Abstract
Increasing evidence suggests that the cytokine transforming growth factor-β (TGF-β) inhibits the development of atherosclerosis. The lipoprotein lipase (LPL) enzyme expressed by macrophages has been implicated in the pathogenesis of atherosclerosis by stimulating the uptake of lipoprotein particles. Unfortunately, the action of TGF-β on the expression of LPL in macrophages remains largely unclear. We show that TGF-β inhibits LPL gene expression at the transcriptional level. Transient transfection assays reveal that the −31/+187 sequence contains the minimal TGF-β-responsive elements. Electrophoretic mobility shift assays show that Sp1 and Sp3 interact with two regions in the −31/+187 sequence. Mutations of these Sp1/Sp3 sites abolish the TGF-β-mediated suppression whereas multimers of the sequence impart the response to a heterologous promoter. TGF-β has no effect on the binding or steady-state polypeptide levels of Sp1 and Sp3. These results, therefore, suggest a novel mechanism for the TGF-β-mediated repression of LPL gene transcription that involves regulation of the action of Sp1 and Sp3.
INTRODUCTION
The enzyme lipoprotein lipase (LPL; EC 3.1.1.34) plays a key role in the overall lipid metabolism and transport by catalysing the hydrolysis of the triacylglycerol component of chylomicrons and very low-density lipoproteins, thereby providing non-esterified fatty acids and 2-monoacylglycerols for tissue utilization (1). LPL is synthesized by several tissues/cell types, with the enzyme expressed by the cells of the vascular wall, particularly macrophages, implicated to play a key role in the pathogenesis of atherosclerosis (1,2). For example, LPL is expressed in the lesion where macrophage-derived foam cells represent the predominant site for the synthesis of the enzyme (3). In addition, inbred murine strains with elevated levels of macrophage LPL show an increased susceptibility to atherosclerosis (4). Furthermore, macrophage LPL expression is increased in patients with diabetes and heterozygous familial hypercholesterolemia (5,6), and this may be responsible, at least in part, for the high incidence of atherosclerosis in such individuals. Moreover, a marked decrease in diet-induced atherosclerosis has been seen in chimeric mice that are deficient for macrophage LPL expression (7–9) and, conversely, macrophage-specific expression of human LPL accelerates atherosclerosis in transgenic apolipoprotein E (apoE)-deficient mice (10). This pro-atherogenic action of LPL involves both its catalytic function and non-catalytic ‘bridging action’ that allows it to interact simultaneously with both specific cell surface proteins/receptors and plasma lipoproteins, thereby increasing the uptake of the latter by macrophages and, thus, leads to foam cell formation (1,2).
The transformation of macrophages into foam cells is inhibited by cytokines such as transforming growth factor-β (TGF-β) by the regulation of key genes that are involved in modulating cholesterol influx and efflux (11–13). For example, TGF-β has been shown to inhibit the expression of genes encoding the scavenger receptors A and CD36 (12–14), which are involved in cholesterol import, and stimulating the expression of a number of genes implicated in mediating cholesterol efflux such as ATP-binding cassette transporter-A1 (ABCA1) and -G1 (ABCG1) (11,12). The anti-atherogenic action of TGF-β is not restricted to the control of macrophage foam cell formation but also extends to other functions. For example, the cytokine has anti-inflammatory properties, as evidenced by a profound inflammatory response reported for TGF-β knockout mice (15). In addition, the cytokine induces the expression of tissue inhibitors of metalloproteinases (16), inhibits nitric oxide and superoxide production (17), and increases the expression of IL-1 receptor antagonist (18). In further support of an anti-atherogenic role of TGF-β, inhibition of its signalling by use of neutralizing antibodies has been shown to accelerate the development of atherosclerotic lesions in apoE-deficient mice (19). It has also been suggested that the protective effect of tamoxifen in the formation of lipid lesions in apoE knockout mice is mediated through increased production of TGF-β in the aorta (20). Additionally, in apoE-deficient mice treated with anti-CD40 antibody, increased levels of TGF-β have been found to be associated with lipid poor lesions (21). More recently, low TGF-β levels have been identified as a risk factor for the high incidence of atherosclerosis in patients with end-stage renal disease (22), and the disruption of TGF-β signalling in T cells has been shown to accelerate atherosclerosis in murine models of the disease (23).
In the light of the TGF-β-regulated expression of several key genes in macrophages that are involved in modulating lipoprotein uptake and foam cell formation, it is essential that a detailed understanding is obtained of the signalling pathway(s) and the transcription factors that are required for such responses. Such studies will not only better our understanding of the molecular basis of foam cell formation and atherogenesis but, in the longer term, may also lead to the identification of potentially novel targets for therapeutic intervention. TGF-β exerts its cellular actions by interacting with a heteromeric complex of transmembrane serine/threonine kinases, the type I and the type II receptors (24,25). In relation with the intracellular signalling pathways, advances have been made in the area of activation of gene transcription by the cytokine (24,25). Thus, on phosphorylation by activated type I receptor, Smad2 or Smad3 form complexes with Smad4. The complex then translocates to the nucleus where it exerts transcriptional activities either directly by interacting with Smad-responsive elements in the regulatory regions of target genes, or indirectly through association with other transcription factors (24,25). Unfortunately, our current understanding of the mechanisms by which TGF-β inhibits gene expression remains enigmatic and this is mainly due to the limited research that has been carried out in this area. We have, therefore, investigated this important aspect using LPL as a model gene. We show here that TGF-β inhibits the expression of the LPL gene in macrophages at the transcriptional level. In addition, we delineate the cis-acting regulatory sequences that are involved in the response and demonstrate a critical role for the Sp1-binding sites in the action of TGF-β. These studies, therefore, provide novel insights into the molecular mechanisms that are required for the TGF-β-mediated inhibition of gene transcription.
MATERIALS AND METHODS
Reagents
The human myeloid leukaemic U937, the mouse macrophage J774.2 and the human hepatoma Hep3B cell lines were from the European Collection of Animal Cell Cultures. All the cell culture reagents were purchased from Autogen-Bioclear, Sigma or Invitrogen. Antisera against Sp1 and Sp3 were from Santa Cruz.
Cell culture
The cell lines were maintained in either DMEM (J744.2 and Hep3B) or RPMI 1640 (U937), which was supplemented with 10% (v/v) heat-inactivated foetal calf serum (HI-FCS; 56°C, 45 min), 2 mM l-glutamine, 100 U/ml penicillin and 100 μg/ml streptomycin (designated as complete medium). The cultures were maintained at 37°C in a humidified atmosphere containing 5% (v/v) CO2 in air. For human monocyte-derived macrophages, blood was collected from healthy volunteers into a syringe containing an equal volume of sterile 2% (w/v) dextran in 1× PBS containing 0.8% (w/v) tri-sodium citrate, and was allowed to stand for 30 min to permit erythrocyte sedimentation. The upper layer was collected, underlayered with Lymphoprep™ (Nycomed Pharmaceuticals) [2:1 (v/v) supernatant/Lymphoprep™] and centrifuged at 800 g for 20 min. The resultant interface was collected and washed 6–8 times with PBS-0.4% (w/v) tri-sodium citrate to remove contaminating platelets. Monocytes were plated out in 75 cm2 flasks in complete medium, as above except containing 5% (v/v) serum, and were allowed to adhere to the surface for 6 h in a humid incubator at 37°C containing 5% (v/v) CO2. The cells were then washed twice with PBS to remove any other mononuclear cells, fresh medium was added and the differentiation allowed to continue for 7 days. The homogeneity of macrophages was verified by morphological analysis. Unless otherwise stated, the cells were preincubated for 4 h in medium containing reduced (0.5%) HI-FCS before stimulation with TGF-β (26–28).
