Abstract
Imbalanced immune responses are a prominent hallmark of cancer and autoimmunity. Myeloid cells can be overly suppressive, inhibiting protective immune responses or inactive not controlling autoreactive immune cells. Understanding the mechanisms that induce suppressive myeloid cells, such as myeloid-derived suppressor cells (MDSCs) and tolerogenic dendritic cells (TolDCs), can facilitate the development of immune-restoring therapeutic approaches. MDSCs are a major barrier for effective cancer immunotherapy by suppressing antitumor immune responses in cancer patients. TolDCs are administered to patients to promote immune tolerance with the intent to control autoimmune disease. Here, we investigated the development and suppressive/tolerogenic activity of human MDSCs and TolDCs to gain insight into signaling pathways that drive immunosuppression in these different myeloid subsets. Moreover, monocyte-derived MDSCs (M-MDSCs) generated in vitro were compared to M-MDSCs isolated from head-and-neck squamous cell carcinoma patients. PI3K-AKT signaling was identified as being crucial for the induction of human M-MDSCs. PI3K inhibition prevented the downregulation of HLA-DR and the upregulation of reactive oxygen species and MerTK. In addition, we show that the suppressive activity of dexamethasone-induced TolDCs is induced by β-catenin–dependent Wnt signaling. The identification of PI3K-AKT and Wnt signal transduction pathways as respective inducers of the immunomodulatory capacity of M-MDSCs and TolDCs provides opportunities to overcome suppressive myeloid cells in cancer patients and optimize therapeutic application of TolDCs. Lastly, the observed similarities between generated- and patient-derived M-MDSCs support the use of in vitro–generated M-MDSCs as powerful model to investigate the functionality of human MDSCs.
Keywords: immunosuppression, MDSC, TolDC, immune tolerance, immunotherapy
In a healthy individual, modulation of immune responses by dampening immune activation is required to maintain immunological homeostasis (1, 2). Perturbation of this immune balance is implicated in diseases such as autoimmunity or cancer. Whereas an overactive immune response can lead to autoimmune disorders, an excessive immunosuppression is a hallmark for cancer (3, 4, 5). Within the immune system, innate immune cells of the myeloid lineage including dendritic cells (DCs), monocytes, and macrophages have been demonstrated to play a dominant role in the initiation and development of pro- and anti-inflammatory immune responses. To this end, myeloid cells employ various mechanisms such as the release of cytokines, growth factors, metabolites, and hormones which modulate the inflammatory environment (6, 7, 8).
Thus, modulating the phenotype and function of myeloid cells has become an attractive strategy as cell-based therapy in diseases such as cancer or autoimmune disorders. Main example of this are the monocyte-derived DCs (MoDCs) that have been used in multiple clinical trials to enhance antitumor immunity in cancer patients (9). But not only vaccines to promote immune activity but also in vitro–generated tolerogenic monocyte-derived dendritic cells (TolDCs) have been developed to promote immune tolerance in vivo for the treatment of autoimmunity (10). This has resulted in the roll out of multiple phase I-II trials to test the therapeutic value of this cell type in patients with a variety of autoimmune disorders (11, 12, 13).
In contrast, during cancer development, myeloid cells are known to acquire an immunosuppressive phenotype supporting a tolerogenic milieu that dampens antitumor immunity (14). In particular, cancer-derived factors are known to favor the development and expansion of a heterogenous group of immune-suppressive immature myeloid cells, called myeloid-derived suppressor cells (MDSCs) (15). Development of MDSCs from both monocytic (M-MDSC) and granulocytic lineage is induced by tumor-derived signals including GM-CSF, IL-6, and PGE-2 (16, 17, 18). MDSCs promote an immunosuppressive tumor microenvironment which facilitates tumor progression and hampers the efficacy of various antitumor therapies such as immune checkpoint blockade (14, 19, 20), leading to a poor prognosis for cancer patients (21). Therefore, targeting the signaling pathways that drive MDSC-mediated immunosuppression in cancer is an attractive opportunity to increase clinical benefit for patients receiving cancer immunotherapy (22).
To his end, further knowledge on the pathways leading to the induction of immune-suppressive myeloid-derived cells is vital for the development of new targeted therapies that could synergize with current therapies. In this study, we have established and validated a robust culture protocol for the in vitro generation of TolDCs and M-MDSCs from the same donor. Using this model, we have identified distinct signal transduction pathways responsible for inducing the suppressive function in M-MDSCs and TolDCs. PI3K-AKT signaling was shown to be crucial in the development of suppressive activity by M-MDSCs, whereas TolDCs rely on β-catenin–dependent Wnt signaling for tolerogenic function. In addition, we compared our in vitro–generated M-MDSC to primary M-MDSCs isolated from head-and-neck squamous cell carcinoma (HNSCC) patients. M-MDSCs generated in vitro were shown to be similar in phenotype and function as M-MDSCs isolated from cancer patients, promoting the use of this model to study human M-MDSCs. Overall, these results give insight into the development of suppressive myeloid cells and identifies potential therapeutic targets to overcome M-MDSCs–mediated immunosuppression in cancer immunotherapy, as well as provides tools for the optimization of TolDC-based therapies in autoimmunity.
Results
M-MDSCs and TolDCs have distinct phenotypes
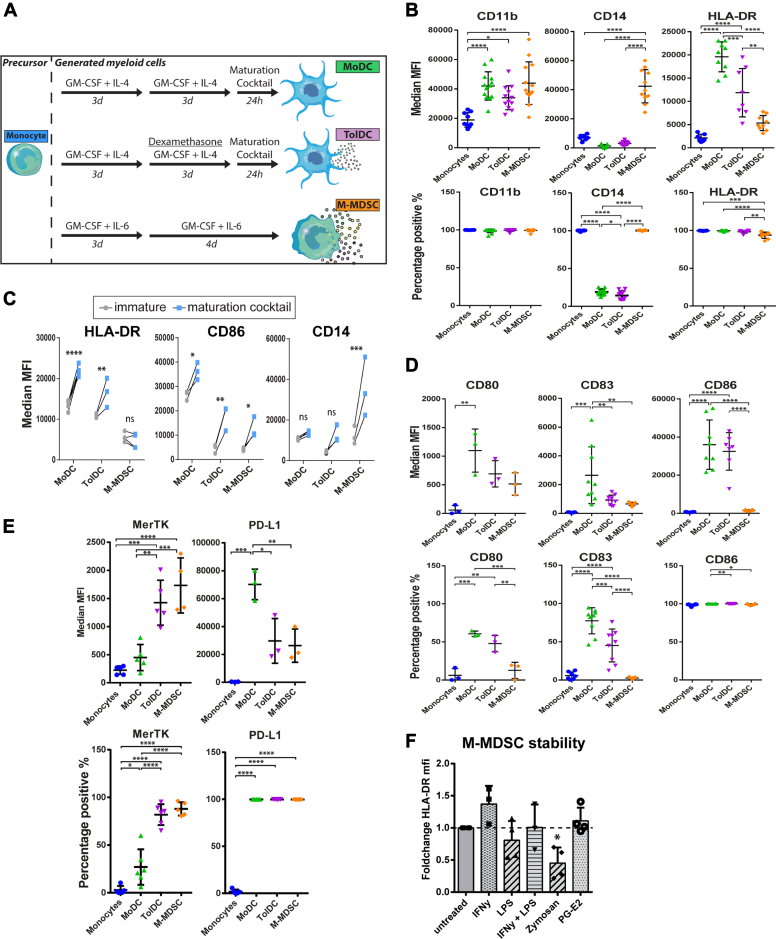
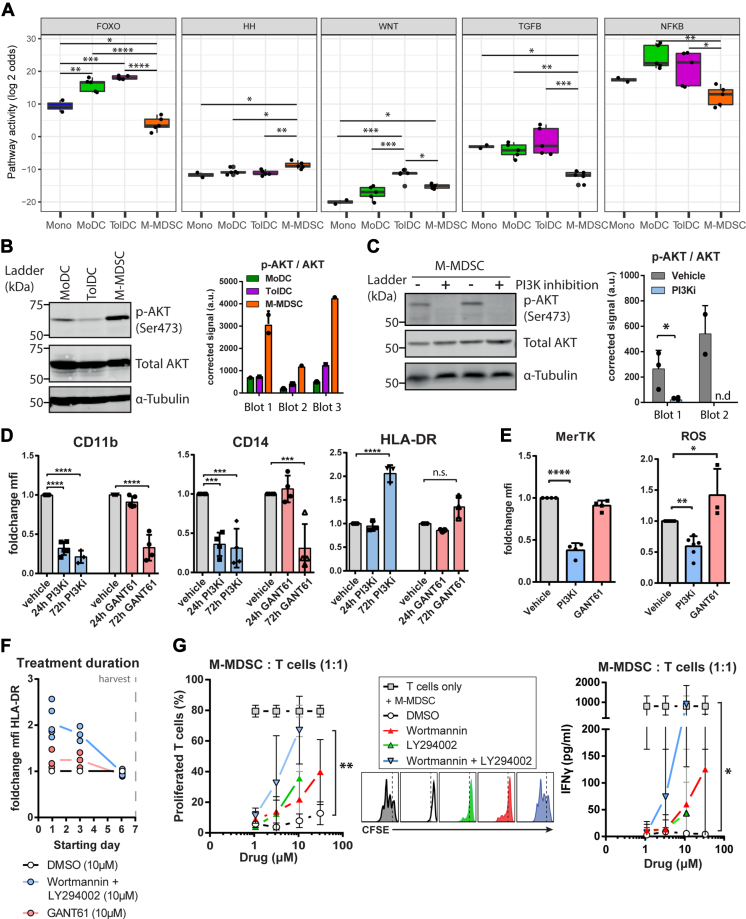
To investigate the phenotypes of suppressive myeloid cells, M-MDSCs were generated in vitro by adapting previous MDSC generation protocols using low dose GM-CSF and IL-6 (16, 17), in parallel with MoDCs and dexamethasone-induced TolDCs following established protocols (23, 24). Phenotypes of in vitro–generated myeloid cells were characterized by flow cytometry, together with their shared precursor monocytes (Figs. 1A and S1A). M-MDSCs had a phenotype of CD11bhigh CD14high HLA-DRlow, similar to immature monocytic myeloid cells (Fig. 1B). Myeloid lineage marker CD11b was expressed by all four cell types and found to be interchangeable with myeloid marker CD33 (Fig. S1B). Both MoDC and TolDC generation protocols require maturation by cytokine stimulation prior to harvest to obtain fully functional DCs (9, 25). In this regard, MoDCs displayed the highest level of maturation markers, TolDCs showed intermediate maturity, and M-MDSCs had the lowest level of maturation markers (Figs. 1B and S1C). Stimulating M-MDSCs with the cytokine-based maturation cocktail did not increase HLA-DR expression, illustrating the persistent immature nature of these cells (Fig. 1C). Expression levels of costimulatory molecules CD80, CD83, and CD86 was highest on MoDCs, intermediate on TolDCs, and low to negative on M-MDSCs (Fig. 1D). Moreover, we looked into the expression of co-inhibitory molecules MerTK and PD-L1, which were reported to distinguish TolDCs from MoDCs and M-MDSCs from monocytes, respectively (19, 26). In line, expression of MerTK on TolDCs discriminated them from MoDCs, whereas M-MDSCs could be separated from monocytes by differential PD-L1 expression (Fig. 1E). However, MerTK and PD-L1 were not exclusively expressed by TolDCs or M-MDSCs but present on both cell types. Of note, the high PD-L1 expression by MoDCs observed here was previously shown to be a transient effect induced by cytokine-induced maturation (27). Lastly, the immature phenotype of M-MDSCs was stable in the absence of GM-CSF/IL-6 and even after 48 h of lipopolysacharide (LPS) stimulation (Fig. S1D). The latter condition further increased the already high expression levels of CD14 by M-MDSCs. Surprisingly, we found that stimulation with various pro-inflammatory agents were unable to upregulate HLA-DR expression by M-MDSCs (Fig. 1F). These data emphasize the ability of low dose GM-CSF and IL-6 to induce immature myeloid cells in vitro with the stable M-MDSC phenotype distinct from TolDCs.
