Abstract
Purpose
A systematic literature review was conducted to estimate the global prevalence of Kirsten rat sarcoma virus gene (KRAS) mutations, with an emphasis on the clinically significant KRAS G12C mutation, and to estimate the prognostic significance of these mutations in patients with colorectal cancer (CRC).
Design
Relevant English-language publications in the Embase, MEDLINE, and the Cochrane Library databases (from 2009 to 2021) and congress presentations (from 2016 to 2021) were reviewed. Eligible studies were those that reported the prevalence and clinical outcomes of the KRAS G12C mutation in patients with CRC.
Results
A total of 137 studies (interventional [n = 8], post hoc analyses of randomized clinical trials [n = 6], observational [n = 122], and longitudinal [n =1]) were reviewed. Sixty-eight studies reported the prevalence of KRAS mutations (KRASm) in 42 810 patients with CRC. The median global prevalence of KRASm was 38% (range, 13.3%-58.9%) and that of the KRAS G12C mutation (KRAS G12C) 3.1% (range, 0.7%-14%). Available evidence suggests that KRASm are possibly more common in tumors that develop on the right side of the colon. Limited evidence suggests a lower objective response rate and inferior disease-free/relapse-free survival in patients with KRAS G12C compared with patients with KRASwt or other KRASm.
Conclusion
Our analysis reveals that KRAS G12C is prevalent in 3% of patients with CRC. Available evidence suggests a poor prognosis for patients with KRAS G12C. Right-sided tumors were more likely to harbor KRASm; however, their role in determining clinical outcomes needs to be investigated further.
Keywords: KRASm, KRAS G12C, prevalence, prognosis, colorectal cancer, systematic literature review, global
This review was conducted to estimate the global prevalence of KRAS mutations, with an emphasis on the clinically significant KRAS G12C mutation, and to estimate the prognostic significance of these mutations in patients with colorectal cancer.
Implications for Practice.
Our systematic literature review of 68 studies from around the globe, reporting on 42 810 patients with colorectal cancer (CRC), revealed a median prevalence of 3.1% (range, 0.7%-14%) for the KRAS G12C mutation. Overall, patients with KRAS G12C had shorter overall survival, progression-free survival, and disease-free survival compared to patients with wild-type KRAS or other non-G12C mutations. The variation in KRAS G12C prevalence across different geographies and the poor prognosis associated with this mutation suggests that KRAS G12C prevalence is an important factor that needs to be considered during patient recruitment in clinical trials of CRC therapies targeting this mutation.
Introduction
Colorectal cancer (CRC) is the third most commonly diagnosed cancer and the second leading cause of cancer-related mortality worldwide.1 In 2020, CRC accounted for around 1.9 million new diagnoses and 935 000 deaths globally.2 The 5-year survival rate for patients with CRC ranges from 91% for those with localized disease to a dismal 15% for those with metastatic disease.3
A number of genes have been implicated in the transformation process from benign neoplasia to invasive carcinoma and metastatic colorectal cancer (mCRC). The development of mutations in the Kirsten rat sarcoma virus gene (KRAS) is an early event in tumorigenesis, which marks the progression from the adenoma to the carcinoma stage.4
KRAS is a proto-oncogene that encodes the 21 kDa guanosine triphosphate (GTP)/guanosine diphosphate (GDP)-binding RAS protein. RAS functions as a molecular switch regulating receptor tyrosine kinase signal transduction by alternating between an active GTP-bound and an inactive GDP-bound states. Mutations in KRAS disrupt this guanine exchange cycle, locking the RAS protein in an active GTP-bound form. The constitutively activated RAS protein can drive uncontrolled cell proliferation, suppress apoptosis, upregulate glucose uptake, promote angiogenesis, and improve cell survival.5 More than 80% of these mutations are located in codon 12 and around 14% are located in codon 13 of KRAS.6,7Mutations in other codons are relatively rare.8-10
For patients with mCRC, first- and second-line treatment approaches typically include chemotherapy combinations with a fluoropyrimidine-based doublet (folinic acid, fluorouracil, and oxaliplatin/folinic acid, fluorouracil and irinotecan [FOLFOX/FOLFIRI]) or triplet (folinic acid, 5-fluorouracil, oxaliplatin, and irinotecan [FOLFOXIRI]) regimen combined with therapies that target either tumor angiogenesis (bevacizumab) or the epidermal growth factor receptor (EGFR) (panitumumab or cetuximab).11-15 However, patients with KRAS mutations (KRASm) or neuroblastoma-RAS (NRAS) mutations are unlikely to benefit from treatment with anti-EGFR therapy and, therefore, have more limited treatment options.16-18 Nonetheless, recently developed KRAS-specific inhibitors offer hope for cancers with certain KRASm. Sotorasib and adagrasib are small-molecule KRAS G12C inhibitors that covalently bind to mutant cysteine residues and lock the G12C-mutated KRAS protein in a non-activated GDP-binding state causing irreversible inhibition of the proliferative activity of the tumor cell.19 Sotorasib and adagrasib have received US FDA approval for the treatment of adult patients with KRAS G12C-mutated locally advanced or metastatic non-small cell lung cancer (NSCLC), and are currently being evaluated in patients with mCRC either as monotherapy or in combination with other therapeutic agents. Other inhibitors that are currently being evaluated for their activity against KRAS G12C-positive solid tumors include JNJ-74699157 (NCT04006301), LY3499446 (NCT04165031), JAB-21822 (NCT05002270), YL-15293 (NCT05119933), GDC-6036 (NCT04449874), BI 1823911 (NCT04973163), and MK-1084 (NCT05067283).
Given the emerging clinical actionability in patients with KRASm, there is a valid need for understanding the prevalence and clinical significance of these mutations in CRC to design appropriate treatment strategies in clinical trials and testing approaches. Additionally, the prognostic significance of these mutations, particularly the “druggable” KRAS G12C mutation in the context of currently approved therapies for CRC has not been definitively established.
The objective of this systematic literature review (SLR) was to assess the prevalence of the KRAS G12C mutation, the prevalence of co-existing mutations, and characterize the clinical outcomes in patients with KRAS G12C-mutated CRC based on robust methodology.
Methods
This SLR was conducted using a standardized approach that was compliant with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses Protocol (PRISMA-P) guidelines.20
Search Strategy
Search strategies were developed around a Population, Intervention, Comparator, Outcomes (PICO) framework in accordance with the Preferred Reporting Items for Systematic reviews and Meta-Analyses (PRISMA) guidelines. The 2 central questions that formed the basis of the literature review were:
What is the prevalence of KRAS G12C in patients with CRC and what are the mutations that have been reported to co-exist with it?
What are the clinical outcomes among patients with CRC who have KRAS G12C-positive tumors?
Searches were carried out in Embase, MEDLINE, and the Cochrane Library databases. A detailed list of the literature sources is provided in Supplementary Table S1. The search timeframe was from January 2009 to July 2021.
Titles and abstracts of identified studies were screened in a double-blind manner by 2 researchers to determine whether they met the predefined inclusion and exclusion criteria (Table 1). Uncertainties regarding the inclusion of studies were resolved by a full-text review carried out in a single-blind manner by the first reviewer, and 10% of these decisions were spot-checked by a second reviewer. All decisions on inclusion and exclusion at title/abstract screening and full-text review, including the reasons for these decisions, were documented. Data were extracted from the identified publications into a data extraction table by the 1st reviewer and independently checked for errors against the original publication by a 2nd reviewer. Discrepancies were resolved through discussion or with the intervention of a 3rd reviewer and the data were qualitatively synthesized. The methodological quality of each individual study was assessed using the Cochrane quality assessment tools. The Risk of Bias (RoB) 2 tool was used to assess the RoB in randomized controlled trials (RCTs), and the Risk of Bias in Non-Randomized Studies—of Interventions (ROBINS-I) tool was used to assess RoB in single-arm trials and observational studies.21,22 Risk of bias assessments was carried out in a single-blind manner by 1 researcher.
Table 1.
PICO framework used for study selection.
| Criteria | Inclusion criteria | Exclusion criteria |
|---|---|---|
| Population | Patients with colorectal cancer (any stage, any line of treatment) carrying a KRAS G12C mutation | Tumor types other than colorectal cancer |
| Intervention | Any anticancer drugs, any line of treatment, or no treatment | Radiotherapy or surgery (unless a relevant comparator arm) |
| Comparator | Any or none | Not applicable |
| Outcome | Outcome reported by KRAS G12C mutation status Epidemiological evidence • Prevalence of KRAS G12C mutation • KRAS mutation and subtypes Clinical evidence • Overall survival • Progression-free survival • Disease-free survival • Adverse events • Objective response rate • Time to response • Duration of response |
Not applicable |
| Study design | Any randomized controlled trial, single-arm trial, observational study | Exclude animal/in vitro studies, case studies, and case reports |
| Date restrictions | • Published since 2009 • Congress abstracts published since 2016 |
Published before 2009 |
| Language restrictions | English language | Non-English language |
| Publication type | All publication types, except editorials and reviews, but including systematic reviews | Editorials and reviews |
| Country | Not restricted | Not applicable |
Abbreviations: KRAS, Kirsten rat sarcoma virus gene; KRAS G12C, KRAS with mutation at codon 12 that results in the substitution of glycine with cysteine; PICO, Population, Intervention, Comparator, Outcome.