RT–PCR
Total cellular RNA was prepared using the RNeasy™ total RNA isolation kit (Qiagen) according to the manufacturer's instructions. Each isolated RNA sample (1 μg) was then subjected to RT–PCR essentially as described before (29–31) except that the annealing temperature and the total number of cycles varied: 21 cycles and 60°C and 24 cycles and 55°C for murine and human LPL, respectively, 9 cycles and 62°C for 28S rRNA, 16 cycles at 60°C for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and 18 cycles at 58°C for β-actin. These conditions were in the exponential phase of amplification and, therefore, provided a direct correlation between the amount of products and the RNA template abundance in the samples. The sequences of the primers were 5′-CATTTACCAGGGGGTCAC-3′ and 5′-AGGCAGAGCCCTTTCTCA-3′ for mouse LPL; 5′-GAGATTTCTCTGTATGGCACC-3′ and 5′-CTGCAAATGAGACACTTTCTC-3′ for human LPL; 5′-CCCTTCATTGACCTCAACTACATGG-3′ and 5′-AGTCTTCTGGGTGGCAGTGATGG-3′ for GAPDH; 5′-TGAACTATGCTTGGGCAGGG-3′ and 5′-AGCGCCATCCATTTTCAGGG-3′ for 28S rRNA; and 5′-TGGAGAAGAGCTATGAGCTGCCTG-3′ and 5′-GTCCCACCAGACACCACTGTGTTG-3′ for β-actin. The PCR products were size-fractionated on a 2% (w/v) agarose gel, photographed using a Syngene gel documentation system (GRI) and quantified using the Gene Tools computer package (Syngene).
RT–PCR based nuclear ‘run-on’ transcription assay
The method employed was a modification of that used by Hildebrandt and Neufer (32). The cells were harvested by scraping them into the medium which was then removed from the flask and centrifuged at 1000 g for 5 min. The resulting cell pellet was washed three times by resuspension in 1 ml of ice-cold PBS and microcentrifugation at 10 000 g for 30 s. After the final wash, the supernatant was discarded and 200 μl of ice-cold buffer A [10 mM HEPES, pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, 0.5 mM phenylmethylsulfonyl fluoride (PMSF), 1 μg/ml pepstatin A, 10 μg/ml aprotinin, 10 μg/ml leupeptin and 10 μg/ml type I-S soybean trypsin inhibitor] was added. The mixture was briefly vortexed and incubated on ice for 5 min. The cells were lysed by pipetting up and down 10 times in a 100 μl Hamilton syringe and centrifuged at 700 g for 10 min at 4°C. The resulting crude nuclear pellet was gently suspended in 10 ml of ice-cold buffer B [15 mM HEPES, pH 7.9, 60 mM KCl, 3 mg/ml BSA, 300 mM sucrose, 0.1 mM EDTA, 0.1 mM EGTA, 1 mM DTT, 0.5 mM spermidine, 0.15 mM spermine, 0.5 mM PMSF, 2 μg/ml leupeptin and 0.5% (v/v) Triton X-100] and was centrifuged at 700 g for 10 min at 4°C. Pelleted nuclei were resuspended in 10 ml of ice-cold buffer C (15 mM HEPES, pH 7.9, 60 mM KCl, 3 mg/ml BSA, 300 mM sucrose, 0.1 mM EDTA, 0.1 mM EGTA, 1 mM DTT, 0.5 mM spermidine, 0.15 mM spermine, 0.5 mM PMSF, 2 μg/ml leupeptin and 5 mM magnesium acetate) and centrifuged as above. Final nuclei pellets were resuspended in 230 μl storage buffer [40% (v/v) glycerol, 75 mM HEPES, pH 7.9, 60 mM KCl, 15 mM NaCl, 5 mM magnesium acetate, 0.1 mM EDTA, 0.1 mM EGTA, 1 mM DTT, 0.5 mM spermidine, 0.15 mM spermine, 2 μg/ml aprotinin and 2 μg/ml leupeptin], quick frozen in liquid nitrogen and stored at −70°C. Nuclear ‘run-on’ reactions were carried out by incubation of nuclei (160 μl), previously thawed on ice, with 2× reaction buffer [20% (v/v) glycerol, 100 mM KCl, 10 mM MgCl2, 4.5 mM DTT, 1.2 mM ATP, 0.6 mM each of CTP, GTP and UTP, 80 U/ml RNasin ribonuclease inhibitor, 0.5 mM spermidine, and 0.15 mM spermine] for 15 min at 22°C. Nuclei were then subjected to DNaseI treatment (20 U) in the presence of 1 mM CaCl2 for 15 min at 37°C. This was followed by digestion of nuclear proteins by the addition of 10× digestion buffer [100 mM Tris–HCl, pH 8.0, 10 mM EDTA, 5% (w/v) SDS and 100 μg/ml proteinase K] and incubation for 30 min at 37°C. Nuclear RNA was extracted in a final volume of 30 μl using the RNeasy total RNA isolation kit (Qiagen). Subsequent RT–PCRs were carried out as described above.
LPL activity assay
Samples for the determination of the heparin-releasable LPL activity were prepared as described previously (27,28). The activity was then determined using ConfluolipTM Continuous Fluorometric Lipase kit (Progen) (33).
Preparation of manipulated LPL promoter–reporter DNA constructs
A series of DNA constructs, containing 5′ truncations of the LPL promoter in the p19Luc vector, were a gift from Dr J. M. Gimble (34). The LPL sequences in these constructs were cloned in front of the luciferase reporter gene in the pGL2-Basic vector (Promega). The preparation of DNA constructs containing mutations of the Sp1 sites (Sp1M44, Sp1M62/65 and Sp1M44/62/65) and those with four copies of the Sp1 sites at position +44, +62 and +65, or the +38 to the +75 region, that are all linked to a heterologous minimal SV40 promoter in the pGL2-promoter vector has been described previously (26). For the preparation of a DNA construct containing the −31 to +9 region linked to a minimal SV40 promoter, the following oligonucleotides were synthesized: 5′-CCGGGTCACTTAAACAGCTGTGCAGTGGAAACAGTGTCAGACTC-3′ and 5′-TCGAGAGTCTGACACTGTTTCCACTGCACAGCTGTTTAAGTGAC-3′. These were designed in such a manner that, following annealing, they had overhangs that allowed their direct cloning into the XmaI and XhoI sites of the pGL2-promoter vector (26,35). The sequences of all the DNA constructs used were verified by DNA sequencing.
Transient transfection assays
U937 or Hep3B cells were transfected using Superfect™ (Qiagen) (26). The cells were harvested by centrifugation (1000 g for 5 min), resuspended in appropriate medium containing 3% (v/v) HI-FCS for U937 (26) and 10% (v/v) HI-FCS for Hep3B (30), subcultured into 6-well plates at a density of 1.5 × 106 cells/well, and incubated for 4 h. The DNA/Superfect™ complex was then prepared by diluting the DNA to 50 μl/μg DNA with appropriate medium. Superfect™ (1.5–4 μl/μg DNA) was added and the mixture was vortexed and incubated at room temperature for 10 min. The complex was then diluted with the appropriate medium (600 μl/μg of DNA) and added to the cells. For Hep3B, the cells were returned to the incubator for 6 h. The medium was then changed and the cells incubated for 24 h in the absence or the presence of 30 ng/ml TGF-β. For U937, the cells were returned to the incubator for a further 3 h before being centrifuged as above and washed with 10 ml of RPMI 1640. The cells were resuspended at a density of 1.5 × 106 cells/ml in RPMI 1640 supplemented with 3% (v/v) FCS and plated out at 2 ml/well in 6-well plates. The cells were then either incubated with phorbol 12-myristate 13-acetate (PMA) (1 μM) in the absence or the presence of TGF-β1 (30 ng/ml) for 12 h (26) or first differentiated with PMA for 12 h and then treated with the cytokine for a further 12 h (cells stimulated for 24 h with PMA were used for comparison in this case). Similar results were obtained with both experimental approaches. The luciferase and the β-galactosidase activities in the cell extracts were then determined using commercially available kits (Promega). The luciferase activity was normalized to the β-galactosidase values or, in some cases the total protein levels, with each transfection being carried out in triplicate and repeated at least three times.