Figure 1.
Generated M-MDSCs have an immature myeloid phenotype that is stable upon stimulation.A, schematic overview of myeloid generation protocols. Phenotype of harvested cells was determined by flow cytometry, shown are (B) MFI and percentage positive cells of lineage markers (n = 7–13), (C) expression of phenotype markers with and without maturation cocktail treatment consisting of IL-1β, TNFα, IL-6, and PGE-2 (n = 3), (D) MFI and percentage positive cells of costimulatory receptors (n = 3–9) and (E) MFI and percentage positive cells of co-inhibitory markers PD-L1 and MerTK (n = 3–7). F, M-MDSCs were stimulated with IFNγ (10 ng/ml), lipopolysacharide (100 ng/ml), Zymosan (1 × 106 beads/ml), or PGE-2 (3,5 μg/ml), and after 48 h, phenotypic maturity was assessed. HLA-DR MFI of treated cells was divided by MFI of untreated cells (n = 3–4). One way ANOVA of unmatched values in all graphs except for the two way ANOVA of unmatched values in panel C. All obtained p-values were adjusted by bonferroni correction. Mean + SD are shown in all graphs ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05, ns = non significant. MDSC, myeloid-derived suppressor cell; MFI, mean fluorescence intensity; M-MDSC, monocyte-derived-MDSC.
M-MDSCs and TolDCs suppress allogeneic and autologous T cell responses
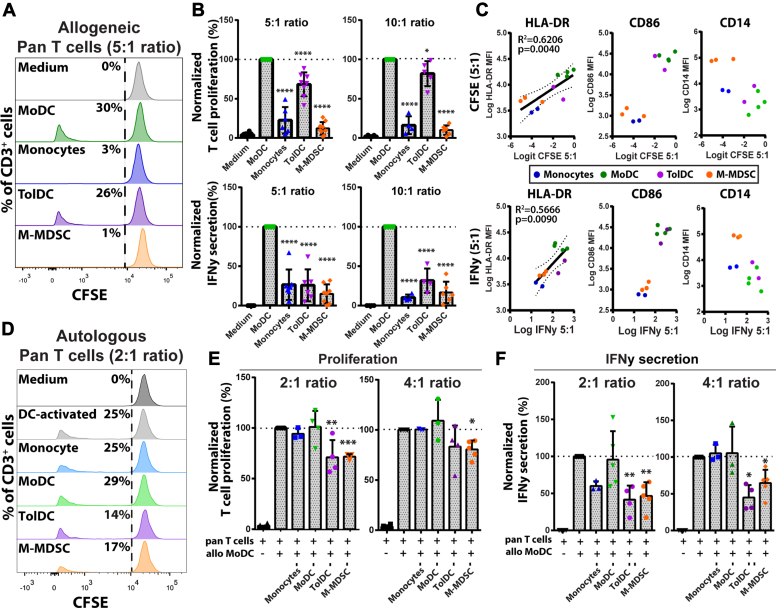
To assess the immunological functions of the myeloid subsets, cocultures with pan T cells were performed and assessed by flow cytometry and ELISA (Fig. S2A). First, myeloid cells were tested for their capacity to induce responses by allogeneic T cells (Fig. 2A). As expected, MoDCs were most potent in inducing allogeneic T cell proliferation and IFNγ secretion (Fig. 2B). In comparison, monocytes and M-MDSCs did not elicit T cell activation, even after stimulating M-MDSCs with the cytokine-based maturation cocktail (Fig. S2B). TolDCs promoted proliferation of allogeneic T cells that were impaired in IFNγ secretion, which nicely shows the induction of immune tolerance. Interestingly, CD4+ T cells were more inhibited in proliferation than CD8+ T cells, in line with reports showing that TolDCs predominantly modulate CD4+ T cells responses (24) (Fig. S2C). The expression of HLA-DR on myeloid cells was found to have a clear linear correlation to their immunostimulatory capacity in the allogeneic T cell cocultures (Fig. 2C). Therefore, the level of HLA-DR expression likely explains a large part of the immunogenic capacity of the myeloid subsets through the number of immunogenic peptides being presented to T cells. Next, the myeloid subsets were tested for their immunosuppressive capacity to inhibit DC-activated autologous T cell responses (Fig. 2D). Proliferation of stimulated T cells was significantly inhibited by coculture with TolDCs and M-MDSCs but not by monocytes and MoDCs in both 2:1 and 4:1 (T cell: Myeloid cell) ratios (Figs. 2E and S2D). This same effect was also seen on the level of IFNγ secretion (Fig. 2F). We concluded that M-MDSCs are less immunogenic than TolDCs with respect to the induction of allogeneic T cell responses, and both cell types are able to suppress ongoing autologous T cell responses.
Figure 2.
Generated M-MDSCs and TolDCs have distinct immunostimulatory and immunosuppressive capacities. Generated myeloid cells were cocultured with allogeneic CD3+ T cells for five days and shown are (A) representative histograms of CFSE-labeled pan T cells after allogeneic coculture, (B) quantified T cell proliferation and IFNγ secretion normalized to MoDC-induced response (n = 6–10). C, biplots of HLA-DR, CD86, and CD14 expression on myeloid cells in relation to induced allogeneic T cell proliferation and IFNγ secretion in the 5:1 cocultures. Generated myeloid cells were cocultured with autologous CD3+ T cells in the presence of allogeneic MoDC for five days and shown are (D) representative histograms of DC-activated CFSE-labeled T cells after autologous coculture, (E) quantified T cell proliferation, and (F) IFN γ secretion normalized to DC-activated T cell response (n = 3–5). One way ANOVA of unmatched values plus bonferroni correction in all figures except for the linear regression in HLA-DR biplots. Mean + SD are shown in all graphs ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05. DC, dendritic cell; MDSC, myeloid-derived suppressor cell; M-MDSC, monocyte-derived-MDSC; MoDC, monocyte-derived DC; TolDC, tolerogenic dendritic cell.
Gene and protein signatures found in M-MDSCs differ to TolDCs and associate to MDSCs from tumor-bearing mice
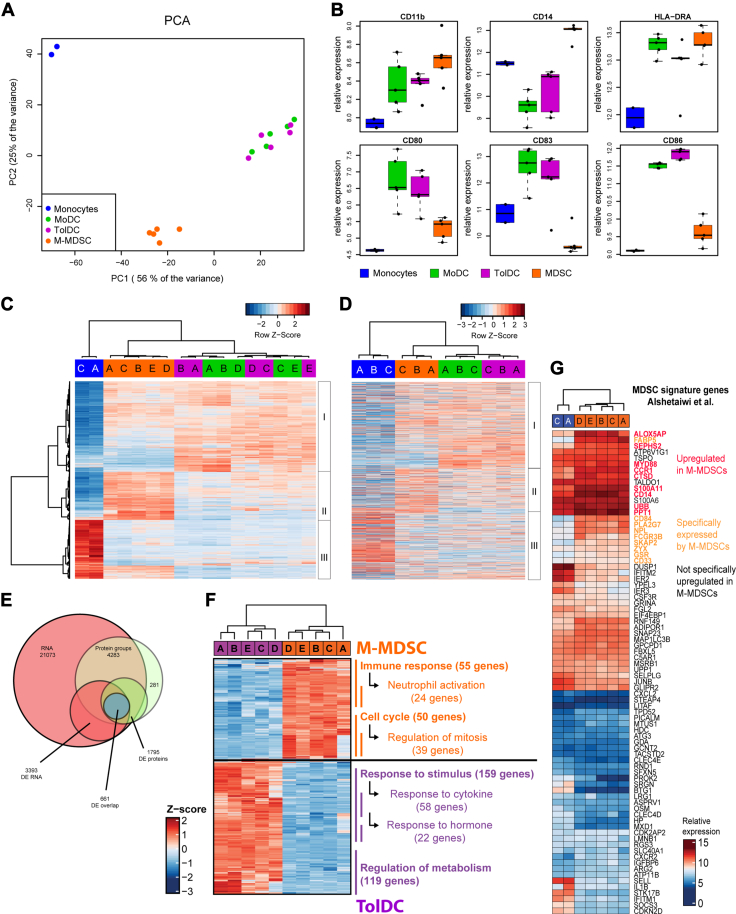
To investigate the biological processes in M-MDSCs and TolDCs, we compared the gene and protein expression profiles associated to these cell types. First, RNA microarray analysis was performed to quantify the expression of 47.000 gene transcripts in the myeloid cells using the Human Genome U133 Plus 2.0 Array probesets. Based on the top 1000 variably expressed probes, all samples clustered closely together per cell type in an unsupervised analysis of principal components discriminating the samples (Fig. 3A). Of note, TolDC and MoDC samples overlapped with each other in one cluster with great distance to M-MDSCs and monocytes. This indicates that all cell types could be clearly discriminated based on their gene expression profile except for MoDCs from TolDCs, which only differed significantly in the expression of 22 genes (Fig. S3A). In addition, we studied the relative expression of myeloid lineage markers and costimulatory molecules within the myeloid cell types and confirmed that these corresponded with the previously measured expression patterns (Fig. 3B). Next, we determined the differentially expressed genes (DEGs) between the samples (p < 0.05 and log2 fold change>2) and performed unsupervised clustering of samples in a heatmap (Fig. 3C). The samples clustered together per cell type and the DEGs clustered together in three separate clusters (I-III) (Fig. 3C). Cluster I was greatly expressed by MoDCs and TolDCs, where cluster II was mainly expressed by M-MDSCs and expression of cluster III was associated to monocytes. To assess the biological relevance of these gene clusters, we determined whether these DEGs were also translated into proteins by performing mass spectrometry on monocytes, MoDCs, TolDCs, and M-MDSCs. Here, we identified the proteins corresponding to most DEGs in panel C and plotted their expression ranked as in panel C but with unsupervised clustering of the samples (Fig. 3D). Based on the expression of these DEG-linked proteins, the samples clustered together per cell type and protein expression patterns revealed three similar protein clusters as initially found for DEGs in panel C. In line, almost all the differentially expressed (p < 0.05 log2 fold change>2) proteins between the samples were also identified as DEGs in the transcriptome analysis (Fig. 3E). This shows that the majority of DEGs are translated into proteins with similar expression patterns and therefore likely of biological relevance. Interestingly, TolDCs could be clearly discriminated from MoDCs based on protein expression, including eight proteins that were also differentially expressed as genes between these two cell types (Fig. S3B). In addition, protein expression of the glucocorticoid receptor was not detected in TolDCs, in line with dexamethasone-induced triggering (28) (Fig. S3C). To investigate which biological processes are differentially active in TolDCs and M-MDSCs, we performed Gene Ontology (GO) term enrichment analysis on the DEGs between these two cell types (Tables S2 and S3). Most upregulated genes in TolDCs were enriched in “response to stimulus” (GO:0050896), including two daughter genesets “response to cytokine” and “response to hormone” (GO:0034097, GO:0009725) or in “regulation of metabolic process” (GO:0019222, Fig. 3F). These first processes likely reflect on the dexamethasone treatment and cytokine-induced maturation endured during the generation of TolDCs. In addition, metabolic activity in DCs is often related to the active uptake, processing, and presentation of molecules by these cells (29, 30). In contrast, two distinctively enriched GO terms for M-MDSCs were “immune response” (GO:0006955) with largest daughter geneset “neutrophil activation” (GO:0042119) and “cell cycle” (GO:0007049) with largest daughter geneset “mitotic cell cycle” (GO:0000278). These processes indicate that M-MDSCs represent activated immature myeloid cells engaged in the promotion of cell survival or proliferation. Surprisingly, the GO-term “neutrophil activation” was also recently reported to be highly enriched in a murine MDSC gene signature shared between both M-MDSCs and granulocyte-derived MDSCs in a model of breast cancer (31). This suggests that genesets historically annotated to activated neutrophils may contain crucial MDSC-associated genes that are also upregulated by the human M-MDSCs generated here in vitro. To investigate this in more detail, we examined the expression pattern of all human ortholog genes of this murine MDSC signature among the human MDSCs and monocytes studied here (Fig. 3G). Although originating from different species, we identified a subset of genes that was also upregulated by human M-MDSCs including MDSC markers S100A11, CD14, CD84, and CD33. Lastly, we also compared our results with those of a previously published TolDC signature (32). We observed that nine out of 10 reported upregulated genes including CD163 and C1Qb were also upregulated in this study (Fig. S3C). Together, the gene and protein signatures identified here demonstrate that the biological processes involved in reacting to inflammation and cytokine stimulation are characteristic to TolDCs. In contrast, the molecular signature of M-MDSCs was related to the activation of immature myeloid cells and compared closely to MDSCs previously found in tumor-bearing mice.
