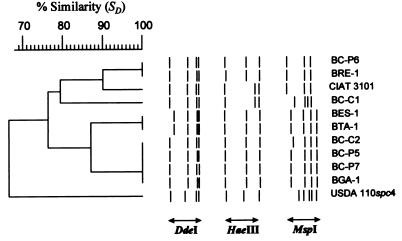
The IGS PCR products were restricted with DdeI, HaeIII, and MspI. Cluster analysis of the combined patterns resolved 11 strains into six genotypes, as shown in Fig. 2. Reference strain USDA 110spc4 was found to constitute an “outgroup” to the other bradyrhizobia, primarily due to its distinctive MspI pattern. Strains CIAT 3101 and BC-C1 of ARDRA cluster B had identical DdeI and HaeIII patterns. However, MspI digestion revealed that the strains had different genotypes. MspI digestion also separated strains BC-P6 and BRE-1 from BC-P7 and BGA-1 of ARDRA cluster A. The last two strains and strains BC-C2 and BC-P5 had identical RFLPs. DdeI restriction resolved strains BES-1 and BTA-1 as having genotypes different from those of strains BC-P5 and BC-P2; all four belong to ARDRA group C. These results clearly indicate the existence of further genotypic diversity within the groups defined by ARDRA.
FIG. 2.
Dice/UPGMA cluster analysis of combined DdeI, HaeIII, and MspI restriction patterns of amplified 16S-23S rDNA IGSs of nodule isolates of endemic woody legumes of the Canary Islands and reference Bradyrhizobium strains (see Table 1). The individual RFLPs are shown as bands defined on the actual restriction fragments. Six genotypes were resolved. Not all groupings are consistent with those obtained by ARDRA.
Cluster analysis of combined 16S rDNA and rDNA IGS RFLPs.
We reasoned that since the 16S rDNA gene and the IGS occupy contiguous DNA regions in the genome, being parts of the same operon, a cluster analysis of their combined restriction patterns should be feasible. The use of this strategy, as shown in Fig. 3, resolved the same six genotypes as did the IGS RFLP analysis and clustered them in accordance with the results obtained by ARDRA.
FIG. 3.
Dice/UPGMA dendrogram based on the combined CfoI, DdeI, MboI, and MspI 16S rDNA RFLPs and 16S-23S rDNA IGS RFLPs of nodule isolates of endemic woody legumes of the Canary Islands and reference Bradyrhizobium strains (see Table 1). The groupings revealed in this cluster analysis are consistent with both the ARDRA and the IGS PCR-RFLP analyses.
Analysis of combined BOX, ERIC, and REP PCR genomic fingerprints.
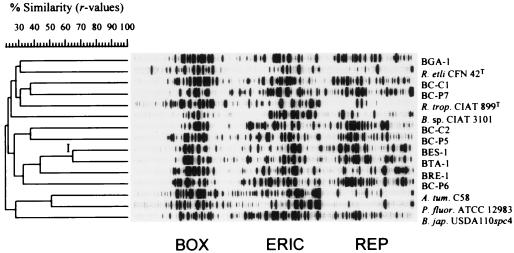
An analysis of the individual and combined BOX, ERIC, and REP PCR patterns revealed a considerable genetic diversity. Almost every Canarian isolate yielded a unique and complex genomic fingerprint with each primer combination, as indicated by the low linkage values in Fig. 4.
FIG. 4.
Product-moment/UPGMA cluster analysis of linearly combined BOX, ERIC, and REP PCR genomic fingerprints of nodule isolates of endemic woody legumes of the Canary Islands and reference strains (see Table 1). The only significant cluster is denoted by I. See the text for explanations. On the scale, r values are expressed as percentages. R. etli, Rhizobium etli; R. trop., Rhizobium tropici; A. tum., Agrobacterium tumefaciens; P. fluor., Pseudomonas fluorescens; B. jap., Bradyrhizobium japonicum.
To analyze the reproducibility of the rep-PCR genomic fingerprinting protocol, five samples were subjected two or three times to rep-PCR. A comparison of the resulting fingerprint patterns yielded similarity coefficients (r values) of ∼0.90 to 0.95 when the patterns were generated in the same PCR experiment and resolved on the same gel, while independent PCR and electrophoresis of repeated samples resulted in r values of ∼0.85 to 0.90 (data not shown). These values are consistent with those from other studies (28a). A relatively high similarity value was found for the single and combined rep-PCR fingerprints of isolates BES-1 and BTA-1 (r > 0.65); these formed the only significant cluster of Canarian isolates (cluster I in Fig. 4). These two strains both grouped in ARDRA cluster B (Fig. 1), illustrating the consistency of results obtained for closely related strains by the combined rep-PCR and the combined ARDRA pattern analysis. Furthermore, these two strains had the same IGS restriction patterns (Fig. 2), this being an independent source of molecular evidence indicating their close genetic relatedness.
