Abstract
The possibilities for low-frequency horizontal transfer of the self-transmissible chlorocatechol degradative genes (clc) from Pseudomonas sp. strain B13 were investigated in activated-sludge microcosms. When the clc genes were transferred into an appropriate recipient bacterium such as Pseudomonas putida F1, a new metabolic pathway for chlorobenzene degradation was formed by complementation which could be selected for by the addition of mono- or 1,4-dichlorobenzene (CB). Under optimized conditions with direct donor-recipient filter matings, very low transfer frequencies were observed (approximately 3.5 × 10−8 per donor per 24 h). In contrast, in matings on agar plate surfaces, transconjugants started to appear after 8 to 10 days, and their numbers then increased during prolonged continuous incubation with CB. In activated-sludge microcosms, CB-degrading (CB+) transconjugants of strain F1 which had acquired the clc genes were detected but only when strain B13 cell densities of more than 105 CFU/ml could be maintained by the addition of its specific growth substrate, 3-chlorobenzoate (3CBA). The CB+ transconjugants reached final cell densities of between 102 and 103 CFU/ml. When strain B13 was inoculated separately (without the designated recipient strain F1) into an activated-sludge microcosm, CB+ transconjugants could not be detected. However, in this case a new 3CBA-degrading strain appeared which had acquired the clc genes from strain B13. The effects of selective substrates on the survival and growth of and gene transfer between bacteria degrading aromatic pollutants in a wastewater ecosystem are discussed.
Horizontal gene transfer between bacteria is a rather general process, leading to the distribution of many traits, such as antibiotic resistance determinants or genes encoding degradative pathways. Transfer occurs by several mechanisms, such as plasmid conjugation, (conjugative) transposition, bacteriophage transduction, and transformation (2, 9, 14). The efficiency and frequency of gene transfer depend on characteristics of the strain and those of the transferred element. In addition, the physiological status of the cell and various environmental parameters play a role (28, 31). In natural environments such as soil or aquatic systems, gene transfer occurs mostly at low frequencies (1, 15, 20, 21, 30, 36).
One factor which has received little attention is the role of a selective environment. Selective constraints by themselves probably do not stimulate gene transfer (a possible exception is the regulation of conjugative transposition in Bacteroides [29]) but determine whether the organisms acquiring new genetic material will multiply and grow. In light of recurring discussions on the risks of gene transfer in the environment, it is important to understand if and how factors considered to be selective actually function. The effect and importance of selective environments are well known from the problems associated with acquired antibiotic resistance in pathogenic bacteria (17, 35). In contrast, the actual selective advantage of, for instance, genes encoding degradative pathways is mostly unknown for natural environments.
Antibiotic resistance genes have been used in numerous studies as markers for gene transfer in microcosms mimicking natural ecosystems (1, 15, 18, 23, 30). Such genetic markers were used because of their easy detection, but they are not an ideal choice for the determination of the actual influence of selective conditions on gene transfer. The addition of antibiotic(s) into a microcosm can prevent growth of nonresistant microorganisms which could act as recipients and may in some cases inhibit conjugational transfer itself. Most studies which used antibiotic resistance genes to determine gene transfer frequencies were performed without any selective (antibiotic) pressure.
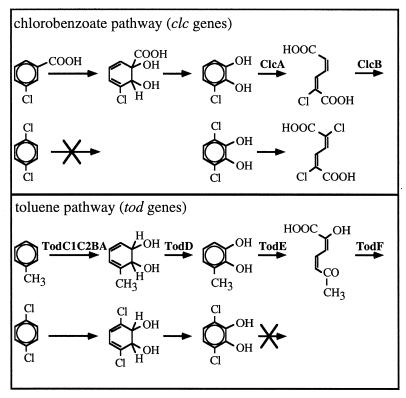
With degradative genes as markers, selective conditions can easily be maintained during gene transfer experiments without interference with the survival of nontarget microorganisms (21). For example, recipient bacteria having acquired new catabolic genes can be selected for by addition of the appropriate substrate. Complications in detecting and enumerating transconjugants arise when donor bacteria are also capable of growing on the specific substrate. Such complications can be circumvented by using donor and recipient bacteria each having one set of complementary degradative genes. For instance, the clc genes for the chlorocatechol pathway can transfer from the donor strain Pseudomonas sp. strain B13 to the recipient strain Pseudomonas putida F1 (22). Although the genetic basis for this transfer has not yet been fully elucidated, the transferred element carrying the clc genes seems to be a conjugative element capable of integrating into the chromosome (25). The resulting F1 transconjugants have the novel capability of metabolizing chlorobenzenes (Fig. 1), a feature which is not found in strain B13 or strain F1.
FIG. 1.
Overview of the pathway complementation for complete CB degradation (shown for 1,4-DCB). Pseudomonas sp. strain B13 possesses the ability to degrade 3-CB via 3-chlorocatechol to 3-oxoadipate. However, CB cannot be transformed to chlorocatechol by the chlorobenzoate pathway. Pseudomonas putida F1 metabolizes toluene via catechol and is also able to convert CB to chlorocatechols. Chlorocatechols were not further converted by the strain unless the genes for the chlorocatechol pathway from B13 were transferred to F1.
Here we used this complementation to study low-frequency gene transfer in activated-sludge microcosms. The selective growth advantage for transconjugants degrading chlorobenzenes enabled the observation of transfer events that would otherwise be below the detection limit. The effect of different donor and/or recipient cell densities on the clc transfer as well as the presence or absence of chlorobenzenes and 3-chlorobenzoate (3CBA) was addressed. Finally, we discuss the extent to which a specific metabolizable substrate imposes selective conditions in a complex ecosystem.
MATERIALS AND METHODS
Bacterial strains and relevant characteristics.
