Abstract
A multicopy plasmid carrying the PDC1 gene (encoding pyruvate decarboxylase; Pdc) was introduced in Saccharomyces cerevisiae CEN.PK113-5D. The physiology of the resulting prototrophic strain was compared with that of the isogenic prototrophic strain CEN.PK113-7D and an empty-vector reference strain. In glucose-grown shake-flask cultures, the introduction of the PDC1 plasmid caused a threefold increase in the Pdc level. In aerobic glucose-limited chemostat cultures growing at a dilution rate of 0.10 h−1, Pdc levels in the overproducing strain were 14-fold higher than those in the reference strains. Levels of glycolytic enzymes decreased by ca. 15%, probably due to dilution by the overproduced Pdc protein. In chemostat cultures, the extent of Pdc overproduction decreased with increasing dilution rate. The high degree of overproduction of Pdc at low dilution rates did not affect the biomass yield. The dilution rate at which aerobic fermentation set in decreased from 0.30 h−1 in the reference strains to 0.23 h−1 in the Pdc-overproducing strain. In the latter strain, the specific respiration rate reached a maximum above the dilution rate at which aerobic fermentation first occurred. This result indicates that a limited respiratory capacity was not responsible for the onset of aerobic fermentation in the Pdc-overproducing strain. Rather, the results indicate that Pdc overproduction affected flux distribution at the pyruvate branch point by influencing competition for pyruvate between Pdc and the mitochondrial pyruvate dehydrogenase complex. In respiratory cultures (dilution rate, <0.23 h−1), Pdc overproduction did not affect the maximum glycolytic capacity, as determined in anaerobic glucose-pulse experiments.
Under most growth conditions, alcoholic fermentation is the predominant mode of sugar dissimilation in Saccharomyces cerevisiae (baker’s yeast). A completely respiratory sugar metabolism is only possible in aerobic cultures grown under sugar limitation and at relatively low specific growth rates (31). This principle is applied in industrial baker’s yeast production, where aerobic fermentation must be avoided because it results in a reduced biomass yield (9).
At high specific growth rates, even aerobic glucose-limited cultures exhibit mixed respirofermentative metabolism (21, 31, 33). This phenomenon is sometimes attributed to a limited respiratory capacity (36, 42). Indeed, models assuming a bottleneck in respiratory glucose dissimilation adequately describe the behavior of sugar-limited chemostat cultures (1, 2). Being purely empirical, such models cannot offer a mechanistic explanation for the onset of respirofermentative metabolism at high specific growth rates. Rate-limiting reactions in respiratory sugar metabolism might reside either in intermediary carbon metabolism (e.g., in the tricarboxylic acid cycle), in mitochondrial electron transport, or in a combination of the two (45).
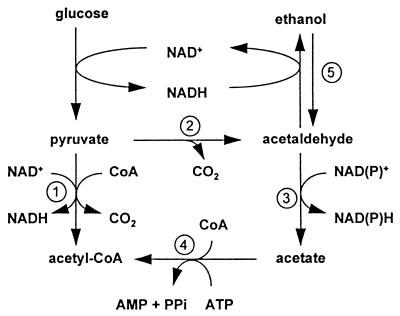
An alternative explanation for the occurrence of aerobic alcoholic fermentation at high specific growth rates is the competition of respiration and fermentation for pyruvate (18, 36, 46), a metabolite located at the branch point between respiration and fermentation (Fig. 1) (35). To be respired, pyruvate must be oxidatively decarboxylated to acetyl coenzyme A (acetyl-CoA); this reaction is catalyzed by the mitochondrial pyruvate dehydrogenase complex. Alcoholic fermentation requires decarboxylation of pyruvate to acetaldehyde by the cytosolic enzyme pyruvate decarboxylase (Pdc). Isolated mitochondria and purified pyruvate dehydrogenase exhibit a much lower Km for pyruvate than does Pdc (19, 25, 33, 46). At low rates of glucose dissimilation during respiratory glucose-limited growth, the intracellular pyruvate concentration is below the Km of pyruvate dehydrogenase (33). Since, in addition to a high Km, Pdc exhibits cooperativity with respect to pyruvate (10, 11, 20), pyruvate will be preferentially metabolized via pyruvate dehydrogenase at low intracellular pyruvate concentrations. In contrast, at high intracellular pyruvate concentrations, pyruvate will be predominantly metabolized via Pdc; in wild-type cells, the later reaction has a much higher Vmax than does mitochondrial pyruvate oxidation (31, 35, 46).
FIG. 1.
Central role of Pdc in respiratory and respirofermentative glucose metabolism in S. cerevisiae. Numbered reactions are catalyzed by the following enzymes: 1, pyruvate dehydrogenase complex; 2, Pdc; 3, acetaldehyde dehydrogenase; 4, acetyl-CoA; 5, alcohol dehydrogenase.
Decarboxylation of pyruvate to acetaldehyde by Pdc does not necessarily lead to alcoholic fermentation. Acetaldehyde may be oxidized to acetyl-CoA by the enzymes of the pyruvate dehydrogenase bypass (Fig. 1) (19, 35). In pyruvate dehydrogenase-negative mutants of S. cerevisiae grown at low specific growth rates in glucose-limited cultures, this bypass enables respiratory glucose dissimilation. This process leads to a reduced biomass yield on glucose due to the consumption of ATP in the acetyl-CoA synthetase reaction (34).
If competition between Pdc and pyruvate dehydrogenase for pyruvate is a physiologically relevant process, it should be possible to manipulate flux distribution at the pyruvate branch point by changing the Vmax and/or the Km of either of these enzymes. In practice, it is difficult to manipulate the kinetic parameters of pyruvate dehydrogenase: this large multienzyme complex is located in the mitochondrial matrix, and genes encoding mitochondrial pyruvate transporters have as yet not been identified (35). In contrast, the structural genes encoding Pdc have been well characterized (17), facilitating genetic modification of Pdc levels.
In this study, the Pdc content of S. cerevisiae was increased by increasing the copy number of the PDC1 structural gene (23). The physiological effects of Pdc overproduction were studied with aerobic glucose-limited chemostat cultures. In addition to the effect on flux distribution at the pyruvate branch point, the effect of Pdc overproduction on the glycolytic capacity of cells pregrown under sugar limitation was investigated.
MATERIALS AND METHODS
Strains and plasmids.
Haploid, prototrophic S. cerevisiae CEN.PK113-7D (MATa MAL2-8c SUC2) and its auxotrophic derivative CEN.PK113-5D (MATa MAL2-8c SUC2 ura3-52) were kindly provided by P. Kötter, Frankfurt, Germany. Precultures of yeast strains were grown to the stationary phase on mineral medium (52) adjusted to pH 6.0 and containing 2% (wt/vol) glucose in shake flasks. After glycerol (30% [vol/vol]) was added, 2-ml aliquots were stored in sterile vials at −70°C. These frozen stock cultures were used to inoculate precultures for chemostat cultivation. Escherichia coli XL1Blue (13) [recA1 endA1 gyrA96 thi-1 hsdR17 supE44 recA1 lac/F′ proAB lacIqZΔM15 Tn10(Tetr)] was used for plasmid amplification. The multicopy plasmid YEplac195 (16), carrying the URA3 marker gene, was a gift from R. D. Gietz (University of Manitoba, Winnipeg, Canada). pRUL321 is a 4.3-kb SphI fragment of the S. cerevisiae PDC1 gene in pBR322 (35a).