The search was carried out in 2 phases: The searches for the 1st SLR (phase I) were performed on July 24, 2019, and covered the years from 2009 to 2019. Searches for phase II of the SLR, which was an update of phase I SLR, were conducted on March 10, 2021, and covered the years from 2019 to 2021. The 2 phases of the search were performed simultaneously for both NSCLC and CRC. The search strings that were used in the different databases are shown in Supplementary Table S2.
Selection Criteria
Studies describing treated (any stage, any anticancer drug or line of treatment with the exception of radiotherapy, or surgery unless a relevant comparator arm was included) or untreated patients with CRC (any stage) and harboring the KRAS G12C mutation were included. Non-English publications, editorials, and reviews were excluded. Articles published between January 2009 and June 2021 and conference abstracts published between January 2016 and June 2021 were included for title and abstract review. RCTs, single-arm trials, and observational studies published since 2009 that met the SLR inclusion criteria were included. The PICO framework and the detailed selection criteria are described in Table 1. Data from publications reporting the incidence and prevalence of KRAS G12C were extracted only when the total study population comprised ≥100 patients, or when the primary aim of the study was to examine the prevalence of KRASm. Data from all publications reporting clinical outcomes for the KRAS G12C were extracted. SLR articles were identified and listed separately; however, data were not extracted from these papers. The reference lists from SLRs were cross-checked against the lists of included references to ensure that no relevant references were omitted.
Studies reporting clinical outcome data, including overall survival (OS), progression-free survival (PFS), objective response rate (ORR), disease-free survival (DFS), relapse-free survival (RFS), and duration of response, were extracted. Epidemiological data extracted included prevalence of KRAS G12C, other KRASm, and co-mutations.
Results
The 2 phases of the search, which were performed simultaneously for both NSCLC and CRC, returned 7549 hits. After removing duplicates and filtering by the pre-specified exclusion criteria, 2038 publications were considered for full manuscript review. Following a full-text review, 185 CRC publications were identified for further review; from these, 48 publications reporting on <100 patients were excluded. Finally, 137 publications were selected for data extraction; of these, 8 were interventional (6 single-arm trials, 2 RCTs), 6 were post hoc analyses, 122 were observational, and 1 was longitudinal. In total, 90 publications reported only epidemiological data, 37 reported epidemiological and clinical outcomes data, and 10 publications reported only clinical outcomes data. Flow diagrams summarizing the study selection process for phases I and II of the SLR are shown in Fig. 1A, 1B, respectively.
Figure 1.
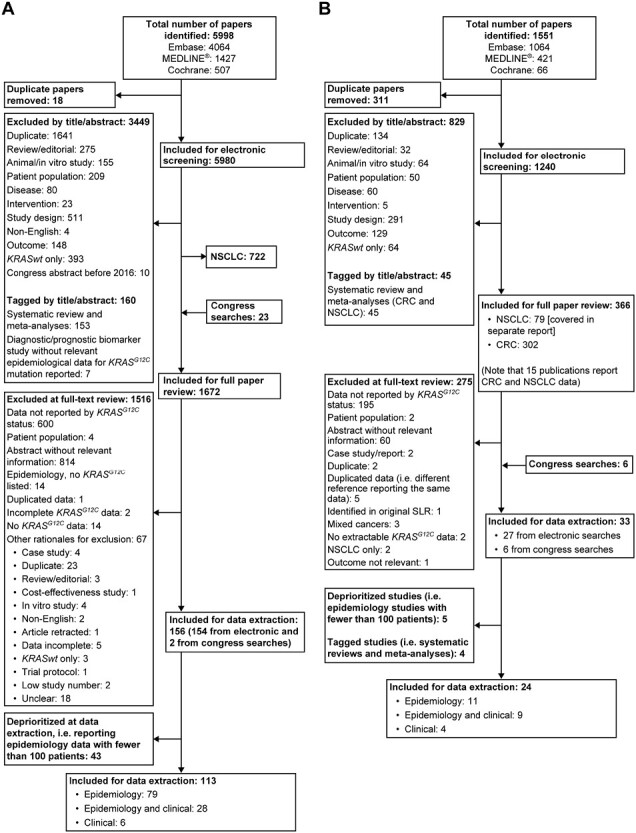
(A) Flow diagram depicting the screening and selection process for study selection in phase I (2009-2019) of the systematic literature search. (B) Flow diagram depicting the screening and selection process for study selection in phase II (2019-2021) of the systematic literature search. Abbreviations: CRC, colorectal cancer; KRASwt, wild-type Kirsten rat sarcoma virus gene; NSCLC, non-small cell lung cancer; SLR, systematic literature review.
Geographical Prevalence of KRASm
Sixty-eight studies that reported on unselected patient populations (participants not chosen for having any particular characteristic other than being adults with CRC) were included. The studies originated from the following countries: China (13 studies); US (8 studies); Italy (6 studies); Japan (4 studies); Iran (4 studies); Australia, South Korea, Spain, Taiwan (3 studies each); Belgium, Brazil, France, India, Mexico, Saudi Arabia (2 studies each); Austria, Denmark, Pakistan, Peru, Russia, Tunisia, Turkey, and the United Kingdom (1 study each). One study reported on mutations from patients from different countries. The country-wise prevalence of KRAS G12C is shown in detail in Supplementary Table S3. Of these, 11 studies had sample sizes > 1000 and 8 had sample sizes 500-1000 (Table 2). Collectively, these 68 studies reported the frequency of KRASm in 42 810 patients.
Table 2.
List of publications reporting the geographical prevalence of the KRAS mutation in >500 patients with colorectal cancer.
| Study | Country | Patients testeda for KRASm (N) | Study type | Patients with the KRAS G12C mutation (n) |
KRAS G12C mutation prevalence within testeda population, % (n/N) |
Median KRAS G12C prevalence, % (n/N) | Patients with KRASm (N1) | KRAS G12C mutation prevalence within KRASm % (n/N1) | Median KRAS G12C prevalence/region % (n/N1) | KRASm prevalence % (N1/N) | Median KRASm prevalence/region KRASm % (N1/N) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| European Union | |||||||||||
| Gvaldin et al (2019)23 | Russia | 744 | Obs/NR | 19 | 2.6 | 3.5 (2.6-5.0) |
238 | 8.0 | 8.9 (8.0-14.0) |
32.0 | 36.5 (32.0-39.7) |
| Palomba et al (2016)24 | Italy | 1284 | Obs/retrospective | 64 | 5 | 457 | 14.0 | 35.6 | |||
| Malapelle et al (2012)25 | Italy | 1691 | Obs/retrospective | 64 | 3.8 | 671 | 9.5 | 39.7 | |||
| Marchetti et al (2011)26 | Italy | 2519 | Obs/prospective | 78 | 3.1 | 940 | 8.3 | 37.3 | |||
| Intercontinental region | |||||||||||
| Gil Ferreira at. (2014)27 | Brazil | 7797 | Obs/retrospective | 206 | 2.6 | 2.6 | 2623 | 7.9 | 7.9 | 33.6 | 33.6 |
| Japan and Asia-Pacific region | |||||||||||
| Price et al (2020)28 | Australia | 1605 | Obs/retrospective | 63 | 3.9 | 2.5 (1.5 to 4.3) |
658 | 9.6 | 5.9 (3.6 to 9.6) |
41.0 | 42.1 (35.9 to 51.5) |
| Wong et al (2020)29 | Australia | 1308 | Obs/prospective | 56 | 4.3 | 674 | 8.3 | 51.5 | |||
| Li et al (2019b)30 | China | 526 | Obs/retrospective | 9 | 1.7 | 253 | 3.6 | 48.1 | |||
| Li et al (2019a)31 | China | 1164 | Obs/retrospective | 29 | 2.5 | 490 | 5.9 | 42.1 | |||
| Loong et al (2020)32 | China | 1114 | Obs/retrospective | 28 | 2.5 | 545 | 5.1 | 48.9 | |||
| Luo et al 202033. | China | 655 | Obs/retrospective | 13 | 2.0 | 305 | 4.3 | 46.6 | |||
| Shen et al, 201334 | China | 674 | Obs/retrospective | 10 | 1.5 | 242 | 4.1 | 35.9 | |||
| Fu et al, 201935 | China | 5495 | Obs/retrospective | 127 | 2.3 | 2070 | 6.1 | 37.7 | |||
| Won et al, 201736 | South Korea | 1092 | Obs/retrospective | 31 | 2.8 | 401 | 7.7 | 36.7 | |||
| North America | |||||||||||
| Nash et al (2010)37 | USA | 531 | Obs/retrospective | 15 | 2.8 | 4.1 (2.8 to 4.2) |
190 | 7.9 | 10.0 (7.9 to 10.4) |
35.8 | 39.0 (35.8 to 39.7) |
| Paulino A (2014)38 | US | 577 | Obs/retrospective | 23 | 4.0 | 229 | 10 | 39.7 | |||
| Maus et al (2014)39 | US | 838 | Obs/retrospective | 34 | 4.1 | 327 | 10.4 | 39.0 | |||
| Nusrat et al (202040 | US | 3469 | Obs/retrospective | 146 | 4.2 | NR | NR | NR | |||
| Greater than 1 geographical region | |||||||||||
| Smith et al (2013b)41 | > 1 | 599 | Obs/retrospective | 17 | 2.8 | 2.8 | 258 | 6.6 | 6.6 | 43.1 | 43.1 |
aPatients with an evaluable test result.