Figure 3.
Transcriptome and proteome analysis reveals clusters of differentially expressed genes and proteins between generated M-MDSCs and TolDCs. Generated myeloid cells (n = 5 donors) were assessed by Affymetrix HGU-133 Plus 2.0 RNA microarray and shown are (A) principal component analysis of top1000 variably expressed probes between subsets, (B) boxplots of genes encoding phenotypic markers, (C) heatmap of all significantly (p < 0.05) and differentially expressed (log2FC > 2) probes between all samples which identifies three clusters of differentially expressed genes (DEGs). Colors indicate cell types, and letters indicate individual donors. D, heatmap showing expression of proteins corresponding to the DEG in panel C as detected by mass spectrometry of generated myeloid cells (n = 3 donors). The same three differentially expressed clusters are also identified. E, Venn diagram showing overlap between the gene and protein expression data including DEG and DEP. F, Geneset enrichment analysis using Gene Ontology terms identifies the biological processes that are differentially activated between M-MDSCs and TolDCs. G, heatmap showing the expression of genes from a previously proposed MDSC signature by Alshetaiwi et al. among the human monocytes and generated M-MDSCs used in this study. Mean + SD in panel B. DEP, differentially expressed protein; MDSC, myeloid-derived suppressor cell; M-MDSC, monocyte-derived-MDSC; TolDC, tolerogenic dendritic cell.
M-MDSCs and TolDCs express distinct immunosuppressive molecules
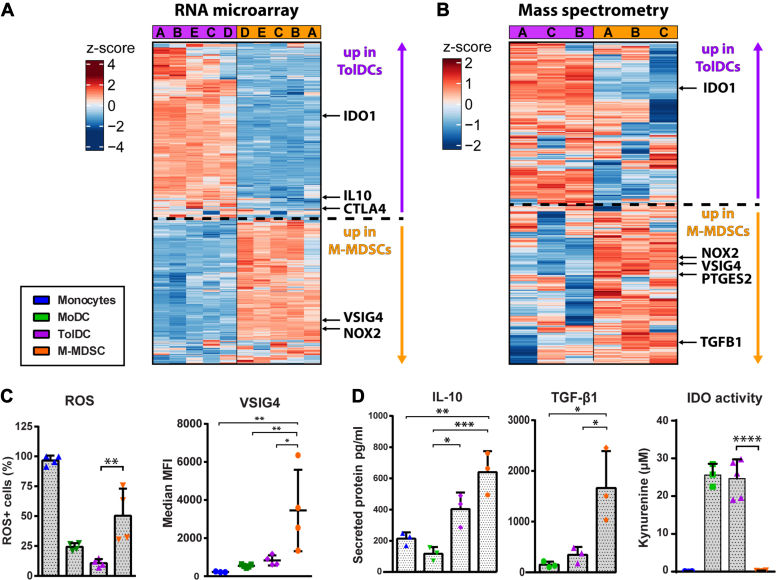
M-MDSCs and TolDCs are both able to suppress activated T cell responses but seem driven by distinct biological processes. Therefore, we set out to identify molecules with known immunosuppressive function that were differentially expressed between TolDCs and M-MDSCs. Based on the RNA microarray, the genes IDO1, IL10, and CTLA4 were identified to be upregulated by TolDCs whereas VSIG4 and NOX2 were specifically expressed by M-MDSCs (Fig. 4A). In addition, the enzyme IDO1 was also found to be upregulated as protein by TolDCs (Fig. 4B). In comparison, NOX2, VSIG4, PGE-2 synthase 2 (PTGES2), and TGF-β1 were specifically upregulated by M-MDSCs. In line with the expression data, M-MDSCs stained positive for reactive oxygen species (ROS), suggesting enzymatic activity by NOX2, and for the co-inhibitory molecule VSIG4 (Fig. 4C). Of note, surface expression of CTLA4 on TolDCs was detected at indistinctively low levels and therefore likely irrelevant to TolDCs function (Fig. S4A). Next, we assessed conditioned media of myeloid cells upon exposure to LPS. While IL-10 was secreted by both M-MDSCs and TolDCs, TGF-β1 was exclusively produced by M-MDSCs (Fig. 4D). TolDCs, in contrast to M-MDSCs, displayed high IDO1 enzymatic activity as evidenced by the accumulation of its product l-kynurenine in TolDC supernatants. Furthermore, high levels of PGE-2 (200pg/ml) were detected in media conditioned by M-MDSCs (Fig. S4B). Taken together, M-MDSCs and TolDCs both produced IL-10 and expressed inhibitory molecules MerTK and PD-L1. However, IDO1 activity was exclusively observed for TolDCs, whereas M-MDSCs expressed a wide variety of immunosuppressive molecules including TGF-β1, ROS, PGE-2, and VSIG4.
Figure 4.
M-MDSC and TolDC express distinct suppressive machinery. Differentially expressed immunosuppression-related molecules between M-MDSCs and TolDCs were identified by RNA microarray and mass spectrometry followed by functional validation. Shown are (A) heatmap of top 1000 variably expressed probes between M-MDSCs and TolDCs, (B) heatmap of top 500 variably expressed proteins between M-MDSCs and TolDCs, (C) expression of ROS and VSIG4 by myeloid cell types determined by flow cytometry (n = 4) and (D) secretion of IL-10 and TGF-β1 plus IDO activity determined in conditioned medium from myeloid cell types (n = 3–5). One way ANOVA of unmatched values plus bonferroni correction in all graphs. Mean + SD in all graphs ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05. MDSC, myeloid-derived suppressor cell; M-MDSC, monocyte-derived-MDSC; ROS, reactive oxygen species; TolDC, tolerogenic dendritic cell.
PI3K-AKT signaling drives the suppressive function of M-MDSCs
To determine which signaling pathways control the immunosuppressive function of M-MDSCs and TolDCs, signaling pathway activities were quantified by Philips signal transduction pathway activity analysis based on the expression of pathway-specific target genes (33). The activities of signaling pathways FOXO, Hedgehog (HH), Wnt, TGF-β, and NFкB were significantly different between TolDCs and M-MDSCs, with every pathway being more active in TolDCs except for HH signaling (Figs. 5A and S5A). The largest difference was observed in FOXO signaling activity which was very low in M-MDSCs as compared to TolDCs. This usually indicates high PI3K signaling activity because of its dominant repressive function on FOXO (34). First, we set out to validate active PI3K signaling in M-MDSCs by assessing the phosphorylation state of downstream kinase AKT/PKB in the myeloid subsets. In line with the PI3K pathway activity score, phosphorylation of AKT was highest in M-MDSCs, suggesting active signaling via the PI3K–AKT axis (Figs. 5B and S5B). Chemical inhibition of PI3K activity by treating M-MDSCs with a combination of Wortmannin and LY294002 (PI3Ki) abrogated the phosphorylation of AKT, confirming the active PI3K–AKT signaling axis (Figs. 5C and S5C). Next, we assessed the phenotype of M-MDSCs treated with PI3Ki and GANT61, a selective inhibitor of Hedgehog pathway transcription factor GLI-1. We observed that PI3Ki treatment had completely reversed the immature phenotype of M-MDSCs, as best evidenced by the increased HLA-DR expression. GANT61-treated M-MDSCs also presented a more mature myeloid phenotype, albeit to a lesser extent than PI3ki-treated cells where reduced CD11b and CD14 expression was observed earlier during treatment and the increase in HLA- DR expression was more substantial (Figs. 5D and S5D). This indicates that PI3Ki-AKT signaling controls the characteristic M-MDSC phenotype. In addition, M-MDSCs treated with PI3Ki had decreased expression of MerTK and ROS suggesting a decreased ability to inhibit T cell responses (Fig. 5E). Of note, starting PI3K inhibition late during M-MDSC generation (4–24 h before harvest) was ineffective in increasing HLA-DR expression as opposed to starting treatment early (3–7 days before harvest) (Fig. 5F). This indicates that PI3K signaling induces an early and irreversible commitment to M-MDSC development. To confirm that PI3K-AKT signaling is essential to the development of suppressive M-MDSCs, monocytes were treated with PI3K inhibitors or vehicle throughout the M-MDSC generation before coculture with CD3/CD28-stimulated autologous T cells (Fig. S5E). The combination of PI3K inhibitors Wortmannin and LY294002 was able to prevent the induction of suppressive activity by M-MDSCs as shown by the dose-dependent rescue of T cell proliferation and IFNγ secretion after coculture (Fig. 5G). Collectively, these observations demonstrate that PI3K-AKT signaling is crucial to the induction of the suppressive phenotype and function of M-MDSCs.
Figure 5.
PI3K-AKT signaling drives M-MDSC suppressive function.A, quantified signaling pathway activities that are significantly different between TolDC and M-MDSC are shown based on Philips signal transduction pathway activity analysis (rules for interpretation are clarified in the Experimental procedures section). B, AKT phosphorylation state in generated myeloid cells was determined, shown are a representative Western blot and semiquantitative p-AKT signal corrected for total AKT signal from three independent blots (n = 3–4 in total). C, PI3K-AKT inhibition (PI3Ki) in M-MDSC was achieved by Wortmannin plus LY294002 (PI3Ki, 10 μM both) treatment for 24 h before harvest, shown are a representative immunoblot and semiquantitative p-AKT signal corrected for total AKT signal of two independent blots (n = 5 in total). D and E, M-MDSCs were treated with PI3Ki or Hedgehog pathway inhibitor GANT61 (10 μM) for indicated periods (72 h in E) before harvest and flow cytometric assessment, shown are foldchange in MFI compared to vehicle-treated cells (n = 3–5). F, M-MDSCs were treated with PI3K inhibitors during 7 days generation period starting at indicated culture days before measuring HLA-DR expression upon harvest to assess the effect of treatment duration on M-MDSC generation (n = 3). G, M-MDSCs were treated with PI3K inhibitors throughout 7 days of generation before coculture with Dynabeads-activated autologous T cells for 3 days, shown are percentage-proliferated T cells with representative CFSE histograms and IFNγ secretion (n = 3–6). One way ANOVA of unmatched values plus Tukey’s HSD in (A). One way ANOVA of unmatched values plus bonferroni in (D, E, and G) Student’s t test in (C). Mean + SD in all graphs. ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05, n.s. = non significant, n.d. = no signal detected. MDSC, myeloid-derived suppressor cell; MFI, mean fluorescence intensity; M-MDSC, monocyte-derived-MDSC; TolDC, tolerogenic dendritic cell.