Pseudomonas sp. strain B13 can grow on 3CBA as the sole source of carbon and energy. The strain carries the self-transmissible clc genes for chlorocatechol degradation (22, 26). The clc genes were initially characterized from P. putida (pAC27) (3, 8) and partially from strain B13 (12). P. putida F1 can use toluene as the sole carbon and energy source. Toluene degradation is encoded by the chromosomally located tod genes (37, 38). Escherichia coli HB101 (pRK2013; tra+ Kmr) (7) and E. coli LE392 (RP1; tra+ Kmr) (4) were used as helper strains in triparental matings.
Media and culturing conditions.
For routine growth of the strains and their enumeration via selective plating, Z3 minimal medium (34) was used with the appropriate aromatic compounds as follows: for strain B13, 3CBA; for F1, toluene; and for chlorobenzene transconjugants, monochlorobenzene (MCB) or 1,4-dichlorobenzene (DCB). Ultrapure agar (Merck AG, Dietikon, Switzerland) was used for selective plating of reactor samples to minimize background growth by indigenous microorganisms. The pH indicator bromothymol blue was added to the agar plates in a concentration of 15 mg/liter to facilitate detection of colonies of strains F1 and of possible chlorobenzene transconjugants, as they then appeared yellowish upon growth on toluene or chlorobenzenes. Growth on toluene or chlorobenzenes was tested by incubating the Z3 agar plates in gas-tight glass jars, with the growth substrates supplied through the vapor phase. 3CBA was added directly into the agar at a concentration of 5 mM. For liquid cultures, toluene, MCB, and 1,4-DCB were supplied in a secondary phase (2,2,4,4,6,8,8-heptamethylnonane [HMN]; Sigma Chemical Co., St. Louis, Mo.). Toluene was dissolved in a ratio of 0.1 (vol/vol) HMN, MCB of 0.04, and 1,4-DCB of 0.02. Per liter of Z3 medium, 400 μl of toluene (346 mg), 400 μl of MCB (443 mg), or 400 mg of 1,4-DCB was added.
Filter, plate, and liquid matings.
Matings were performed with cultures pregrown on Luria-Bertani (LB) medium which were washed and resuspended in Z3 mineral medium prior to the experiments. Filter matings were performed on 0.45-μm-pore-size cellulose nitrate filters (Sartorius AG, Göttingen, Germany). Cells were transferred to the filter with a syringe, and the filter was incubated on an LB agar plate at 30°C for 24 h. Afterwards, the bacteria were resuspended from the filter by vortexing in 1 ml of Z3 medium and plating appropriate dilutions of the cell suspension on selective media. Plate matings between Pseudomonas sp. strain B13 and P. putida F1 were performed on Z3 agar plates in the presence of either MCB or 1,4-DCB as the sole source of carbon and energy. Different amounts of cells per plate (diameter, 9 cm) ranging from approximately 104 to 108 were tested. For liquid matings, the cells were resuspended in 200 ml of Z3 medium in a 2-liter Erlenmeyer flask and incubated on a shaker with 1,4-DCB as the only substrate. Samples of approximately 1 ml were taken regularly to determine cell numbers by selective plating and to measure chloride concentration. The chloride concentration in samples was measured with a Chlor-o-Counter according to the instructions given by the manufacturer (Flohr Instruments, Nieuwegein, The Netherlands). Preparation of cell extracts (from transconjugants and parent strains) and activity measurements of catechol 1,2-dioxygenase and catechol 2,3-dioxygenase were performed as described elsewhere (12, 22).
Microcosm experiments.
Microcosms consisted of 1.0-liter glass reactors and were operated with a volume of approximately 0.5 liter of culture medium and at a dilution rate of 0.04 h−1. The medium was sterile synthetic wastewater (SWW, based on DIN 38412 T24, German Industry Norm, 1981) whose total dissolved organic carbon content we increased to 375 mg/liter by taking threefold quantities of each of the carbon substrates (meat extract and peptone). The microcosms were maintained at 30°C and a pH of between 7.4 and 7.6 by automatic temperature and pH control. The mixed microbial community was obtained from an activated-sludge treatment process. For each experiment, the reactors were inoculated with 50 ml of sludge freshly obtained from the Kloten/Opfikon municipal sewage treatment plant near Zürich, Switzerland. The sludge culture was then incubated in the reactor for at least 48 h (approximately 2 volume changes) before inoculation of B13 and/or F1 and before any of the specific substrates was supplied (see below). The microcosms were constantly and vigorously aerated with air passing through a sterile filter at a flow rate of 100 to 120 ml/min. Stirring at 400 rpm was used to ensure complete mixing. Pseudomonas sp. strain B13 and P. putida F1 were pregrown overnight on a rich medium (LB or nutrient broth). In all experiments, the strains were inoculated to a final cell density of approximately 107 CFU/ml in the microcosm. An overview of the different experimental setups is given in Table 1. 3CBA was added directly into the SWW medium before autoclaving to a final concentration of 0.7 mM (109 mg/liter). Toluene was added into the medium stream at 10 μl/h (8.7 mg/h) via a syringe pump shortly before the medium entered the reactor. 1,4-DCB was added through the air, by directing an adjustable portion of the inflowing air through a vessel with crystalline 1,4-DCB. Air concentrations of 1,4-DCB were approximately 100 μg/liter. Samples of a 10-ml volume were taken from the reactors and processed to determine concentrations of aromatic compounds, optical density at 600 nm, and cell numbers of the applied strain(s). Cell numbers were derived by selective plating. The detection limit for the different strains in the reactors was 10 CFU/ml. Selective plating in combination with colony hybridization was used to verify the presence of the specific degradative genes.
TABLE 1.
Setup of the activated-sludge microcosm experiments
| Microcosm no. | Straina
|
Substrate
|
|||
|---|---|---|---|---|---|
| B13 | F1 | 3CBA | Toluene | 1,4-DCB | |
| I | + | + | − | − | − |
| II | + | + | − | − | + |
| III | + | + | + | + | − |
| IV | + | + | + | + | + |
| V | + | − | + | − | + |
Strains inoculated after 2 volume changes of sludge growth (the reactor volume was 500 ml). Total sludge density was maintained at an A600 of ≈0.8.