Nucleic acid manipulation and plasmid construction.
Plasmid DNA was isolated from E. coli by the ammonium acetate method (26). Restriction endonucleases (Boehringer) and T4 DNA ligase (Pharmacia) were used according to the suppliers’ recommendations. A 3,484-bp SphI-XhoI fragment from pRUL321 containing the PDC1 gene was cloned in YEplac195 digested with SphI and SalI, giving pRUL178. This construct, as well as the vector YEplac195, was used to transform S. cerevisiae CEN.PK113-5D by electroporation (6), selecting for uracil prototrophy. This procedure resulted in S. cerevisiae GG393 (containing the PDC1 construct) and GG393 (containing the empty vector).
Media.
E. coli was grown in LB medium (37). Yeast cultures for genetic experiments were grown on either YPD (40) or MY (55) medium supplemented as required with uracil (20 mg · liter−1). For physiological characterization of wild-type and recombinant yeast strains, a defined mineral medium containing vitamins (52) was used. For chemostat cultivation, the glucose concentration in reservoir media was 7.5 g · liter−1 (0.25 mol of carbon per liter).
Chemostat cultivation in fermentors.
Aerobic chemostat cultivation was performed at 30°C with laboratory fermentors (Applikon, Schiedam, The Netherlands) at a stirrer speed of 800 rpm. All chemostat cultivation runs were started at a dilution rate (D, equal to specific growth rate in steady-state cultures) of 0.10 h−1. After steady states had been established at higher dilution rates, the culture was brought back to a D of 0.10 h−1 to check for hysteresis effects. These were not found (data not shown). A steady state was defined as a situation in which at least five volume changes had passed after the last change in growth conditions and in which the biomass concentration, as well as the specific rates of carbon dioxide production and oxygen consumption, had remained constant (<2% variation) for at least two volume changes. The working volume of the cultures was kept at 1.0 liter by a peristaltic effluent pump coupled to an electrical level sensor. This setup ensured that under all growth conditions, biomass concentrations in samples taken directly from the culture differed by <1% from biomass concentrations in samples taken from the effluent line (30). The exact working volume was measured after each experiment. The pH was kept at 5.0 ± 0.1 by an ADI 1030 biocontroller via the automatic addition of 2 mol of KOH liter−1. The fermentors were flushed with air at a flow rate of 0.5 liter · min−1 by use of a Brooks 5876 mass flow controller. The dissolved oxygen concentration was continuously monitored with an oxygen electrode (model 34 100 3002; Ingold) and remained above 60% air saturation. Steady-state data are reported for cultures without detectable oscillations in oxygen consumption and carbon dioxide production rates. Chemostat cultures were routinely checked for purity by phase-contrast microscopy and by plating on YPD agar. The minor loss of ethanol in the exhaust gas due to evaporation at high dilution rates (49) and the dilution of cultures as a result of alkali titration were not taken into account for calculating carbon recoveries of steady-state chemostat cultures. These calculations were based on a carbon content of dry yeast biomass of 48%.
Gas analysis.
The exhaust gas was cooled in a condenser (2°C) and dried with a Perma Pure dryer (type PD-625-12P). O2 and CO2 concentrations were determined with a Servomex type 1100A analyzer and a Beckman model 864 infrared detector, respectively. The off-gas flow rate was measured as described previously (54). Specific rates of CO2 production (qCO2) and O2 consumption (qO2) were calculated according to van Urk et al. (45).
Determination of culture dry weight.
Culture samples (10 ml) were filtered over preweighed nitrocellulose filters (pore size, 0.45 μm; Gelman Sciences). After the removal of medium, the filters were washed with demineralized water, dried in a Sharp type R-4700 microwave oven for 20 min at a 360-W output, and weighed. Duplicate determinations varied by <1%.
Determination of maximum glycolytic capacity.
Samples containing exactly 100 mg (dry weight) of biomass from a chemostat culture were harvested by centrifugation at 10,000 × g for 5 min, washed once, and resuspended in 5 ml of 0.9% (wt/vol) NaCl solution. The cell suspension was then immediately introduced into a thermostat-controlled (30°C) vessel containing 10 ml of fivefold-concentrated mineral medium (pH 5.6). The volume was adjusted to 40 ml with demineralized water. After 10 min of incubation, a 10-ml glucose pulse (100 g · liter−1) was applied, and samples (1 ml) were taken at appropriate times. The working volume was 50 ml, with a 10-ml headspace, which was continuously flushed with CO2 gas at a flow rate of ca. 10 ml · min−1. Ethanol production in sample supernatants was determined according to Verduyn et al. (50) with alcohol oxidase from Hansenula polymorpha (a kind gift from Bird Engineering, Schiedam, The Netherlands). Specific rates of ethanol production are expressed as millimoles of ethanol · gram of dry yeast biomass−1 · h−1.
Metabolite analysis.
Glucose in reservoir media and supernatants was determined with a glucose oxidase kit (Merck kit 14144; detection limit, ca. 5 μM). Ethanol, glycerol, and pyruvate were determined by high-pressure liquid chromatography with an HPX-87H Aminex ion-exchange column (300 by 7.8 mm; Bio-Rad) at 60°C. The column was eluted with 5 mM H2SO4 at a flow rate of 0.6 ml · min−1. Pyruvate was detected at 214 nm with a Waters 441 UV meter coupled to a Waters 741 data module. Ethanol and glycerol were detected with an ERMA type ERC-7515A refractive-index detector coupled to a Hewlett-Packard type 3390A integrator. Acetate was determined with Boehringer test kit 148261 (detection limit, ca. 0.1 mM).
Preparation of cell extracts.
For preparation of cell extracts, culture samples were harvested by centrifugation, washed twice with 10 mM potassium phosphate buffer (pH 7.5) containing 2 mM EDTA, concentrated fourfold, and stored at −20°C. Before being assayed, the samples were thawed, washed, and resuspended in 100 mM potassium phosphate buffer (pH 7.5) containing 2 mM MgCl2 and 1 mM dithiothreitol (sonication buffer). Extracts were prepared by sonication with 0.7-mm-diameter glass beads at 0°C in an MSE sonicator (150-W output, 7-μm peak-to-peak amplitude) for 3 min at 0.5-min intervals. Unbroken cells and debris were removed by centrifugation at 4°C (20 min at 36,000 × g). The clear supernatant was used as the cell extract.
Enzyme analysis.
Enzyme assays were performed by use of a Hitachi model 100-60 spectrophotometer at 30°C and 340 nm (E340 of reduced pyridine dinucleotide cofactors, 6.3 mM−1) with freshly prepared extracts. All enzyme activities are expressed as micromoles of substrate converted · minute−1 · milligram of protein−1. When necessary, extracts were diluted in sonication buffer. All assays were performed in duplicate with two concentrations of cell extract. Specific activities in these duplicate experiments differed by <10%.