Abbreviations: KRAS, Kirsten rat sarcoma virus gene; KRASm, KRAS mutation(s); KRAS G12C, KRAS with mutation at codon 12 that results in the substitution of glycine with cysteine; NR, not reported; Obs, observational; UK, United Kingdom; US, United States of America.
The global median prevalence of KRAS G12C in patients with CRC was 3.1% (range, 0.7% to 14.0%) (Fig. 2A). The reported median prevalence was 3.8% (range, 1.3% to 7.8%) in Europe, 2.9% in the intercontinental region (range, 1.0% to 11.3%) (Middle East: 2.3% [range, 1.0% to 3.6%]; South America: 3.4% [range, 1.1% to 11.3%]), and 4.0% in North America (range, 0.8% to 9.2%). The median of reported prevalence estimates in Japan and the Asia-Pacific region (JAPAC) was 2.5% (range, 0.7% to 14.0%). When a study describing a large outlying prevalence of 14.0% in Indian patients with CRC42 was removed from the analysis, the median prevalence was 2.3% (0.7% to 6.9%).
Figure 2.
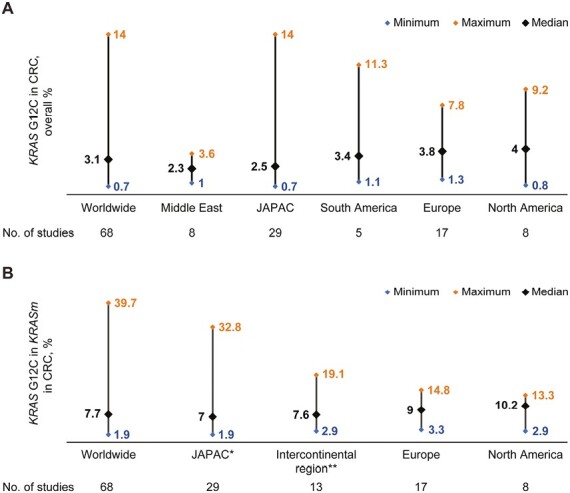
(A) Worldwide prevalence of KRAS G12C mutations in the entire cohort of included populations with colorectal cancer. (B) Worldwide prevalence of KRAS G12C mutations within the KRASm population in patients with colorectal cancer. *JAPAC, Japan & Asia-Pacific regions; **Intercontinental region: South America, Middle East. Abbreviation: KRASm, Kirsten rat sarcoma virus gene mutations.
The reported median prevalence of KRASm in CRC in the included studies was 38.0% (range, 13.3% to 58.9%), with the median prevalence on a regional level ranging from 38.0% (range, 17.0% to 55.2%) in Europe to 40.7% (range, 13.3% to 51.5%) in JAPAC. The median prevalence of KRASm in CRC in studies from the intercontinental region was 38.2% (range, 16.7% to 58.9%) (Middle East: 37.3% [range, 26.0% to 50.9%]; South America: 41.4% [range, 16.7% to 58.9%]), and 37.4% in North America (range, 23.1% to 50.0%).
Among patients with KRASm, the median global prevalence of KRAS G12C in patients with CRC was 7.7% (range, 1.9% to 39.7%) (Fig. 2B). The median regional prevalence was 9.0% in Europe (range, 3.3% to 14.8%), 10.0% in North America (range, 2.9% to 13.3%), 7.0% in JAPAC (range, 1.9% to 32.8%), although this was reduced to 6.5% (range, 1.9% to 21.2%) when a study with a large outlier prevalence42 was excluded, and 7.6% in the intercontinental region (range, 2.9% to 19.1%) (Middle East: 5.1% [range, 2.9% to 8.6%]; South America: 7.9% [range, 6.7% to 19.1%])
Tumor sidedness, KRAS mutations, and outcomes
Five studies that reported on tumor sidedness noted that tumors on the right side of the colon were more likely to have KRASm than those that developed on the left side of the colon.43-47 Three of the five studies reported KRASm prevalence in the rectum separately,45-47 even though the rectum is anatomically considered a part of the left colon.48 Two studies did not specify which parts of the colon were included for assessing tumor sidedness.43,44
Multivariate analysis from one study reported that survival was significantly lower in patients with right-sided tumors, however, the association between the presence of KRASm and tumor location was not studied.49 Multivariate analyses from another study reported that patients with tumors on the left side had a lower risk of death compared with those with tumors on the right side (hazard ratio [HR], 0.699; 95% CI: 0.350, 1.398; P = .312) although the risk of death between the groups was more similar if the left-sided tumors were positive for KRAS G12C (HR: 0.812; 95% CI: 0.131, 5.053; P = .823).50 However, none of these findings was statistically significant.
Clinical outcomes with KRAS mutations
In total, 31 publications reporting clinical outcomes with KRAS G12C were included and comprised 6 single-arm studies (from 4 clinical trials), 4 post hoc analyses from RCTs, and 21 observational studies. No publication reported on time to response, depth of response, or treatment discontinuation in patients with KRAS G12C.
OS and PFS
Seventeen publications reported OS (n = 16) and/or PFS (n = 9) outcomes in patients with KRASm-positive CRC (Table 3). Most studies (11 of 17 studies) reported shorter OS and/or PFS for patients with KRAS G12C-positive tumors than for those with tumors harboring KRASwt or other KRASm. Of the remaining 6 studies, 2 reported outcomes in individual patients, 2 reported outcomes only for patients with KRAS G12C-positive tumors in studies that did not have a comparator arm. Two studies reported a higher OS rate in KRAS G12C-positive patients compared with those with KRASwt or other KRASm.
Table 3.
List of Publications Reporting Overall Survival and Progression-free Survival Data in Patients With KRAS G12C -positive Colorectal Cancers.
| Study | N | Patients with the KRAS G12C mutation (n) | Metastatic | Study type | Treatment LoT |
OS KRASwt/KRASm mOS, months (95% CI) |
OS KRAS G12C mOS, months (95% CI) |
PFS KRASwt/ KRASm mPFS, months (95% CI) |
PFS KRAS G12C mPFS, months (95% CI) |
|---|---|---|---|---|---|---|---|---|---|
| Wong et al, 202029 | 1,308 | 56 | Yes (63%) | Obs | None Prior LoT: 1, 2, or ≥ 3 |
NR | KRAS G12C, 31.7 | NR | NR |
| Modest et al, 201651 | 1,239 | 28 | Yes | Posthoc from RCT | FUFIRI, mIROX FOLFIRI + BEV CAP + OXA + BEV; CAP + IRI + BEV; FP + BEV, BEV; XELOX/FUFOX LoT: 1L |
b
KRASwt, 26.9 (25.2, 28.5) KRASm 21.0 (18.5, 23.5) |
KRAS G12C, 16.8 (15.6, 18.0) |
b
KRASwt, 10.3 (9.7, 10.8) KRASm 9.5 (8.9, 10.1) |
KRAS G12C, 10.1 (6.4, 13.8) |
| De Roock et al, 201052a | 886 | 24 | Yes | Obs |
LoT: ≥ 2
CET + CHT Prior LoT 1: n = 84 (12.9%); 2: n = 320 (49.3%); 3: n = 156 (24.0%); 4: n = 60 (9.2%); ≥ 5: n = 25 (3.9%); unknown: n = 4 (0.6%) |
Reported for each patient individually | Reported for each patient individually | Reported for each patient individually | Reported for each patient individually |
| Schirripa et al, 202053 | 839 | 145 Study cohort |
Yes | Obs | None | Other KRASm, 36.7 (32.2, 41.5) | KRAS G12C, 28.9 (24.0, 35.2) | NR | NR |
| 329 | 57 External validation cohort |
NR | Other KRASm, 35.8 (31.0, 42.8) | KRAS G12C, 25.9 (17.2, 37.3) | NR | NR | |||
| Ottaiano et al, 202054 | 446 | 13 | Yes | Obs | None Prior LoT: 1 or ≥ 2 OS: not defined |
OS not reported for KRASm but for individual codon 12 mutations | KRAS G12C, C 7.3 (1.6, 12.6) | NR | NR |
| Jones et al, 201710 | 392 | 15 | Yes | Obs | Curative surgery and/or FOLFOX as 1 LoTc |
b
KRASwt 35.1 KRASm 25.8 (P = .006) |
KRAS G12C, 24.9; (P < .02 vs KRASwt) | NR | NR |
| Fiala et al, 201655 | 358 | 9 | Yes | Obs | BEV in combination with FOLFOX, FOLFIRI, 5-FU/FA, XELOX, XELIRI or CAP, OXA or IRI alone LoT: 1 or ≥ 2 |
b
KRASwt, 29.2 (26.3, 32.1) KRASm, 22.8 (P = .003) |
KRAS G12C, 27.4 (10.0, 44.8) |
b
KRASwt, 10.8 (9.2, 12.3) KRASm, 9.2 (P = .30) |
KRAS G12C 10.6 (6.8, 14.3) |
| George et al, 202056 | 273 | 25 | Yes | Obs | None | All, 26.4 KRASm, 25.8 KRASm (non- KRAS G12C), 27.1 |
KRAS G12C, 23 (P < .001; vs other non- KRAS G12C KRASm) |
NR | NR |