Beta-catenin–dependent Wnt signaling drives the tolerogenic function of TolDCs
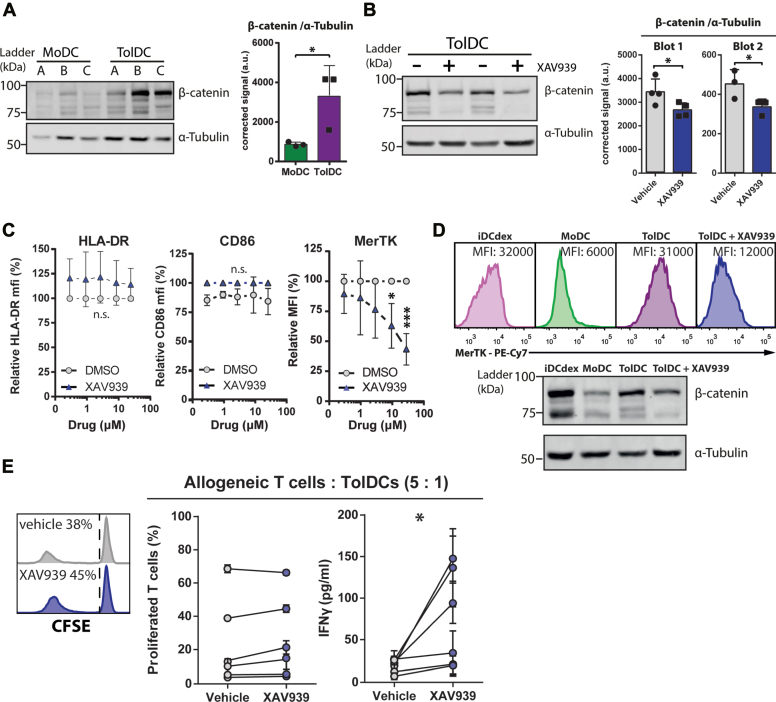
Only the Wnt signaling pathway was more active in TolDCs than both M-MDSCs and MoDCs according Philips signal transduction pathway activity analysis (Fig. 5A). Therefore, we hypothesized that active Wnt signaling was specifically involved in the suppressive function of TolDCs. To validate this, we first assessed the accumulation of β-catenin in TolDCs, which mediates signaling via the canonical Wnt pathway (35). In line with the measured Wnt pathway activity score, we observed more β-catenin accumulation in TolDCs than in MoDCs, suggestive of active signaling by the Wnt pathway (Fig. 6A). Next, we treated TolDCs with the Wnt pathway antagonist XAV939 which promotes the degradation of cytosolic β-catenin (36). Indeed, XAV939-treated TolDCs showed a modest (35%) but significant reduction in β-catenin expression (Figs. 6B and S6A). Of note, nondegraded β-catenin is inhibited in translocating to the nucleus by XAV939, further antagonizing signaling (36). To assess whether inhibition of β-catenin signaling resulted in immunologically relevant alterations of TolDCs, we assessed the phenotype of monocytes treated with increasing concentrations of XAV939 throughout TolDC generation. We observed that the expression of HLA-DR and CD86 remained stable whereas the expression of co-inhibitory molecule MerTK was drastically decreased by XAV939 treatment in a dose-dependent manner (Figs. 6C and S6B). This indicates that Wnt pathway signaling specifically controls the tolerogenic phenotype of TolDCs in a manner that is dependent on β-catenin. Moreover, we observed that the expression pattern of MerTK among immature TolDCs (iDCdex), MoDCs, TolDCs, and XAV939-treated TolDCs closely followed β-catenin expression, suggesting that expression of MerTK may be induced by β-catenin signaling either directly or indirectly (Figs. 6D and S6A). To confirm that Wnt signaling is involved in the tolerogenic function of TolDCs, we cocultured XAV939-treated TolDCs with allogeneic pan T cells and measured T cell proliferation and IFNγ secretion after 5 days. In particular, IFNγ secretion by allogeneic T cells activated by XAV939-treated TolDCs was greatly increased compared to vehicle-treated TolDCs, indicating a reduced ability to induce tolerogenic immune responses (Fig. 6E). Taken together, these data show that β-catenin–dependent Wnt signaling is active in dexamethasone-induced TolDCs and controls its tolerogenic function.
Figure 6.
Beta catenin–dependent Wnt signaling drives tolerogenic function of TolDCs.A, β-catenin accumulation was determined in MoDC and TolDC, shown are a representative Western blot and semiquantitative β-catenin signal corrected for α-tubulin signal from three independent donors. B, inhibition of β-catenin was achieved by treating TolDCs with 10 μM XAV939 for 3 days before harvest. Shown are a representative blot and semiquantitative β-catenin signal corrected for α-tubulin signal of two independent blots (n = 7 in total). C, TolDCs were treated with XAV939 using indicated concentrations throughout generation period before harvest and flow cytometric assessment, shown are foldchange in MFI (n = 4). D, representative histograms of MerTK expression combined with representative blot of β-catenin expression in generated myeloid cells. Ten micromolars of XAV939 was added at day 1 of generation in treated condition. E, TolDCs were treated with 10 μM XAV939 3 days before harvest before coculture with allogeneic T cells for 5 days, shown are percentage-proliferated T cells with representative CFSE histograms and IFNγ secretion. Unpaired t test in A and paired t test in B and E; one way ANOVA in C. Mean + SD in all graphs. ∗p < 0.05, n.s. = non significant. MFI, mean fluorescence intensity; MoDC, monocyte-derived DC; TolDC, tolerogenic dendritic cell.
In vitro–generated M-MDSCs resemble M-MDSCs from HNSCC patients
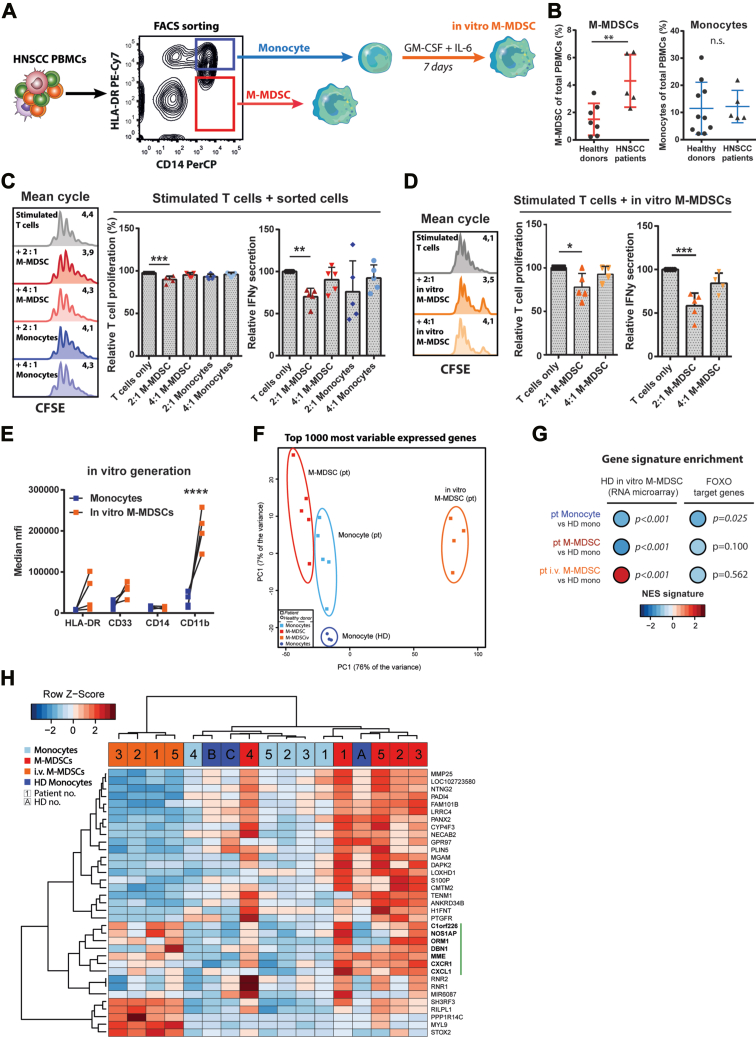
To validate our findings, we set out to compare in vitro–generated M-MDSCs to M-MDSCs from cancer patients. To this end, we collected peripheral blood from patients with squamous cell tumors of the head and neck (Table 1). HNSCC patients were selected because MDSCs, including CD14+ M-MDSCs, were shown to be prominent in this patient group and constituted a major obstruction to applied immunotherapies (37, 38, 39, 40, 41, 42, 43). M-MDSCs and monocytes were FACS-sorted out of HNSCC patient peripheral blood mononuclear cells (PBMCs) after which part of the sorted monocytes were used to generate in vitro M-MDSCs using low dose GM-CSF and IL-6 (Fig. 7A). PBMCs of patients and healthy donors were assessed by flow cytometry to quantify the frequencies of M-MDSCs and monocytes (Fig. S7A). We observed that M-MDSC but not monocyte frequencies were increased in HNSCC patients compared to healthy donors, which is in line with previous reports (38) (Fig. 7B). M-MDSCs and monocytes isolated from HNSCC patients were co-cultured ex vivo with CD3/CD28-stimulated T cells from healthy donors to determine their suppressive capacity (Fig. 7C). M-MDSCs were able to inhibit proliferation and IFNγ secretion of stimulated T cells in a 2:1 ratio (T cell: M-MDSC). In contrast, monocytes isolated from HNSCC patients did not inhibit stimulated T cell responses. However, M-MDSCs generated in vitro from patient monocytes were again potently able to suppress stimulated T cell responses in the 2:1 ratio, showing functional similarity to M-MDSCs directly isolated from patients (Fig. 7D). In vitro–generated M-MDSCs had retained similar expression levels of HLA-DR, CD33, and CD14 compared to its precursor monocytes, indicating a shared immature myeloid phenotype between the three cell types (Figs. 7E and S7B). Of note, M-MDSCs generated in vitro were larger in size than patient-derived myeloid cell types and had greatly increased expression of CD11b and PD-L1 (Fig. S7C), possibly induced by adhesion to culture plates (44, 45, 46). To compare the molecular features between the three patient-derived subsets and monocytes from healthy donors, we examined gene and protein expression profiles through RNAseq and mass spectrometry (Fig. S7D). In an unsupervised analysis of principal components using the top 1000 variably expressed genes, all samples clustered together as four distinct cell types, with in vitro–generated M-MDSCs being most distant from the other cell types (Fig. 7F). This is most likely explained by the in vitro culture needed to obtain the latter cell type in contrast to the freshly isolated cell types. To determine whether these M-MDSCs generated in vitro from cancer patient monocytes were comparable to the M-MDSCs generated in vitro from healthy donor (HD) monocytes, we performed a geneset enrichment using the in vitro M-MDSC signature obtained earlier by RNA microarray (M-MDSC versus HD monocyte). It became clear that patient-derived in vitro–generated M-MDSCs were significantly enriched for this in vitro M-MDSC signature whereas patient monocytes and M-MDSCs were not (Figs. 7G and S7E). In line with these findings, enrichment scores of FOXO target genes were negative in all patient-derived myeloid subsets indicative of PI3K activity, but this was only significant for patient monocytes. This suggests that monocytes from cancer patients have been susceptible to tumor-derived signals (e.g. GM-CSF and IL-6) in comparison to the PI3K-naive healthy donor monocytes. Finally, to investigate the difference between patient M-MDSCs and patient monocytes, the DEG (p < 0.05 and log2 FC > 2) between these two cell types were calculated (Fig. 7H). A cluster of genes was identified with relative high expression in patient M-MDSCs and in in vitro–generated M-MDSCs that are associated to immunological function, as indicated by the vertical green bar. These included NOS1AP, a positive regulator of nitric oxide, and ORM1, which was previously shown to induce immunosuppression in macrophages (47). Lastly, GSEA was performed using the Hallmark database containing a large set of pathways (Table 2). Here, we observed that ‘TNFα signaling via NF-кB’ and ‘Hypoxia’ were significantly enriched in patient M-MDSCs compared to patient monocytes, suggestive of increased tumor-imprinting on the former cell type. Of interest, although not significant, the pathways ‘Mitotic spindle’, ‘PI3K-AKT-mTOR signaling,’ and ‘Reactive oxygen species pathway’ were also found enriched in patient M-MDSCs in contrast to patient monocytes. This is comparable to the results previously obtained comparing in vitro–generated M-MDSCs to HD monocytes (Figs. 3F, 4C, and 5A). Taken together, these results show clear phenotypic and functional similarity between patient M-MDSCs and in vitro–generated M-MDSCs but no significant overlap in gene expression profiles was found, likely due to the in vitro culture period of the latter cell type. The differences in gene expression between patient M-MDSCs and patient monocytes are comparable to the processes discriminating in vitro–generated M-MDSCs from HD monocytes.
Table 1.