Hexokinase (EC 2.7.1.1) was assayed according to Postma et al. (32). Phosphoglucose isomerase (EC 5.3.1.9) was assayed according to Bergmeyer (7) with minor modifications. The assay mixture contained the following: Tris-HCl buffer (pH 8.0), 50 mM; MgCl2, 5 mM; NADP+, 0.4 mM; glucose-6-phosphate dehydrogenase (Boehringer), 1.8 U · ml−1; and cell extract. The reaction was started with 2 mM fructose-6-phosphate. Phosphofructokinase (EC 2.7.1.11) was assayed according to de Jong-Gubbels et al. (14) with minor modifications. The assay mixture contained the following: imidazole-HCl (pH 7.0), 50 mM; MgCl2, 5 mM; NADH, 0.15 mM; fructose-2,6-diphosphate, 0.10 mM; fructose-1,6-diphosphate aldolase (Boehringer), 0.5 U · ml−1; glycerol-3-phosphate dehydrogenase (Boehringer), 0.6 U · ml−1; triosephosphate isomerase, 1.8 U · ml−1 (Boehringer); and cell extract. The endogenous activity was measured after the addition of 0.25 mM fructose-6-phosphate. The reaction was started with 0.5 mM ATP. Fructose-1,6-diphosphate aldolase (EC 4.1.2.13) was assayed according to van Dijken et al. (44). Triosephosphate isomerase (EC 5.3.1.1) was assayed according to Bergmeyer (7) with minor modifications. The assay mixture contained the following: triethanolamine-HCl buffer (pH 7.6), 100 mM; NADH, 0.15 mM; glycerol-3-phosphate dehydrogenase, 8.5 U · ml−1 (Boehringer); and cell extract. The reaction was started with 6 mM glyceraldehyde-3-phosphate. Glyceraldehyde-3-phosphate dehydrogenase (EC 1.2.1.12) was assayed according to Bergmeyer (7) with minor modifications. The assay mixture contained the following: triethanolamine-HCl buffer (pH 7.6), 100 mM; ATP, 1 mM; EDTA, 1 mM; MgSO4, 1.5 mM; NADH, 0.15 mM; phosphoglycerate kinase, 22.5 U · ml−1 (Boehringer); and cell extract. The reaction was started with 5 mM 3-phosphoglycerate. The assay of phosphoglycerate kinase (EC 2.7.2.3) was identical to that of glyceraldehyde-3-phosphate dehydrogenase except that phosphoglycerate kinase was replaced by glyceraldehyde-3-phosphate dehydrogenase at 8.0 U · ml−1 (Boehringer). Phosphoglycerate mutase (EC 2.7.5.3) was assayed according to Bergmeyer (7). Enolase (EC 4.2.1.11) was assayed according to Bergmeyer (7) with minor modifications. The assay mixture contained the following: triethanolamine-HCl buffer (pH 8.0), 100 mM; MgSO4, 1.5 mM; NADH, 0.15 mM; ADP, 10 mM; pyruvate kinase, 26.3 U · ml−1 (Sigma); lactate dehydrogenase, 11.3 U · ml−1 (Boehringer); and cell extract. The reaction was started with 1 mM 2-phosphoglycerate. Pyruvate kinase (EC 2.7.1.40) was assayed according to de Jong-Gubbels et al. (14) with minor modifications. The assay mixture contained the following: cacodylic acid-KOH (pH 6.2), 100 mM; KCl, 100 mM; ADP, 10 mM; fructose-1,6-diphosphate, 1 mM; MgCl2, 25 mM; NADH, 0.15 mM; lactate dehydrogenase (Boehringer), 11.25 U · ml−1; and cell extract. The reaction was started with 2 mM phosphoenolpyruvate. Pdc (EC 4.1.1.1) and alcohol dehydrogenase (EC 1.1.1.1) were assayed according to Postma et al. (33). Pdc was also assayed without the addition of thiamine pyrophosphate as a control for the overproduction of Pdc in the presence of low concentrations of this cofactor extant in situ (8).
Protein determinations.
The protein content of whole cells was estimated by a modified biuret method (51). Protein concentrations in cell extracts were determined by the Lowry method. Dried bovine serum albumin (fatty acid free; Sigma) was used as a standard.
RESULTS
Growth in shake-flask cultures.
For an initial characterization, strains CEN.PK113-7D (prototrophic wild type), GG392 (CEN.PK113-5D host transformed with an empty vector), and GG393 (CEN.PK113-5D host with a multicopy PDC1 vector) were grown on glucose in shake-flask cultures. In the two plasmid-containing strains, the uracil auxotrophy of the CEN.PK113-5D host was complemented by the URA3 gene on the YEplac195 vector. Therefore, a defined medium lacking uracil could be used for all three strains.
In cell extracts prepared from exponentially growing cultures, the Pdc activities in the prototrophic wild-type strain and the empty-vector reference strain were virtually the same (Table 1). The Pdc activity in cell extracts of strain GG393 was 3.4-fold higher than that in the reference strains. This result confirms an earlier report (38) that the presence of multiple copies of the PDC1 gene leads to a substantial increase in Pdc activity in batch cultures on glucose.
TABLE 1.
Effect of Pdc overexpression on the maximum specific growth ratea
| Strain | μmax (h−1) | Pdc activity (U · mg of protein−1) |
|---|---|---|
| CEN.PK113-7D (wild type) | 0.44 ± 0.01 | 1.9 ± 0.05 |
| GG392 (empty vector) | 0.43 ± 0.01 | 1.8 ± 0.04 |
| GG393 (PDC1 construct) | 0.39 ± 0.01 | 6.2 ± 0.2 |
Determined for exponentially growing shake-flask cultures on mineral medium containing 2% (wt/vol) glucose. Dat are presented as the average ± standard deviation for two independent experiments.
The presence of the YEplac195 empty vector had no significant effect on the specific growth rate on glucose. However, the specific growth rate of the Pdc-overproducing strain was ca. 10% lower than that of the two reference strains (Table 1). A similar small negative effect of PDC1 overexpression on specific growth rate was found previously (38).
Overexpression of PDC1 in glucose-limited chemostat cultures (D, 0.10 h−1).
For aerobic glucose-limited chemostat cultures of strain GG393 (D, 0.10 h−1), the presence of multiple copies of the PDC1 gene resulted in 14-fold-higher Pdc activity in cell extracts relative to that for the two isogenic reference strains (Table 2). Since episomal vectors carrying auxotrophic markers may be unstable in chemostat cultures (12, 28), Pdc activity in cell extracts of chemostat cultures was monitored for over 100 generations at a D of 0.10 h−1. No significant loss of Pdc activity was observed in either the reference strains or the PDC1-overexpressing strain (data not shown).
TABLE 2.