DNA isolation, DNA manipulations, and hybridizations.
Total DNA isolations from P. putida F1, Pseudomonas sp. strain B13, and putative transconjugants and Southern hybridizations were carried out as described elsewhere (11, 16). The DNA probe for the clc genes was a 4.5-kb PstI fragment containing the clcRAB genes from Ralstonia sp. strain JS705 (33). The probe for the tod genes was a 1.9-kb PstI fragment containing the todC2BA region (37). As a probe for detecting the 16S ribosomal DNA (rDNA) genes, we used a 700-bp cloned 16S rDNA fragment of Ralstonia sp. strain JS705 (33). For amplification of the 16S rRNA gene, the following eubacterial primers were used: V1.1, 5′GCG.GCG.TGC.CTA.ATA.CAT.GC 3′ (E. coli 16S rDNA bp 41 to 60), and V3.2, 5′ATC.TAC.GCA.TTT.CAC.CGC.TAC 3′ (705 to 685); or 970404, 5′GTG.CTG.CAG.GGT.TAC.CTT.GTT.ACG.ACT 3′ (E. coli 1510 to 1483), and 970405, 5′GGA.GAG.TTA.GAT.CTT.GGC.TCA.G 3′ (E. coli 6 to 27). Amplification was performed by using the PCR with an annealing temperature of 50°C (primers 970404 plus 970405) or 58°C (primers V1.1 plus V3.2) and for 35 cycles. Amplified DNAs were cloned into pGEM-T Easy (Promega Corporation, Madison, Wis.). DNA sequencing was performed as described elsewhere (11).
Chemical analysis.
For analysis of the 3CBA microcosm, samples were centrifuged to remove the biomass and 20 μl of 1 M phosphoric acid was added per ml to lower the pH. Concentrations of 3CBA were measured with a Gynko high-pressure liquid chromatograph (Gynkotek AG, Regensdorf, Switzerland) equipped with a Waters Nova-Pak C18 reversed-phase column (Machery-Nagel AG, Oensingen, Switzerland) and UV/VIS detector set at 206 nm. The mobile phase was a solution of 50% methanol and 50% NaH2PO4 (50 mM [pH 3.0]) at a flow rate of 1 ml/min. For 1,4-DCB analyses, microcosm samples were extracted with 2 volumes of hexane (Fluka, Buchs, Switzerland), and the extracted samples were stored in sealed vials at −20°C until they were analyzed. 1,4-DCB was analyzed on a Hewlett-Packard 5890 series II gas chromatograph equipped with an ECD detector.
RESULTS
Transfer of an element containing the clc genes from Pseudomonas sp. strain B13 to P. putida F1.
In filter matings on LB medium agar plates with strains B13 and F1, MCB+ transconjugants appeared at a low frequency of approximately 3.5 × 10−8 per donor per 24 h. The number of transconjugants was determined from the number of colonies growing on chlorobenzene within 5 days. A similar frequency was observed in triparental matings with strains B13 and F1 and a mobilizing strain [either E. coli HB101(pRK2013) or E. coli LE392 (RP1)].
In matings with donor and recipient cells streaked on the surface of mineral agar plates (incubated in the presence of 1,4-DCB or MCB), transconjugants able to utilize both 1,4-DCB and MCB were easily obtained, especially with high concentrations of parent cells. The transconjugant colonies in this case started to appear after 8 to 10 days of incubation with MCB or 1,4-DCB. It was difficult to enumerate transconjugant colonies and frequencies of transfer, since a mixture of small and large colonies which steadily increased with time was observed. The number of MCB+ colonies obtained after 17 days of incubation with different starting cell densities of the parent strains is shown in Table 2. On control plates with only F1 or B13, no MCB+ colonies were detected.
TABLE 2.
Numbers of MCB+ transconjugants obtained after 17 days in agar plate surface matings between Pseudomonas sp. strain B13 and P. putida F1 at different cell densities
| Cell densitya for indicated strain
|
No. of transconjugantsa,b | |
|---|---|---|
| B13 | F1 | |
| 1.4 × 108 | 2.6 × 108 | >500 |
| 1.4 × 107 | 2.6 × 107 | 106 ± 14 |
| 1.4 × 106 | 2.6 × 106 | 42 ± 8 |
| 1.4 × 105 | 2.6 × 105 | 25 ± 6 |
| 1.4 × 104 | 2.6 × 104 | 7 ± 2 |
CFU per plate.
Mean data from an experiment performed in triplicate.
Once selected, transconjugants capable of metabolizing chlorobenzenes grew faster on agar plates with MCB. Easily visible colonies formed within 3 to 5 days, indicating that we would be able to distinguish later in the microcosm experiments between chlorobenzene transconjugants which had arisen in the reactor and those that arose on the selective plates (not visible for the first 6 days). Therefore, in all other mating experiments, transconjugant formation was determined by the number of MCB+ colonies that grew to an easily visible size (≥1 mm) within 5 days.