Cluster analysis of combined 16S rDNA RFLPs and rep-PCR genomic fingerprints.
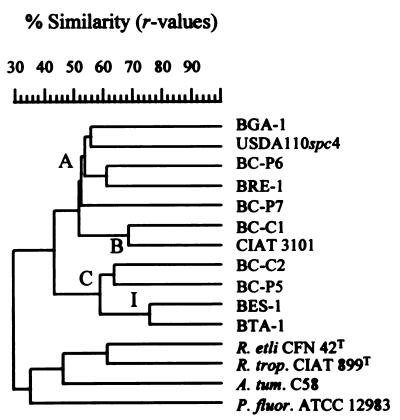
As noted above, the great diversity of rep-PCR patterns obtained resulted in low linkage values of the fingerprints, yielding only one significant cluster. Therefore, a cluster analysis was performed on the combination of four 16S rDNA restriction patterns and the three rep-PCR genomic fingerprints (Fig. 5). The resulting dendrogram, here referred to as BERCDMM (for BOX, ERIC, and REP PCR plus ARDRA with restriction enzymes CfoI, DdeI, MboI, and MspI) was found to reflect the overall diversity and phylogenetic positions of the environmental isolates analyzed in this study (see Table 3).
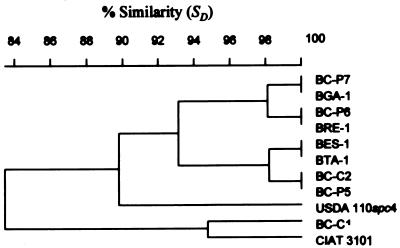
FIG. 5.
Product-moment/UPGMA cluster analysis of linearly combined BOX, ERIC, and REP PCR genomic fingerprints and CfoI, DdeI, MboI, and MspI restriction patterns of amplified 16S rDNA of nodule isolates of endemic woody legumes of the Canary Islands and reference strains (see Table 1). Clusters A, B, and C contain all the Canarian isolates and the two reference Bradyrhizobium strains. On the scale, r values are expressed as percentages. For species abbreviations, see the legend for Fig. 4.
TABLE 3.
Summary of fingerprinting and sequencing results
| Bradyrhizobium strain | Single and combined types of 16S rDNA and IGS restriction patternsa
|
Type of rep-PCR fingerprintb
|
Combined ARDRA and rep-PCR fingerprint and sequence typec
|
|||||
|---|---|---|---|---|---|---|---|---|
| CDMM | DHM | CDMM-DHM | BOX | ERIC | REP | BERCDMM | 16S | |
| BC-C1 | BBAA | 3 | BBAA-3 | U | U | U | U-BBAA | S2 |
| CIAT 3101 | BBAA | 2 | BBAA-2 | U | U | U | U-BBAA | S2 |
| USDA 110 | BBAB | 6 | BBAB-6 | U | U | U | U-BBAB | S4 |
| BC-P6 | BBAB | 1 | BBAB-1 | U | U | U | U-BBAB | S3 |
| BC-P7 | BBAB | 5 | BBAB-5 | U | U | U | U-BBAB | S3 |
| BGA-1 | BBAB | 5 | BBAB-5 | U | U | U | U-BBAB | S3 |
| BRE-1 | BBAB | 1 | BBAB-1 | U | U | U | U-BBAB | S3 |
| BC-C2 | AAAB | 5 | AAAB-5 | U | U | U | U-AAAB | S1 |
| BC-P5 | AAAB | 5 | AAAB-5 | U | U | U | U-AAAB | S1 |
| BES-1 | AAAB | 4 | AAAB-4 | I | I | I | I-AAAB | S1 |
| BTA-1 | AAAB | 4 | AAAB-4 | I | I | I | I-AAAB | S1 |
A and B denote the types of restriction pattern for the restriction enzyme as follows. The CDMM column indicates the restriction patterns obtained, respectively, with the tetrameric endonucleases CfoI, DdeI, MboI, and MspI used for ARDRA. The DHM column indicates the types of restriction patterns obtained with DdeI, HaeIII, and MspI, respectively, used in IGS PCR-RFLP analysis. The CDMM-DHM column indicates the genotypes resolved by combined 16S rDNA and 16S-23S rDNA restriction analysis with the enzymes indicated above. The sizes of the resulting restriction fragments in each pattern are listed in Table 2.
U and I stand, respectively, for unique and type I rep-PCR patterns generated with the indicated rep-PCR primers.
Letters in the BERCDMM column indicate the type of combined 16S rDNA restriction patterns and rep-PCR fingerprints. In the 16S column, S1 to S4 are the sequence types obtained for the Canarian and reference bradyrhizobia, as indicated in Fig. 6.