| Garrido-Laguna et al, 201245 | 238 | 11 | Yes | Obs | CET or PAN, n = 122 PI3K/Akt/mTOR inhibitors, n = 80 Prior therapies: adjuvant CHT, anti-EGFR |
b
KRASm, 57.5 (50.0, 64.8) KRASwt, 89.5 (63.5, 120.1) (P = .007) |
KRAS G12C, 50.0 (14.3, 59.3) |
Patients treated with anti-EGFR
b
KRASm, 15 weeks KRASwt, 22 weeks (P = .01) |
NR |
| Dadduzio et al, 201657 | 218 | 11 | Yes | Obs | FOLFOX/XELOX ± IRI FOLFOX/XELOX ± IRI± BEV Prior lines of systemic therapies: 0 to ≥ 2 |
b
KRASm (codon 12) 32.0 (26.3, 37.7) KRASm (codon 13) 31.0 (24.3, 37.8) |
KRAS G12C, 24.4 (10.6, 38.2) |
KRASm,
(codon 12) 1L: 10.8 2L: 5.9 3L: 4.6b |
NR |
| Ucar et al, 202058 | 191 | 7.4% in multiple mutation group | Yes | Obs |
LoT: 1 mFOLFOX6 or FOLFIRI |
b
Single KRASm, 22.7 Multiple KRASm, 40.7 (P = .01) |
KRAS G12C, 28.4 | Single KRASm, 8.8 Multiple KRASm, 12.8 (P = .05) |
NR |
| Renaud et al, 201559 | 180 | 9 | Yes | Obs | Thoracic metastasectomy Neoadjuvant and adjuvant therapy based on 5-FU |
b
KRASwt,
98.00 (74.21, 121.78) KRASm, 55.00 (28.69, 81.31) |
KRAS G12C, 37 (15.09,58.91) | NR | NR |
| Modest et al, 201260a | 119 | 13 | Yes | Posthoc analyses from RCT | CET + XELIRI CET + FOLFIRI CET + FOLFOX6 LoT: 1L (CET-based) |
OS not reported for KRASm collectively, only for individual codon 12 mutations |
KRAS G12C, 14.3 (6.7, 21.9) |
PFS not reported for KRASm collectively, only for individual codon 12 mutations |
KRAS G12C, 4.9 (3.7, 6.2) |
| Cushman-Vokoun et al, 201361 | 111 | 4 | Yes (Stage IV 25%) | Obs | None | 2-year OS ratea KRASwt, 84.5% (75.2, 95.0) |
2-year OS rate KRAS G12C, 100% (100.0, 100.0) |
NR | NR |
| Andre et al, 201362 | 65 | 1 | Yes | Int Single arm |
PAN + IRI in patients refractory to CHT LoT: 2 or 3 |
b
KRASwt, 11.9 (6.8, 18.2) |
KRAS G12C, 4.5 (1 patient) |
KRASwt 6.3 (3.7, 8.7) |
KRAS G12C, 3.1 (1 patient) |
| Hong et al, 202063 | 42 | 42 | Yes, or locally advanced | Int Single arm |
Sotorasib Prior LoT: ≥ 2 (98%) |
NR | NR | NR | KRAS G12C, 4.0 (0, 11.1) |
| Cercek et al, 201464 | 17 | 1 | Yes | Int Single arm |
Ganetespib LoT: ≥ 3 |
KRASwt (n = 6): median OS not calculated; individual patient OS: 26, 15, 16, 113, 11, 63 weeks |
KRAS G12C, 22 weeks (1 patient) |
KRASwt, (n = 6): median PFS not calculated, individual patient PFS: 5, 6, 4, 9, 2, 15 weeks |
KRAS G12C, 12 weeks (1 patient) |
aStudies included prior or concomitant treatment with anti-EGFR therapies combined with oxaliplatin, which have been associated with either no benefit or worse outcomes in KRASm patients with CRC (62).
bOther KRASm subtypes and/or mutations reported.
cUnclear whether chemotherapy was used as neoadjuvant/adjuvant therapy.
Abbreviations: BEV: bevacizumab; CAP: capecitabine; CET, cetuximab; CHT: chemotherapy; EGFR: epidermal growth factor receptor; FA: folinic acid; FOLFIRI: IRI + 5-FU/FA; FOLFOX: OXA + 5-FU/FA; FP: fluoropyrimidine; 5-FU: 5-fluorouracil; FUFIRI: infusional 5-FU; FUFOX: OXA + 5-FU (bolus)/FA; Int: interventional; IRI: irinotecan; KRAS: Kirsten rat sarcoma viral oncogene homolog; KRASm: KRAS mutation(s); KRASG12C: KRAS with mutation at codon 12 that results in the substitution of glycine with cysteine; KRASwt: wild-type KRAS; 1L, 2L, 3L: first, second, third LoT; LoT: line of therapy; mFOLFOX: modified FOLFOX; mIROX: IRI + OXA; mOS: median overall survival; mPFS: median progression-free survival; mTOR: mammalian target of rapamycin; NR: not reported; Obs: observational; OS: overall survival; OXA: oxaliplatin; PAN: panitumumab; PFS: progression-free survival; PI3K: phosphatidylinositol-3-kinase; RCT: randomized controlled trial; XELIRI: CAP + IRI; XELOX : CAP + OXA.
A study of patients with stage IV mCRC reported a significantly shorter median OS in patients with KRAS G12C (28.9 months [range, 24.0, 35.2]) than in those with other KRASm (36.7 months [range, 32.2, 41.5]).53 Similar results were observed in an external validation cohort (patients with KRASm-positive CRC who were treated in a medical oncology unit of another hospital in the same timeframe as this study) that was analyzed as part of this study (median OS: KRAS G12C 25.9 months [range, 17.2, 37.3] vs other KRASm 35.8 months [range, 31.0, 42.8]). A retrospective review of genomic profiling data from patients with mCRC reported significantly shorter OS for patients with KRAS G12C versus patients with other KRASm (23 vs 27.1 months, P < .001).56
In a post hoc analysis of data from 3 clinical trials of 119 patients with mCRC who were treated with cetuximab-based first-line regimens, patients with KRAS G12C tumors had the shortest survival times compared with patients with other analyzed KRASm (KRASG12D/V/A/S/R).60 The median OS for patients with KRAS G12C was 14.3 (95% CI: 6.7, 21.9) months and the median PFS was 4.9 (95% CI: 3.7, 6.2) months. For patients with other KRASm, the median OS ranged from 15.2 months (KRAS G12S) to 23.3 months (KRAS G12D) months and the median PFS from 5.3 months (KRAS G12R) to 9.8 months (KRAS G12A).
A study of patients with mCRC who were treated with standard chemotherapy, with or without an anti-angiogenic agent, reported a significantly worse OS for patients with KRAS G12C (7.3 months) and KRAS G12S (5.0 months) compared with patients harboring other KRASm who exhibited a median survival time ranging from 11.6 to 27.0 months.54
A post hoc analysis of pooled clinical data from 1239 patients from 5 randomized trials, in which patients with mCRC were treated with chemotherapy with or without bevacizumab as first-line treatment (67%), reported that patients harboring KRAS G12C had the shortest median OS (16.8 [95% CI: 15.6, 18.0] months).51 Patients with other KRASm, including KRAS G12A/D/S/V and KRAS G13D, had OS times ranging from 17.6 to 25.2 months.51 The median PFS for patients with KRAS G12C was 10.1 (95% CI: 6.4, 13.8) months and ranged from 8.8 to 10.5 months in patients with other KRASm. In an observational, retrospective study in Italian patients with stage IV mCRC treated with chemotherapy (most [65%] of whom had also received bevacizumab as first-line treatment), the median OS for patients with KRAS G12C was 24.4 months (95% CI: 10.6, 38.2) and ranged from 5.7 to 39.1 months among patients with other KRASm.57
In a retrospective analysis of data from 404 patients with mCRC who were treated with bevacizumab (80% received bevacizumab as first-line treatment), the median OS in patients with KRAS G12C was shorter (27.4 [95% CI: 10.0, 44.8] months) than in patients with KRAS G12D (28.7 months) and KRAS G12S (32.7 months), but longer than in those with KRAS G12A, KRAS G12V, and KRAS G13D (range, 16.1 to 22.8 months).55 The median PFS was 10.6 (95% CI: 6.8, 14.3) months in patients with KRAS G12C, while the median PFS ranged from 3.5 to 15.1 months with other KRASm. In this study, the PFS and OS of patients with KRAS G12C tumors were comparable to those with other KRASm tumors (with the exception of KRAS G12V and KRAS G12A tumors which were associated with significantly shorter survival outcomes) and to those with KRASwt tumors.