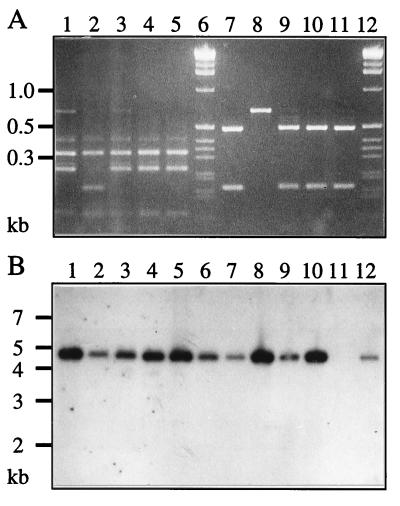
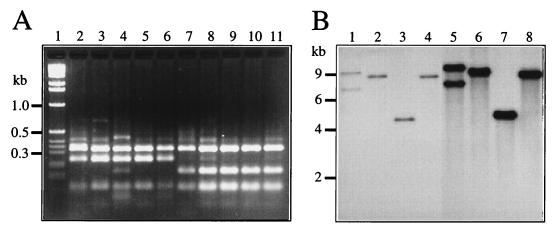
Individual 1,4-DCB+ transconjugant colonies obtained in the plate matings were all able to grow on 1,4-DCB, MCB, 3CBA, or toluene. Measurements of activities of catechol 1,2-dioxygenase and catechol 2,3-dioxygenase in cell extracts of the transconjugant grown on the different aromatic carbon sources indicated a mixed pattern of activity characteristic for both strain F1 and strain B13 (Table 3). Part of the 16S rDNA was amplified from total DNA by PCR, and the PCR products were digested with Sau3AI or HaeIII (Fig. 2A). The resulting banding patterns were identical for F1 and the transconjugants but different for B13. This is evidence that F1 was the recipient and B13 was the donor strain. In addition, total DNA of individual transconjugants and of strains B13 and F1 was isolated, digested with BglII or HindIII, separated by agarose gel electrophoresis, and hybridized with a probe containing the clc genes. In this case, Pseudomonas sp. strain B13 and all 10 analyzed transconjugants revealed an identical hybridization pattern with the clc gene probe. The clc gene cluster was present on a 4.5-kb BglII fragment (Fig. 2B) and on a 9-kb HindIII fragment (data not shown). This indicated that the clc genes had been transferred intact from strain B13 to strain F1.
TABLE 3.
Enzyme activities of chlorocatechol 1,2-dioxygenase (CC-1,2-D) and catechol 2,3-dioxygenase (C-2,3-D) in cell extracts of strains B13 and F1 and a transconjugant of F1 (RR1) capable of growth on CB
| Strain | Growth substrate | Sp acta
|
|
|---|---|---|---|
| CC-1,2-Db | C-2,3-Dc | ||
| B13 | 3CBA | 451 ± 133d | 0 |
| F1 | Toluene | 0 | 98 ± 11 |
| RR1 | Acetate | 0 | 0 |
| RR1 | 3CBA | 469 ± 150 | 0 |
| RR1 | Toluene | 0 | 116 ± 3 |
| RR1 | MCB | 289 ± 106 | 45 ± 5 |
Expressed in nanomoles per minute per milligram of protein.
The substrate for chlorocatechol 1,2-dioxygenase was 3-chlorocatechol.
The substrate for the catechol 2,3-dioxygenase was 3-methylcatechol.
Mean data from measurements of three independently grown cultures.
FIG. 2.
Confirmation of the transconjugants as being of F1 origin and carrying the clc genes. (A) Sau3AI (lanes 1 to 5) and HaeIII digestions (lanes 7 to 11) of PCR products obtained from different transconjugants, F1 and B13 with the eubacterial 16S rDNA primers V1.1 plus V3.2. Lanes: 1 and 7, F1; 2 and 8, B13; 3 to 5 and 9 to 11, three transconjugants; 6 and 12, molecular size marker. (B) Autoradiogram of hybridization with the clc probe to total DNAs digested with BglII. Lanes: 1 to 10, different independently derived transconjugants; 11, F1; 12, B13. Size markers are indicated on the left in kilobase pairs.
Transfer of the clc genes from Pseudomonas sp. strain B13 to P. putida F1 in liquid cultures.
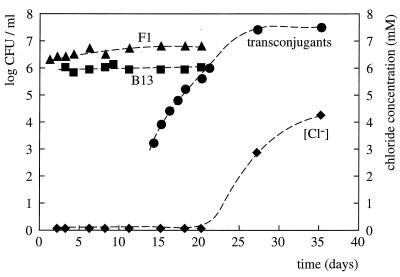
In mixtures of strains B13 and F1, each at approximately 5 × 106 CFU/ml, incubated in a flask with Z3 mineral medium and with 1,4-DCB as the sole carbon and energy source, CB+ transconjugants appeared after 14 days (Fig. 3). The transconjugants grew and stabilized at a final cell density of approximately 5 × 107 CFU/ml within 10 days after their first appearance. Since the transconjugant strain possessed all three phenotypes (3CBA+, toluene+, and 1,4-DCB+) used for selective enumeration, donor and recipient strains could no longer be enumerated at high transconjugant numbers. The transconjugants completely mineralized the 1,4-DCB in the medium, as observed from the formation of chloride (Fig. 3). In cultures operated in parallel with strain B13 or F1 only, no CB+ transconjugants or formation of chloride could be observed (data not shown). Interestingly, both parent strains remained viable throughout the experiment, although no growth substrate was available to them. This experiment was repeated with various lower initial cell densities of donor and recipient (104 to 106 CFU/ml). It was not possible to maintain such low cell densities, because both populations increased again to densities of approximately 106 CFU/ml, possibly due to traces of carbon substrate in the medium.
FIG. 3.
Increase of CB-metabolizing transconjugants in a two-strain mixture of F1 and B13, incubated in mineral medium with 1,4-DCB as the sole substrate. Symbols: diamonds, chloride concentration; circles, cell numbers derived on CB plates (transconjugants); squares, cell numbers derived on 3-CBA (strain B13); triangles, cell numbers on toluene (strain F1).
Survival of and gene transfer between Pseudomonas sp. strain B13 and P. putida F1 in activated-sludge microcosms.