The overall topology of the dendrogram was found to be very similar to that constructed for the combined 16S rDNA restriction patterns. A cluster comprising exclusively the Bradyrhizobium strains was again found and was resolved consistently into subgroups A, B, and C previously defined by ARDRA.
The relatedness of strains BC-C1 and CIAT 3101, revealed by ARDRA but not apparent by rep-PCR genomic fingerprint analysis, is again evident (r > 0.67) in the BERCDMM tree (Fig. 5). This highlights the effect of 16S rDNA RFLP patterns on clustering rep-PCR genomic fingerprints of highly diverse strains.
Strains BES-1 and BTA-1 (both originating from Tenerife) form a subgroup (I) with a high degree of similarity (r > 0.76) within cluster C of BERCDMM. This refined the clustering of these two strains as revealed by ARDRA in a way consistent with rep-PCR and rDNA IGS RFLP data, reflecting their high degree of genotypic similarity at the strain level.
These results show that the strategy of combining rep-PCR genomic fingerprints and 16S rDNA PCR-RFLPs in computer-assisted pattern analysis yields phylogenetically sound groupings for such disparate taxa as Rhizobium and Bradyrhizobium strains.
Partial 16S rDNA sequence analysis.
When subjected to PCR with primer pair Y1/Y2 or its derivatives, all Canarian isolates and reference strains Bradyrhizobium sp. strain (Centrosema) CIAT 3101 and B. japonicum USDA 110spc4 yielded a 264-bp product, after subtracting the terminal primer sequences, corresponding to positions 44 to 337 in the E. coli 16S rRNA sequence (4, 48). Three different sequences (S1 to S3) were found among the Canarian strains (Fig. 6 and Table 3). Cluster S1 contains isolates BC-C2, BC-P5, BES-1, and BTA-1 and corresponds to ARDRA cluster C (Fig. 1 and Table 3). Members of the S1 cluster lacked a 100% homolog in the GenBank sequence database and thus represent a new sequence type for the genus Bradyrhizobium. Strains BC-C1 and CIAT 3101 (ARDRA group B) have the same rDNA sequence (cluster S2), which is identical to that of phototrophic Bradyrhizobium sp. strain (Aeschynomene) BTAi1 (GenBank accession no. M55492) (48).
Isolates BC-P6, BC-P7, BGA-1, and BRE-1 (ARDRA group A) displayed a 100% sequence identity to each other and to strain USDA 6T and form cluster S3. Based on the available sequence information strain USDA 6T is expected to yield a related but unique 16S rDNA restriction pattern that would be designated BAAB (Tables 2 and 3).
The sequence data are in full agreement with those obtained by ARDRA, but analysis of the combined 16S rDNA-plus-IGS RFLP or ARDRA-plus-rep-PCR patterns resulted in a much finer taxonomic resolution than that achieved by partial rDNA sequencing.
Differences between the sequences of partial rDNA clones and of PCR products, as has been reported recently (13) for some Acacia nodulating Sinorhizobium strains, were not found, suggesting that no sequence microheterogeneity is found in the 16S rRNA genes of these strains. This is in good agreement with the results of Kündig et al. (17), who reported that most Bradyrhizobium strains have a single copy of the rRNA operon.
Analysis of host range.
Nodulation experiments were carried out to examine the host range of the strains. The observed lack of nodulation on two soybean varieties, the modern commercial cultivar McCall and the primitive cultivar Peking, indicates that the Canarian isolates have a different host range than typical B. japonicum strains (Table 4), despite their close phylogenetic relatedness as revealed by ARDRA and partial 16S rDNA sequencing (Fig. 1 and 6). Conversely, B. japonicum USDA 110spc4 did not nodulate tagasaste. Interestingly, Bradyrhizobium sp. strain (Centrosema) CIAT 3101, a Colombian isolate, induces fully effective nodules on A. pendula, C. proliferus, and V. unguiculata. None of the Canarian isolates nodulated the last species effectively, but all of them formed N2-fixing nodules on M. atropurpureum and A. pendula (Table 4). Experiments with L. corniculatus showed that the nodulation patterns of the Canarian isolates are not identical; BC-P6 was found to be the only strain capable of inducing effective nodulation on L. corniculatus.
TABLE 4.