Preparation of whole cell extracts
Whole cell extracts were prepared essentially as described previously (26,29,30,35). Protease inhibitors (0.5 mM PMSF, 2 mM benzamidine, 1 μg/ml pepstatin A, 10 μg/ml aprotinin and 2.5 μg/ml leupeptin) and DTT (0.5 mM) were added to all buffers before use. The concentration of proteins in extracts was determined using the microBCA protein assay kit according to the manufacturer's instructions (Pierce).
Electrophoretic mobility shift assay (EMSA)
EMSA was carried out essentially as reported previously (26,29,30,35). The sequences of the oligonucleotides used for EMSA were as follows: −31/+8, 5′-GTCACTTAAACAGCTGTGCAGTGGAAACAGTGTCAG-3′ and 5′-AGTCTGACACTGTTTCCACTGCACAGCTGTTTAAGT-3′; +9/+49, 5′-CTCGATTTCTCCTCCTACTCCTCCTCCGAGGAATTCT-3′ and 5′-GGGCAGAATTCCTCGGAGGAGGAGTAGGAGGAGAAAT-3′; +46/+90, 5′-GCCCCCTGTAACTGTTCTGCCCTCCCCTTTAAAGGTTGACTT-3′ and 5′-GGCAAGTCAACCTTTAAAGGGGAGGGCAGAACAGTTACAGGG-3′; +90/+118, 5′-GCCCTACGGCGCTCCACCGCGCTCCAGT-3′ and 5′-AGGACTGGAGCGCGGTGGAGCGCCGTAG-3′; +119/+160, 5′-CTTGCGCCTCCTGCTCAACCCGCTCCTGACTGCCCACGC-3′ and 5′-GCGGCGTGGGCAGTCAGGAGCGGGTTGAGCAGGAGGCG-3′; and +159/+195, 5′-CGCGTAGTTCCAGCAGCAAAGCAGAAGGGTGCA-3′ and 5′-CCGGTGCACCCTTCTGCTTTGCTGCTGGAACTA-3′.
SDS–PAGE and western-blot analysis
Samples (10 μg whole cell extracts) were size-fractionated under reducing conditions using 7.5–15% (w/v) polyacrylamide gels containing SDS (26–30). For western-blot analysis, the proteins on the gel were transferred onto Immobilon-P PVDF membranes (Millipore) by blotting (26–30). The blotted membranes were incubated first for 1 h at room temperature in blocking solution [1× Tris-buffered saline (TBS) containing 5% (w/v) non-fat milk powder and 0.05% (w/v) Tween-20] in order to reduce any non-specific interaction of the antiserum with the membrane. Following three washes of 5 min each with 1× TBS containing 0.05% (v/v) Tween-20, the membrane was incubated with the primary antibody for 1 h in 1× TBS containing 5% (w/v) non-fat milk powder and 0.1% (v/v) Tween-20. After washes as above, the membrane was incubated with secondary horseradish peroxidase (HRP)-conjugated antibodies (goat anti-rabbit IgG or mouse anti-goat IgG) for 30 min at room temperature in 1× TBS containing 5% (w/v) non-fat milk powder and 0.05% (v/v) Tween-20. After washing once again as above, the membranes were developed using an enhanced chemiluminescence detection kit (Amersham) and XAR sensitive film (Kodak). The sizes of the proteins were determined by comparison with Rainbow molecular weight markers (Amersham) that had been subjected to electrophoresis and blotting on the same gel as the test samples.
RESULTS
TGF-β inhibits LPL gene expression in macrophages at the transcriptional level
We have previously employed the murine J774.2 cell line to investigate the regulation of LPL gene expression in macrophages by cytokines (26–28). The work, both in our laboratory and others have shown that the regulation of LPL and a number of other genes in this cell line correlates with what is seen in primary macrophage cultures [(26–28) and references therein]. For the initial experiments, the cells were incubated with 10 ng/ml of TGF-β for 20 h, a period when maximal changes to a range of factors in LPL expression are seen (26–28), and RNA was used for RT–PCR with primers against LPL and the control β-actin. A similar experiment using PDGF, which has been shown previously to induce LPL mRNA expression in macrophages (36), was included for comparative purposes. As expected, PDGF increased LPL mRNA expression (Figure 1A). On the other hand, incubation of the cells with TGF-β led to a marked decrease in LPL mRNA expression (Figure 1A). Further experiments showed that TGF-β decreased LPL mRNA expression in a time-dependent manner, with a maximal reduction of ∼60% produced at 20 h (data not shown). The profile was similar to that seen previously for LPL in response to a number of other factors (27). In order to investigate the dose–response of TGF-β action, the cells were treated for 20 h with different concentrations of cytokine. The range of concentrations used was based on an extensive survey of TGF-β-mediated regulation of gene expression. As shown in Figure 1B, a concentration-dependent reduction of LPL mRNA expression was observed with maximal decrease of ∼70% seen with 30 ng/ml of the cytokine. Finally, in order to rule out the possibility that the TGF-β-mediated decrease in LPL mRNA expression was a peculiar property of the J774.2 cell line, RT–PCR analysis was carried out using primary cultures of human monocyte-derived macrophages. As expected, TGF-β decreased LPL mRNA expression with maximal reduction of ∼70% seen at 12 h (Figure 1C).
Figure 1.
TGF-β decreases LPL mRNA expression in macrophages. (A) J774.2 macrophages were either left untreated (C) or stimulated for 20 h with PDGF-BB (PDGF; 10 ng/ml) or 10 ng/ml TGF-β1 (TGF-β); (B) J774.2 macrophages were exposed to the indicated concentration of the cytokine for 20 h; and (C) primary cultures of human monocyte-derived macrophages were incubated for the indicated period of time with 30 ng/ml of TGF-β1. Total RNA was isolated and subjected to RT–PCR using primers for LPL, β-actin or GAPDH as shown. The amplification products were size-fractionated by agarose gel electrophoresis. The data shown are representative of at least two independent experiments.
We next investigated whether the TGF-β-mediated reduction of LPL mRNA expression in J774.2 macrophages was accompanied by a corresponding decrease in the level of protein and enzymatic activity. As shown in Figure 2A, western-blot analysis showed that the cytokine produced a dramatic decrease in LPL protein levels. Similarly a decrease in the functional, heparin-releasable LPL enzymatic activity was seen (Figure 2B). The LPL activity assay was extended to primary cultures of human monocyte-derived macrophages. As shown in Figure 2C, TGF-β also decreased LPL activity in these cells.
Figure 2.
The action of TGF-β on LPL gene expression is mediated at the level of gene transcription. (A) Western-blot analysis was carried out using whole cell extracts from J774.2 macrophages that were either left untreated (C) or exposed to TGF-β1 (TGF-β; 30 ng/ml) for 20 h, and probed with antiserum against LPL. (B and C) J774.2 cells (B) or primary cultures of human monocyte-derived macrophages (C) were either left untreated (Control) or stimulated for 20 h with TGF-β1 (30 ng/ml) as indicated. The heparin-releasable LPL activity was then determined as described under Materials and Methods. The LPL activity from cytokine-stimulated cells is represented as a percentage of the activity of untreated control cells (assigned as 100%) Asterisks show significant reduction compared to untreated cells (**P < 0.005, *P < 0.05). (D) J774.2 macrophages were either left untreated (C) or stimulated with TGF-β1 (TGF-β) for 24 h. Following nuclear ‘run-on’ transcription assay, nuclear RNA was extracted and subjected to RT–PCR using primers against LPL or GAPDH.
The TGF-β-mediated decrease in LPL mRNA expression could be due to changes in either gene transcription or mRNA stability. RT–PCR-based nuclear ‘run-on’ transcription assays showed that the action of TGF-β on LPL mRNA expression was mediated at the level of gene transcription (Figure 2D). This conclusion was supported further by a similar profile of decay of LPL mRNA in the absence or the presence of TGF-β following inhibition of gene transcription using actinomycin D (data not shown). We next determined the regulatory sequences and the transcription factors that are required for the response.