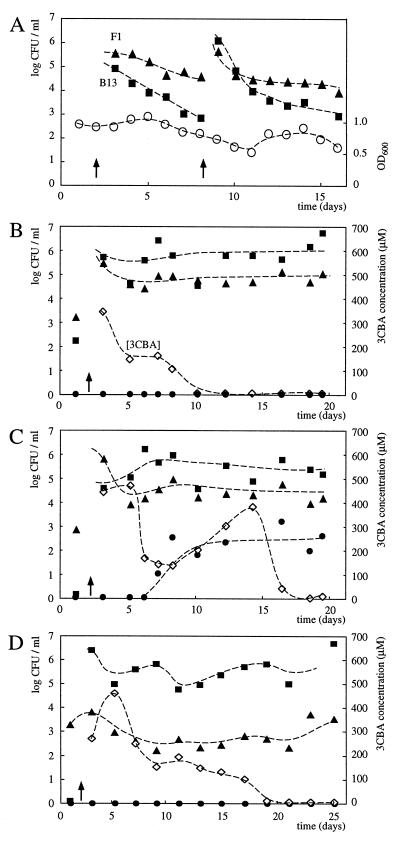
Pseudomonas sp. strain B13 and P. putida F1 were both inoculated into two microcosms (I and II, Table 1) operated in parallel. In this and the following experiments, each of the strains was inoculated to a final cell density of approximately 107 CFU/ml. No transconjugants appeared during the 2-week monitoring period, either in the reactor to which 1,4-DCB had been added or in the one without 1,4-DCB. The survival of strain F1 and especially that of strain B13 was poor. The cell numbers of these strains in the reactors decreased within a couple of days to below 105 CFU/ml (Fig. 4A). Therefore, the two strains were again inoculated after 6 days. Even after reinoculation, there was no improved survival of strain B13, whereas strain F1 stabilized between 104 and 105 CFU/ml. There was no difference in the population sizes for B13 and F1 between the two parallel microcosms (with and without the addition of 1,4-DCB).
FIG. 4.
Survival of strains B13 and F1 and increase of CB transconjugants in activated-sludge microcosms. (A) Microcosms I and II, operated without 3CBA or toluene but microcosm number II with 1,4-DCB. (B) Microcosm III operated with 3CBA and toluene but without 1,4-DCB. (C) Microcosm IV, similar to III but with 1,4-DCB. (D) Microcosm V inoculated with B13 only. The day of inoculation after the microcosm was started with sludge is indicated by a vertical arrow. Symbols: squares, cell numbers derived on 3CBA plates (mostly B13); triangles, those derived on toluene plates (mostly F1); closed circles, those derived on plates incubated with CB (transconjugants of F1 with strain B13 clc genes); open circles, optical density of the total sludge population (OD600); open diamonds, 3CBA concentration (micromolar). The data from the sludge microcosms are typical for the outcome of the gene transfer process but not for the exact moment at which the transconjugants appeared.
In a further experiment, two microcosms (III and IV, Table 1) were operated in parallel, now both with the addition of 3CBA and toluene and one with the addition of 1,4-DCB. The addition of these substrates started upon inoculation of the strains B13/F1 (any initial increase in the 3CBA concentration was due to wash-in). Indeed, the survival of both strains was improved, an effect which was most pronounced with regard to the population size of Pseudomonas sp. strain B13 (Fig. 4B). Compared to the previous experiment, the cell densities of strain B13 remained 2 to 3 orders of magnitude higher (around 106 CFU/ml). The survival of both Pseudomonas sp. strain B13 and P. putida F1 was slightly lower with 1,4-DCB than without it (Fig. 4C). In addition, degradation of 3CBA proceeded faster and more constantly in the microcosm without 1,4-DCB. Interestingly, CB+ bacteria started to appear from day 6 onward in the microcosm to which 1,4-DCB was added but not in the control without 1,4-DCB (Fig. 4C). The chlorobenzene degraders reached a density of between 102 and 103 CFU/ml.
To verify the nature of these chlorobenzene degraders, we isolated their total DNA and total DNA from strain F1, digested these with EcoRI or HindIII, and performed Southern hybridizations with a 16S rDNA probe. Identical banding patterns were observed for all isolated DNAs, indicating that strain F1 had also been the recipient strain in the reactors (data not shown). In addition, we verified the presence of the clc genes of strain B13 as described above, which indicated that the element containing the clc genes had been transferred (data not shown).
Finally, a microcosm (V, Table 1) into which only strain B13 was inoculated was operated. The substrate 3CBA (at a concentration of 0.7 mM) was added to maintain a relatively high population density of strain B13. In addition, 1,4-DCB was flushed through the reactor continuously to select for possible CB+ transconjugants which could arise due to transfer of the clc gene cluster from B13 to indigenous recipients. Indigenous toluene-metabolizing bacteria which hybridized to the tod gene probe were detected in the sludge at levels of around 103 CFU/ml (Fig. 4D). However, Southern hybridizations with the 16S rDNA probe performed on isolated total DNA of such strains, digested with EcoRI or HindIII, showed different banding patterns than those of strain F1 (data not shown), indicating that none of them was identical to F1.
Strain B13 survived well in microcosm number V, and 3CBA was degraded to low levels (Fig. 4D). Despite the presence of possible indigenous recipients and good survival of strain B13, no CB+ transconjugants were detected during this experiment. Interestingly, though, on selective agar plates with 3CBA as the carbon source, a strain different from B13 appeared in samples taken from the reactor after 9 days. Before 9 days, this strain had not been detected on 3CBA-selective plates. First of all, it could be distinguished from strain B13 on the basis of colony morphology. The strain reached numbers similar to those of strain B13 at the end of the experiment; however, we did not perform an exact enumeration of its population size. PCR amplification of the 16S rDNA from such colonies and subsequent restriction enzyme digestion showed that they were all identical to each other but different from B13 (Fig. 5A). Total DNA was isolated from the new 3CBA+ strain, named S11, and from B13. Southern hybridization with the clc gene probe revealed identical restriction patterns for S11 and B13 (Fig. 5B), suggesting that the clc genes had been transferred from strain B13. To further confirm the presence of the clc element from B13 in strain S11, a plate mating was performed between S11 and P. putida F1. MCB-degrading transconjugants were also obtained in this case. In Southern hybridizations of XbaI-digested total DNAs with the clc gene probe, the banding patterns of the new F1 transconjugants were identical (data not shown) to those obtained previously from matings between B13 and F1 (25). The DNA sequence of a part of the 16S rDNA from the strain S11 was most similar (98.2% identity in 1,496-nt overlap) to that of Ralstonia eutropha.