HNSCC patient characteristics
| Code | Age | Gender | Localization primary | T stage | N stage | HPV status |
|---|---|---|---|---|---|---|
| 1 | 59 | v | unknown | T0 | N1 | unknown |
| 2 | 68 | v | oropharynx | T1 | N1 | HPV P16 + |
| 3 | 68 | m | hypopharynx | T4a | N2b | n.a. |
| 4 | 61 | m | oropharynx | T4 | N2 | unknown |
| 5 | 65 | m | oropharynx | T4 | N2 | HPV P16 + |
Figure 7.
In vitro–generated M-MDSCs recapitulate phenotype and function of M-MDSCs isolated from HNSCC patients.A, schematic overview of monocytes, M-MDSCs, and in vitro M-MDSCs obtained from HNSCC patients (pt). B, quantification of M-MDSCs and monocytes among total PBMCs (n = 5–9). C and D, FACS-sorted monocytes, M-MDSCs plus in vitro–generated M-MDSCs from HNSCC patients were cocultured with dynabeads-stimulated T cells from healthy donors (HD) for 3 days; shown are representative CFSE histograms, relative T cell proliferation (mean cycle), and IFNγ secretion (n = 4). E, phenotype of in vitro–generated M-MDSCs were compared to their precursor monocytes within HNSCC patients. F, PCA of top 1000 variably expressed genes between samples as measured by RNAseq. G, geneset enrichment analysis using an in vitro (generated) M-MDSC signature and a FOXO signature. H, heatmap showing the expression of significantly upregulated genes (p < 0.05, logFC > 2) in patient M-MDSCs versus patient monocytes. Vertical green bar indicates gene cluster of interest. Unpaired t test in B. One way ANOVA of unmatched values plus bonferroni correction in C and D. Two way ANOVA of matched values plus bonferroni correction in E. Mean + SD in all graphs. ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05, n.s. = non significant. Pt = patient-derived, i.v. = in vitro generated and HD = healthy donor-derived. HNSCC, head-and-neck squamous cell carcinoma; MDSC, myeloid-derived suppressor cell; M-MDSC, monocyte-derived-MDSC; PBMC, peripheral blood mononuclear cell.
Table 2.
Hallmark pathway enrichment patient MDSCs versus patient Mono (RNAseq input)
| Hallmark pathway | Size | ES | NES | NOM_pval | FDR q-val | FWER p-val |
|---|---|---|---|---|---|---|
| 1. Hallmark_TNFA_SISIGNALLING_VIA_NFKB | 193 | 0.38 | 1.45 | 0.003 | 0.254 | 0.271 |
| 2. HALLMARK_HYPOXIA | 174 | 0.36 | 1.31 | 0.035 | 0.382 | 0.611 |
| 3. HALLMARK_WNT_BETA_CATENIN_SIGNALLING | 37 | 0.43 | 1.25 | 0.149 | 0.398 | 0.777 |
| 4. HALLMARK_MYOGENESIS | 150 | 0.32 | 1.19 | 0.131 | 0.468 | 0.902 |
| 5. HALLMARK_P53_PATHWAY | 191 | 0.31 | 1.18 | 0.088 | 0.383 | 0.910 |
| 6. HALLMARK_MITOTIC_SPINDLE | 196 | 0.31 | 1.17 | 0.108 | 0.368 | 0.941 |
| 7. HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION | 159 | 0.30 | 1.12 | 0.182 | 0.431 | 0,977 |
| 8. HALLMARK_APICAL_JUNCTION | 156 | 0.30 | 1.09 | 0.266 | 0.462 | 0.988 |
| 9. HALLMARK_PI3K_AKT_MTOR_SIGNALLING | 95 | 0.29 | 0.97 | 0.518 | 0.820 | 1.00 |
| 10. HALLMARK_REACTIVE_OXYGEN_SPECIES_PATHWAY | 47 | 0.31 | 0.95 | 0.553 | 0.783 | 1.00 |
| 11. HALLMARK_KRAS_SIGNALLING_DN | 108 | 0.26 | 0.91 | 0.702 | 0.858 | 1.00 |
| 12. HALLMARK_TGF_BETA_SIGNALLING | 53 | 0.28 | 0.86 | 0.739 | 0.932 | 1.00 |
| 13. HALLMARK_NOTCH_SIGNALLING | 30 | 0.30 | 0.82 | 0.757 | 0.940 | 1.00 |
| 14. HALLMARK_ANGIOGENESIS | 27 | 0.30 | 0.78 | 0.775 | 0.927 | 1.00 |
Discussion
In depth understanding of the molecular mechanisms that initiate and control immunosuppression by myeloid cells is essential to overcome the myeloid-induced disbalance of the immune system observed in patients with cancer and autoimmune disorders. Myeloid cells are implicated in aggravated suppression of immunity as seen in cancer as well as in defective modulation of immune responses as observed in autoimmunity. Therefore, signaling pathways controlling immunomodulation by myeloid cells are an attractive target to potentially restore immune homeostasis and increase clinical benefit for a large number of patients. Here, we investigated the mechanisms involved in the development of suppressive M-MDSCs and TolDCs derived from nonsuppressive monocytes of healthy human subjects. First, we characterized these cell types using flow cytometry, functional assays, RNA microarray, and mass spectrometry. We show that in vitro–generated M-MDSCs expressed high levels of CD11b, CD33, CD14 and low levels of HLA-DR, CD80, CD83, and CD86, consistent with the immature myeloid phenotype of M-MDSCs observed in cancer patients (48). TolDCs were characterized by a semimature phenotype as a result of the combined dexamethasone treatment and cytokine-induced maturation. Both suppressive phenotypes were complemented by the expression of co-inhibitory molecules MerTK and PD-L1. Interestingly, stimulation with various pro-inflammatory agents including a cocktail of cytokines, LPS, and IFNγ did not induce the expression of costimulatory molecules, demonstrating the resilient nature of M-MDSCs. In addition, M-MDSCs were unable to initiate allogeneic T cell responses, whereas TolDCs induced proliferation of IFNγlow T cells, characteristic for a tolerogenic response. As expected, stimulated autologous T cell responses were suppressed by M-MDSCs that were found to express a variety of immunosuppressive molecules including IL-10, TGF-β1, ROS, PGE-2, and VSIG4. TolDCs were equally able to inhibit autologous T cells and displayed distinctive IDO1 activity in addition to IL-10 secretion. Transcriptome and proteome analysis determined that in vitro–generated human M-MDSCs expressed genes previously associated to activated immature myeloid cells and MDSCs from tumor-bearing mice such as S100A11, CD33, and unique MDSC marker CD84 (31). When in vitro–generated M-MDSCs were compared to M-MDSCs isolated from HNSCC patients, these were similar in phenotype and immunosuppressive capacity. Taken together, these findings support the value of in vitro–generated M-MDSCs as experimental model for M-MDSCs induced by tumors in vivo.
PI3K-AKT signaling was identified to drive the induction of the suppressive phenotype and function of M-MDSCs, based on the repressed expression of FOXO target genes, increased phosphorylation of AKT, and chemical inhibition studies using PI3K inhibitors. Signaling via PI3K, in particular the class Ib isoform PI3Kγ, has been shown to control immunosuppression by myeloid cells during inflammation and cancer (49, 50). GM-CSF and IL-6 were previously shown to induce PI3K signaling through their cognate receptors GM-CSFR and (soluble) IL-6R (51, 52). This suggests that treating monocytes with GM-CSF and IL-6 in vitro resembles PI3K-driven expansion and activation of M-MDSCs in cancer patients. In line with these observations, we observed similar processes distinguishing in vitro–generated M-MDSCs from healthy donor monocytes as well as between patient M-MDSCs and patient monocytes. In addition, expression of VSIG4 detected here on in vitro–generated M-MDSCs was previously reported to induce PI3K-AKT signaling as well, which impaired LPS-induced activation and maturation of macrophages (53). This might explain the refractory nature of the immature phenotype observed here for in vitro–generated M-MDSCs upon various stimuli including LPS and possibly also for resilient M-MDSCs in cancer patients. Indeed, we observed that inhibition of PI3K activity late during M-MDSC generation in vitro was unable to prevent suppressive activity. This suggests that PI3K inhibitors, such as the PI3Kδ/γ-inhibitor IPI-545 currently in a phase 1 trial to enhance anti-PD-1 therapy (NCT02637531), do not modulate existing M-MDSCs but rather prevent the generation of de novo M-MDSCs to overcome immunosuppression. Together, these observations support the combination of PI3K inhibitors with immunotherapy to revert MDSC-mediated immunosuppression and increase response rates in cancer patients. Future studies are needed to understand how GM-CSF and IL-6 precisely induce PI3K-driven M-MDSC formation and the dynamics of inhibiting this process in vivo.
β-catenin–dependent Wnt pathway signaling was identified to drive the suppressive function induced by dexamethasone in TolDCs. Previously, Wnt signaling has been suggested to explain the CD163 expression found in dexamethasone-induced TolDCs and engagement of this pathway has been shown to induce tolerance in MoDCs and naturally occurring DCs (8, 32, 54). Culture protocols aimed to generate suppressive monocyte-derived cell types have been extensively studied as potential strategy to treat autoimmune disease (10). Potent anti-inflammatory and tolerogenic activity which is needed to overcome autoreactive effector cells were evidenced in TolDCs induced by in vitro dexamethasone treatment but the precise mechanisms controlling these functions are unknown (55). Here, we show, to the best of our knowledge for the first time, that Wnt pathway activity is essential to the suppressive function of dexamethasone-induced TolDCs. In addition, inhibition of β-catenin signaling decreased the expression of negative T cell regulator MerTK and subsequent tolerogenic capacity of TolDCs. Interestingly, the effect of β-catenin antagonist XAV939 on TolDC functionality was larger than the effect on β-catenin abundance levels, suggesting that the level of β-catenin activity, not expression, is responsible for tolerogenic activity. This is supported by the observation that β-catenin peptides were detected at similar levels between MoDCs and TolDCs (data not shown). These data warrant new studies into manipulating Wnt signaling in TolDCs with a focus on enhancing the efficacy of TolDC-based therapy for patients with an autoimmune disease.
In conclusion, we have demonstrated that human M-MDSCs generated in vitro highly resemble M-MDSCs isolated from cancer patients and therefore can be used as a model to study their functional behavior. The development of M-MDSCs from monocytes was dependent on PI3K-AKT signaling highlighting the potential of PI3K inhibition to overcome immunosuppression in cancer patients. In contrast, TolDCs generated in vitro using dexamethasone were dependent on β-catenin signaling for suppressive activity, showing new opportunities to be exploited for their therapeutic application in autoimmunity.
Experimental procedures
Healthy donor blood
PBMCs were isolated from healthy donor blood (Sanquin) by density centrifugation using Lymphoprep (Axis-Shield). Monocytes and T cells were isolated using CD14 positive magnetic beads and Pan T Cell Isolation kit, respectively (Miltenyi Biotec). Purity was determined by flow cytometry and accepted at >95% CD14+ and >98% CD3+. Isolated cells were resuspended in culture medium X-VIVO 15 (Lonza) containing 2% human male AB serum (Sigma) and either subjected to a culture protocol and incubated at 37 °C in 5% CO2 or cryopreserved in culture medium + 10% DMSO (WAK-Chemie) to be stored in liquid nitrogen.
HNSCC patients
Blood collection of patients was approved by the Committee on Research involving Human Subjects Arnhem-Nijmegen and written informed consent was obtained from all patients. Five HNSCC patients with lymph node metastases, but without distant metastases, were included. Blood samples were collected from patients before start of chemoradiotherapy and without prior systemic treatment or radiotherapy. PBMCs were isolated from blood by density gradient centrifugation using Lymphoprep (Axis-Shield). T cells were depleted from PBMCs using the Pan T cell Isolation Kit and LS column (Miltenyi Biotec) according to manufacturer’s instructions. The T cell negative fraction was stained with antibodies against CD14 and HLA-DR (Table S1) in PBS with 0.1% bovine serum albumin and 0.4% ethylenediaminetetraacetic acid for 30 min. CD14+ HLA-DRneg/low (M-MDSCs) and CD14+ HLA-DRplus/high were sorted with a FACSAria (BD Biosciences) using BD FACSDiva software (https://www.bdbiosciences.com/en-us/products/instruments/flow-cytometers/research-cell-sorters/bd-facsaria-iii). Sorted cells were cocultured with 2.5 × 104 carboxyfluorescein succinimidyl ester (CFSE)-labeled pan T cells from healthy donors in 200 μl culture medium. To minimize donor-donor variation, the same patient-donor (Myeloid-T cell) combinations were used. T cells were stimulated by Dynabeads (1 bead per 5 T cells) and after 3 days, supernatants were taken for IFNγ ELISA and proliferation of T cells was measured by quantifying mean cycle of proliferation by flow cytometry. Mean cycle was calculated per condition as the log2 of the geometric mean fluorescence intensity (MFI) of CFSE from unstimulated T cells divided by the geometric MFI of CFSE from all T cells in the conditions.