Identification of the minimal regulatory region in the LPL promoter that is required for the TGF-β-mediated inhibition of LPL gene transcription
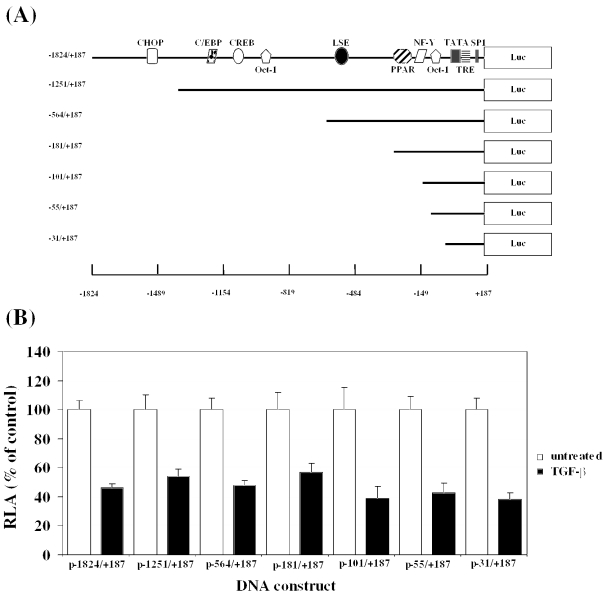
We have previously delineated the regulatory sequences that are required for the inhibition of LPL gene transcription by interferon γ (IFN-γ) via transient transfection of a range of 5′ LPL promoter deletion constructs into the human U937 myeloid leukaemic cell line (26). Transfection of macrophages is relatively difficult and U937 cells, which have been employed widely to delineate the sequence elements that are involved in the regulation of macrophage gene transcription, yielded the highest transfection efficiency from a range of monocyte/macrophage cell lines examined (J774.2 macrophages could not be transfected efficiently) [(26) and references therein]. Thus, U937 monocytes were transfected with the LPL promoter–luciferase DNA constructs, and luciferase activity in cell extracts from differentiated macrophages, in the absence or the presence of TGF-β, was then determined and normalized to that of the β-galactosidase control. A schematic representation of the LPL promoter–luciferase DNA constructs used along with binding sites for some key transcription factors (1,2) are shown in Figure 3A. When the cells were transfected using a DNA construct containing the largest LPL promoter fragment (−1824/+187), ∼50% reduction in the relative luciferase activity was obtained (Figure 3B). Such a reduction was comparable with that seen at the level of LPL mRNA expression, protein level and enzymatic activity (Figures 1 and 2). A similar reduction in the relative luciferase activity of ∼50% was seen with all the LPL promoter deletion constructs (Figure 3B). This suggests that the −31 to +187 LPL promoter region contains the minimal TGF-β responsive elements (TGF-βREs) and was, therefore, investigated in detail. However, instead of mapping further the precise sequences that are involved in the TGF-β response by analysing the effect of finer deletions or specific mutations in the −31/+187 region, we decided to first investigate the interaction of proteins with this promoter region using EMSA. Further studies could then focus on those regions that interact with DNA-binding proteins.
Figure 3.
The −31/+187 region contains the minimal TGF-βREs. (A) A schematic representation of the LPL promoter–luciferase DNA constructs showing key sites identified by bioinformatic or functional analysis (1,2): C/EBP, CCAAT/enhancer binding protein; CHOP, C/EBP homologous protein; CREB, cAMP response element binding protein; Oct-1, octamer binding protein-1; LSE, LPL silencing element; PPAR, peroxisome-proliferator activated receptor; NF-Y, nuclear factor Y; TATA, TATA box; TRE, thyroid responsive element; and Sp1, specificity protein-1. ‘+1’ refers to the transcription initiation site. (B) Transient transfection assays with the indicated LPL promoter–luciferase DNA constructs were carried out as described in Materials and Methods. The luciferase activity was normalized to the β-galactosidase activity and is represented as relative luciferase activity (RLA). For each construct, the RLA from untreated cells has been assigned as 100% (open histogram) with that from TGF-β cells being represented to this value (filled histogram). Each value represents mean ± SD from four independent experiments. The TGF-β-response was statistically significant in each case (P < 0.05).
Further analysis of the −31/+187 region by EMSA and transfection assays
EMSA was carried out using the six double-stranded oligonucleotides that spanned the −31/+187 region (−31/+8, +9/+49, +46/+90, +90/+118, +119/+160 and +159/+195) and extracts from J774.2 macrophages that were either left untreated or stimulated with TGF-β. Using the +90/+118, +119/+160 and +159/+195 oligonucleotides, no specific DNA–protein complexes could be detected even after long autoradiographic exposures (Figure 4A). In contrast, specific DNA–protein complexes were seen with the other three oligonucleotides, thereby implicating the corresponding sequences in the TGF-β response (Figure 4A). When the interaction of DNA-binding proteins to these three sites with extracts from untreated and TGF-β-stimulated cells was examined, an increase in binding to the −31/+8 region was observed (Figure 4A). In contrast, no reproducible changes in the binding of the proteins to the +9/+49 and +46/+90 region could be seen in independent experiments (Figure 4A).
Figure 4.
The Sp1- and Sp3-binding sites in the −31/+187 region contains TGF-βREs (A) EMSA was carried out using radiolabelled oligonucleotides against the sequences shown and extracts from J774.2 cells that were either left untreated (UT) or exposed to TGF-β1 (30 ng/ml) for 24 h (TGF-β). (B) Transient transfection assays were carried out using the −31/+9 and +38/+75 region of the LPL 5′ sequence linked to the heterologous minimal SV40 promoter. The relative luciferase activity (RLA) in the absence or the presence of TGF-β in differentiated macrophages (open and filled histograms, respectively) was determined as described in Materials and Methods. The RLA from cells transfected with the p+38/+75 construct in untreated cells has been assigned as 100% with the remainder shown relative to this value. Asterisks show significant reduction compared to untreated cells (P < 0.05). (C) Alignment of the +9 to +90 sequence of mouse LPL (Mo) with the corresponding sequence from the human (Hu) and the rat (R) counterpart. The three Sp1/Sp3 sequences [+44 to +51 (antisense strand), +62 to +67 (antisense strand) and +65 to +71 (sense strand)] are shown in upper case. The corresponding sequences in the three mutants in these Sp1/Sp3 sites (see Figure 5A) are also shown with the mutated base in boldface and underlined. (D) Antibody supershift experiments were carried out using radiolabelled +9/+49 or +46/+90 oligonucleotides and extracts from J774.2 macrophages exposed to TGF-β1 (30 ng/ml) for 24 h in the absence (–) or the presence of antisera against Sp1 (+Sp1) or Sp3 (+Sp3), or, the non-immune serum (+NIS). C and SS represent DNA–protein and antibody–DNA–protein supershift complex(es), respectively.
The +9/+90 region contains three recognition sequences for the transcription factors Sp1 and Sp3 [a single site at position +44 to +51 (antisense strand) and a dual site at position +62 to +67 (antisense strand) and +65 to +71 (sense strand)] that are highly conserved among the mouse, rat and the human gene (Figure 4C). We have previously shown that these sites are required for the IFN-γ-mediated reduction of LPL gene transcription (26). We next evaluated the activity of DNA constructs containing either a fragment with all these three Sp1/Sp3 sites (+38/+75) or the −31/+9 region, to which a marginal increase in binding was seen when extracts from TGF-β-treated cells were used (Figure 4A), when placed in front of the heterologous, minimal SV40 promoter linked to a luciferase reporter gene. The reporter gene activity from the −31/+9 region in untreated macrophages was only marginal compared with that produced by the +38/+75 region, thereby demonstrating the major contribution by the Sp1/Sp3 sites to the constitutive transcription of the LPL gene (Figure 4B). More importantly, a marked, and statistically significant, TGF-β-mediated reduction in reporter gene activity was seen with the +38/+75 sequence but not with the −31/+9 region (Figure 4B). Our further studies, therefore, focused on these Sp1 and Sp3 sites in the +38/+75 region.