Specific activities of glycolytic enzymes in cell extracts of aerobic glucose-limited chemostat cultures (D, 0.10 h−1) of various S. cerevisiae strainsa
| Enzyme | Sp act (U · mg of protein−1) in:
|
||
|---|---|---|---|
| CEN.PK113-7D (wild type) | GG293 (empty vector) | GG393 (Pdc overproducer) | |
| Hexokinase (EC 2.7.1.1) | 1.7 ± 0.1 | 1.8 ± 0.1 | 1.6 ± 0.2 |
| Phosphoglucose isomerase (EC 5.3.1.9) | 2.8 ± 0.2 | 2.9 ± 0.2 | 2.4 ± 0.1 |
| Phosphofructokinase (EC 2.7.1.11) | 0.3 ± 0.04 | 0.4 ± 0.04 | 0.3 ± 0.04 |
| Fructose-1,6-diphosphate aldolase (EC 4.1.2.13) | 1.0 ± 0.02 | 1.0 ± 0.1 | 0.8 ± 0.03 |
| Triosephosphate isomerase (EC 5.3.1.1) | 52.5 ± 4.1 | 57.7 ± 1.4 | 45.3 ± 2.3 |
| Glyceraldehyde-3-phosphate dehydrogenase (EC 1.2.1.12) | 5.6 ± 0.3 | 5.3 ± 0.2 | 4.4 ± 0.1 |
| Phosphoglycerate kinase (EC 2.7.2.3) | 7.4 ± 0.1 | 6.5 ± 0.2 | 6.7 ± 0.4 |
| Phosphoglycerate mutase (EC 2.7.5.3) | 6.8 ± 0.4 | 6.5 ± 0.3 | 5.9 ± 0.4 |
| Enolase (EC 4.2.1.11) | 0.7 ± 0.02 | 0.7 ± 0.01 | 0.6 ± 0.03 |
| Pyruvate kinase (EC 2.7.1.40) | 2.9 ± 0.1 | 2.6 ± 0.1 | 2.4 ± 0.2 |
| Pdc (EC 4.1.1.1) | 0.6 ± 0.1 | 0.6 ± 0.03 | 8.4 ± 0.5 |
| Alcohol dehydrogenase (EC 1.1.1.1) | 9.1 ± 0.4 | 9.6 ± 0.5 | 6.1 ± 0.1 |
Data are presented as the average ± standard deviation for duplicate enzyme assays on two independent cultures. Glycolytic flux for hexose and triose intermediates was estimated from the specific ethanol production rate in an off-line fermentation assay. Calculations of the flux were based on a soluble protein content of 33% (33). For strains CEN.PK113-7D, GG293, and GG393, glycolytic flux values for hexose and triose intermediates were 0.37, 0.38, and 0.39 U · mg of protein−1 and 0.75, 0.76, and 0.78 U · mg of protein−1, respectively.
To check whether PDC1 overexpression affected levels of other key enzymes of glucose dissimilation, the 12 enzyme activities of glycolysis and alcoholic fermentation were assayed in cell extracts of chemostat-grown cells. The presence of the YEPlac195 vector in strain GG392 did not cause significant changes in enzyme activities compared to those in the prototrophic wild-type strain CEN.PK113-7D (Table 2). However, the activities of the glycolytic enzymes upstream of Pdc were 10 to 15% lower in the PDC1-overexpressing strain GG393 than in the two reference strains (Table 2). The only enzyme tested that exhibited a larger difference in activity was NAD-dependent alcohol dehydrogenase, an enzyme downstream of Pdc. Its activity was ca. 35% lower in the PDC1-overexpressing strain than in the reference strains (Table 2).
Physiology of wild-type and PDC1-overexpressing strains in chemostat cultures.
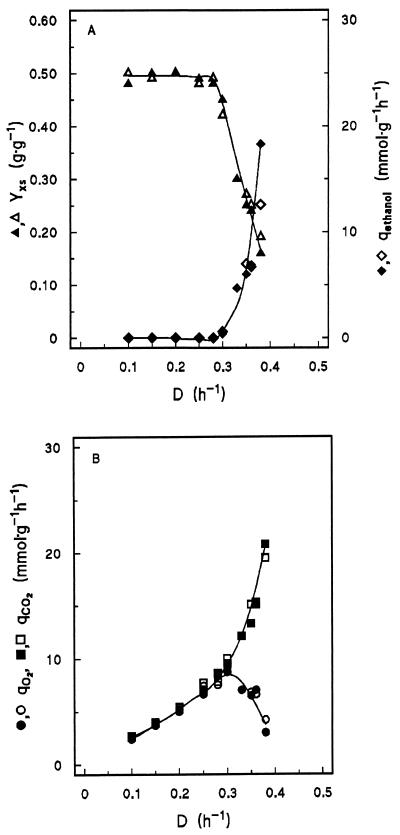
In aerobic glucose-limited chemostat cultures of the reference strains CEN.PK113-7D and GG392 grown at a D of <0.30 h−1, over 98% of the substrate carbon was recovered as biomass and CO2. The biomass yield of 0.49 g · g of glucose−1 (Fig. 2A) was similar to that of respiratory cultures of other wild-type S. cerevisiae strains (48). Completely respiratory metabolism was further evident from the absence of ethanol and other typical fermentation products in culture supernatants. In these respiratory cultures, the specific rates of O2 consumption and CO2 production (Fig. 2B) increased linearly with D.
FIG. 2.
Effect of D on biomass yield (Yxs; grams of dry yeast biomass · gram of glucose−1) and the specific rate of ethanol production (qethanol) (A) and on the specific rates of oxygen consumption (qO2) and carbon dioxide production (qCO2) (B) in aerobic glucose-limited chemostat cultures of prototrophic wild-type S. cerevisiae CEN.PK113-7D (closed symbols) and the empty-vector control strain (GG392; open symbols). Fluxes are expressed in millimoles · g of dry yeast biomass−1 · h−1.
At a D of 0.30 h−1, the specific oxygen consumption rate for the reference strains reached a maximum of 8.8 mmol · g of dry yeast biomass−1 · h−1. At higher Ds, glucose metabolism became respirofermentative: the specific rate of CO2 production increased sharply, while the specific rate of O2 consumption decreased (Fig. 2B). In addition to ethanol, respirofermentative cultures produced acetate and pyruvate, albeit at low rates (<0.5 mmol · g−1 · h−1). A low rate of glycerol production (<0.7 mmol · g−1 · h−1) was only detected at a D of 0.38 h−1. Aerobic fermentation was accompanied by a decreased biomass yield on glucose (Fig. 2A).
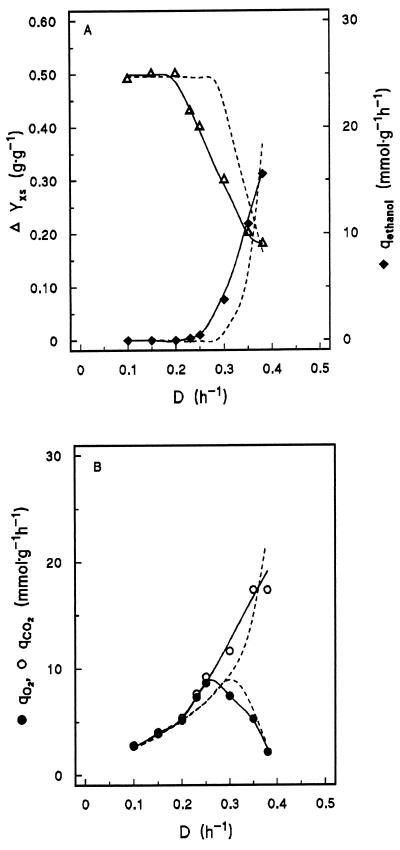
At low Ds, the Pdc-overproducing strain exhibited respiratory growth, with virtually the same biomass yield on glucose (0.50 g · g−1) as the reference strains (Fig. 3A). At Ds of up to 0.20 h−1, the profiles of the specific rate of O2 consumption and the specific rate of CO2 production were not significantly different from those of the reference strains (Fig. 3B). In these cultures, over 98% of the glucose carbon was recovered as biomass and carbon dioxide. However, the D at which aerobic alcoholic fermentation set in (0.23 h−1) was much lower in the PDC1-overexpressing strain than in the two reference strains. In addition, the specific rates of O2 consumption and CO2 production both increased steeply at a D of ca. 0.20 h−1 (Fig. 3B). The specific rate of O2 consumption reached a maximum of 8.6 mmol · g of dry yeast biomass−1 at a D of 0.25 h−1. This maximum specific rate of O2 consumption was the same as that of the reference strains. However, in contrast to the reference strains, the Pdc-overproducing strain reached its maximum specific rate of O2 consumption in a culture that already produced ethanol (Fig. 3). The pattern of formation of other metabolites was comparable to that in the two reference strains (data not shown).