Host range analysis of the Canarian isolates and selected reference strains on several legume hosts
| Rhizobial strain | Nodule characteristics ofa:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| A. pendula | C. prolif. | G. max | G. soja | L. leuco. | L. cornic. | M. atrop. | V. unguic. | |
| BC-C1 | F | F | 0 | 0 | 0 | N | F | N |
| BC-C2 | F | F | 0 | 0 | 0 | N | F | N |
| BC-P5 | F | F | 0 | 0 | 0 | NS | F | N |
| BC-P6 | F | F | 0 | 0 | 0 | F | F | N |
| BC-P7 | F | F | 0 | 0 | 0 | N | F | N |
| BES-1 | F | F | 0 | 0 | 0 | NS | F | N |
| BGA-1 | F | F | 0 | 0 | 0 | NS | F | N |
| BRE-1 | F | F | 0 | 0 | 0 | N | F | N |
| BTA-1 | F | F | 0 | 0 | 0 | NS | F | N |
| CIAT 3101 | F | F | 0 | 0 | ND | B | F | F |
| USDA 110spc4 | ND | 0 | F | F | ND | B | ND | F |
| R. tropici CIAT 899T | ND | B | ND | ND | F | ND | ND | ND |
F, nodules contained leghemoglobin, as determined visually under the binocular on longitudinally cut nodules; N, nodules lacked leghemoglobin; NS, nodules lacked leghemoglobin and additionally contained visibly darkened and senescing cells; B, bumps or empty pseudonodules; 0, no nodules; ND, not determined. C. prolif., C. proliferus; G. soja, Glycine soja; L. leuko., L. leucocephala; L. cornic., Lotus corniculatus; M. atrop., M. atropurpureum; V. unguic., Vigna unguiculata. Cultivars of these species are those specified in Materials and Methods.
DISCUSSION
Woody legumes from the tropics and subtropics are nodulated by a diverse set of rhizobia, including both fast- and slow-growing strains (6, 13, 50). Dommergues et al. (6) have proposed that N2-fixing trees and shrubs should be divided into three broad groups, based on their nodulation patterns with fast-, fast- and slow-, and slow-growing rhizobia. With one possible exception (31), all strains isolated so far from endemic shrub or tree legumes from the Canary Islands are slow-growing strains, phenotypically resembling Bradyrhizobium spp. (21, 31, 41a). The available data on the nodulation patterns of these legumes are scarce but indicate that nodulation with bradyrhizobia seems to predominate in the Canaries. Gault et al. (11) have reported that in southeastern Australia tagasaste plants are nodulated effectively both by fast- and slow-growing rhizobia compatible with several Lotus species and propose a symbiotic relationship between the genera Chamaecytisus and Lotus. Indeed, all Canarian strains were able to nodulate L. corniculatus in our experiments, but only BC-P6 induced N2-fixing nodules. Taken together, these data suggest that tagasaste belongs to group two, as defined by Dommergues et al. (6).
León-Barrios et al. (21) have reported that strains BES-1, BGA-1, BRE-1, and BTA-1 nodulate two taxa in the C. proliferus taxonomic complex as well as Teline canariensis, Teline stenopetala, and M. atropurpureum, but not G. max cv. Amsoy, Lupinus spp., or Lotus campylocladus. The last is an endemic Lotus species from the pine wood belts of the Canaries, frequently found associated with C. proliferus stands. Our nodulation experiments with the advanced commercial soybean cultivar McCall or the primitive cultivar Peking confirm the findings of León-Barrios et al. (21) and suggest that these isolates are not typical B. japonicum strains with regard to host range. Conversely, the well-known soybean strain B. japonicum USDA 110spc4 did not nodulate tagasaste in our experiments. In a later study, Santamaría et al. (31) reported that strains BGA-1 and BTA-1 nodulate Cajanus cajan. The results reported here extend the host range database for the Canarian isolates by providing data on the nodulation of another four legume species (Table 4).
The taxonomy of the genus Bradyrhizobium needs to be further developed, especially with regard to isolates from tree or shrub legumes, since it still relies predominantly on the soybean nodulation phenotype. Currently three Bradyrhizobium species are validly described: B. japonicum (15), B. elkanii (18), and B. liaoningense (47), all nodulating soybeans. The taxonomic positions of most of the tree-associated bradyrhizobia not nodulating soybeans remain uncertain, and isolates of this type are generically referred to as Bradyrhizobium sp. strains (host genus).
For the Canarian strains isolated so far from woody legumes only scarce phenotypic data are available (21, 31), and no cluster analysis based on phenotypic traits has been carried out. Several reports have stressed the lack of correlation between phenotype- and genotype-based Bradyrhizobium classification methods (32, 40). For this reason, we chose genotypic typing methods to survey bradyrhizobial biodiversity.
Our 16S rDNA PCR-RFLP and sequencing analysis grouped the Canarian isolates within the B. japonicum/Rhodopseudomonas palustris rDNA cluster, lending strong support to the classification of each of these strains as a Bradyrhizobium sp. This in turn is consistent with their slow growth, alkalization of culture media and NAD-dependent 6-phosphogluconate dehydrogenase activity (15, 21, 31, 41a).
ARDRA with four tetrameric restriction enzymes revealed the existence of three well-defined groups of bradyrhizobia within our sample set (Table 3), indicating the existence of intrageneric diversity within the small collection of strains analyzed. A recent computer-based study (25) evaluated the efficacy of selected tetrameric restriction enzymes for rDNA RFLP phylogenetic analysis of natural isolates, showing that a minimum of three had to be used to detect more than 99% of the operational taxonomic units within a model data set of over 100 proximally and distally related full-length rDNA sequences. Our choice of enzymes was based on this work and the results of Laguerre et al. (19).