In vitro generation of monocyte-derived cell types
M-MDSCs were generated in vitro by culturing 1 × 106 monocytes/well of a 6-well plate in 2 ml culture medium supplemented by GM-CSF and IL-6 (both 10 ng/ml; Cell Genix) for 7 days. Cytokines were refreshed at day 3 by adding 0.5 ml of culture medium supplemented with 5× cytokine concentrations. Immature MoDCs were generated by culturing 1 × 106 monocytes/well of a 6-well plate in 2 ml culture medium supplemented with GM-CSF (450U/ml) and IL-4 (300U/ml; Cell Genix) for 6 days. At day 3, cytokines were refreshed in the same manner as described for the M-MDSC protocol. On day 6, a maturation cocktail was added containing PGE-2 (10 μg/ml; Pfizer), TNFα (500 U/ml), IL-1β (1000 U/ml), and IL-6 (1000 U/ml; all Cell Genix) for 24 h. Generated cells were harvested at day 7 by incubating plates at 4 degrees for 1 h before collection by means of gentle scraping. For the generation of TolDCs, dexamethasone (10−6 M) was added at day 3 in the MoDC protocol. In the stimulation assays, the following stimuli were used: LPS (100 ng/ml, Sigma), IFNγ (10 ng/ml, Thermo Fisher Scientific), Zymosan (1 × 106 beads/ml, Sigma), and PGE-2 (3.5 μg/ml, Pfizer).
Flow cytometry
To determine the expression of phenotypic markers, 2.5 to 5 × 104 myeloid cells/well were plated in 96-well v-bottom plates and washed with PBS. Cells were first stained for cell viability using Fixable Viability Dye eFluor506 (1:2000 dilution; eBioscience) in PBS for 20 min. Extracellular staining was performed at 4 °C in PBA (PBS + 1% bovine serum albumin + 0.05% sodium azide) in the dark. The antibody details are provided in Table S1. Intracellular ROS staining was performed using CM-H2DCFDA (Thermo Fisher Scientific) using manufacturer’s instructions. Myeloid cells stained with viability dye and CD11b only were used to set gates for marker positivity. To quantify T cell proliferation, all cells were transferred from assay plates to 96 wells v-bottom plates and washed with PBS. T cells were stained for cell viability, CD3 and CD8/CD4 markers using Fixable Viability Dye eFluor780 (1:2000 dilution, eBioscience) and the antibodies listed in Table S1. Unlabeled T cells were included to set the gate for CFSE positivity, and CD11b staining was used to exclude myeloid cells from analysis. Stained cells were analyzed using a FACSVerse flow cytometer (BD Biosciences). Quality control of the flow cytometer's performance and coefficient of variation values were monitored on a day-to-day basis using CS&T beads (BD Biosciences). Data was analyzed using FlowJo V10 software (https://www.flowjo.com/solutions/flowjo/downloads) (Treestar). In this manuscript, the MFI. The median was used in every experiment to represent the MFI.
Allogeneic T cell activation assay
The ability of myeloid cells to induce immune responses by allogeneic T cells was determined in a coculture of mixed donors in 96 wells round bottom culture plates. 1 × 105 thawed allogeneic pan T cells labeled with CFSE (5 μM, Invitrogen) were plated/well in at least triplicates with myeloid cells in indicated ratios in 200 μl assay medium IMDM (Gibco) containing 10% heat-inactivated and filtered human serum (AB male, Sigma) + 1% antimycotic/antibiotic (Gibco). T cells left unstimulated or stimulated with 0.2 μl CD3/CD28 Dynabeads (Gibco) were used as negative and positive controls, respectively. After 5 days, supernatants were taken for IFNγ ELISA and cells were analyzed for proliferation by quantifying the CFSE dilution by flow cytometry.
DC-activated autologous T cell suppression assay
The suppression of autologous T cell responses by myeloid cells was determined in a coculture of three cell types in 96 wells round bottom culture plates. 5 × 104 thawed autologous pan T cells labeled with CFSE were stimulated by 5 × 103 allogeneic MoDCs and plated in at least triplicates to which myeloid cells (autologous to T cells) were added in indicated ratios to T cells in assay medium. T cells left unstimulated or stimulated with 0.2 μl CD3/CD28 Dynabeads were used as negative and positive controls, respectively. After 5 days, supernatants were taken for IFNγ ELISA and cells were analyzed for proliferation by quantifying the CFSE dilution by flow cytometry.
RNA sequencing
Cells were washed twice with PBS before snap-freezing dry pellets using liquid nitrogen and storage at −80 °C. Total RNA was extracted from cell pellets using RNeasy Plus Micro Kit (Qiagen) according to manufacturer’s instructions. Per sample, 30 ng of RNA was used for preparation of RNA-seq libraries using the KAPA RNA HyperPrep Kit with RiboErase (KAPA Biosystems). Fragmentation and priming were performed at 85 °C for 6 min. For adapter ligation, 1.5 μM adapter stocks (8 bp NEXTflex DNA barcodes, Bioo Scientific) were used and sufficient library yield was achieved by 15 cycles of PCR. Library amplification cleanup was performed, and library size was determined using the High Sensitivity DNA bioanalyzer kit, and concentration was measured using the dsDNA High Sensitivity Assay (Denovix). Paired-end sequencing reads of 50 bp were generated using an Illumina NextSeq 500. Sequence reads were processed using the seq2science pipeline (https://github.com/vanheeringen-lab/seq2science). Differential expressed genes were identified using DeSeq2 (56).
RNA microarray
Cells were washed twice with PBS before snap-freezing dry pellets using liquid nitrogen and storage at −80 °C. Total RNA was extracted from cell pellets using Quick-RNA MiniPrep Plus Kit (Zymo Research) according to manufacturer’s instructions, and RNA concentration and purity were measured with a NanoDrop (Thermo Fisher Scientific). RNA integrity number was assessed and RNA integrity number ≥ 7 was considered of sufficient quality. Total RNA was labeled and hybridized to the GeneChip Human Genome U133 Plus 2.0 (Affymetrix). Quality control was performed on Affymetrix data of each individual sample based on twelve different quality parameters following Affymetrix recommendations and previously published literature. In summary, these parameters include the average value of all probe intensities, presence of negative or extremely high (>16 bit) intensity values, poly-A RNA (sample preparation spike-ins) and labeled cRNA (hybridization spike ins) controls, GAPDH and ACTB 3’/5′ ratio, the center of intensity and values of positive and negative border controls determined by affyQCReport package in R, and an RNA degradation value determined by the AffyRNAdeg function from the Affymetrix package in R. Samples that failed quality control were removed prior to data analysis. Packages and libraries developed for hgu133plus2.0 affymetrix data analysis were obtained from the Bioconductor Project (www.bioconductor.org) and assessed in Rstudio (Version v2021.09.1+372, RStudio Team 2021).
Philips transduction pathway analysis
Tests to quantitatively measure functional activity of PI3K, Wnt, NFκB, TGFβ, HH, AP1, AR, ER, JAK-STAT, and Notch signal transduction pathways on Affymetrix Human Genome U133 Plus 2.0 expression microarrays data have been described before (57). In brief, the pathway assays are based on the concept of a Bayesian network computational model which calculates from mRNA levels of a selected set, usually between 20 and 30, target genes of the pathway-associated transcription factor, a probability score for pathway activity, which is translated to a log2 value of the transcription factor odds, that is, a log2odds score scaling with pathway activity. The models have all been calibrated on a single cell type and were subsequently frozen and validated on a number of other cell and tissue types without further adaptations of the models. The range (minimum-maximum pathway activity) on the log2odds scale is different for each signaling pathway. While the signaling pathway assays can be used on all cell types, the log2odds score range may vary per cell/tissue type. Of note, measurement of the activity of the PI3K pathway assay is based on the inverse inference of activity of the PI3K pathway from the measured activity of the FOXO transcription factor, in the absence of cellular oxidative stress (34). For this reason, the FOXO activity score is presented in the figures instead of PI3K pathway activity. In cell culture experiments in vitro, PI3K pathway activity can be directly (inversely) inferred from FOXO activity.
General rules for interpretation of signal transduction pathway activity scores
An important and unique advantage of the pathway activity assays is that they can in principle be performed on each cell type. Important considerations for interpretation of log2 odds pathway activity scores are.
-
(1)
on the same sample, log2 odds pathway activity scores cannot be compared between different signaling pathways, since each of the signaling pathways has its own range in log2 odds activity scores;
-
(2)
the log2 odds range for pathway activity (minimum-maximum activity) may vary depending on cell type. Once the range has been defined using samples with known pathway activity, on every new sample, the absolute value can be directly interpreted against that reference. If the range has not been defined, only differences in log2 odds activity score between samples can be interpreted;
-
(3)
pathway activity scores are highly quantitative, and even small differences in log2 odds can be reproducible and meaningful;
-
(4)
a negative log2 odds ratio does not mean that the pathway is inactive.
Mass spectrometry
Cells were washed twice with PBS before snap-freezing dry pellets using liquid nitrogen and storage at −80 °C. Cell pellets were lysed in SDS lysis buffer (4% SDS, 1 mM DTT, 100 mM Tris pH 7.5) and sonicated for five cycles (30 s on/30 s off, Diagenode Bioruptor Pico). Protein yield was measured using Pierce BCA kit (Thermo Fisher Scientific). Protein lysates (∼20 μg) were digested with trypsin using Filter Aided Sample-Preparation (58). Peptides were desalted and stored on C18-StageTips prior to mass spectrometry analysis (59). Samples were applied to on-line Easy-nLC 1000 (Thermo Fisher Scientific) separation using 2 hour gradients. Mass spectra were collected on an Orbitrap Exploris 480 (Thermo Fisher Scientific) in data-dependent top 20 mode with dynamic exclusion set at 45s. Raw mass spectra were analyzed in MaxQuant 1.6.0.1 (60), with match between runs and label-free quantification enabled. The fragmentation spectra were searched against the Uniprot human protein database (downloaded 2017–06). Potential contaminants, reverse sequences, and proteins identified by only one peptide were excluded from the analysis using Perseus software (https://maxquant.net/perseus/). Proteins quantified in all triplicates of at least one sample group were considered for downstream analysis. Next, missing LFQ values were imputed for statistical analysis using ‘replace missing values from normal distribution’ function with default settings.
IL-10, TGF-β, PGE-2, and IFNγ ELISA
1 × 105 cells/well were cultured in 200 μl of culture medium in a 96 wells round bottom culture plate for detection of IL-10, TGF-β, and PGE-2. Conditioned medium was collected after 24 h (IL-10 and TGF-β) or 48 h (PGE-2) of culture and clarified by centrifugation. For IL-10 quantification, cells were stimulated with 1 μg/ml LPS (Sigma Aldrich) and supernatants were processed using the IL-10 ELISA kit (Invitrogen). For TGF-β1 quantification, cells were cultured without serum, and supernatants were activated and processed using the TGF-β1 ELISA kit (R&D Systems) following the manufacturer’s instructions. For PGE-2 quantification, supernatants were processed using the PGE-2 ELISA KIT (R&D Systems). IFNγ content was measured in supernatants of myeloid–T cell cocultures at indicated timepoints and processed using the IFNγ ELISA kit (Invitrogen) following manufacturer’s instructions. Absorption was measured using an iMark Microplate Reader (Bio-Rad).