FIG. 3.
Effect of D on biomass yield (Yxs; grams of dry yeast biomass · gram of glucose−1) and the specific rate of ethanol production (qethanol) (A) and on the specific rates of oxygen consumption (qO2) and carbon dioxide production (qCO2) (B) in aerobic glucose-limited chemostat cultures of the Pdc-overproducing strain S. cerevisiae GG393. Fluxes are expressed in millimoles · gram of dry yeast biomass−1 · h−1. The broken lines represent data for the wild-type and control strains as depicted in Fig. 2.
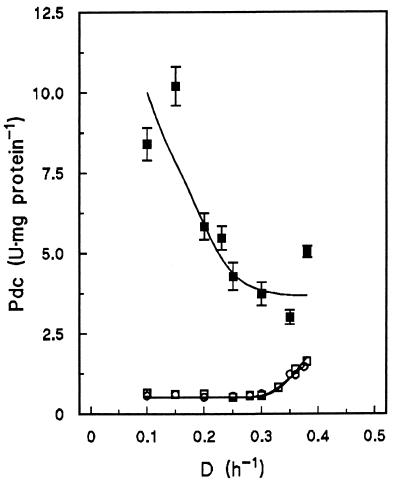
The Pdc level in cell extracts of the reference strains was ca. 0.6 U · mg of protein−1 during respiratory growth and increased during respirofermentative growth to a value of 1.6 U · mg of protein−1 at a D of 0.38 h−1 (Fig. 4). In contrast, the Pdc level in cell extracts of the Pdc-overproducing strain decreased with increasing D, down to 5.0 U · mg of protein−1 at a D of 0.38 h−1. Nevertheless, at all Ds studied, the Pdc activity in cell extracts of the Pdc-overproducing strain was at least 3.5-fold higher than that in cell extracts of the reference strains (Fig. 4).
FIG. 4.
Pdc activities in cell extracts of aerobic glucose-limited chemostat cultures of the prototrophic wild-type strain (S. cerevisiae CEN.PK113-7D; □), empty-vector reference strain (GG392; ○) and Pdc-overproducing strain (GG393; ▪) as a function of D. Values are presented as the averages ± standard deviations for two independent enzyme assays.
In the two reference strains, the protein content of the biomass increased with increasing D in respiratory cultures, from ca. 42% at a D of 0.10 h−1 to ca. 49% at a D of 0.30 h−1. At higher Ds, the protein content decreased again. In the Pdc-overproducing strain, the protein content was ca. 47% in respiratory cultures and, as in the reference strains, decreased at higher Ds.
Effect of Pdc overproduction on maximum glycolytic capacity.
To investigate whether Pdc overproduction affected the maximum glycolytic (fermentative) capacity of chemostat-grown cells, culture samples were washed, made anaerobic, and exposed to excess glucose. During the first 30 min after glucose addition, the increase in biomass concentration was negligible and the increase in ethanol concentration was linear with time and proportional to the amount of biomass present (data not shown).
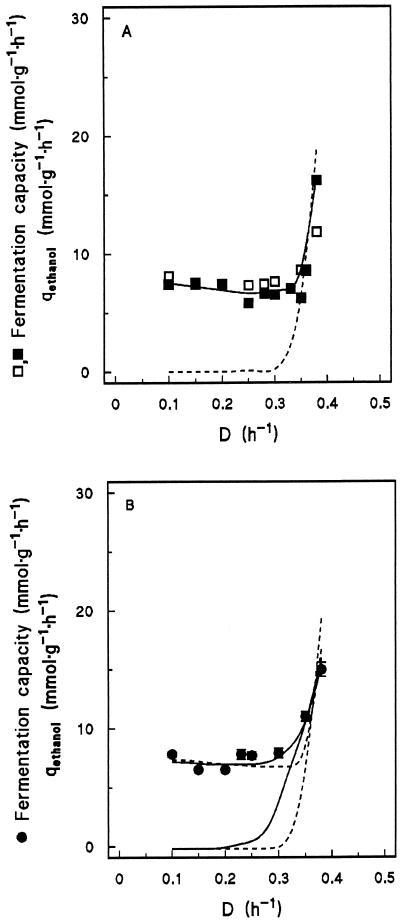
Cells from respiratory cultures already exhibited substantial fermentative capacity (ca. 7.5 mmol of ethanol · g of dry yeast biomass−1 · h−1) when incubated under anaerobic conditions in the presence of excess glucose (Fig. 5A). This fermentative capacity, which was approximately constant in respiratory cultures, increased only in cells pregrown at high Ds at which high in situ rates of alcoholic fermentation occurred in aerobic chemostat cultures. At the highest specific D studied (0.38 h−1), the fermentative capacity was 16.2 mmol · g of dry yeast biomass−1 · h−1 (Fig. 5A).
FIG. 5.
Effect of D on the maximum specific rate of ethanol production (determined off line under anaerobic conditions with excess glucose) in aerobic glucose-limited chemostat cultures of S. cerevisiae wild-type and empty-vector control strains (A) and Pdc-overproducing strain (B) (compared to the data presented in panel A). The broken line in panel A represents the specific rate of ethanol production (qethanol) during glucose-limited growth of the reference strains as presented in Fig. 2A. The solid line in panel B represents the qethanol of the Pdc-overproducing strain as plotted in Fig. 3A. Fluxes are expressed in millimoles · g of dry yeast biomass−1 · h−1. Values for the Pdc-overproducing strain are presented as the average ± standard deviation for two independent off-line fermentation assays.
The fermentative capacity of respiratory cultures of the Pdc-overproducing strain was not significantly different from that of respiratory cultures of the two reference strains (Fig. 5B). As observed with the reference cultures, the fermentative capacity of the Pdc-overproducing culture increased only at high Ds at which alcoholic fermentation occurred in chemostat cultures (Fig. 5B).
DISCUSSION
Pdc overproduction.
Our data confirm the conclusion of Schaaff et al. (38) that Pdc by itself does not control the specific rate of growth of S. cerevisiae on glucose. Instead, a 3.4-fold overproduction of Pdc in batch cultures resulted in a ca. 10% decrease in the specific growth rate (Table 1). In the interpretation of this effect, it should be considered that in wild-type S. cerevisiae, Pdc is already an abundant protein. Based on a specific activity of purified Pdc of 54 U · mg−1 (4), the Pdc activity in cell extracts of batch cultures of the two reference strains (1.85 U · mg of protein−1) would correspond to 3.4% of the soluble cell protein. In the overproducing strain, this value would increase to an estimated 11.5% of the soluble protein (Table 1).