FIG. 5.
Verification of the indigenous transconjugants (Ralstonia sp. strain S11) to which the clc genes of B13 were transferred. (A) Sau3AI digestions of PCR-amplified (primers V1.1 plus V3.2) 16S rDNAs from 3CBA degraders from microcosm V. Lanes: 2 to 6, strain S11; 7 to 11, B13. (B) Southern hybridization of total DNAs of S11 (lanes 1 to 4) and B13 (lanes 5 to 8) with the clc gene probe. DNAs digested with BamHI (lanes 1 and 5), HindIII (lanes 2 and 6), NotI (lanes 3 and 7), and SmaI (lanes 4 and 8) are shown.
DISCUSSION
Gene transfer by bacterial conjugation occurs most efficiently when high numbers of donor and recipient bacteria are present in mating aggregates. This is achieved in the laboratory by defined filter or agar plate surface matings. In environments where cell densities are much lower or many different species are present, conjugation frequencies are several orders of magnitude lower. For instance, when studying transfer of the RP4p plasmid between P. fluorescens strains in filter matings and in soil microcosms, Smit (30) found 100-fold-lower transfer frequencies in soil. Similar observations were made for transfer of the plasmid pJP4 from Alcaligenes eutrophus to Variovorax paradoxus in soil microcosms (20) and for the mercury resistance plasmid pQKH6 between P. putida strains in pilot-scale percolating filter sewage treatment systems (1). In the study of gene transfer in natural environments, such observations are important. However, these observations are dependent upon direct detection of the transconjugant bacteria. In many other cases, such as transfer of chromosomal markers (30) or transfer of plasmids to uncharacterized recipient bacteria, frequencies will be too low to be detected directly. Instead of focusing on determining transfer frequencies, researchers should therefore concentrate on the possibilities of transconjugants to grow and increase their population size; it is this aspect which is often neglected in risk-associated studies.
Here we used transfer of the clc element of Pseudomonas sp. strain B13 to P. putida F1 to obtain a system suitable for studying very low gene transfer frequencies under selective conditions. The clc genes of strain B13 are very likely located on a conjugative transposable element capable of integrating into the chromosome of various other host strains (25). The estimated frequency of this conjugation and integration from B13 into F1 was rather low: 3.5 × 10−8 per donor in filter matings. Some of our observations with plate matings suggest that the transfer may become more efficient or triggered after prolonged incubation on mineral agar plates with MCB or 1,4-DCB. Transfer of the clc genes from B13 to F1 could be easily detected because F1 transconjugants expressed a complete pathway for CB degradation (22). Since very few bacterial species efficiently degrade CBs, selective conditions could be maintained for the transconjugants by adding CB to our experimental systems.
Despite the low transfer frequencies observed in filter matings and the small likelihood of donor-recipient contact in shaking flasks and wastewater microcosms, we could detect transfer of the clc element from B13 to F1. As expected, the CB transconjugant population size increased after an initial lag period. This period probably reflects the chance for transconjugants to arise at such relative low donor and recipient cell densities. Mathematical approaches have been made to investigate kinetics of plasmid transfer in liquid matings (13, 27). However, the mass action model proposed by Levin et al. is not applicable for matings with resting cells, such as in our shaking- flask experiments (13). In the wastewater microcosms it is difficult to calculate actual transfer frequencies, since the growth rate of the transconjugants in the microcosms would have to be known, and the presence of flocs complicates the estimation of cell-to-cell contacts (27). Therefore, transfer frequencies for the clc element in the liquid matings or wastewater microcosms were not calculated. The rise of the transconjugants appeared to be dependent on the donor and recipient cell population sizes. This was concluded mainly from plate matings, and from the microcosm experiments, in which transconjugants were only found when donor and recipient cell densities could be maintained around 105 CFU/ml or above. In the B13/F1 resting cell mixtures in shaking flasks, it was not possible to perform matings with cell densities lower than 106 CFU/ml.
The question remains whether the presence of a unique carbon source is sufficient to create conditions which select for the growth of a specific transconjugant. The two-culture liquid mating experiment clearly demonstrated the transconjugant’s growth advantage when CB was the only substrate available, since it was the only bacterium capable of utilizing CB in that system. In the activated-sludge microcosms not just one specific substrate was added; a combination of up to three substrates (3CBA, toluene, or 1,4-DCB), mixed with undefined other carbon sources (from peptone and meat extract), was added. In addition, many more species were present in the microcosms.
Without specific substrates added to the microcosms (3CBA and toluene), strains B13 and F1 maintained themselves, but at relatively low population levels. No transconjugants could be detected without or with the addition of 1,4-DCB. When 3CBA and toluene were added simultaneously, both B13 and F1 survived much better, indicating the selective effects of these substrates. However, their population sizes were still 100-fold lower than expected from biomass calculations. For example, with a 3CBA concentration of 0.7 mM in the incoming medium, about 106 CFU of B13 per ml were maintained. Yield calculations predict a population size of around 108 CFU/ml (at μmax = 0.13 h−1, Ks 3CBA = 0.05 mM, Ymax = 61.9 mg [dry weight]/mmol [32], and a mean dry weight per cell of 1.4 × 10−10 mg of C [19]). Similarly, the transconjugants’ population size (5 × 102 CFU/ml) reached after addition of 1,4-DCB was 200-fold lower than expected. The added amount of 1,4-DCB (calculated from the flux of 1,4-DCB from the vapor phase into the water phase, resulting in an available dissolved concentration of 0.8 mg/liter) would have been sufficient for sustaining a population size of 105 CFU/ml (24). Our observations of the relatively low maintenance of introduced bacteria in activated-sludge microcosms are similar to those described by others (21).