In contrast to these reports, 2 studies noted that the clinical outcomes in patients with KRAS G12C were either similar or superior to the outcomes observed in patients with other KRASm. A retrospective review from Turkey reported no difference in the OS for patients with KRAS G12C and patients with other KRASm.58 In another retrospective study, in which KRAS was identified through allele-specific polymerase chain reaction on paraffin-embedded tumor specimens, patients with KRAS G12C and KRAS G12S had the highest 2-year OS rates (both 100%); the OS rates were lower in those with other KRASm (OS range, 34% to 87%) and KRASwt (OS, 85%).61
The small sample sizes in each study, the difference in treatment protocols, and number of prior lines of therapy made a direct comparison of these outcomes difficult.
Objective Response Rate
At the time of the conduct of the SLR, 2 publications had reported the ORR in patients with KRAS G12C-positive mCRC and other gastrointestinal cancers and treated with sotorasib.63,65 A phase I trial of sotorasib in 42 patients with CRC reported an overall ORR of 7.1% (95% CI: 1.5, 19.5) across all doses tested.63 An ORR of 12.0% was reported for patients with CRC and other GI cancers treated with 960 mg/day of oral sotorasib.65 However, this study lacked a comparator arm, precluding a comparison of outcomes.
In a post hoc analysis of outcomes in patients who received cetuximab in combination with FOLFOX, FOLFIRI, or capecitabine/oxaliplatin/irinotecan (XELOXIRI) as first-line treatment for mCRC, patients with KRAS G12C tumors had a worse ORR (17.0%) compared with patients with KRAS G12D (46.0%), KRAS G12V (44.0%), and KRAS G12A (42.0%) tumors, but a better ORR than patients with KRAS G12S (13.0%) and KRAS G12R (0%) tumors.60
Disease-Free Survival and Relapse-Free Survival
Three studies reported DFS outcomes. Overall, patients with KRAS G12C tumors appeared to have inferior DFS rates than those with KRASwt tumors. Among patients with stage III adenocarcinoma of the colon who received adjuvant FOLFOX monotherapy or combination therapy with cetuximab, the combined 3-year DFS rate in both arms for patients with KRAS G12C tumors was 61.0% (95% CI: 50.0%, 73.0%) with an HR of 1.66 (95% CI: 1.14, 2.41; P = .008) compared with a 3-year DFS rate of 77% (95% CI: 75%, 80%) in patients with KRASwt tumors.66 Among patients with stage II CRC, a significantly shorter DFS was noted in patients with KRAS G12C than in those with KRASwt (HR 2.03: P = .006).43 However, it should be noted that data from only 6 patients with KRAS G12C tumors were available for this analysis. In another study of patients treated with cetuximab, an odds ratio of 0.95 (95% CI: 0.18, 5.15; P = .95) for DFS was reported for patients with KRAS G12C; however, the comparator was not clearly defined.67
Two studies reported RFS in patients with KRASm. An analysis of outcomes in Japanese patients who had undergone curative surgical resection for stage I-III CRC revealed a significantly worse 3-year RFS rate in patients with KRAS G12C than in patients with KRASwt (33.3% vs. 81.9%; HR 6.57 [95% CI: 1.90, 17.7; P < .001]).68 In a post hoc analysis of 1404 patients with stage II or stage III colon cancer treated with irinotecan added to fluorouracil/leucovorin as adjuvant, neither KRAS G12C or any other KRASm had a prognostic value for RFS.44
Discussion
In this SLR, studies describing the global prevalence of the KRASm in patients with CRC, mutation prevalence by primary tumor sidedness, and the clinical significance of KRASm were identified and described. A worldwide median prevalence of 38.0% for KRASm was estimated based on the included studies. The observed prevalence of KRASm varied widely across studies, although the median prevalence in regions with the lowest prevalence (North America, 37.4%; range, 23.1% to 50.0%) and the highest prevalence (JAPAC, 40.7%; range, 13.3% to 51.5%) were not widely different and had overlapping ranges. The majority of prevalence studies for which the RoB could be estimated (46/51, 90%) had a low RoB. While it is possible that these percentages reflect actual differences in the prevalence of KRASm, the contribution of heterogeneity in study design and size in causing these variations cannot be discounted, even though this analysis was limited to unselected studies with > 100 patients and to studies that specifically analyzed the prevalence of this mutation in the population. The prevalence of KRASm noted in this SLR is comparable to that reported in a recent analysis of real-world data from 6477 adult patients with mCRC (3.7% for KRAS G12C and 45.5% for other KRASm),69 as well as to that reported in other published databases, such as the AACR Project GENIE, which currently reports a KRAS G12C prevalence of 2.9% in 12 187 cases of colorectal adenocarcinoma.70 The median prevalence of KRAS G12C as reported in the entire cohorts of the included populations in this study was 3.1%. The prevalence of KRAS G12C in CRC was much lower than that in NSCLC, where a prevalence of 14.0% has been reported.71
Limited evidence was available to assess any link between the prevalence of KRAS G12C and primary tumor sidedness. The available literature indicated that tumors that develop on the right side of the colon were more likely to harbor KRASm. Published evidence indicates that tumors that develop on the right side of the colon respond poorly to anti-EGFR therapies compared with left-sided colorectal tumors.72,73 Indeed, the National Comprehensive Cancer Network (NCCN) guidelines recommend treatment with anti-EGFR therapies (eg, cetuximab and panitumumab) only for patients with left-sided tumors. The increased frequency of CpG island methylation in the EGFR promoter on right-sided primary tumors leading to loss of EGFR expression (and the consequent lack of response to anti-EGFR therapies) and the hypermutations observed in right-sided colorectal carcinomas may provide a possible explanation for this phenomenon.74,75 It will be important to assess if patients with right-sided tumors, who are known to experience poorer outcomes and also have a limited range of therapeutic options, can benefit from currently available KRAS-targeting therapies.
When clinical outcome data were analyzed with respect to KRASm, most studies reported inferior survival and prognostic outcomes for patients with KRAS G12C. However, these studies could not be compared directly because of considerable heterogeneity in study populations, treatments, and the number of prior lines of therapy. It should also be noted that only a few studies reported prognostic data in patients with KRAS G12C and that these findings were inconsistent.
Studies published since the completion of this SLR continue to provide contradictory evidence regarding the prognostic significance of KRAS G12C. Outcome analysis from a population-based registry of Australian patients with mCRC and a real-life and population-based cohort of Nordic patients with mCRC or locally advanced untreatable CRC did not reveal any difference between patients with KRAS G12C and those with other KRASm.76,77 In contrast, 4 recent studies from Japan, US, and China reported poorer survival rates and significantly inferior PFS in patients with KRAS G12C than in those with other KRASm.69,78-80 These conflicting findings have been attributed to the difference in average patient age, disease stage, and data sources (selected patient populations vs population-based/real-life registries).76 Given these findings, a deeper investigation into the biological and mechanistic role of KRAS G12C (vis-à-vis other KRASm) may possibly provide better insights into the clinical significance of this mutation.
Limited evidence was available regarding prognostic outcomes with KRAS-targeting therapies. However, given the number of targeted therapies that are currently in the developmental pipeline for CRC, it is expected that there will be a considerable increase in our knowledge base regarding clinical outcomes with KRAS G12C-positive tumors in the near future. For example, results from a recent phase I/II study of adagrasib, revealed an ORR of 19% in patients receiving adagrasib monotherapy for untreatable or metastatic tumors with KRAS G12C mutations.81 The phase II CodeBreaK 100 study, the results of which were published after the conduct of this SLR, reported an ORR of 9.7% among patients who received sotorasib monotherapy for KRAS G12C-mutated mCRC and had progressed following fluoropyrimidine, oxaliplatin, and irinotecan treatment.82 Recent studies have shown that treatment approaches that combine KRAS inhibitors with anti-EFGR antibodies yield considerably enhanced ORRs in patients with refractory mCRC.83,84 These findings complement studies in preclinical models which had indicated that EGFR blockade could potentially overcome the EGFR-driven adaptive resistance to KRAS inhibitors—a phenomenon which is more pronounced in CRC in comparison to NSCLC.85
A major strength of this study was the inclusion of a large number of studies which enabled a robust estimate of the prevalence of KRASm in > 40 000 patients from different geographical regions. This information can be used to support future testing and treatment strategies. The limitations of this SLR include the analysis of data from studies that used a wide variety of techniques to estimate the prevalence of KRASm. This may have contributed to the outlier prevalence observed in studies from certain geographical regions. The use of standardized molecular methods to identify KRASm may help clarify some of these discrepancies. Another limitation was the availability of relatively few studies that investigated the role of KRASm with respect to tumor sidedness or studies that compared response rates and duration of response in patients with different KRASm. KRASm have not been routinely assessed in clinical trials or in real-world practice as they were considered “undruggable” until recently. Consequently, there is a limited amount of published data describing outcomes in patients with KRAS G12C mutations. This literature review was, therefore, not designed to definitively answer the proposed research questions but rather to summarize existing data and provide context for the interpretation of the multiple sources of existing data derived from next-generation sequencing.
Conclusions
To summarize, the prevalence of KRAS G12C in CRC varied widely across studies. Limited evidence was available to assess any association between the KRAS G12C mutation and tumor sidedness. The prognostic significance of KRASm could not be definitively assessed as the studies were highly variable in terms of the patient population and interventional therapies used; however, overall, the reviewed evidence suggests shorter OS, PFS, and DFS in patients with KRAS G12C than in those with patients with KRASwt and other KRASm. Well-designed studies with clearly delineated research questions are needed to assess the differences in clinical outcomes in patients with KRASm and KRASwt and to investigate the mechanistic link between the KRAS G12C mutation and tumor sidedness.