HPLC kynurenine detection
1 × 105 cells/well were cultured in 200 μl of culture medium in a 96 wells round bottom culture. Conditioned medium was collected after 24 h of incubation and clarified by centrifugation. The supernatants (100 μl) were diluted with a solution 3-nitro-l-tyrosine (10 μM, Sigma-Aldrich) in PBS (100 μl) (internal standard). Then proteins were precipitated by adding a solution of trichloroacetic acid (2 M, Sigma-Aldrich) in MilliQ (25 μl). The samples were spun down for 5 min at 13.800 rcf. The supernatants (150 μl) were transferred to HPLC vials and measured on the auto-sampling HPLC device (Shimadzu) with XSelect Peptide CSHTM C18 column (130 Å, 3.5 μm, 4.6 mm × 100 mm, Waters). First l-kynurenine (Sigma-Aldrich) standards were run to generate a standard curve, followed by the samples. Chromatographic peak areas were determined by manual integration of each peak at 360 nm. The peak area of the 3-nitro-l-tyrosine was measured and concentrations of l-kynurenine were determined by extrapolation into the linear standard curve.
Western blot analysis
Cells were lysed in RIPA buffer and 30 μg protein was loaded onto 8% acrylamide gels for protein separation by SDS PAGE followed by protein transfer onto polyvinylidene difluorid membranes (Immobilon-FL). Blots were blocked with block buffer in PBS (Intercept) at room temperature for 1 h and incubated with primary antibodies overnight at 4 °C (Table S1). Secondary antibodies were incubated for 1 h at room temperature. Signal was detected with Odyssey Clx (LI-COR) and assessed and quantified using Image Studio (LI-COR) and ImageJ (National Institute of Health).
PI3K inhibition assays
1 × 105 monocytes were plated in 200 μl culture medium supplemented with GM-CSF and IL-6 in a 96 wells round bottom culture plate for 7 days to generate M-MDSCs as described above. PI3K inhibitors Wortmannin (Invivogen), LY294002 (Invivogen), and vehicle DMSO were added at day 0 and day 3 in indicated concentrations. At day 7, plates were harvested for phenotype assessment or spun down to discard supernatant, washed with PBS, and resuspended the cells in 100 μl fresh culture medium. 1 × 105 CFSE-labeled autologous T cells stimulated with Dynabeads (1 bead per 5 T cells) were added in culture medium to the treated M-MDSCs (1:1 T cell: M-MDSC ratio). After 3 days, supernatants were taken for IFNγ ELISA and proliferation of T cells was measured by quantifying CFSE signal by flow cytometry.
Beta-catenin inhibition assays
In phenotyping assays, 1 × 105 monocytes were plated in 200 μl culture medium supplemented with GM-CSF and IL-4 in a 96 wells round bottom culture plate and treated as described above to generate TolDCs. β-catenin inhibitor XAV939 (Sigma-Aldrich) and vehicle DMSO were added at day 0 and day 3 in indicated concentrations before harvest and phenotype assessment. In functional assays, TolDCs were generated in 6-well plates as described above and 10 μM of XAV939 or vehicle DMSO were added at day 3. After harvest, cells were washed and replated in 2 × 104 per well in 96 wells round bottom culture plates in 100 μl culture medium. 1 × 105 CFSE-labeled allogeneic pan T cells were added to the treated TolDCs (5: 1 T cell: TolDC ratio) in 100 μl culture medium. After 5 days, supernatants were taken for IFNγ ELISA and proliferation of T cells was measured by quantifying CFSE signal by flow cytometry.
Statistical analysis
Data was analyzed using Prism software version 5.0 (https://www.graphpad.com/support/prism-5-updates/) (GraphPad Software). Data transformation was performed to meet normal distribution assumption of linear regressions in HLA-DR biplots. Applied statistical tests to determine significant differences, error bars, and sample sizes are stated in the figure legends.
For Figure 5, D and E, the absolute data of all experimental groups was first normalized to the control group (vehicle-treated) within each experiment. For visualization and space-saving purposes, the data of PI3Ki and GANT61 experiments are shown combined into a limited number of figures. Statistical analyses were done using GraphPad software (https://www.graphpad.com/support/prism-5-updates/). Because Gaussian distributions are assumed, the ordinary one Way ANOVA was selected without matching or pairing the data. Due to the combined visualization of the PI3Ki and GANT61 data, a pre-selection of the pairwise columns to be compared was setup to make sure that all PI3Ki columns were compared to each other (and vehicle) but not the GANT61 columns and that GANT61 columns were compared to each other (and vehicle) and not to PI3Ki columns. Lastly, Bonferroni method was selected to correct for multiple comparisons.
For Figure 5G, the absolute data is shown (no normalization to control group). The data of interest selected here for statistical comparison are all experimental groups at a concentration of 10 μM, including vehicle (DMSO). As Gaussian distributions are assumed, the ordinary one Way ANOVA was selected without matching or pairing the data. The mean of every experimental group at 10 μM was compared to the mean of control group (DMSO at 10 μM). Lastly, Bonferroni method was selected to correct for multiple comparisons.
Compliance with ethical standards
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional research committees (Committee on Research involving Human Subjects Arnhem-Nijmegen) and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Data availability
The RNA microarray and RNAseq datasets have been deposited in NCBI’s Gene Expression Omnibus under the super-series accession number GSE183855. All proteomics data are available on the PRIDE repository (PXD027715).
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
The authors would like to thank Rob Woestenenk and the Radboud Technology Center Flow Cytometry for their assistance in FACS sorting and Michiel Kerkkamp for patient inclusion and patient care; Simone Hins-deBree for overseeing clinical patient information; and Inge Wortel for their assistance in linear regression analysis.
Author contributions
G. F. v. W., I. J. M. d. V., C. G. F., and G. F.-G. methodology; G. F. v. W., S. S., I. J. M. d. V., C. G. F., and G. F.-G. formal analysis; G. F. v. W., J. C.-E., S. S., J. B., E. P., I. R.-T., Y. W.-R., and A. v. d. S. investigation; Y. W.-R. and A. v. d. S. data curation; S. F. and C. M. L. v. H. resources; G. F. v. W. writing–original draft; S. K. H., A. v. d. S., M. V., I. J. M. d. V., C. G. F., and G. F.-G. writing–review and editing.
Funding and additional information
G. F. v. W. was supported by The Dutch Cancer Society (grant 10673); J. C.-E. was supported by Health Holland, Top Sector Life Sciences & Health grant LSHM18056-SGF; Y. W.-R. was full-time employed by Molecular Pathway Diagnostics, Philips; A. v. d. S. was a full-time regular employee of Philips (Philips Research); I. J. M. d. V. received Health Holland, Top Sector Life Sciences & Health grant LSHM18056-SGF; C. G. F. received ERC Adv Grant ARTimmune (834618); G. F.-G. was supported by Health Holland, Top Sector Life Sciences & Health grant LSHM18056-SGF. The Figdor lab and Vermeulen lab are part of the Oncode Institute, which is partly funded by The Dutch Cancer Society.
Reviewed by members of the JBC Editorial Board. Edited by Clare E. Bryant
Supporting information
References
- 1.Audiger C., Rahman M.J., Yun T.J., Tarbell K.V., Lesage S. The importance of dendritic cells in maintaining immune tolerance. J. Immunol. 2017;198:2223–2231. doi: 10.4049/jimmunol.1601629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.ElTanbouly M.A., Noelle R.J. Rethinking peripheral T cell tolerance: checkpoints across a T cell’s journey. Nat. Rev. Immunol. 2021;21:257–267. doi: 10.1038/s41577-020-00454-2. [DOI] [PubMed] [Google Scholar]
- 3.Takaba H., Takayanagi H. The mechanisms of T cell selection in the Thymus. Trends Immunol. 2017;38:805–816. doi: 10.1016/j.it.2017.07.010. [DOI] [PubMed] [Google Scholar]
- 4.Proekt I., Miller C.N., Lionakis M.S., Anderson M.S. Insights into immune tolerance from AIRE deficiency. Curr. Opin. Immunol. 2017;49:71–78. doi: 10.1016/j.coi.2017.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 6.Mantovani A., Cassatella M.A., Costantini C., Jaillon S. Neutrophils in the activation and regulation of innate and adaptive immunity. Nat. Rev. Immunol. 2011;11:519–531. doi: 10.1038/nri3024. [DOI] [PubMed] [Google Scholar]
- 7.Galli S.J., Borregaard N., Wynn T.A. Phenotypic and functional plasticity of cells of innate immunity: macrophages, mast cells and neutrophils. Nat. Immunol. 2011;12:1035–1044. doi: 10.1038/ni.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suryawanshi A., Hussein M.S., Prasad P.D., Manicassamy S. Wnt signaling cascade in dendritic cells and regulation of anti-tumor immunity. Front. Immunol. 2020;11:122. doi: 10.3389/fimmu.2020.00122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bol K.F., Aarntzen E.H.J.G., Pots J.M., Olde Nordkamp M.A.M., van de Rakt M.W.M.M., Scharenborg N.M., et al. Prophylactic vaccines are potent activators of monocyte-derived dendritic cells and drive effective anti-tumor responses in melanoma patients at the cost of toxicity. Cancer Immunol. Immunother. 2016;65:327–339. doi: 10.1007/s00262-016-1796-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morante-Palacios O., Fondelli F., Ballestar E., Martínez-Cáceres E.M. Tolerogenic dendritic cells in autoimmunity and inflammatory diseases. Trends Immunol. 2021;42:59–75. doi: 10.1016/j.it.2020.11.001. [DOI] [PubMed] [Google Scholar]
- 11.Jauregui-Amezaga A., Cabezón R., Ramírez-Morros A., España C., Rimola J., Bru C., et al. Intraperitoneal administration of autologous tolerogenic dendritic cells for refractory crohn’s disease: a phase I study. J. Crohns. Colitis. 2015;9:1071–1078. doi: 10.1093/ecco-jcc/jjv144. [DOI] [PubMed] [Google Scholar]
- 12.Zubizarreta I., Flórez-Grau G., Vila G., Cabezón R., España C., Andorra M., et al. Immune tolerance in multiple sclerosis and neuromyelitis optica with peptide-loaded tolerogenic dendritic cells in a phase 1b trial. Proc. Natl. Acad. Sci. U. S. A. 2019;116:8463–8470. doi: 10.1073/pnas.1820039116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kurochkina Y., Tikhonova M., Tyrinova T., Leplina O., Sizikov A., Sulutian A., et al. SAT0212 the safety and tolerability of intra-articular injection of tolerogenic dendritic cells in patients with rheumatoid arthritis: the preliminary results. Annals. Rheumatic Diseases. 2018 doi: 10.1136/annrheumdis-2018-eular.2880. [DOI] [Google Scholar]
- 14.Messmer M.N., Netherby C.S., Banik D., Abrams S.I. Tumor-induced myeloid dysfunction and its implications for cancer immunotherapy. Cancer Immunol. Immunother. 2015;64:1–13. doi: 10.1007/s00262-014-1639-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Veglia F., Sanseviero E., Gabrilovich D.I. Myeloid-derived suppressor cells in the era of increasing myeloid cell diversity. Nat. Rev. Immunol. 2021;21:485–498. doi: 10.1038/s41577-020-00490-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Casacuberta-Serra S., Parés M., Golbano A., Coves E., Espejo C., Barquinero J. Myeloid-derived suppressor cells can be efficiently generated from human hematopoietic progenitors and peripheral blood monocytes. Immunol. Cell Biol. 2017;95:538–548. doi: 10.1038/icb.2017.4. [DOI] [PubMed] [Google Scholar]