As part of our previous studies on the regulation of LPL gene transcription by IFN-γ, we have confirmed by using competition EMSA and antibody supershift assay that Sp1 and Sp3 in extracts from untreated macrophages interact specifically with these three recognition sequences in the +9/+90 region (26). However, in contrast to the action of TGF-β (Figure 4A), exposure of macrophages to IFN-γ leads to a dramatic decrease in the binding of factors to these sites (26). In the light of this major difference, we decided to confirm that Sp1 and Sp3 also bound to the sites when extracts from TGF-β-stimulated cells were used. For this, antibody supershift assays were carried out using extracts from macrophages that had been treated with TGF-β for 24 h and antisera against Sp1 and Sp3. As shown in Figure 4D, an inhibition of specific DNA–protein complexes and the appearance of slower migrating antibody–DNA–protein supershift complex was seen when antisera against Sp1 and Sp3, but not the non-immune serum, was included, thereby confirming the interaction of these two factors.
The Sp1/Sp3 sites are crucial for the TGF-β response
In order to evaluate the role of the identified Sp1/Sp3 recognition sequences in the TGF-β response, the effect of mutations of the single site at position +44, dual site at position +62/+65 and all three sites in the −31/+187 context was investigated by transfection assays. As expected, the relative luciferase activity, normalized for the β-galactosidase internal control, from the wild-type −31/+187 region was reduced by ∼60% when the cells were treated with TGF-β (Figure 5A). Mutations of the Sp1 sites have a marked effect on the promoter activity of untreated cells (Figure 5A), thereby indicating that the Sp1 sites are also necessary for constitutive transcription. It is interesting to note that while the activity of the DNA construct with mutations of the sites at position +62 and +65 is only marginally more than that of the promoterless pGL2-Basic vector, the activity of the construct with mutations of all three sites (positions +44, +62 and +65) was substantially greater (Figure 5A). Although the exact reason for this remains to be determined, it is likely that this may be due to some form of interaction between the different Sp1/Sp3 sites. More importantly, however, the TGF-β response was abolished with mutations of the dual site at positions +62 and +65 or mutations of all three sites (Figure 5A). In contrast, a statistically significant TGF-β response was seen with mutation of the site at position +44 although the reduction was less extensive compared to that seen with the −31/+187 region that contains all three Sp1/Sp3 sites (Figure 5A). The TGF-β response with the construct containing the mutation at position +44 was most probably due to the presence of intact Sp1/Sp3 sites at positions +62 and +65. Overall, therefore, these results suggest that the Sp1/Sp3 sites are required for the TGF-β-mediated decrease in LPL promoter activity with the contribution from the +62 and +65 sites being more extensive than that from the site at position +44.
Figure 5.
Investigation of the importance of the Sp1/Sp3 sites in the −31/+187 sequence on the TGF-β response. Transient transfection assays were carried out using the following DNA constructs: The promoterless pGL2-Basic vector (pGL2 Basic), −31/+187 region of the LPL 5′ sequence linked to the luciferase gene either intact (p−31/+187) or containing mutations of the Sp1/Sp3 sites at position +44 (pSp1 M44), +62 and +65 (pSp1 M62/M65), or +44, +62 and +65 (pSp1 M44/62/65) (see Figure 4C for the sequences of the bases substituted) (A); the +38/+75 region of the LPL 5′ sequence (p+38/+75) or four copies of the Sp1/Sp3 sites at position +44 or the dual site at position +62/+65 (pSp1+44×4 and pSp1+62/+65×4, respectively) all linked to the heterologous minimal SV40 promoter (B and C); and the TGF-β inducible p3TP-Lux plasmid (C). These were transfected into either U937 cells (A and B) or Hep3B cells (C), and the relative luciferase activity (RLA) in the absence or presence of TGF-β (open and filled histograms, respectively) was determined as described in Materials and Methods. In (A), the RLA from cells transfected with the p−31/+187 construct in untreated cells has been assigned as 100% with the remainder shown relative to this value. For (B and C), the activity of each construct in untreated cells has arbitrarily been assigned as 100% with that from TGF-β-stimulated cells being represented as a percentage of this value. Asterisks show significant change compared with untreated control cells (**, P < 0.005; *, P < 0.05).
In order to investigate the role of the Sp1 sites further, the ability of multimers of the sites to confer the TGF-β response to a heterologous promoter was investigated. For this, we used DNA constructs containing four copies of the Sp1/Sp3 site at position +44 or the dual site at positions +62 and +65 that are linked to the minimal SV40 promoter in the pGL2-promoter vector along with that containing the +38/+75 region (i.e. all three Sp1/Sp3 sites). These constructs were transfected into U937 cells and the relative luciferase activity was determined in macrophages that were either left untreated or exposed to TGF-β. As shown in Figure 5B, a TGF-β-mediated reduction in reporter gene activity was obtained with all the LPL regulatory sequences used. Thus, the Sp1/Sp3 sites at positions +44 and +62/+65 are able to confer the TGF-β response to a heterologous promoter.
In order to rule out the possibility that the TGF-β-mediated repression through the Sp1/Sp3 recognition sequences was a peculiar property of macrophages, representative experiments were repeated in the human hepatoma Hep3B cell line. A TGF-β-inducible plasmid 3T3-Lux (37) was also included for comparative purposes. Similar to U937 cells, a TGF-β-mediated reduction of reporter gene activity was seen with DNA constructs containing the +38/+75 region of the LPL gene (all three Sp1/Sp3 sites) or four copies of the dual site at position +62/+65, that are linked to the minimal SV40 promoter (Figure 5C). In contrast, and as expected, TGF-β induced the activity of the 3T3-Lux plasmid (Figure 5C).
The action of TGF-β on LPL expression is independent of any change in the steady-state levels of Sp1 and Sp3
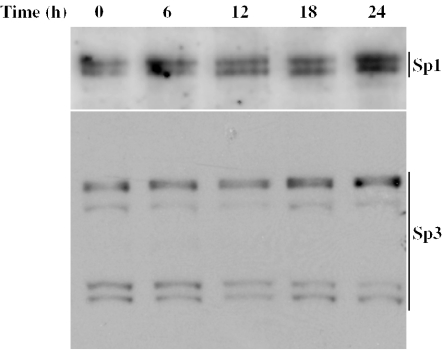
EMSA showed that TGF-β had little effect on the binding of Sp1 and Sp3 to regulatory sequences in the LPL gene (Figure 4A). This suggested that the cytokine probably had no effect on the steady-state levels of Sp1 and Sp3 polypeptides. To confirm this assumption, J774.2 macrophages were treated with TGF-β for 0, 6, 12, 18 and 24 h. Samples of proteins from these time points were subjected to western-blot analysis using antisera against Sp1 and Sp3. The Sp1 antiserum detected two closely migrating proteins with a molecular mass of ∼112 kDa (Figure 6). It is likely that one of these represents a post-translational variant of the other. In the case of Sp3, at least three polypeptides are known to be produced from the same mRNA by alternate use of translational initiation codons (26,38). In the present study, at least four polypeptides were seen that formed two closely migrating doublets with approximate molecular mass of 115 and 70 kDa (Figure 6). The expression of either the Sp1 or the Sp3 polypeptides was not affected by incubation of the cells with TGF-β (Figure 6).
Figure 6.
The effect of TGF-β on steady-state levels of Sp1 and Sp3 polypeptides. Western-blot analysis was carried out using whole cell extracts from J774.2 macrophages exposed to TGF-β1 (30 ng/ml) for the times indicated. The blotted membranes were probed with antiserum against Sp1 or Sp3, and the antigen–antibody complexes were detected as described in Materials and Methods. The position of the Sp1 and Sp3 polypeptides are shown on the right-hand side of the figure. The data shown are representative of three independent experiments.