Negative effects of protein overproduction on specific growth rate (protein burden effects) have been reported for many homologous and heterologous proteins (3, 24, 41). Adverse effects are not always due to specific catalytic or regulatory properties of an overproduced protein. Even overproduction of a metabolically inert protein will reduce the specific growth rate when it represents a substantial fraction of the cell protein and thus dilutes enzyme activities that (together) control specific growth rate (41). The abundance of Pdc in S. cerevisiae can adequately explain the small reduction in the specific growth rate of the overproducing strain.
The 14-fold overproduction of Pdc in glucose-limited chemostat cultures (D, 0.10 h−1; Table 2) resulted in the highest Pdc activities in S. cerevisiae reported to date. In similar cultures of the two reference strains, the Pdc activity of 0.6 U · mg of protein−1 (Table 2) corresponded to 1.1% of the soluble cell protein (calculated as described above). In the overproducing strain, this value increased to ca. 16%, leading to a dilution of other enzymes by ca. 15%. This result may explain the small (ca. 10 to 15%) decrease in most other glycolytic enzyme levels in cell extracts (Table 2). The 35% decrease in alcohol dehydrogenase activity cannot be explained by dilution alone. This decrease possibly represented an effect of changed metabolite levels (e.g., acetaldehyde) on alcohol dehydrogenase biosynthesis.
The very high Pdc levels achieved by the introduction of multiple copies of the PDC1 gene in combination with aerobic sugar-limited cultivation may be of applied significance, in particular for whole-cell bioconversions in which Pdc is the key catalyst (e.g., production of phenylacetyl carbinol [27]).
The extent of Pdc overproduction decreased with increasing D. A plausible explanation for this result might be a decreasing copy number of the episomal vector with increasing specific growth rate. In addition, a decreasing residence time (resulting in a shorter duration of Pdc accumulation in the cells) and/or unknown regulatory phenomena associated with the presence of multiple copies of the PDC1 promoter may have contributed to this effect. Although Pdc overproduction had significant effects on maximum specific growth rate and levels of other key enzymes, the magnitude of these effects was small. It is therefore unlikely that effects on critical D, which are discussed below, were due to aspecific protein burden effects.
Flux distribution at the pyruvate branch point.
In aerobic glucose-limited chemostat cultures, Pdc overproduction caused a substantial decrease in the D at which aerobic fermentation set in (the so-called critical D; Fig. 4). The most straightforward explanation for this observation is that the increased level of Pdc allows the enzyme to compete more effectively with the mitochondrial pyruvate dehydrogenase complex. As mentioned above, decarboxylation by Pdc does not necessarily commit pyruvate to alcoholic fermentation, as acetaldehyde can also be channelled into respiratory dissimilation via the pyruvate dehydrogenase bypass (Fig. 1) (19, 35). This pathway plays an essential role in S. cerevisiae, probably by providing cytosolic acetyl-CoA for biosynthesis (15, 35, 43). In wild-type, glucose-limited chemostat cultures, respiratory dissimilation of pyruvate via acetyl-CoA predominantly occurs via the pyruvate dehydrogenase complex (34). However, in pyruvate dehydrogenase-negative mutants, respiratory pyruvate dissimilation can be completely redirected via the bypass at low specific growth rates. This process results in a 15% decrease in the biomass yield on glucose due to consumption of ATP in the acetyl-CoA synthetase reaction (34).
In chemostat cultures, the biomass yield of the Pdc-overproducing strain had already started to decrease at a D of ca. 0.20 h−1, at which ethanol formation was still absent (Fig. 4). At a D of 0.23 h−1, at which ethanol formation started, the biomass yield was 14% lower than in the respiratory cultures grown at lower Ds. In this range of Ds, the specific rate of oxygen consumption also increased sharply, providing further indication for decreased growth efficiency. This result suggests that, before alcoholic fermentation sets in, respiratory pyruvate metabolism is redirected via the respiratory pyruvate dehydrogenase bypass. Then, as the bypass enzymes become saturated, acetaldehyde is reduced to ethanol. Alternatively, the switch from respiratory acetaldehyde metabolism to alcoholic fermentation may be due to a limited capacity for respiratory reoxidation of NAD(P)H. However, in the Pdc-overproducing strain, the specific rate of oxygen consumption reached its maximum at a D at which alcoholic fermentation had already set in. This phenomenon, which is not observed in wild-type strains (5, 31, 33, 53), argues against a major role of NAD(P)H reoxidation in the switch to respirofermentative metabolism in the Pdc-overproducing strain.
Our results demonstrate that flux distribution at the pyruvate branch point can be manipulated by overproduction of Pdc. This finding does not necessarily imply that competition of pyruvate-metabolizing enzymes is responsible for aerobic alcoholic fermentation in wild-type cells. In addition to pyruvate, mitochondrial respiration competes with alcoholic fermentation for the NADH produced in glycolysis. To further investigate the relevance of competition between respiration and fermentation for pyruvate, it would be of interest to study strains with reduced Pdc levels. We have tried to do this by growing S. cerevisiae strains expressing only the PDC5 structural gene. Unfortunately, although these strains exhibited much reduced Pdc levels at low Ds, PDC5 was induced at higher specific growth rates, thus obscuring any effect on critical D (14a).
Maximum glycolytic (fermentative) capacity.
Various authors have investigated the relationship between Pdc activity and glycolytic flux. In batch cultures of S. cerevisiae mutants with different levels of PDC1 expression, fermentation rates exhibited a linear correlation with a broad range of Pdc activities in cell extracts (39). A similar correlation was found in a comparison of fermentation rates and Pdc activities in different yeast species (47).
In their study on the overproduction of glycolytic enzymes in S. cerevisiae, Schaaff et al. (38) demonstrated that a fourfold overproduction of Pdc did not enhance alcoholic fermentation rates in glucose-grown batch cultures. However, results from batch cultures cannot necessarily be extrapolated to other growth conditions. Of particular interest is the effect of Pdc overproduction on the fermentative capacity of cells grown under conditions resembling the industrial aerobic and sugar-limited production of baker’s yeast, for which fermentative capacity is one of the major quality parameters (9, 35).
Overproduction of Pdc had no effect on the fermentative capacity of respiratory cultures grown at low Ds (Fig. 5). This result indicates that, as previously found for batch cultures, Pdc is not the major rate-controlling enzyme determining fermentative capacity in glucose-limited cultures. It is now generally accepted that fluxes through metabolic pathways can hardly ever be described in terms of a single enzyme that limits the overall rate (22, 29). Therefore, our results do not rule out the possibility that overproduction of Pdc (in combination with other enzymes) may be required for improving the fermentative capacity of baker’s yeast. In that case, a trade-off situation will occur with respect to, on the one hand, biomass productivity in the aerobic production process (which is negatively affected by Pdc overproduction due to the reduced critical specific growth rate at which aerobic fermentation sets in) and, on the other hand, fermentative capacity in the dough application.
ACKNOWLEDGMENTS
We thank our colleagues at Delft University of Technology, Leiden University, and Gist-Brocades B.V. for stimulating discussions and Saskia Cooman for technical support.