Competition with indigenous bacteria for the same specific substrates is certainly one explanation for a lower than maximally achievable population size, but this factor seemed to be limited to toluene. 3CBA and 1,4-DCB are not readily metabolized by the indigenous sludge bacteria, although part of these substrates may have been lost by incomplete degradation. The relative flux of a specific substrate compared to those of other metabolizable compounds has also been implicated in the selective maintenance of bacterial populations (5). However, if true, this would imply that part of the 3CBA and 1,4-DCB was not metabolized at all, since B13 and F1 transconjugants would prefer the carbon substrates from peptone and meat extract. The concept of relative fluxes is important, though, to explain that the concentration of a specific substrate per se does not lead to outgrowth of introduced or transconjugant bacteria. For example, in contrast to the limited maintenance of B13 in activated sludge plus the high input of 3CBA, 1 μM 3CBA was sufficient to promote growth of transconjugants carrying plasmid pBR60 in flow-through lake microcosms (10). In these lake microcosms, the total amount of metabolizable carbon was sufficient only for a total population size of between 105 and 106 CFU/ml. In the activated-sludge microcosms, limitation of a trace element (e.g., iron) may have been another reason for the low yield, based on utilization of specific substrate by the introduced or transconjugant strains. During competition with the indigenous microorganisms, part of the metabolizable carbon may be wasted by synthesizing and excreting iron chelators (6).
In summary, our results showed that although some genetic elements transfer at very low frequencies, their transfer occurs even under nonoptimal conditions such as those in sewage sludge. The transconjugants had in all cases obtained the clc element from the donor strain B13, and even without any specific recipient added to the system, the clc element ended up in an indigenous recipient. Most importantly, selection and growth of the introduced strains and transconjugants were dependent on the presence of sufficiently high concentrations of specific substrates (toluene, 3CBA, or 1,4-DCB). The detection of F1 transconjugants’ acquisition of the clc genes from strain B13 was only possible due to their selective advantage of utilizing 1,4-DCB, thereby increasing the population’s size to detectable densities. This demonstrates how the evolution of a new metabolic pathway can be based on rare transfer events and weak selective forces, as it most likely occurs under environmental conditions.
ACKNOWLEDGMENTS
We thank Thomas Fleischmann and Michael Nay for technical assistance and Thomas Egli for critically reading the manuscript.
This work was supported by grant 5002-038279 from the Swiss Priority Program Biotechnology.
REFERENCES
- 1.Ashelford K E, Fry J C, Learner M A. Plasmid transfer between strains of Pseudomonas putida, and their survival, within a pilot scale percolating-filter sewage treatment system. FEMS Microbiol Ecol. 1995;18:15–25. [Google Scholar]
- 2.Bertram J, Strätz M, Dürre P. Natural transfer of conjugative transposon Tn916 between gram-positive and gram-negative bacteria. J Bacteriol. 1991;173:443–448. doi: 10.1128/jb.173.2.443-448.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coco W M, Rothmel R K, Henikoff S, Chakrabarty A M. Nucleotide sequence and initial functional characterization of the clcR gene encoding a LysR family activator of the clcABD chlorocatechol operon in Pseudomonas putida. J Bacteriol. 1993;175:417–427. doi: 10.1128/jb.175.2.417-427.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Datta N, Hedges R W, Shaw E J, Sykes R B, Richmond M H. Properties of an R factor from Pseudomonas aeruginosa. J Bacteriol. 1971;108:1244–1249. doi: 10.1128/jb.108.3.1244-1249.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Egli T. The ecological and physiological significance of the growth of heterotrophic microorganisms with mixtures of substrates. Adv Microb Ecol. 1995;14:305–386. [Google Scholar]
- 6.Egli T. Multiple-nutrient-limited growth of microorganisms: what are the consequences for bioremediation processes. Presented at the International Symposium “Environmental Biotechnology,” Oostende, Belgium, 21 to 23 April 1997. 1997. [Google Scholar]
- 7.Figurski D H, Helinski D R. Replication of an origin-containing derivate of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frantz B, Chakrabarty A M. Organization and nucleotide sequence determination of a gene cluster involved in 3-chlorocatechol degradation. Proc Natl Acad Sci USA. 1987;84:4460–4464. doi: 10.1073/pnas.84.13.4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frost L S. Bacterial conjugation—everybody’s doin’ it. Can J Microbiol. 1992;38:1091–1096. doi: 10.1139/m92-179. [DOI] [PubMed] [Google Scholar]
- 10.Fulthorpe R R, Wyndham R C. Survival and activity of a 3-chlorobenzoate-catabolic genotype in a natural system. Appl Environ Microbiol. 1989;55:1584–1590. doi: 10.1128/aem.55.6.1584-1590.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knobel H R, Egli T, van der Meer J R. Cloning and characterization of the genes encoding nitrilotriacetate monooxygenase of Chelatobacter heintzii ATCC 29600. J Bacteriol. 1996;178:6123–6132. doi: 10.1128/jb.178.21.6123-6132.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leveau J H J, van der Meer J R. The tfdR gene product can successfully take over the role of the insertion element-inactivated TfdT protein as a transcriptional activator of the tfdCDEF gene cluster, which encodes chlorocatechol degradation in Ralstonia eutropha JMP134(pJP4) J Bacteriol. 1996;178:6824–6832. doi: 10.1128/jb.178.23.6824-6832.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levin B R, Stewart F M, Rice V A. The kinetics of conjugative plasmid transmission: fit of a simple mass action model. Plasmid. 1979;2:247–260. doi: 10.1016/0147-619x(79)90043-x. [DOI] [PubMed] [Google Scholar]