DISCUSSION
TGF-β has recently been shown to play a key anti-atherogenic role (11–23). Consistent with such a role, we show here that the cytokine inhibits the expression in macrophages of LPL (Figures 1 and 2), which has been implicated in the uptake of lipid/lipoproteins and the transformation of macrophages into foam cells (1,2). This finding also correlates well with previous studies showing that the cytokine inhibits foam cell formation by suppressing the expression of scavenger receptor A and CD36, which are involved in lipid/lipoprotein uptake, and stimulating the expression of ABCA1 and ABCG1, which have been implicated in enhancing cholesterol efflux (11–14). We show that the TGF-β-mediated inhibition of macrophage LPL gene expression is mediated at the transcriptional level and that the −31/+187 region contains the TGF-βREs (Figure 3). We demonstrate that three binding sites for Sp1 and Sp3 in this region are essential for the response (Figures 4 and 5).
Sp1 and Sp3 belong to the zinc finger class of transcriptional regulators, which bind to an identical recognition sequence and tend to be co-expressed in several tissues and cell types, including macrophages (26,39). While these proteins were originally implicated in the constitutive expression of several genes, they have subsequently been shown to be involved in inducible gene expression that includes responses triggered by TGF-β (39). Although the Smads are largely responsible for the transcriptional activation of these genes, other factors have also been found to play an important role, including Sp1/Sp3 (40–45). For example, Sp1/Sp3 have been implicated in the TGF-β-induced expression of tenascin-C, adenine nucleotide translocator 1, endoglin, α2(I) collagen, p15 (Ink4B) cyclin-dependent kinase inhibitor, urokinase type plasminogen activator receptor and several others (40–45). In many cases Sp1 acts in conjunction with Smads (40,41,43,44). TGF-β is also known to inhibit the expression of a number of genes (24,25). However, in contrast to gene activation, the mechanisms responsible for transcriptional inhibition by the cytokine have been identified for only a few genes, and include the Smad3-mediated stimulation of expression of repressors that then interact with their recognition sequence in the promoter region of target genes (e.g. ATF-3 for Id1, SIP1 for E-cadherin and high-mobility group protein-IY for nitric oxide synthase 2) (46–48). In the case of Sp1 and Sp3, although Sp1 acts mainly as an activator of gene transcription, Sp3 contains a transcriptionally repressive domain and can act as an activator or a repressor, depending on the promoter and cell type (39). Interestingly, the TGF-β-mediated downregulation of human type II collagen expression in articular chondrocytes occurs through changes in the Sp3/Sp1 ratio (49). It was found that TGF-β increased the expression of Sp3 in articular chondrocytes without significant changes to its binding level, but repressed both the synthesis and binding activity of Sp1 (49). A similar transcriptional regulation of TGF-β1 receptor is also dependent upon the Sp1/Sp3 ratio (50). The mechanism for the regulation of the LPL gene, however, appears to be different from these cases given that there are no changes in both the expression of Sp1 and Sp3 in macrophages and their binding to recognition sequences in the regulatory region (Figures 4 and 6). Finally, TGF-β is also known to inhibit differentiation of myoblasts, osteoblasts and adipocytes through Smad3-mediated functional repression of key transcription factors involved in the differentiation programme (e.g. Runx2/CBFA1, MyoD and C/EBPs for the differentiation of osteoblasts, myoblasts and adipocytes, respectively) (51–53). A TGF-β-mediated modulation of the trans-activation potential of Sp1 and Sp3 remains the most likely mechanism for the regulation of LPL gene expression. This could involve post-translational modification of Sp1/Sp3, limited availability of co-activators shared with factors whose expression is induced by TGF-β, or a cytokine-mediated interaction(s) with specific co-repressors.
A role for Sp1 and Sp3 in the constitutive expression of LPL has been reported previously (54). This site (position −91 to −83) coincides with a naturally occurring −73T/G substitution that is associated with decreased promoter activity (54). Sp1 and Sp3 in nuclear extracts from THP-1 macrophages were shown to specifically interact with this site (54). This site, however, is clearly not necessary for the TGF-β response as its deletion does not affect the cytokine-mediated inhibition of LPL promoter activity (Figure 3).
Although binding sites for Sp1 members are present in the regulatory regions of a large number of genes, they are subject to differential regulation by TGF-β, with the cytokine either inducing or inhibiting the expression or having no effect. The precise reason(s) for such differential regulation of gene promoters by the Sp1 family remain currently unclear. The consensus sequence for binding by Sp1 is GGGGCGGGG (GC-rich boxes) or GGTGTGGGG (GT/CACC boxes) (39). However, Sp1 members have also been found to interact with sequences that diverge from this consensus (39). It is interesting to note that the Sp1 recognition sequences identified in the LPL gene (Figure 4B) diverge from this consensus. It is, therefore, possible that such variations in the Sp1/Sp3-binding sites between different regulatory regions could be responsible for the differential actions of TGF-β. An alternative explanation for the differential action could be the involvement of sites for two or more transcription factors in the response. Indeed, as detailed above, the Sp1 sites have been shown to act in conjunction with sites for other transcription factors (40,41,43,44).
TGF-β and IFN-γ often have opposite effects in a range of cellular functions (55). It has been shown that IFN-γ induces the expression of Smad7, an antagonistic Smad, which prevents the interaction of Smad3 with the TGF-β receptor (55). In contrast to such findings, both IFN-γ and TGF-β inhibit LPL gene transcription and the responses are mediated through Sp1 and Sp3 [(26–28); this study]. However, the mechanisms by which IFN-γ and TGF-β mediate the response difference. Thus, in contrast to TGF-β, IFN-γ reduces the binding of Sp1 and Sp3 by decreasing the DNA-binding activity of Sp1 and the steady-state polypeptide levels of Sp3 (26).
In conclusion, we have identified a novel pathway by which TGF-β inhibits LPL gene transcription that involves modulation of the action of Sp1 and Sp3. Future studies should seek to identify the signal transduction pathways involved in the response and the molecular mechanisms by which TGF-β modulates Sp1 and Sp3.
Acknowledgments
We thank Jeffrey Gimble for the LPL promoter–luciferase DNA constructs, Joan Massagué for the p3TP-Lux plasmid and the Wellcome Trust and the British Heart Foundation for financial support. Funding to pay the Open Access publication charges for this article was provided by the Wellcome Trust.