- 17.Lechner M.G., Liebertz D.J., Epstein A.L. Characterization of cytokine-induced myeloid-derived suppressor cells from normal human peripheral blood mononuclear cells. J. Immunol. 2010;185:2273–2284. doi: 10.4049/jimmunol.1000901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mao Y., Poschke I., Wennerberg E., Pico de Coaña Y., Egyhazi Brage S., Schultz I., et al. Melanoma-educated CD14+ cells acquire a myeloid-derived suppressor cell phenotype through COX-2-dependent mechanisms. Cancer Res. 2013;73:3877–3887. doi: 10.1158/0008-5472.CAN-12-4115. [DOI] [PubMed] [Google Scholar]
- 19.Pico de Coaña Y., Wolodarski M., van der Haar Àvila I., Nakajima T., Rentouli S., Lundqvist A., et al. PD-1 checkpoint blockade in advanced melanoma patients: NK cells, monocytic subsets and host PD-L1 expression as predictive biomarker candidates. Oncoimmunology. 2020;9:1786888. doi: 10.1080/2162402X.2020.1786888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krieg C., Nowicka M., Guglietta S., Schindler S., Hartmann F.J., Weber L.M., et al. High-dimensional single-cell analysis predicts response to anti-PD-1 immunotherapy. Nat. Med. 2018;24:144–153. doi: 10.1038/nm.4466. [DOI] [PubMed] [Google Scholar]
- 21.Shipp C., Speigl L., Janssen N., Martens A., Pawelec G. A clinical and biological perspective of human myeloid-derived suppressor cells in cancer. Cell. Mol. Life Sci. 2016;73:4043–4061. doi: 10.1007/s00018-016-2278-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Henau O., Rausch M., Winkler D., Campesato L.F., Liu C., Cymerman D.H., et al. Overcoming resistance to checkpoint blockade therapy by targeting PI3Kγ in myeloid cells. Nature. 2016;539:443–447. doi: 10.1038/nature20554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao J.X., Madrenas J., Zeng W., Zhong R., Grant D. Generation of dendritic cell-like antigen-presenting cells in long-term mixed leucocyte culture: phenotype and functional studies. Immunology. 1997;91:135–144. doi: 10.1046/j.1365-2567.1997.00220.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Naranjo-Gómez M., Raïch-Regué D., Oñate C., Grau-López L., Ramo-Tello C., Pujol-Borrell R., et al. Comparative study of clinical grade human tolerogenic dendritic cells. J. Transl. Med. 2011;9 doi: 10.1186/1479-5876-9-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cabezón R., Ricart E., España C., Panés J., Benitez-Ribas D. Gram-negative Enterobacteria induce tolerogenic maturation in dexamethasone conditioned dendritic cells. PLoS One. 2012;7 doi: 10.1371/journal.pone.0052456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cabezo´n R., Carrera-Silva E.A., Flórez-Grau G., Errasti A.E., Calderón-Gómez E., Lozano J.J., et al. MERTK as negative regulator of human T cell activation. J. Leukoc. Biol. 2015;97:751–760. doi: 10.1189/jlb.3A0714-334R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hobo W., et al. Dendritic cell mediated reactivation of functionally impaired memory T cells to improve graftversus-tumor responses after stem cell transplantation. Eur. J. Immunol. 2009;39:S522. [Google Scholar]
- 28.Rosewicz S., McDonald A.R., Maddux B.A., Goldfine I.D., Miesfeld R.L., Logsdon C.D. Mechanism of glucocorticoid receptor down-regulation by glucocorticoids. J. Biol. Chem. 1988;263:2581–2584. [PubMed] [Google Scholar]
- 29.Basit F., Mathan T., Sancho D., De Vries J.M. Human dendritic cell subsets undergo distinct metabolic reprogramming for immune response. Front. Immunol. 2018;9:2489. doi: 10.3389/fimmu.2018.02489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wculek S.K., Khouili S.C., Priego E., Heras-Murillo I., Sancho D. Metabolic control of dendritic cell functions: digesting information. Front. Immunol. 2019;10:775. doi: 10.3389/fimmu.2019.00775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alshetaiwi H., Pervolarakis N., McIntyre L.L., Ma D., Nguyen Q., Rath J.A., et al. Defining the emergence of myeloid-derived suppressor cells in breast cancer using single-cell transcriptomics. Sci. Immunol. 2020;5 doi: 10.1126/sciimmunol.aay6017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Navarro-Barriuso J., Mansilla M.J., Naranjo-Gómez M., Sánchez-Pla A., Quirant-Sánchez B., Teniente-Serra A., et al. Comparative transcriptomic profile of tolerogenic dendritic cells differentiated with vitamin D3, dexamethasone and rapamycin. Sci. Rep. 2018;8:14985. doi: 10.1038/s41598-018-33248-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van de Stolpe A., Holtzer L., van Ooijen H., de Inda M.A., Verhaegh W. Enabling precision medicine by unravelling disease pathophysiology: quantifying signal transduction pathway activity across cell and tissue types. Sci. Rep. 2019;9:1603. doi: 10.1038/s41598-018-38179-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Ooijen H., Hornsveld M., Dam-de Veen C., Velter R., Dou M., Verhaegh W., et al. Assessment of functional phosphatidylinositol 3-kinase pathway activity in cancer tissue using forkhead box-O target gene expression in a knowledge-based computational model. Am. J. Pathol. 2018;188:1956–1972. doi: 10.1016/j.ajpath.2018.05.020. [DOI] [PubMed] [Google Scholar]
- 35.Pai S.G., Carneiro B.A., Mota J.M., Costa R., Leite C.A., Barroso-Sousa R., et al. Wnt/beta-catenin pathway: modulating anticancer immune response. J. Hematol. Oncol. 2017;10:101. doi: 10.1186/s13045-017-0471-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang S.M.A., Mishina Y.M., Liu S., Cheung A., Stegmeier F., Michaud G.A., et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature. 2009;461:614–620. doi: 10.1038/nature08356. [DOI] [PubMed] [Google Scholar]
- 37.Young M.R.I., Wright M.A., Matthews J.P., Collins S.L., Petruzzelli G.J., Young M.R. Mechanisms of immune suppression in patients with head and neck cancer: presence of CD34+ cells which suppress immune functions within cancers that secrete granulocyte-macrophage colony-stimulating factor. Clin. Cancer Res. 1995;1:95–103. [PubMed] [Google Scholar]
- 38.Vasquez-Dunddel D., Pan F., Zeng Q., Gorbounov M., Albesiano E., Fu J., et al. STAT3 regulates arginase-i in myeloid-derived suppressor cells from cancer patients. J. Clin. Invest. 2013;123:1580–1589. doi: 10.1172/JCI60083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weed D.T., Vella J.L., Reis I.M., De la Fuente A.C., Gomez C., Sargi Z., et al. Tadalafil reduces myeloid-derived suppressor cells and regulatory t cells and promotes tumor immunity in patients with head and neck squamous cell carcinoma. Clin. Cancer Res. 2015;21:39–48. doi: 10.1158/1078-0432.CCR-14-1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davis R.J., Van Waes C., Allen C.T. Overcoming barriers to effective immunotherapy: MDSCs, TAMs, and Tregs as mediators of the immunosuppressive microenvironment in head and neck cancer. Oral Oncol. 2016;58:59–70. doi: 10.1016/j.oraloncology.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Davis R.J., Moore E.C., Clavijo P.E., Friedman J., Cash H., Chen Z., et al. Anti-PD-L1 efficacy can be enhanced by inhibition of myeloid-derived suppressor cells with a selective inhibitor of PI3Kd/g. Cancer Res. 2017;77:2607–2619. doi: 10.1158/0008-5472.CAN-16-2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Greene S., Robbins Y., Mydlarz W.K., Huynh A.P., Schmitt N.C., Friedman J., et al. Inhibition of MDSC trafficking with SX-682, a CXCR1/2 inhibitor, enhances NK-cell immunotherapy in head and neck cancer models. Clin. Cancer Res. 2020;26:1420–1431. doi: 10.1158/1078-0432.CCR-19-2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Younis R.H., Han K.L., Webb T.J. Human head and neck squamous cell carcinoma–associated semaphorin 4D induces expansion of myeloid-derived suppressor cells. J. Immunol. 2016;196:1419–1429. doi: 10.4049/jimmunol.1501293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Petit-Bertron A.-F., Fitting C., Cavaillon J.M., Adib-Conquy M. Adherence influences monocyte responsiveness to interleukin-10. J. Leukoc. Biol. 2003;73:145–154. doi: 10.1189/jlb.0802388. [DOI] [PubMed] [Google Scholar]
- 45.Miyazawa A., Ito S., Asano S., Tanaka I., Sato M., Kondo M., Hasegawa Y. Regulation of PD-L1 expression by matrix stiffness in lung cancer cells. Biochem. Biophys. Res. Commun. 2018;495:2344–2349. doi: 10.1016/j.bbrc.2017.12.115. [DOI] [PubMed] [Google Scholar]
- 46.Kwak T., Wang F., Deng H., Condamine T., Kumar V., Perego M., et al. Distinct populations of immune-suppressive macrophages differentiate from monocytic myeloid-derived suppressor cells in cancer. Cell Rep. 2020;33:108571. doi: 10.1016/j.celrep.2020.108571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakamura K., Ito I., Kobayashi M., Herndon D.N., Suzuki F. Orosomucoid 1 drives opportunistic infections through the polarization of monocytes to the M2b phenotype. Cytokine. 2015;73:8–15. doi: 10.1016/j.cyto.2015.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bronte V., Brandau S., Chen S.H., Colombo M.P., Frey A.B., Greten T.F., et al. Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nat. Commun. 2016;7 doi: 10.1038/ncomms12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kaneda M.M., Messer K.S., Ralainirina N., Li H., Leem C.J., Gorjestani S., et al. PI3Kγ 3 is a molecular switch that controls immune suppression. Nature. 2016;539:437–442. doi: 10.1038/nature19834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Foubert P., Kaneda M.M., Varner J.A. PI3Kγ activates integrin α4 and promotes immune suppressive myeloid cell polarization during tumor progression. Cancer Immunol. Res. 2017;5:957–968. doi: 10.1158/2326-6066.CIR-17-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Van De Laar L., Coffer P.J., Woltman A.M. Regulation of dendritic cell development by GM-CSF: molecular control and implications for immune homeostasis and therapy. Blood. 2012;119:3383–3393. doi: 10.1182/blood-2011-11-370130. [DOI] [PubMed] [Google Scholar]
- 52.Wegiel B., Bjartell A., Culig Z., Persson J.L. Interleukin-6 activates PI3K/Akt pathway and regulates cyclin A1 to promote prostate cancer cell survival. Int. J. Cancer. 2008;122:1521–1529. doi: 10.1002/ijc.23261. [DOI] [PubMed] [Google Scholar]
- 53.Li J., Diao B., Guo S., Huang X., Yang C., Feng Z., et al. VSIG4 inhibits proinflammatory macrophage activation by reprogramming mitochondrial pyruvate metabolism. Nat. Commun. 2017;8:1322. doi: 10.1038/s41467-017-01327-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Valencia J., Hernández-López C., Martínez V.G., Hidalgo L., Zapata A.G., Vicente Á., et al. Wnt5a skews dendritic cell differentiation to an unconventional phenotype with tolerogenic features. J. Immunol. 2011;187:4129–4139. doi: 10.4049/jimmunol.1101243. [DOI] [PubMed] [Google Scholar]
- 55.Anderson A.E., Swan D.J., Wong O.Y., Buck M., Eltherington O., Harry R.A., et al. Tolerogenic dendritic cells generated with dexamethasone and vitamin D3 regulate rheumatoid arthritis CD4+ T cells partly via transforming growth factor-β1. Clin. Exp. Immunol. 2017;187:113–123. doi: 10.1111/cei.12870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:1–21. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van de Stolpe A., Verhaegh W., Blay J.Y., Ma C.X., Pauwels P., Pegram M., et al. RNA based approaches to profile oncogenic pathways from low quantity samples to drive precision oncology strategies. Front. Genet. 2021;11:598118. doi: 10.3389/fgene.2020.598118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wiśniewski J.R., Zougman A., Nagaraj N., Mann M. Universal sample preparation method for proteome analysis. Nat. Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 59.Rappsilber J., Mann M., Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2007;2:1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 60.Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA microarray and RNAseq datasets have been deposited in NCBI’s Gene Expression Omnibus under the super-series accession number GSE183855. All proteomics data are available on the PRIDE repository (PXD027715).