Comparative sequencing of a short stretch of the 16S rRNA gene of each of the Bradyrhizobium strains listed in Table 1 provided an independent confirmation of the ARDRA-based classification. For this purpose we chose primer pair Y1/Y2 (48), since it amplifies a hypervariable region of the 16S rDNA, thus allowing phylogenetic comparisons of close relatives. Furthermore, this region has been analyzed in several other studies for the genotypic characterization of Rhizobium and Bradyrhizobium isolates (13, 32, 40, 48). As expected, all three types of sequences obtained (S1 to S3) were most similar to those of strains in the B. japonicum/R. palustris branch of the alpha subclass of the Proteobacteria (32, 40, 48). Interestingly, S1 was found to be a new sequence representing a line of descent not previously described within the Bradyrhizobium/R. palustris rDNA complex.
To analyze the diversity and phylogenetic relationships of the Canarian isolates at finer taxonomic resolution, 16S-23S rDNA IGS PCR-RFLPs and rep-PCR genomic fingerprints with BOX, ERIC, and REP primers were generated from all environmental and reference Bradyrhizobium strains. IGS RFLP with three enzymes resolved six genotypes. The clustering of these RFLPs was not entirely consistent with groupings revealed by ARDRA. These inconsistencies were overcome by combining ARDRA and IGS RFLP pattern analyses, which yielded a dendrogram consistent with the separately obtained groupings. The resolution obtained by this approach was significantly higher than that achieved by partial sequencing of the 16S rDNA region.
An even finer taxonomic resolution was found in the highly complex BOX, ERIC, and REP PCR genomic fingerprints. Only strains BTA-1 and BES-1 were found to have similar rep-PCR patterns, forming cluster I in Fig. 4 (see also Table 3). It has been shown previously (16) that rep-PCR genomic fingerprints have a greater discrimination power than serotyping, allowing the determination of the phylogenetic relationships of phenotypically nearly identical B. japonicum strains in seroclusters 123, 127, and 129. To maximize strain discrimination by rep-PCR genomic fingerprinting, Judd et al. (16) analyzed combined ERIC-plus-REP fingerprints manually. This is the first study on rhizobial isolates that fully exploits the taxonomic resolution of rep-PCR by combining BOX, ERIC, and REP PCR genomic fingerprints in computer-assisted pattern analysis, maximizing strain discrimination and the phylogenetic coherency of the obtained clusters (28). The great diversity (low correlation) of rep-PCR banding patterns obtained was associated with very low linkage levels. Thus, we explored the value of combining the three rep-PCR genomic fingerprints with the four 16S rDNA restriction patterns. Cluster analysis using the product-moment correlation coefficients (12) of the resulting combined gel yielded a dendrogram (Fig. 5) integrating the phylogenetic information of ARDRA and rep-PCR genomic fingerprinting. This approach allows for the illustration of phylogenetic relationships between isolates with a dynamic range from the genus to the strain level (Fig. 5). Presently, this is difficult to achieve with any other single technique available.
The great genotypic diversity revealed by our genotypic analysis is in good agreement with the great phenotypic diversity reported by Santamaría et al. based on serotyping and electrophoretic profiling of lipopolysaccharides (31). These authors detected 21 profiles within the 27 Bradyrhizobium isolates studied and reported that of the strains included in the present work only BC-P5 (BTA-8) and BC-P6 (BTA-9) have the same lipopolysaccharide profiles. This result is not consistent with our genotypic data (Table 3), which show that these two strains belong to different 16S rDNA homology groups and have very different IGS and rep-PCR fingerprints. Conversely, strains we have found to have identical rDNA genotypes do not display similar lipopolysaccharide profiles. Moreover, Santamaría et al. (31) have reported eight serogroups on the basis of indirect enzyme-linked immunosorbent assay tests; the serogroups were determined by serological cross-reaction to polyclonal antisera raised against strains BGA-1 and BTA-1. Almost no correlation could be found between our genotypic groupings and the serogroups defined by Santamaría et al. (31), further illustrating the lack of correlation between phenotype- and genotype-based methods in grouping Bradyrhizobium strains.
Together, our ARDRA, partial 16S rDNA sequencing, IGS RFLP, and rep-PCR data indicate a large genotypic diversity from intrageneric to strain levels, and provide the first analysis of the phylogenetic position and genotypic diversity of Bradyrhizobium strains nodulating endemic woody legumes from the Canarian archipelago. The type of combinational analysis of fingerprinting data presented here also reveals the power and suitability of computer-assisted pattern analysis for rapid, accurate, and thorough surveys of bacterial diversity.