This project was financially supported by Gist-Brocades B.V., Delft, The Netherlands; the Dutch Ministry of Economic Affairs (via the ABON program Metabolic Fluxes in Yeasts and Fungi); and the European Community (DG XII Framework IV Program on Cell Factories, project From Gene to Product in Yeast, a Quantitative Approach).
REFERENCES
- 1.Alexander M A, Jeffries T W. Respiratory efficiency and metabolite partitioning as regulatory phenomena in yeasts. Enzyme Microb Technol. 1990;12:2–19. [Google Scholar]
- 2.Auberson L C M, von Stockar U. A unified stoichiometric model for oxidative and oxidoreductive growth of yeasts. Biotechnol Bioeng. 1992;40:1243–1255. doi: 10.1002/bit.260401014. [DOI] [PubMed] [Google Scholar]
- 3.Bailey J E. Host-vector interactions in Escherichia coli. Adv Biochem Eng Biotechnol. 1993;48:29–52. doi: 10.1007/BFb0007195. [DOI] [PubMed] [Google Scholar]
- 4.Barburina I, Gao Y, Hu Z, Jordan F, Hohmann S, Furey W. Substrate activation of brewers’ yeast pyruvate decarboxylase is abolished by mutation of cysteine 221 to serine. Biochemistry. 1994;33:5630–5635. doi: 10.1021/bi00184a035. [DOI] [PubMed] [Google Scholar]
- 5.Barford J P, Hall R J. An examination of the Crabtree effect in Saccharomyces cerevisiae: the role of respiratory adaptation. J Gen Microbiol. 1979;114:267–275. [Google Scholar]
- 6.Becker D M, Guarente L. High-efficiency transformation of yeast by electroporation. Methods Enzymol. 1991;194:182–187. doi: 10.1016/0076-6879(91)94015-5. [DOI] [PubMed] [Google Scholar]
- 7.Bergmeyer H U. Methods of enzymatic analysis. 2nd ed. Vol. 1. New York, N.Y: Academic Press, Inc.; 1974. [Google Scholar]
- 8.Bergmeyer H U. Methods of enzymatic analysis. 3th ed. Vol. 7. Weinheim, Germany: VCH Verlagsgesellschaft; 1985. pp. 457–467. [Google Scholar]
- 9.Beudeker R F, van Dam H W, van der Plaat J B, Vellenga K. Developments in baker’s yeast production. In: Verachtert H, De Mot R, editors. Yeast biotechnology and biocatalysis. New York, N.Y: Marcel Dekker, Inc.; 1990. pp. 103–146. [Google Scholar]
- 10.Boiteux A, Hess B. Allosteric properties of yeast pyruvate decarboxylase. FEBS Lett. 1970;9:293–296. doi: 10.1016/0014-5793(70)80381-7. [DOI] [PubMed] [Google Scholar]
- 11.Boles E, Heinisch J, Zimmermann F K. Different signals control the activation of glycolysis in the yeast Saccharomyces cerevisiae. Yeast. 1993;9:761–770. doi: 10.1002/yea.320090710. [DOI] [PubMed] [Google Scholar]
- 12.Bugeja V C, Kleinman M J, Stanbury P F, Gingold E B. The segregation of the 2mu-based yeast plasmid pJDB248 breaks down under conditions of slow, glucose-limited growth. J Gen Microbiol. 1989;135:2891–2897. doi: 10.1099/00221287-135-11-2891. [DOI] [PubMed] [Google Scholar]
- 13.Bullock W O, Fernandez J M, Short J M. XL1Blue: a high efficiency plasmid transforming recA Escherichia coli strain with β-galactosidase selection. BioTechniques. 1987;5:376–379. [Google Scholar]
- 14.de Jong-Gubbels P, Vanrolleghem P, Heijnen J J, van Dijken J P, Pronk J T. Regulation of carbon metabolism in chemostat cultures of Saccharomyces cerevisiae grown on mixtures of glucose and ethanol. Yeast. 1995;11:407–418. doi: 10.1002/yea.320110503. [DOI] [PubMed] [Google Scholar]
- 14a.Flikweert, M. T. Unpublished data.
- 15.Flikweert M T, van der Zanden L, Janssen W M T M, Steensma H Y, van Dijken J P, Pronk J T. Pyruvate decarboxylase: an indispensable enzyme for growth of Saccharomyces cerevisiae on glucose. Yeast. 1996;12:247–257. doi: 10.1002/(SICI)1097-0061(19960315)12:3%3C247::AID-YEA911%3E3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 16.Gietz R D, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 17.Hohmann S. Characterization of PDC6, a third structural gene for pyruvate decarboxylase in Saccharomyces cerevisiae. J Bacteriol. 1991;173:7963–7969. doi: 10.1128/jb.173.24.7963-7969.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holzer H. Regulation of carbohydrate metabolism by enzyme competition. Cold Spring Harbor Symp Quant Biol. 1961;26:277–288. doi: 10.1101/sqb.1961.026.01.034. [DOI] [PubMed] [Google Scholar]
- 19.Holzer H, Goedde H W. Zwei Wege von Pyruvat zu Acetyl-Coenzym A in Hefe. Biochem Z. 1957;329:175–191. [PubMed] [Google Scholar]
- 20.Hübner G, Weidhase R, Schellenberger A. The mechanism of substrate activation of pyruvate decarboxylase: a first approach. Eur J Biochem. 1978;92:175–181. doi: 10.1111/j.1432-1033.1978.tb12735.x. [DOI] [PubMed] [Google Scholar]
- 21.Käppeli O. Regulation of carbon metabolism in Saccharomyces cerevisiae and related yeasts. Adv Microb Physiol. 1986;28:181–209. doi: 10.1016/s0065-2911(08)60239-8. [DOI] [PubMed] [Google Scholar]
- 22.Kell D B, Westerhoff H V. Metabolic control theory: its role in microbiology and biotechnology. FEMS Microbiol Rev. 1986;39:305–320. [Google Scholar]
- 23.Kellerman E, Seeboth P G, Hollenberg C P. Analysis of the primary structure and promoter function of a pyruvate decarboxylase gene (PDC1) from Saccharomyces cerevisiae. Nucleic Acids Res. 1986;14:8963–8977. doi: 10.1093/nar/14.22.8963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koch A L. The protein burden of lac operon products. J Mol Evol. 1983;19:455–462. doi: 10.1007/BF02102321. [DOI] [PubMed] [Google Scholar]
- 25.Kresze G B, Ronft H. Pyruvate dehydrogenase complex from bakers’ yeast. 1. Purification and some kinetic and regulatory properties. Eur J Biochem. 1981;119:573–579. doi: 10.1111/j.1432-1033.1981.tb05646.x. [DOI] [PubMed] [Google Scholar]
- 26.Lee S, Suraiya R. A simple procedure for maximum yield of high-quality plasmid DNA. BioTechniques. 1990;9:676–679. [PubMed] [Google Scholar]
- 27.Long A, Ward O P. Biotransformation of benzaldehyde by Saccharomyces cerevisiae: characterization of the fermentation and toxicity effects of substrates and products. Biotechnol Bioeng. 1989;34:933–941. doi: 10.1002/bit.260340708. [DOI] [PubMed] [Google Scholar]