Supplementary Material
Acknowledgment
Medical writing support at the manuscript drafting stage was provided by Sukanya Raghuraman, PhD, of Cactus Life Sciences (part of Cactus Communications) and funded by Amgen Inc.
Contributor Information
John H Strickler, Department of Medicine, Duke University Medical Center, Durham, NC, USA.
Takayuki Yoshino, Department of Gastroenterology and Gastrointestinal Oncology, National Cancer Center Hospital East, Kashiwa, Japan.
Kendall Stevinson, Health Economics and Outcomes Researc, Amgen Inc., Thousand Oaks, CA, USA.
Christian Stefan Eichinger, Value Demonstration Practice, Oxford Pharmagenesis, Oxford, UK.
Christina Giannopoulou, Health Economics and Outcomes Research, Amgen (Europe) GmbH, Rotkreuz, Switzerland.
Marko Rehn, Global Medical Affairs, Amgen Inc., Thousand Oaks, CA, USA.
Dominik Paul Modest, Department for Hematology, Oncology and Cancer Immunology (CVK), Charité Universitätsmedizin Berlin, Berlin, Germany.
Conflict of Interest
John Strickler reports grants or contracts to research institution from AstraZeneca, Bayer, Seagen, Amgen Inc., Daiichi-Sankyo, Nektar, Abbvie, Erasca, and Gossemer Bio, AStar D3, Curegenix, Sanofi, Roche/ Genentech, and Silverback Therapeutics, consulting fees from Abbvie, Amgen Inc., AstraZeneca, Bayer, Inivata, Natera, GlaxoSmithKline(GSK), Mereo Biopharma, Pionyr Immunotherapeutics, Seagen, Pfizer, Silverback Therapeutics, and Viatris, support for attending meetings and/or travel from Seagen, and participation on a Data Safety Monitoring Board or Advisory Board for Abbvie and Pionyr Immunotherapeutics. Takayuki Yoshino, reports research funding from Ono Pharmaceutical Co., Ltd., Sanofi K.K., Daiichi Sankyo Co., Ltd., PAREXEL International Inc., Pfizer Japan Inc., Taiho Pharmaceutical Co., Ltd., MSD K.K., Amgen K.K., Genomedia Inc., Sysmex Corporation, Chugai Pharmaceutical Co., Ltd., and Nippon Boehringer Ingelheim Co., Ltd., and honoraria from Taiho Pharmaceutical Co., Ltd., Chugai Pharmaceutical Co., Ltd., Eli Lilly Japan K.K., Merck Biopharma Co., Ltd., Bayer Yakuhin, Ltd., Ono Pharmaceutical Co., Ltd., and MSD K.K. Kendall Stevinson was employed by Amgen Inc. at the time of conduct of the study and has stock ownership in Amgen Inc., is currently employed by Iovance Biotherapeutics, reports research funding from Amgen Inc. and Iovance Biotherapeutics, and travel, accommodation, and expense reimbursement from Iovance Biotherapeutics. Christian Stefan Eichinger is an employee of Oxford Pharmagenesis which received funding for the conduct of this systematic literature review. Christina Giannopoulou is an employee of Amgen (Europe) and reports stock ownership in Amgen (Europe). Marko Rehn, is an employee of Amgen Inc. and reports stock ownership in Amgen Inc.. Dominik Paul Modest reports consultancy (including expert testimony) for Amgen Inc., Servier, AstraZeneca, Merck Sharp & Dohme, Bristol-Myers Squibb, PierreFabre, Sanofi, Lilly, Merck, Cureteq, Onkowissen.de, G1, Incyte, Taiho, Takeda, and COR2ED, research funding to institution from Amgen Inc., and Servier, honoraria from Amgen, Servier, AstraZeneca, Merck Sharp & Dohme, Bristol-Myers Squibb, PierreFabre, Sanofi, Lilly, Merck, Cureteq, Onkowissen.de, G1, Incyte, Taiho, Takeda, and COR2ED.
Author Contributions
Conception/design: K.S., C.S.E., M.R. Provision of study material or patients: K.S., C.S.E., M.R. Collection and/or assembly of data: K.S., C.S.E. Data analysis and interpretation: all authors. Final approval of manuscript: all authors.
Funding
This study was funded by Amgen Inc.
Data Availability
Qualified researchers may request data from Amgen clinical studies. Complete details are available at http://www.amgen.com/datasharing
References
- 1. World Health Organization. Fact sheets: cancer. 2021. Accessed October 20, 2021.https://www.who.int/news-room/fact-sheets/detail/cancer.
- 2. Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209-249. 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 3. Cancer.Net. Colorectal Cancer: Statistics. 2021. Accessed October 05, 2021.https://www.cancer.net/cancer-types/colorectal-cancer/statistics).
- 4. Pritchard CC, Grady WM.. Colorectal cancer molecular biology moves into clinical practice. Gut. 2011;60(1):116-129. 10.1136/gut.2009.206250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Huang L, Guo Z, Wang F, Fu L., Wang F and Fu L. KRAS mutation: from undruggable to druggable in cancer. Signal Transduct Target Ther. 2021;6(1):386. 10.1038/s41392-021-00780-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Andreyev HJ, Norman AR, Cunningham D, Oates JR, Clarke PA.. Kirsten ras mutations in patients with colorectal cancer: the multicenter “RASCAL” study. J Natl Cancer Inst. 1998;90(9):675-684. 10.1093/jnci/90.9.675. [DOI] [PubMed] [Google Scholar]
- 7. Hobbs GA, Der CJ, Rossman KL.. RAS isoforms and mutations in cancer at a glance. J Cell Sci. 2016;129(7):1287-1292. 10.1242/jcs.182873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Normanno N, Tejpar S, Morgillo F, et al. Implications for KRAS status and EGFR-targeted therapies in metastatic CRC. Nat Rev Clin Oncol. 2009;6(9):519-527. 10.1038/nrclinonc.2009.111. [DOI] [PubMed] [Google Scholar]
- 9. Imamura Y, Lochhead P, Yamauchi M, et al. Analyses of clinicopathological, molecular, and prognostic associations of KRAS codon 61 and codon 146 mutations in colorectal cancer: cohort study and literature review. Mol Cancer. 2014;13(1):135. 10.1186/1476-4598-13-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jones RP, Sutton PA, Evans JP, et al. Specific mutations in KRAS codon 12 are associated with worse overall survival in patients with advanced and recurrent colorectal cancer. Br J Cancer. 2017;116(7):923-929. 10.1038/bjc.2017.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Loree JM, Kopetz S.. Recent developments in the treatment of metastatic colorectal cancer. Ther Adv Med Oncol. 2017;9(8):551-564. 10.1177/1758834017714997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hashiguchi Y, Muro K, Saito Y, et al. Japanese Society for Cancer of the Colon and Rectum (JSCCR) guidelines 2019 for the treatment of colorectal cancer. Int J Clin Oncol. 2020;25(1):1-42. 10.1007/s10147-019-01485-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Biller LH, Schrag D.. Diagnosis and treatment of metastatic colorectal cancer: a review. JAMA. 2021;325(7):669-685. 10.1001/jama.2021.0106. [DOI] [PubMed] [Google Scholar]
- 14. National Comprehensive Cancer Network. Rectal Cancer (Version 1.2022). 2022. Accessed May 16, 2022.https://www.nccn.org/professionals/physician_gls/pdf/rectal.pdf.
- 15. National Comprehensive Cancer Network. Colon Cancer (Version 1.2022). 2022. Accessed May 16, 2022. https://www.nccn.org/professionals/physician_gls/pdf/colon.pdf.