- 14.Lorenz M G, Wackernagel W. Bacterial gene transfer by natural genetic transformation in the environment. Microbiol Rev. 1994;58:563–602. doi: 10.1128/mr.58.3.563-602.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mancini P, Fertels S, Nave D, Gealt M A. Mobilization of plasmid pHSV106 from Escherichia coli HB101 in a laboratory-scale waste treatment facility. Appl Environ Microbiol. 1987;53:665–671. doi: 10.1128/aem.53.4.665-671.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marmur J. A procedure for the isolation of deoxyribo-nucleic acid from microorganisms. J Mol Biol. 1961;3:208–218. [Google Scholar]
- 17.Moellering R C. Interaction between antimicrobial consumption and selection of resistant bacterial strains. Scand J Infect Dis Suppl. 1990;70:18–24. [PubMed] [Google Scholar]
- 18.Naik G A, Bhat L N, Chopade B A, Lynch J M. Transfer of broad-host-range antibiotic resistance plasmids in soil microcosms. Curr Microbiol. 1994;28:209–215. [Google Scholar]
- 19.Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C: American Society for Microbiology; 1987. [Google Scholar]
- 20.Neilson J W, Josephson K L, Pepper I L, Arnold R B, Di Giovanni G D, Sinclair N A. Frequency of horizontal gene transfer of a large catabolic plasmid (pJP4) in soil. Appl Environ Microbiol. 1994;60:4053–4058. doi: 10.1128/aem.60.11.4053-4058.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nüsslein K, Maris D, Timmis K, Dwyer D F. Expression and transfer of engineered catabolic pathways harbored by Pseudomonas spp. introduced into activated sludge microcosms. Appl Environ Microbiol. 1992;58:3380–3386. doi: 10.1128/aem.58.10.3380-3386.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oltmanns R H, Rast H G, Reineke W. Degradation of 1,4-dichlorobenzene by enriched and constructed bacteria. Appl Microbiol Biotechnol. 1988;28:609–616. [Google Scholar]
- 23.Pukall R, Tschäpe H, Smalla K. Monitoring the spread of broad host and narrow host range plasmids in soil microcosms. FEMS Microbiol Ecol. 1996;20:53–66. [Google Scholar]
- 24.Ravatn, R. Unpublished results.
- 25.Ravatn, R., S. Studer, D. Springael, A. J. B. Zehnder, and J. R. van der Meer. Chromosomal integration, tandem amplification and deamplification in Pseudomonas putida F1 of a 105-kilobase genetic element containing the chlorocatechol degradative genes from Pseudomonas sp. strain B13. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 26.Reineke W, Wessels S W, Rubio M A, Latorre J, Schwien U, Schmidt E, Schlömann M, Knackmuss H J. Degradation of monochlorinated aromatics following transfer of genes encoding chlorocatechol catabolism. FEMS Microbiol Lett. 1982;14:291–294. [Google Scholar]
- 27.Reuss M, Dössereck C. Reaction engineering aspects of conjugation in biodegradation processes. Ann NY Acad Sci. 1994;721:428–439. doi: 10.1111/j.1749-6632.1994.tb47414.x. [DOI] [PubMed] [Google Scholar]
- 28.Rochelle P A, Fry J C, Day M J. Factors affecting conjugal transfer of plasmids encoding mercury resistance from pure cultures and mixed natural suspensions of epilithic bacteria. J Gen Microbiol. 1989;135:409–424. doi: 10.1099/00221287-135-2-409. [DOI] [PubMed] [Google Scholar]
- 29.Salyers A A, Shoemaker N B, Stevens A M, Li L Y. Conjugative transposons: an unusual and diverse set of integrated gene transfer elements. Microbiol Rev. 1995;59:579–590. doi: 10.1128/mr.59.4.579-590.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smit E. Conjugal gene transfer between bacteria in soil and rhizosphere. Ph.D. thesis. Wageningen, The Netherlands: University of Wageningen; 1994. [Google Scholar]
- 31.Top E, Mergeay M, Springael D, Verstraete W. Gene escape model: transfer of heavy metal resistance genes from Escherichia coli to Alcaligenes eutrophus on agar plates and in soil samples. Appl Environ Microbiol. 1990;56:2471–2479. doi: 10.1128/aem.56.8.2471-2479.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tros M E, Bosma T N P, Schraa G, Zehnder A J B. Measurement of minimum substrate concentration (Smin) in a recycling fermentor and its prediction from the kinetic parameters of Pseudomonas sp. strain B13 from batch and chemostat cultures. Appl Environ Microbiol. 1996;62:3655–3661. doi: 10.1128/aem.62.10.3655-3661.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van der Meer, J. R. Unpublished results.
- 34.van der Meer J R, Roelofsen W, Schraa G, Zehnder A J B. Degradation of low concentrations of dichlorobenzenes and 1,2,4-trichlorobenzene by Pseudomonas sp. strain P51 in nonsterile soil columns. FEMS Microbiol Ecol. 1987;45:333–341. [Google Scholar]
- 35.Vandenbroucke-Grauls C M. The threat of multiresistant microorganisms. Eur J Clin Microbiol Infect Dis. 1993;12:27–30. doi: 10.1007/BF02389874. [DOI] [PubMed] [Google Scholar]
- 36.Zhou J Z, Tiedje J M. Gene transfer from a bacterium injected into an aquifer to an indigenous bacterium. Mol Ecol. 1995;4:613–618. doi: 10.1111/j.1365-294x.1995.tb00261.x. [DOI] [PubMed] [Google Scholar]
- 37.Zylstra G J, Gibson D T. Toluene degradation by Pseudomonas putida F1. Nucleotide sequence of the todC1C2BADE genes and their expression in Escherichia coli. J Biol Chem. 1989;264:14940–14946. [PubMed] [Google Scholar]
- 38.Zylstra G J, McCombie W R, Gibson D T, Finette B A. Toluene degradation by Pseudomonas putida F1: genetic organization of the tod operon. Appl Environ Microbiol. 1988;54:1498–1503. doi: 10.1128/aem.54.6.1498-1503.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]