REFERENCES
- 1.Mead J.R., Irvine S.A., Ramji D.P. Lipoprotein lipase: structure, function, regulation and role in disease. J. Mol. Med. 2002;80:753–769. doi: 10.1007/s00109-002-0384-9. [DOI] [PubMed] [Google Scholar]
- 2.Mead J.R., Ramji D.P. The pivotal role of lipoprotein lipase in atherosclerosis. Cardiovasc. Res. 2002;55:261–269. doi: 10.1016/s0008-6363(02)00405-4. [DOI] [PubMed] [Google Scholar]
- 3.O'Brien K.D., Gordon D., Deeb S., Ferguston M., Chait A. Lipoprotein lipase is synthesized by macrophage-derived foam cells in human coronary atherosclerotic plaques. J. Clin. Invest. 1992;89:1544–1550. doi: 10.1172/JCI115747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Renier G., Skamene E., DeSanctis J.B., Radzioch D. High macrophage lipoprotein lipase expression and secretion are associated in inbred murine strains with susceptibility to atherosclerosis. Arterioscler. Thromb. 1993;13:190–196. doi: 10.1161/01.atv.13.2.190. [DOI] [PubMed] [Google Scholar]
- 5.Sartippour M.R., Renier G. Upregulation of macrophage lipoprotein lipase in patients with type 2 diabetes: role of peripheral factors. Diabetes. 2000;49:597–602. doi: 10.2337/diabetes.49.4.597. [DOI] [PubMed] [Google Scholar]
- 6.Beauchamp M.-C., Letendre E., Renier G. Macrophage lipoprotein lipase expression is increased in patients with heterozygous familial hypercholesterolemia. J. Lipid Res. 2002;43:215–222. [PubMed] [Google Scholar]
- 7.Babaev V.R., Fazio S., Gleaves L.A., Carter K.J., Semenkovich C.F., Linton M.F. Macrophage LPL promotes foam cell formation and atherosclerosis in vivo. J. Clin. Invest. 1999;103:1697–1705. doi: 10.1172/JCI6117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Babaev V.R., Patel M.B., Semenkovich C.F., Fazio S., Linton M.F. Macrophage lipoprotein lipase promotes foam cell formation and atherosclerosis in low density lipoprotein receptor-deficient mice. J. Biol. Chem. 2000;275:26293–26299. doi: 10.1074/jbc.M002423200. [DOI] [PubMed] [Google Scholar]
- 9.Van Eck M., Zimmermann R., Groot P.H., Zechner R., Van Berkel T.J. Role of macrophage-derived lipoprotein lipase in lipoprotein metabolism and atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2000;20:53–62. doi: 10.1161/01.atv.20.9.e53. [DOI] [PubMed] [Google Scholar]
- 10.Wilson K., Fry G.L., Chappell D.A., Sigmund C.D., Medh J.D. Macrophage-specific expression of human lipoprotein lipase accelerates atherosclerosis in transgenic apolipoprotein E knockout mice but not in C57BL/6 mice. Arterioscler. Thromb. Vasc. Biol. 2001;21:1809–1815. doi: 10.1161/hq1101.097805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Panousis C.G., Evans G., Zuckerman S.H. TGF-β increases cholesterol efflux and ABC-1 expression in macrophage-derived foam cells: opposing the effects of IFN-γ. J. Lipid Res. 2001;42:856–863. [PubMed] [Google Scholar]
- 12.Argmann C.A., Van Den Diepstraten C.H., Sawyez C.G., Edwards J.Y., Hegele R.A., Wolfe B.M., Huff M.W. Transforming growth factor-β1 inhibits macrophage cholesteryl ester accumulation induced by native and oxidized VLDL remnants. Arterioscler. Thromb. Vasc. Biol. 2001;21:2011–2018. doi: 10.1161/hq1201.099426. [DOI] [PubMed] [Google Scholar]
- 13.Bottalico L.A., Wager R.E., Agellon L.B., Assoian R.K., Tabas I. Transforming growth factor-β1 inhibits scavenger receptor activity in THP-1 human macrophages. J. Biol. Chem. 1991;266:22866–22871. [PubMed] [Google Scholar]
- 14.Draude G., Lorenz R.L. TGF-β1 downregulates CD36 and scavenger receptor A but upregulates LOX-1 in human macrophages. Am. J. Physiol. Heart Circ. Physiol. 2000;278:H1042–H1048. doi: 10.1152/ajpheart.2000.278.4.H1042. [DOI] [PubMed] [Google Scholar]
- 15.Shull M.M., Ormsby I., Kier A.B., Pawlowski S., Diebold R.L., Yin M., Allen R., Sidman C., Proetzel G., Calvin D., Annunziata N., Doetschman T. Targeted disruption of mouse transforming growth factor-β1 gene results in multifocal inflammatory disease. Nature. 1992;359:693–699. doi: 10.1038/359693a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edwards D.R., Murphy G., Reynolds J.J., Whitman S.E., Dochertt A.J., Angel P., Heath J.K. Transforming growth factor-β modulates the expression of collagenase and metalloproteinase inhibitor. EMBO J. 1987;6:1899–1904. doi: 10.1002/j.1460-2075.1987.tb02449.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ding A., Nathan C.F., Graycar J., Derynk R., Stuehr D.J., Srimal S. Macrophage deactivating factor and transforming growth factor-β1, -β2 and -β3 inhibit induction of macrophage nitric oxide synthesis by IFN-γ. J. Immunol. 1990;145:940–944. [PubMed] [Google Scholar]
- 18.Di Febbo C., Baccante G., Reale M., Castellani M.L., Angelini A., Cuccurullo F., Porreca E. Transforming growth factor β1 induces IL-1 receptor antagonist production and gene expression in rat vascular smooth muscle cells. Atherosclerosis. 1998;136:377–382. doi: 10.1016/s0021-9150(97)00240-2. [DOI] [PubMed] [Google Scholar]
- 19.Mallat Z., Gojova A., Marchiol-Fournigault C., Esposito B., Kamate C., Merval R., Fradelizi D., Tedgui A. Inhibition of transforming growth factor-β signaling accelerates atherosclerosis and induces an unstable plaque phenotype in mice. Circ. Res. 2001;89:930–934. doi: 10.1161/hh2201.099415. [DOI] [PubMed] [Google Scholar]
- 20.Reckless J., Metcalf J., Grainger D.J. Tamoxifen decreases cholesterol sevenfold and abolishes lipid lesion development in apolipoprotein E knockout mice. Circulation. 1997;95:1542–1548. doi: 10.1161/01.cir.95.6.1542. [DOI] [PubMed] [Google Scholar]
- 21.Lutgens E., Cleutjens K.B., Heeneman S., Koteliansky V.E., Burkly L.C., Daemen M.J. Both early and delayed anti-CD40L antibody treatment induces a stable plaque phenotype. Proc. Natl Acad. Sci. USA. 2000;97:7464–7469. doi: 10.1073/pnas.97.13.7464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stefoni S., Cianciolo G., Donati G., Dormi A., Silvestri M.G., Coli L., Pascalis A.D., Lannelli S. Low TGF-β1 serum levels are a risk factor for atherosclerosis disease in ESRD patients. Kidney Int. 2002;61:324–335. doi: 10.1046/j.1523-1755.2002.00119.x. [DOI] [PubMed] [Google Scholar]
- 23.Robertson A.K., Rudling M., Zhou X., Gorelik L., Flavell R.A., Hansson G.K. Disruption of TGF-β signaling in T cells accelerates atherosclerosis. J. Clin. Invest. 2003;112:1342–1350. doi: 10.1172/JCI18607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Derynck R., Zhang Y.E. Smad-dependent and Smad-independent pathways in TGF-β family signalling. Nature. 2003;425:577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- 25.Shi Y., Massague J. Mechanisms of TGF-β signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- 26.Hughes T.R., Tengku-Muhammad T.S., Irvine S.A., Ramji D.P. A novel role of Sp1 and Sp3 in the interferon-γ-mediated suppression of macrophage lipoprotein lipase gene transcription. J. Biol. Chem. 2002;277:11097–11106. doi: 10.1074/jbc.M106774200. [DOI] [PubMed] [Google Scholar]
- 27.Tengku-Muhammad T.S., Hughes T.R., Cryer A., Ramji D.P. Differential regulation of lipoprotein lipase in the macrophage J774.2 cell line by cytokines. Cytokine. 1996;8:525–533. doi: 10.1006/cyto.1996.0071. [DOI] [PubMed] [Google Scholar]
- 28.Tengku-Muhammad T.S., Cryer A., Ramji D.P. Synergism between IFN-γ and TNF-α in the regulation of lipoprotein lipase in the macrophage J774.2 cell line. Cytokine. 1998;10:38–48. doi: 10.1006/cyto.1997.0254. [DOI] [PubMed] [Google Scholar]
- 29.Mead J.R., Hughes T.R., Irvine S.A., Singh N.N., Ramji D.P. Interferon-γ stimulates the expression of the inducible cAMP early repressor in macrophages through the activation of casein kinase 2: a potentially novel pathway for IFN-γ-mediated inhibition of gene transcription. J. Biol. Chem. 2003;278:17741–17751. doi: 10.1074/jbc.M301602200. [DOI] [PubMed] [Google Scholar]