ACKNOWLEDGMENTS
We thank Milagros León-Barrios and Cristina Bolaños for providing strains and unpublished data, Javier Francisco-Ortega for information on C. proliferus, and Steven G. Pueppke for reading the manuscript, providing plant germplasm, and help with nodulation assays.
This work was supported by the Deutsche Forschungsgemeinschaft through the SFB 395, the DOE (DE FG 0290ER20021), the NSF Center for Microbial Ecology (DIR 8809640), Heinz Inc., Roger Seeds Co., and the Consortium for Plant Biotechnology Research (DE-FC05-02OR22072). P.V. was the recipient of a scholarship from the Deutscher Akademischer Austauschdienst.
REFERENCES
- 1.Acebes-Ginovés J R, del Arco-Aguilar M, Wildpret de la Torre W. Revisión taxonómica de Chamaecytisus proliferus (L.Fil) Link en Canarias. Vieraea. 1991;20:191–202. [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Smit J A, Seidman J C, Struhl K S, editors. Current protocols in molecular biology, section 2.4. New York, N.Y: John Wiley and Sons; 1994. [Google Scholar]
- 3.Bramwell D. Endemism in the flora of the Canary Islands. In: Valentine D H, editor. Taxonomy, phytogeography and evolution. London, United Kingdom: Academic Press; 1972. pp. 141–159. [Google Scholar]
- 4.Brosius J, Dull T J, Sleeter D D, Noller H F. Gene organisation and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981;148:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- 5.de Bruijn F J. Use of repetitive (repetitive extragenic palindromic and enterobacterial repetitive intergeneric consensus) sequences and the polymerase chain reaction to fingerprint the genomes of Rhizobium meliloti isolates and other soil bacteria. Appl Environ Microbiol. 1992;58:2180–2187. doi: 10.1128/aem.58.7.2180-2187.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dommergues Y R, Diem H G, Gauthier D L, Dreyfus B L, Cornet F. Nitrogen-fixing trees in the tropics: potentialities and limitations. In: Veeger C, Newton W E, editors. Advances in nitrogen fixation research. Proceedings of the 5th International Symposium on Nitrogen Fixation. Wageningen, The Netherlands: Martinus Nijhoff/Dr. W. Junk Publishers; 1984. pp. 7–8. [Google Scholar]
- 7.Fox G E, Wisotzkey J D, Jurtshuk P. How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int J Syst Bacteriol. 1992;42:166–170. doi: 10.1099/00207713-42-1-166. [DOI] [PubMed] [Google Scholar]
- 8.Francisco-Ortega J, Jackson M T, Santos-Guerra A, Fernández Galván M. Historical aspects of the origin and distribution of tagasaste [Chamaecytisus proliferus (L.fil.) Link ssp. palmensis (Christ) G. Kunkel], a fodder tree from the Canary Islands. J Adel Bot Gard. 1991;14:67–76. [Google Scholar]
- 9.Francisco-Ortega J, Newbury J H, Ford-Lloyd B V. Numerical analysis of RAPD data highlights the origin of cultivated tagasaste (Chamecytisus proliferus ssp. palmensis) in the Canary Islands. Theor Appl Genet. 1993;87:264–270. doi: 10.1007/BF00223775. [DOI] [PubMed] [Google Scholar]
- 10.Francisco-Ortega J, Jackson M T, Santos-Guerra A, Fernández-Galván M, Ford-Lloyd B V. The phytogeography of Chamaecytisus proliferus (L.fil.) Link complex (Fabaceae: Genisteae) in the Canary Islands: a multivariate analysis. Vegetatio. 1994;110:1–17. [Google Scholar]
- 11.Gault R R, Pilka A, Hebb D M, Brockwell J. Nodulation studies on legumes exotic to Australia: symbiotic relationships between Chamaecytisus palmensis (tagasaste) and Lotus spp. Austr J Exp Agric. 1994;34:385–394. [Google Scholar]
- 12.Häne B G, Jäger K, Drexler H. The Pearson product-moment correlation coefficient is better suited for identification of DNA fingerprint profiles than band matching algorithms. Electrophoresis. 1993;14:967–972. doi: 10.1002/elps.11501401154. [DOI] [PubMed] [Google Scholar]
- 13.Haukka K, Lindström, Young J P W. Diversity of partial 16S rRNA sequences among and within strains of African rhizobia isolated from Acacia and Prosopis. System Appl Microbiol. 1996;19:352–359. [Google Scholar]
- 14.Heyndrickx M, Vauterin L, Vandamme P, Kersters K, De Vos P. Applicability of combined amplified ribosomal DNA restriction analysis (ARDRA) patterns in bacterial phylogeny and taxonomy. J Microbiol Methods. 1996;26:247–259. [Google Scholar]