- 16. Lièvre A, Bachet J-B, Le Corre D, et al. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006;66(8):3992-3995. 10.1158/0008-5472.CAN-06-0191. [DOI] [PubMed] [Google Scholar]
- 17. Douillard J-Y, Oliner KS, Siena S, et al. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med. 2013;369(11):1023-1034. 10.1056/NEJMoa1305275. [DOI] [PubMed] [Google Scholar]
- 18. Karapetis CS, Khambata-Ford S, Jonker DJ, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359(17):1757-1765. 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 19. Liu J, Kang R, Tang D.. The KRAS-G12C inhibitor: activity and resistance. Cancer Gene Ther. 2022;29(7):875-878. 10.1038/s41417-021-00383-9. [DOI] [PubMed] [Google Scholar]
- 20. Shamseer L, Moher D, Clarke M, et al. ; PRISMA-P Group. Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015: elaboration and explanation. BMJ. 2015;350(1756-1833):g7647. 10.1136/bmj.g7647. [DOI] [PubMed] [Google Scholar]
- 21. Sterne JA, Hernán MA, Reeves BC, et al. ROBINS-I: a tool for assessing risk of bias in non-randomised studies of interventions. BMJ. 2016;355:i4919. 10.1136/bmj.i4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sterne JAC, Savović J, Page MJ, et al. RoB 2: a revised tool for assessing risk of bias in randomised trials. BMJ. 2019;366:l4898. 10.1136/bmj.l4898. [DOI] [PubMed] [Google Scholar]
- 23. Gvaldin DY, Kit OI, Omelchuk EP, et al. Frequency of somatic mutations in the KRAS gene in patients of the South Russia diagnosed with colorectal cancer. J Clin Oncol. 2019 (Abstract e15081);37(15_suppl):e15081-e15081. 10.1200/jco.2019.37.15_suppl.e15081. [DOI] [Google Scholar]
- 24. Palomba G, Doneddu V, Cossu A, et al. Prognostic impact of KRAS, NRAS, BRAF, and PIK3CA mutations in primary colorectal carcinomas: a population-based study. J. 2016;14(1479-5876):292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Malapelle U, Bellevicine C, Salatiello M, et al. Sanger sequencing in routine KRAS testing: a review of 1720 cases from a pathologist’s perspective. J Clin Pathol. 2012;65(10):940-944. 10.1136/jclinpath-2012-200773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Marchetti A, Pinto C, Taddei GL, et al. KRAS mutations in colorectal cancer patients in Italy: Results from the KRAS aKtive program. J Clin Oncol. 2011 (Abstract e14000);29(15_suppl):e14000-e14000. 10.1200/jco.2011.29.15_suppl.e14000. [DOI] [Google Scholar]
- 27. Gil Ferreira C, Aran V, Zalcberg-Renault I, et al. KRAS mutations: variable incidences in a Brazilian cohort of 8,234 metastatic colorectal cancer patients. BMC Gastroenterol. 2014;14(1471-230X):73. 10.1186/1471-230X-14-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Price TJ, Piantadosi C, Townsend AR, et al. Prognostic differences of RAS mutations: Results from South Australian (SA) metastatic colorectal (mCRC) registry. J Clin Oncol. 2020 (Abstract 4067);38(15_suppl):4067-4067. 10.1200/jco.2020.38.15_suppl.4067. [DOI] [Google Scholar]
- 29. Wong H-L, Cui W, Loft M, et al. Assessing real-world outcomes in metastatic colorectal cancer with KRASG12C mutation. J Clin Oncol. 2020 (Abstract e16072);38(15_suppl):e16072-e16072. 10.1200/jco.2020.38.15_suppl.e16072. [DOI] [Google Scholar]
- 30. Li W, Qiu T, Guo L, Ying J, Zhou A.. NGS-based oncogenic mutations analysis in advanced colorectal cancer patients improves targeted therapy prediction. Pathol Res Pract. 2019;215(3):483-489. 10.1016/j.prp.2018.12.037. [DOI] [PubMed] [Google Scholar]
- 31. Li W, Liu Y, Cai S, et al. Not all mutations of KRAS predict poor prognosis in patients with colorectal cancer. Int J Clin Exp Pathol. 2019;12(3):957-967. [PMC free article] [PubMed] [Google Scholar]
- 32. Loong H-F, Du N, Cheng C, et al. KRAS G12C mutations in Asia: a landscape analysis of 11,951 Chinese tumor samples. Transl Lung Cancer Res. 2020;9(2226-4477):1759-1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Luo Q, Chen D, Fan X, Fu X, Ma T, Chen D., Ma T and Chen D. KRAS and PIK3CA bi-mutations predict a poor prognosis in colorectal cancer patients: a single-site report. Transl Oncol. 2020;13(12):100874. 10.1016/j.tranon.2020.100874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shen Y, Wang J, Han X, et al. Effectors of epidermal growth factor receptor pathway: the genetic profiling of KRAS, BRAF, PIK3CA, NRAS mutations in colorectal cancer characteristics and personalized medicine. PLoS One. 2013;8(12):e81628. 10.1371/journal.pone.0081628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fu X, Huang Y, Fan X, et al. Demographic trends and KRAS/BRAFV600E mutations in colorectal cancer patients of South China: a single-site report. Int J Cancer. 2019;144(9):2109-2117. 10.1002/ijc.31973. [DOI] [PubMed] [Google Scholar]
- 36. Won DD, Lee JI, Lee IK, et al. The prognostic significance of KRAS and BRAF mutation status in Korean colorectal cancer patients. BMC Cancer. 2017;17(1):403. 10.1186/s12885-017-3381-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nash GM, Gimbel M, Cohen AM, et al. KRAS mutation and microsatellite instability: two genetic markers of early tumor development that influence the prognosis of colorectal cancer. Ann Surg Oncol. 2010;17(2):416-424. 10.1245/s10434-009-0713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Paulino A. KRAS mutation in colorectal carcinoma: analysis of 577 cases. Am J Clin Pathol. 2014 (Abstract 81SA);142(suppl_1):A242-A242. 10.1093/ajcp/142.suppl1.242. [DOI] [Google Scholar]
- 39. Maus MKH, Grimminger PP, Mack PC, et al. KRAS mutations in non-small-cell lung cancer and colorectal cancer: implications for EGFR-targeted therapies. Lung Cancer. 2014;83(2):163-167. 10.1016/j.lungcan.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 40. Nusrat M, Roszik J, Holla V, et al. Therapeutic vulnerabilities among KRAS G12C mutant (mut) advanced cancers based on co-alteration (co-alt) patterns. J Clin Oncol. 2020 (Abstract 3625);38(15_suppl):3625-3625. 10.1200/jco.2020.38.15_suppl.3625. [DOI] [Google Scholar]
- 41. Smith JC, Brooks L, Hoff PM, et al. KRAS mutations are associated with inferior clinical outcome in patients with metastatic colorectal cancer, but are not predictive for benefit with cediranib. Eur J Cancer. 2013;49(10):2424-2432. 10.1016/j.ejca.2013.02.023. [DOI] [PubMed] [Google Scholar]
- 42. Veldore VH, Rao MR, Prabhudesai SA, et al. Prevalence of KRAS mutations in metastatic colorectal cancer: a retrospective observational study from India. Indian J Cancer. 2014;51(4):531-537. 10.4103/0019-509X.175371. [DOI] [PubMed] [Google Scholar]
- 43. Deschoolmeester V, Boeckx C, Baay M, et al. KRAS mutation detection and prognostic potential in sporadic colorectal cancer using high-resolution melting analysis. Br J Cancer. 2010;103(10):1627-1636. 10.1038/sj.bjc.6605959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Roth AD, Tejpar S, Delorenzi M, et al. Prognostic role of KRAS and BRAF in stage II and III resected colon cancer: results of the translational study on the PETACC-3, EORTC 40993, SAKK 60-00 trial. J Clin Oncol. 2010;28(3):466-474. 10.1200/JCO.2009.23.3452. [DOI] [PubMed] [Google Scholar]
- 45. Garrido-Laguna I, Hong DS, Janku F, et al. KRASness and PIK3CAness in patients with advanced colorectal cancer: outcome after treatment with early-phase trials with targeted pathway inhibitors. PLoS One. 2012;7(5):e38033. 10.1371/journal.pone.0038033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Karagkounis G, Torbenson MS, Daniel HD, et al. Incidence and prognostic impact of KRAS and BRAF mutation in patients undergoing liver surgery for colorectal metastases. Cancer. 2013;119(23):4137-4144. 10.1002/cncr.28347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chang Y-Y, Lin J-K, Lin T-C, et al. Impact of KRAS mutation on outcome of patients with metastatic colorectal cancer. Hepatogastroenterology. 2014;61(135):1946-1953. [PubMed] [Google Scholar]
- 48. Colorectal Cancer Alliance. Colorectal Cancer Info. Biomarkers. Sidedness. Accessed September 21, 2022. https://www.ccalliance.org/colorectal-cancer-information/biomarkers/biomarkers-sidedness.