- 28.Meinander N Q, Hahn-Hägerdal B. Fed-batch xylitol production with two recombinant Saccharomyces cerevisiae strains expressing XYL1 at different levels, using glucose as a cosubstrate: a comparison of production parameters and strain stability. Biotechnol Bioeng. 1997;54:391–394. doi: 10.1002/(SICI)1097-0290(19970520)54:4<391::AID-BIT12>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 29.Niederberger P, Prasad R, Miozzari G, Kacser H. A strategy for increasing an in vivo flux by genetic manipulations. The tryptophan system of yeast. Biochem J. 1992;287:473–479. doi: 10.1042/bj2870473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Noorman H J, Baksteen J, Heijnen J J, Luyben K C A M. The bioreactor overflow device: an undesired selective separator in continuous cultures? J Gen Microbiol. 1991;137:2171–2177. [Google Scholar]
- 31.Petrik M, Käppeli O, Fiechter A. An expanded concept for glucose effect in the yeast Saccharomyces uvarum: involvement of short and long-term regulation. J Gen Microbiol. 1983;129:43–49. [Google Scholar]
- 32.Postma E, Scheffers W A, van Dijken J P. Adaptation of the kinetics of glucose transport to environmental conditions in the yeast Candida utilis CBS 621: a continuous-culture study. J Gen Microbiol. 1988;134:1109–1116. [Google Scholar]
- 33.Postma E, Verduyn C, Scheffers W A, van Dijken J P. Enzymatic analysis of the Crabtree effect in glucose-limited chemostat cultures of Saccharomyces cerevisiae. Appl Environ Microbiol. 1989;55:468–477. doi: 10.1128/aem.55.2.468-477.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pronk J T, Wenzel T J, Luttik M A H, Klaassen C C M, Scheffers W A, Steensma H Y, van Dijken J P. Energetic aspects of glucose metabolism in a pyruvate-dehydrogenase-negative mutant of Saccharomyces cerevisiae. Microbiology. 1994;140:601–610. doi: 10.1099/00221287-140-3-601. [DOI] [PubMed] [Google Scholar]
- 35.Pronk J T, Steensma H Y, van Dijken J P. Pyruvate metabolism in Saccharomyces cerevisiae. Yeast. 1996;12:1607–1633. doi: 10.1002/(sici)1097-0061(199612)12:16<1607::aid-yea70>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 35a.Pronk, J. T., and H. Y. Steensma. Unpublished data.
- 36.Rieger M, Käppeli O, Fiechter A. The role of limited respiration in the incomplete oxidation of glucose by Saccharomyces cerevisiae. J Gen Microbiol. 1983;129:653–661. [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 38.Schaaff I, Heinisch J, Zimmermann F K. Overproduction of glycolytic enzymes in yeast. Yeast. 1989;5:285–290. doi: 10.1002/yea.320050408. [DOI] [PubMed] [Google Scholar]
- 39.Schmitt H D, Zimmermann F K. Genetic analysis of the pyruvate decarboxylase reaction in yeast glycolysis. J Bacteriol. 1982;151:1146–1152. doi: 10.1128/jb.151.3.1146-1152.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sherman F, Fink G R, Hicks J B. Laboratory course manual for methods in yeast gene genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1986. [Google Scholar]
- 41.Snoep J L, Yomano L P, Westerhoff H V, Ingram L O. Protein burden in Zymomonas mobilis: negative flux and growth control due to overproduction of glycolytic enzymes. Microbiology. 1995;141:2329–2337. [Google Scholar]
- 42.Sonnleitner B, Käppeli O. Growth of Saccharomyces cerevisiae is controlled by its limited respiratory capacity: formulation and verification of a hypothesis. Biotechnol Bioeng. 1986;28:927–937. doi: 10.1002/bit.260280620. [DOI] [PubMed] [Google Scholar]
- 43.van den Berg M A, Steensma H Y. ACS2, a Saccharomyces cerevisiae gene encoding acetyl-coenzyme A synthetase, essential for growth on glucose. Eur J Biochem. 1995;231:704–713. doi: 10.1111/j.1432-1033.1995.tb20751.x. [DOI] [PubMed] [Google Scholar]
- 44.van Dijken J P, Harder W, Beardsmore A J, Quayle J R. Dihydroxyacetone: an intermediate in the assimilation of methanol by yeasts? FEMS Microbiol Lett. 1978;4:97–102. [Google Scholar]
- 45.van Urk H, Mak P R, Scheffers W A, van Dijken J P. Metabolic responses of Saccharomyces cerevisiae CBS 8066 and Candida utilis CBS 621 upon transition from glucose limitation to glucose excess. Yeast. 1988;4:283–291. doi: 10.1002/yea.320040406. [DOI] [PubMed] [Google Scholar]
- 46.van Urk H, Schipper D, Breedveld G J, Mak P R, Scheffers W A, van Dijken J P. Localization and kinetics of pyruvate-metabolizing enzymes in relation to aerobic alcoholic fermentation in Saccharomyces cerevisiae CBS 8066 and Candida utilis CBS 621. Biochim Biophys Acta. 1989;992:78–86. doi: 10.1016/0304-4165(89)90053-6. [DOI] [PubMed] [Google Scholar]
- 47.van Urk H, Voll W S L, Scheffers W A, van Dijken J P. Transient-state analysis of metabolic fluxes in Crabtree-positive and Crabtree-negative yeasts. Appl Environ Microbiol. 1990;56:281–287. doi: 10.1128/aem.56.1.281-287.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Verduyn C. Physiology of yeasts in relation to growth yields. Antonie Leeuwenhoek. 1991;60:325–353. doi: 10.1007/BF00430373. [DOI] [PubMed] [Google Scholar]
- 49.Verduyn C. Energetic aspects of metabolic fluxes in yeasts. Ph.D. thesis. Delft, The Netherlands: Delft University of Technology; 1992. [Google Scholar]
- 50.Verduyn C, van Dijken J P, Scheffers W A. Colorimetric alcohol assays with alcohol oxidase. J Microbiol Methods. 1984;2:15–25. [Google Scholar]
- 51.Verduyn C, Postma E, Scheffers W A, van Dijken J P. Physiology of Saccharomyces cerevisiae in anaerobic glucose-limited chemostat cultures. J Gen Microbiol. 1990;136:395–403. doi: 10.1099/00221287-136-3-395. [DOI] [PubMed] [Google Scholar]
- 52.Verduyn C, Postma E, Scheffers W A, van Dijken J P. Effect of benzoic acid on metabolic fluxes in yeasts: a continuous culture study on the regulation of respiration and alcoholic fermentation. Yeast. 1992;8:501–517. doi: 10.1002/yea.320080703. [DOI] [PubMed] [Google Scholar]
- 53.Von Meyenburg H K. Energetics of the budding cycle of Saccharomyces cerevisiae during glucose-limited aerobic growth. Arch Mikrobiol. 1969;66:289–303. doi: 10.1007/BF00414585. [DOI] [PubMed] [Google Scholar]
- 54.Weusthuis R A, Luttik M A H, Scheffers W A, van Dijken J P, Pronk J T. Is the Kluyver effect in yeasts caused by product inhibition? Microbiology. 1994;140:1723–1729. doi: 10.1099/13500872-140-7-1723. [DOI] [PubMed] [Google Scholar]
- 55.Zonneveld B J M. Cheap and simple yeast media. J Microbiol Methods. 1986;4:287–291. [Google Scholar]