- 15.Jordan D C. Family III. Rhizobiaceae Conn 1938. In: Krieg N R, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 1. Baltimore, Md: The Williams & Wilkins Co.; 1984. pp. 234–244. [Google Scholar]
- 16.Judd A K, Schneider M, Sadowsky M J, de Bruijn F J. Use of repetitive sequences and the polymerase chain reaction technique to classify genetically related Bradyrhizobium japonicum serocluster 123 strains. Appl Environ Microbiol. 1993;59:1702–1708. doi: 10.1128/aem.59.6.1702-1708.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kündig K, Beck C, Hennecke H, Göttfert M. A single rRNA gene region in Bradyrhizobium japonicum. J Bacteriol. 1995;177:5151–5154. doi: 10.1128/jb.177.17.5151-5154.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuykendall L D, Saxena B, Devine T E, Udell S E. Genetic diversity in Bradyrhizobium japonicum Jordan 1982 and a proposal for Bradyrhizobium elkanii sp. nov. Can J Microbiol. 1992;38:501–505. [Google Scholar]
- 19.Laguerre G, Allard M R, Revoy F, Amarger N. Rapid identification of rhizobia by restriction fragment length polymorphism analysis of PCR-amplified 16S rRNA genes. Appl Environ Microbiol. 1994;60:56–63. doi: 10.1128/aem.60.1.56-63.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laguerre G, Mavingui P, Allard M R, Charnay M P, Louvrier P, Mazurier S I, Rigottier-Gois L, Amarger N. Typing of rhizobia by PCR and PCR-restriction fragment length polymorphism analysis of chromosomal and symbiotic gene regions: application to Rhizobium leguminosarum and its different biovars. Appl Environ Microbiol. 1996;62:2029–2036. doi: 10.1128/aem.62.6.2029-2036.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.León-Barrios M, Gutiérrez-Navarro A M, Pérez-Galdona R, Corzo J. Characterization of Canary Island isolates of Bradyrhizobium sp. (Chamaecytisus proliferus) Soil Biol Biochem. 1991;23:487–489. [Google Scholar]
- 22.Lindström K, Laguerre G, Normand P, Rasmussen U, Heulin T, Jarvis B D W, de Lajudie P, Martínez.Romero E, Chen W-X. Taxonomy and phylogeny of diazotrophs. In: Elmerich C, Kondorosi A, Newton W E, editors. Biological nitrogen fixation for the 21st century. Proceedings of the 11th International Congress on Nitrogen Fixation, Paris, July 1997. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. [Google Scholar]
- 23.Martínez-Romero E, Caballero-Mellado J. Rhizobium phylogenies and bacterial genetic diversity. Crit Rev Plant Sci. 1996;15:113–140. [Google Scholar]
- 24.Messing J. M13, the universal primer and the polylinker. Focus (BRL) 1988;10:21–26. [Google Scholar]
- 25.Moyer C L, Tiedje J M, Dobbs F C, Karl D M. A computer-simulated restriction fragment length polymorphism analysis of bacterial small-subunit rRNA genes: efficacy of selected tetrameric restriction enzymes for studies of microbial diversity in nature. Appl Environ Microbiol. 1996;62:2501–2507. doi: 10.1128/aem.62.7.2501-2507.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ovalle C, Longeri L, Aronson J, Herrera A, Avendaño J. N2-fixation, nodule efficiency and biomass accumulation after two years in three Chilean legume trees and tagasaste Chamaecytisus proliferus subsp. palmensis. Plant Soil. 1996;179:131–140. [Google Scholar]
- 27.Oyaizu H, Matsumoto S, Minamisawa K, Gamou T. Distribution of rhizobia in leguminous plants surveyed by phylogenetic identification. J Gen Appl Microbiol. 1993;39:339–354. [Google Scholar]
- 28.Rademaker J L W, de Bruijn F J. Characterization and classification of microbes by rep-PCR genomic fingerprinting and computer assisted pattern analysis. In: Caetano-Anollés G, Gresshoff P M, editors. DNA markers: protocols, applications and overviews. New York, N.Y: John Wiley and Sons, Inc.; 1997. pp. 151–171. [Google Scholar]
- 28a.Rademaker J L W, Louws F J, de Bruijn F J. Characterization of the diversity of ecologically important microbes by rep-PCR fingerprinting. In: Akkermans A D L, van Elsas J D, de Bruijn F J, editors. Molecular microbial ecology manual, suppl. 3. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. pp. 1–26. [Google Scholar]
- 29.Regunath B R, Francisco-Ortega J, Newbury H J, Ford-Lloyd B V. Methods for increasing the efficiency of seed germination in the fodder legumes tagasaste and escobon [Chamaecytisus proliferus (L.fil.) Link sensu lato] Seed Sci. Technol. 1993;21:225–235. [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T A. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 31.Santamaría M, Corzo J, León-Barrios M, Gutiérrez-Navarro A M. Characterization and differentiation of indigenous rhizobia isolated from Canarian shrub legumes of agricultural and ecological interest. Plant Soil. 1997;190:143–152. [Google Scholar]