- 49. Aldiab A, Al Khayal KA, Al Obaid OA, et al. Clinicopathological features and predictive factors for colorectal cancer outcome in the Kingdom of Saudi Arabia. Oncology (Huntingt). 2017;92(2):75-86. 10.1159/000450857. [DOI] [PubMed] [Google Scholar]
- 50. Usón PLS Jr, Bugano DDG, Moura F, Carvalho RS, Maluf FC.. Worst outcomes according to RAS mutation variants: an analysis in patients with metastatic colorectal adenocarcinoma. J BUON. 2018;23(4):925-935. [PubMed] [Google Scholar]
- 51. Modest DP, Ricard I, Heinemann V, et al. Outcome according to KRAS-, NRAS- and BRAF-mutation as well as KRAS mutation variants: pooled analysis of five randomized trials in metastatic colorectal cancer by the AIO colorectal cancer study group. Ann Oncol. 2016 (Abstract 3551);27(9):1746-1753. 10.1093/annonc/mdw261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. De Roock W, Claes B, Bernasconi D, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010;11(8):753-762. 10.1016/S1470-2045(10)70130-3. [DOI] [PubMed] [Google Scholar]
- 53. Schirripa M, Nappo F, Cremolini C, et al. KRAS G12C metastatic colorectal cancer: Specific features of a new emerging target population. Clin Colorectal Cancer. 2020;19(3):219-225. 10.1016/j.clcc.2020.04.009. [DOI] [PubMed] [Google Scholar]
- 54. Ottaiano A, Normanno N, Facchini S, et al. Study of Ras mutations’ prognostic value in metastatic colorectal cancer: STORIA analysis. Cancers (Basel). 2020;12(7):1919. 10.3390/cancers12071919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Fiala O, Buchler T, Mohelnikova-Duchonova B, et al. G12V and G12A KRAS mutations are associated with poor outcome in patients with metastatic colorectal cancer treated with bevacizumab. Tumour Biol. 2016;37(5):6823-6830. 10.1007/s13277-015-4523-7. [DOI] [PubMed] [Google Scholar]
- 56. George B, Taylor BW, Lasowski M, et al. Prognostic effect of specific RAS/BRAF mutations in patients (pts) with metastatic colorectal cancer (mCRC). J Clin Oncol. 2020 (Abstract 4050);38(15_suppl):4050-4050. 10.1200/jco.2020.38.15_suppl.4050. [DOI] [Google Scholar]
- 57. Dadduzio V, Basso M, Rossi S, et al. KRAS exon 2 mutations as prognostic indicators in advanced colorectal cancer in clinical practice: a mono-institutional study. Mol Diagn Ther. 2016;20(1):65-74. 10.1007/s40291-015-0178-8. [DOI] [PubMed] [Google Scholar]
- 58. Ucar G, Ergun Y, Aktürk Esen S, et al. Prognostic and predictive value of KRAS mutation number in metastatic colorectal cancer. Medicine (Baltim). 2020;99(39):e22407. 10.1097/MD.0000000000022407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Renaud S, Romain B, Falcoz P-E, et al. KRAS and BRAF mutations are prognostic biomarkers in patients undergoing lung metastasectomy of colorectal cancer. Br J Cancer. 2015;112(4):720-728. 10.1038/bjc.2014.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Modest DP, Brodowicz T, Stintzing S, et al. Impact of the specific mutation in KRAS codon 12 mutated tumors on treatment efficacy in patients with metastatic colorectal cancer receiving cetuximab-based first-line therapy: a pooled analysis of three trials. Oncology (Huntingt). 2012;83(5):241-247. 10.1159/000339534. [DOI] [PubMed] [Google Scholar]
- 61. Cushman-Vokoun AM, Stover DG, Zhao Z, et al. Clinical utility of KRAS and BRAF mutations in a cohort of patients with colorectal neoplasms submitted for microsatellite instability testing. Clin Colorectal Cancer. 2013;12(3):168-178. 10.1016/j.clcc.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. André T, Blons H, Mabro M, et al. Panitumumab combined with irinotecan for patients with KRAS wild-type metastatic colorectal cancer refractory to standard chemotherapy: a GERCOR efficacy, tolerance, and translational molecular study. Ann Oncol. 2013;24(2):412-419. 10.1093/annonc/mds465. [DOI] [PubMed] [Google Scholar]
- 63. Hong DS, Fakih MG, Strickler JH, et al. KRAS(G12C) inhibition with sotorasib in advanced solid tumors. N Engl J Med. 2020;383(13):1207-1217. 10.1056/NEJMoa1917239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Cercek A, Shia J, Gollub M, et al. Ganetespib, a novel Hsp90 inhibitor in patients with KRAS mutated and wild type, refractory metastatic colorectal cancer. Clin Colorectal Cancer. 2014;13(4):207-212. 10.1016/j.clcc.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Strickler JH, Fakih M, Price TJ, et al. AMG 510, a novel small molecule inhibitor of KRAS(G12C), for patients (pts) with advanced gastrointestinal (GI) cancers: Results from the CodeBreaK100 phase I trial. Ann Oncol. 2020 (Abstract 83MO);31(suppl_6):S1274-S1275. 10.1016/j.annonc.2020.10.103. [DOI] [Google Scholar]
- 66. Yoon HH, Tougeron D, Shi Q, et al. KRAS codon 12 and 13 mutations in relation to disease-free survival in BRAF-wild-type stage III colon cancers from an adjuvant chemotherapy trial (N0147 alliance). Clin Cancer Res. 2014;20(11):3033-3043. 10.1158/1078-0432.CCR-13-3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Pang X-L, Li Q-X, Ma Z-P, et al. Association between clinicopathological features and survival in patients with primary and paired metastatic colorectal cancer and KRAS mutation. Onco Targets Ther. 2017;10(1178-6930 ):2645-2654. 10.2147/OTT.S133203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hayama T, Hashiguchi Y, Okamoto K, et al. G12V and G12C mutations in the gene KRAS are associated with a poorer prognosis in primary colorectal cancer. Int J Colorectal Dis. 2019;34(8):1491-1496. 10.1007/s00384-019-03344-9. [DOI] [PubMed] [Google Scholar]
- 69. Fakih M, Tu H, Hsu H, et al. Real-world study of characteristics and treatment outcomes among patients with KRAS p.G12C-mutated or other KRAS mutated metastatic colorectal cancer. Oncologist. 2022;27(8):663-674. 10.1093/oncolo/oyac077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Consortium APG. AACR Project GENIE Consortium. AACR Project GENIE: Powering precision medicine through an international consortium. Cancer Discov. 2017;7(2159-8290):818-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Nassar AH, Adib E, Kwiatkowski DJ.. Distribution of KRAS (G12C) somatic mutations across race, sex, and cancer type. N Engl J Med. 2021;384(2):185-187. 10.1056/NEJMc2030638. [DOI] [PubMed] [Google Scholar]
- 72. Chen K-H, Shao Y-Y, Chen H-M, et al. Primary tumor site is a useful predictor of cetuximab efficacy in the third-line or salvage treatment of KRAS wild-type (exon 2 non-mutant) metastatic colorectal cancer: a nationwide cohort study. BMC Cancer. 2016;16(1471-2407):327. 10.1186/s12885-016-2358-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Moretto R, Cremolini C, Rossini D, et al. Location of primary tumor and benefit from anti-epidermal growth factor receptor monoclonal antibodies in patients with RAS and BRAF wild-type metastatic colorectal cancer. Oncologist. 2016;21(8):988-994. 10.1634/theoncologist.2016-0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. The Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Scartozzi M, Bearzi I, Mandolesi A, et al. Epidermal growth factor receptor (EGFR) gene promoter methylation and cetuximab treatment in colorectal cancer patients. Br J Cancer. 2011;104(11):1786-1790. 10.1038/bjc.2011.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Alawawdeh A, Piantadosi C, Townsend AR, et al. Prognostic differences of RAS mutations: results from the South Australian metastatic colorectal registry. Target Oncol. 2022;17(1):35-41. 10.1007/s11523-021-00856-9. [DOI] [PubMed] [Google Scholar]
- 77. Osterlund E, Ristimäki A, Kytölä S, et al. KRAS-G12C mutation in one real- life and three population-based Nordic cohorts of metastatic colorectal cancer. Front Oncol. 2022;12(826073):2234-943X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Chida K, Kotani D, Masuishi T, et al. The prognostic impact of KRAS G12C mutation in patients with metastatic colorectal cancer: a multicenter retrospective observational study. Oncologist. 2021;26(10):845-853. 10.1002/onco.13870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Henry JT, Coker O, Chowdhury S, et al. Comprehensive clinical and molecular characterization of KRAS (G12C)-mutant colorectal cancer. JCO Precis Oncol. 2021;5(2473-4284). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Lin Z, Liu Y, Cai S, et al. Not all Kirsten rat sarcoma viral oncogene homolog mutations predict poor survival in patients with unresectable colorectal liver metastasis. Technol Cancer Res Treat. 2021;20(1533-0338):15330338211039131. 10.1177/15330338211039131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Yaeger R, Weiss J, Pelster MS, et al. Adagrasib with or without cetuximab in colorectal cancer with mutated KRAS G12C. N Engl J Med. 2023;388(1):44-54. 10.1056/NEJMoa2212419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Fakih MG, Kopetz S, Kuboki Y, et al. Sotorasib for previously treated colorectal cancers with KRAS(G12C) mutation (CodeBreaK100): a prespecified analysis of a single-arm, phase 2 trial. Lancet Oncol. 2022;23(1):115-124. 10.1016/S1470-2045(21)00605-7. [DOI] [PubMed] [Google Scholar]
- 83. Klempner SJ, Weiss J, Pelster M, et al. KRYSTAL-1: Updated efficacy and safety of adagrasib (MRTX849) with or without cetuximab in patients with advanced colorectal cancer (CRC) harboring a KRASG12C mutation. Ann Oncol. 2022 (Abstract LBA24);33(suppl_7):S1391S808-S1391SS69. 10.1016/j.annonc.2022.08.020. [DOI] [Google Scholar]
- 84. Kuboki Y, Yaeger R, Fakih M, et al. Sotorasib in combination with panitumumab in refractory KRAS G12C-mutated colorectal cancer: safety and efficacy for phase Ib full expansion cohort. Ann Oncol. 2022 (Abstract 3150);33(suppl_7):S680S136-S680S681. 10.1016/j.annonc.2022.07.453. [DOI] [Google Scholar]
- 85. Amodio V, Yaeger R, Arcella P, et al. EGFR blockade reverts resistance to KRAS(G12C) inhibition in colorectal cancer. Cancer Discov. 2020;10(8):1129-1139. 10.1158/2159-8290.CD-20-0187. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Qualified researchers may request data from Amgen clinical studies. Complete details are available at http://www.amgen.com/datasharing