- 30.Foka P., Irvine S.A., Kockar F., Ramji D.P. Interleukin-6 represses the transcription of the CCAAT/enhancer binding protein-α gene in hepatoma cells by inhibiting its ability to autoactivate the proximal promoter region. Nucleic Acids Res. 2003;31:6722–6732. doi: 10.1093/nar/gkg861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kousteni S., Kockar F.T., Sweeney G.E., Ramji D.P. Characterisation and developmental regulation of the Xenopus laevis CCAAT-enhancer binding protein-β gene. Mech. Dev. 1998;77:143–148. doi: 10.1016/s0925-4773(98)00128-2. [DOI] [PubMed] [Google Scholar]
- 32.Hildebrandt A.L., Neufer P.D. Exercise attenuates the fasting-induced transcriptional activation of metabolic genes in skeletal muscle. Am. J. Physiol. 2000;278:E1078–E1086. doi: 10.1152/ajpendo.2000.278.6.E1078. [DOI] [PubMed] [Google Scholar]
- 33.Duque M., Graupner M., Stutz H., Wicher I., Zechner R., Paltauf F., Hermetter A. New fluorogenic triacylglycerol analogs as substrates for the determination and chiral discrimination of lipase activities. J. Lipid Res. 1996;37:868–876. [PubMed] [Google Scholar]
- 34.Gimble J.M., Hua X., Wanker F., Morgan C., Robinson C., Hill M.R., Nadon N. In vitro and in vivo analysis of murine lipoprotein lipase gene promoter: tissue-specific expression. Am. J. Physiol. 1995;268:E213–E218. doi: 10.1152/ajpendo.1995.268.2.E213. [DOI] [PubMed] [Google Scholar]
- 35.Kockar F.T., Foka P., Hughes T.R., Kousteni S., Ramji D.P. Analysis of the Xenopus laevis CCAAT-enhancer binding protein α gene promoter demonstrates species-specific differences in the mechanisms for both auto-activation and regulation by Sp1. Nucleic Acids Res. 2001;29:362–372. doi: 10.1093/nar/29.2.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Inaba T., Kawamura M., Gotoda T., Harada K., Shimada M., Ohsuga J., Shimano H., Akanuma Y., Yazaki Y., Yamada N. Effects of platelet-derived growth factor on the synthesis of lipoprotein lipase in human monocyte-derived macrophages. Arterioscler. Thromb. Vasc. Biol. 1995;15:522–528. doi: 10.1161/01.atv.15.4.522. [DOI] [PubMed] [Google Scholar]
- 37.Wrana J.L., Attisano L., Carcamo J., Zentella A., Doody J., Laito M., Wang X.-F., Massagué J. TGF-β signals through a heteromeric protein kinase receptor complex. Cell. 1992;71:1003–1014. doi: 10.1016/0092-8674(92)90395-s. [DOI] [PubMed] [Google Scholar]
- 38.Kennett S.B., Udvadia A.J., Horowitz J.M. Sp3 encodes multiple proteins that differ in the capacity to stimulate or repress transcription. Nucleic Acids Res. 1997;25:3110–3117. doi: 10.1093/nar/25.15.3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Philipsen S., Suske G. A tale of three fingers: the family of mammalian Sp/XKLF transcription factors. Nucleic Acids Res. 1999;27:2991–3000. doi: 10.1093/nar/27.15.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jinnin M., Ihn H., Asano Y., Yamane K., Trojanowska M., Tamaki K. Tenascin-C upregulation by transforming growth factor-β in human dermal fibroblasts involves Smad3, Sp1, and Ets1. Oncogene. 2004;23:1656–1667. doi: 10.1038/sj.onc.1207064. [DOI] [PubMed] [Google Scholar]
- 41.Law A.K., Gupta D., Levy S., Wallace D.C., McKeon R.J., Buck C.R. TGF-β1 induction of the adenine nucleotide translocator 1 in astrocytes occurs through Smads and Sp1 transcription factors. BMC Neurosci. 2004;5:1. doi: 10.1186/1471-2202-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Botella L.M., Sanchez-Elsner T., Rius C., Corbi A., Bernabeu C. Identification of a critical Sp1 site within the endoglin promoter and its involvement in the transforming growth factor-β stimulation. J. Biol. Chem. 2001;276:34486–34494. doi: 10.1074/jbc.M011611200. [DOI] [PubMed] [Google Scholar]
- 43.Inagaki Y., Nemoto T., Nakao A., Dijke P.T., Kobayashi K., Takehara K., Greenwel P. Interaction between GC box binding factors and Smad proteins modulates cell lineage-specific α2 (I) collagen gene transcription. J. Biol. Chem. 2001;276:16573–16579. doi: 10.1074/jbc.M010485200. [DOI] [PubMed] [Google Scholar]
- 44.Feng X.H., Lin X., Derynck R. Smad2, Smad3 and Smad4 cooperate with Sp1 to induce p15 (Ink4B) transcription in response to TGF-β. EMBO J. 2000;19:5178–5193. doi: 10.1093/emboj/19.19.5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park I.K., Lyu M.A., Yeo S.J., Han T.H., Kook Y.H. Sp1 mediates constitutive and transforming growth factor-β-inducible expression of urokinase type plasminogen activator receptor gene in human monocyte-like U937 cells. Biochim. Biophys. Acta. 2000;1470:302–310. doi: 10.1016/s0167-4781(99)00246-8. [DOI] [PubMed] [Google Scholar]
- 46.Kang Y., Chen C.-R., Massagué J. A self-enabling TGF-β response coupled to stress signaling: Smad engages stress response factor ATF3 for Id1 repression in epithelial cells. Mol. Cell. 2003;11:915–926. doi: 10.1016/s1097-2765(03)00109-6. [DOI] [PubMed] [Google Scholar]
- 47.Comijn J., Berx G., Vermassen P., Verschueren K., van Grunsven L., Bruyneel E., Mareel M., Huylebroeck D., van Roy F. The two-handed E box binding zinc finger protein SIP1 downregulates E-cadherin and induces invasion. Mol. Cell. 2001;7:1267–1278. doi: 10.1016/s1097-2765(01)00260-x. [DOI] [PubMed] [Google Scholar]
- 48.Pellacani A., Wiesel P., Razavi S., Vasilj V., Feinberg M.W., Chin M.T., Reeves R., Perrella M.A. Down-regulation of high mobility group-I(Y) protein contributes to the inhibition of nitric-oxide synthase 2 by transforming growth factor-β1. J. Biol. Chem. 2001;276:1653–1659. doi: 10.1074/jbc.M008170200. [DOI] [PubMed] [Google Scholar]
- 49.Chadjichristos C., Ghayor C., Herrouin J.-F., Ala-Kokko L., Suske G., Pujol J.-P., Galéra P. Down-regulation of human type II collagen gene expression by transforming growth factor-β1 (TGF-β1) in articular chondrocytes involves SP3/SP1 ratio. J. Biol. Chem. 2002;277:43903–43917. doi: 10.1074/jbc.M206111200. [DOI] [PubMed] [Google Scholar]
- 50.Ammanamanchi S., Brattain M.G. Sp3 is a transcriptional repressor of transforming growth factor-β receptors. J. Biol. Chem. 2001;276:3348–3352. doi: 10.1074/jbc.M002462200. [DOI] [PubMed] [Google Scholar]
- 51.Alliston T., Choy L., Ducy P., Karsenty G., Derynck R. TGF-β-induced repression of CBFA1 by Smad3 decreases cbfa1 and osteocalcin expression and inhibits osteoblast differentiation. EMBO J. 2001;20:2254–2272. doi: 10.1093/emboj/20.9.2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu D., Black B.L., Derynck R. TGF-β inhibits muscle differentiation through functional repression of myogenic transcription factors by Smad3. Genes Dev. 2001;15:2950–2966. doi: 10.1101/gad.925901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Choy L., Derynck R. Transforming growth factor-β inhibits adipocyte differentiation by Smad3 interacting with CCAAT/enhancer-binding protein (C/EBP) and repressing C/EBP transactivation function. J. Biol. Chem. 2003;278:9609–9619. doi: 10.1074/jbc.M212259200. [DOI] [PubMed] [Google Scholar]
- 54.Yang W.-S., Deeb S.S. Sp1 and Sp3 transactivate the human lipoprotein lipase gene promoter through binding to a CT element: synergy with the sterol regulatory element binding protein and reduced transactivation of a naturally occurring promoter variant. J. Lipid Res. 1998;39:2054–2064. [PubMed] [Google Scholar]
- 55.Ulloa L., Doody J., Massagué J. Inhibition of transforming growth factor-β/SMAD signalling by the interferon-γ/STAT pathway. Nature. 1999;397:710–713. doi: 10.1038/17826. [DOI] [PubMed] [Google Scholar]