- 32.So R B, Ladha J K, Young J P W. Photosynthetic symbionts of Aeschynomene spp. form a cluster with bradyrhizobia on the basis of fatty acid and rRNA analyses. Int J Syst Bacteriol. 1994;44:392–403. doi: 10.1099/00207713-44-3-392. [DOI] [PubMed] [Google Scholar]
- 33.Sokal R R, Michener C D. A statistical method for evaluating systematic relationships. Univ Kans Sci Bull. 1958;38:1409–1438. [Google Scholar]
- 34.Somasegaran P, Hoben H J. Handbook for rhizobia—methods in legume-Rhizobium technology. Heidelberg, Germany: Springer-Verlag; 1994. [Google Scholar]
- 35.Streit W, Kosch K, Werner D. Nodulation competitiveness of Rhizobium leguminosarum bv. phaseoli and Rhizobium tropici strains measured by glucuronidase (gus) gene fusion. Biol Fertil Soils. 1992;14:140–144. [Google Scholar]
- 36.Townsend R J, Radcliffe J E. Tagasaste forage production systems. N Z J Agric Res. 1990;33:627–634. [Google Scholar]
- 37.Triplett E W, Sadowsky M J. Genetics of competition for nodulation of legumes. Annu Rev Microbiol. 1992;46:399–428. doi: 10.1146/annurev.mi.46.100192.002151. [DOI] [PubMed] [Google Scholar]
- 38.Tyler K D, Wang G, Tyler S D, Johnson W M. Factors affecting reliability and reproducibility of amplification-based DNA fingerprinting of representative bacterial pathogens. J Clin Microbiol. 1997;35:339–346. doi: 10.1128/jcm.35.2.339-346.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Rhijn P, Vanderleyden J. The Rhizobium-plant symbiosis. Microbiol Rev. 1995;59:124–142. doi: 10.1128/mr.59.1.124-142.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Rossum D, Schuurmans F P, Gillis M, Muyotcha A, van Verseveld H W, Stouthamer A H, Boogerd F C. Genetic and phenetic analyses of Bradyrhizobium strains nodulating peanut (Arachis hypogaea L.) roots. Appl Environ Microbiol. 1995;61:1599–1609. doi: 10.1128/aem.61.4.1599-1609.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Versalovic J, Schneider M, de Bruijn F J, Lupski J R. Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Methods Mol Cell Biol. 1994;5:25–40. [Google Scholar]
- 41a.Vinuesa, P. Unpublished results.
- 42.Vinuesa-Fleischmann P, Negrín M A, Jaizme-Vega M C, Hernández-Moreno J M. Arbuscular mycorrhizal tagasaste plants mediate transformations on the P pools in an andisol. In: Azcón-Aguila C, Barea J M, editors. Proceedings of the 4th European Symposium on Mycorrhizas 1994. Brussels, Belgium: Office for Official Publications of the European Communities; 1996. pp. 599–602. [Google Scholar]
- 43.Weisburg W G, Barns S M, Pelletier D A, Lane D J. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Welsh J, McClelland M. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res. 1990;18:7213–7218. doi: 10.1093/nar/18.24.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Werner D, Wilcockson J, Zimmermann E. Adsorption and selection of rhizobia by ion exchange papers. Arch Microbiol. 1975;105:27–32. doi: 10.1007/BF00447108. [DOI] [PubMed] [Google Scholar]
- 46.Willems A, Collins M D. Phylogenetic analysis of rhizobia and agrobacteria based on 16S rRNA gene sequences. Int J Syst Bacteriol. 1993;43:305–313. doi: 10.1099/00207713-43-2-305. [DOI] [PubMed] [Google Scholar]
- 47.Xu L M, Ge C, Cui Z, Li J, Fan H. Bradyrhizobium liaoningense sp. nov., isolated from the root nodules of soybeans. Int J Syst Bacteriol. 1995;45:706–711. doi: 10.1099/00207713-45-4-706. [DOI] [PubMed] [Google Scholar]
- 48.Young J P W, Downer H L, Eardly B D. Phylogeny of the phototrophic Rhizobium strain BTAi1 by polymerase chain reaction-based sequencing of a 16S rRNA gene segment. J Bacteriol. 1991;173:2271–2277. doi: 10.1128/jb.173.7.2271-2277.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Young J P W, Haukka K E. Diversity and phylogeny of rhizobia. New Phytol. 1996;133:87–94. [Google Scholar]
- 50.Zhang X, Harper R, Karsito M, Lindström K. Diversity of Rhizobium bacteria isolated from the root nodules of leguminous trees. Int J Syst Bacteriol. 1991;41:104–113. [Google Scholar]