Abstract
Background
In glioblastoma (GBM), the effects of altered glycocalyx are largely unexplored. The terminal moiety of cell coating glycans, sialic acid, is of paramount importance for cell-cell contacts. However, sialic acid turnover in gliomas and its impact on tumor networks remain unknown.
Methods
We streamlined an experimental setup using organotypic human brain slice cultures as a framework for exploring brain glycobiology, including metabolic labeling of sialic acid moieties and quantification of glycocalyx changes. By live, 2-photon and high-resolution microscopy we have examined morphological and functional effects of altered sialic acid metabolism in GBM. By calcium imaging we investigated the effects of the altered glycocalyx on a functional level of GBM networks.
Results
The visualization and quantitative analysis of newly synthesized sialic acids revealed a high rate of de novo sialylation in GBM cells. Sialyltrasferases and sialidases were highly expressed in GBM, indicating that significant turnover of sialic acids is involved in GBM pathology. Inhibition of either sialic acid biosynthesis or desialylation affected the pattern of tumor growth and lead to the alterations in the connectivity of glioblastoma cells network.
Conclusions
Our results indicate that sialic acid is essential for the establishment of GBM tumor and its cellular network. They highlight the importance of sialic acid for glioblastoma pathology and suggest that dynamics of sialylation have the potential to be targeted therapeutically.
Keywords: glioblastoma, sialic acid, sialidases, glioma circuits, cancer neuroscience
Key Points.
GBM tumors and cell lines exhibit highly dynamic sialylation and desialylation.
Network connectivity of GBM depends on the level of sialylation on GBM cells.
Sialylation of GBM cells is modified by chemical inhibitors suggesting drugability.
Importance of the Study.
Cancer cells have been shown to exhibit altered composition of the glycocalyx. They are known to overexpress sialic acid moieties that terminate glycan branches. Recent studies have indicated that aberrant sialylation is vital for tumor cells to escape immune surveillance and retain malignance. However, the role of sialic acid turnover in glioblastoma (GBM) is under-investigated. Here we show, for the first time, that sialylation dynamics determine electric activity and network formation of GBM cells. Our study highlights that sialic acid is essential for electric signaling in GBM. Intra- and transcellular activity of GBM cells is lost upon the inhibition of sialic acid synthesis. Meanwhile, the inhibition of desialylation leads to overexcitability and compromises the hierarchy of electric network signaling of GBM cells. Our results highlight the importance of sialic acid for glioblastoma pathology. In the future, sialylation dynamics have the potential to be targeted therapeutically.
Glioblastoma (GBM)—the most malignant brain tumor—is hallmarked by an aggressive growth with infiltration of tumor cells into surrounding brain causing tremendous therapeutic challenges.1 Recent breakthrough in GBM research revealed exceptional intratumoral and intertumoral heterogeneity on transcriptional and genomic level. Transcriptionally, 4 different cellular states were described recapitulating distinct brain cell types: neural precursor cell (NPC)-like, oligodendrocyte precursor cell (OPC)-like, astrocyte (AC)-like, and mesenchymal (MES)-like, with varying proportions across patients.2 These cellular states revealed a high grade of dynamic adaptation in response to microenvironmental alterations and contribute to the plasticity and malignancy of GBM.2,3 In many cancers, an important molecular contributor to the malignancy of cancer cells is a dense layer of multifunctional glycans on the cellular surface, named glycocalyx.4,5 The surface of tumor cells is abundantly glycosylated and exhibit unique properties of glycan structure, including increased sialylation and fucosylation, truncated O-glycans, and increased branching of N- and O-linked glycans.6–8 Excessive sialylation of cancer cells was demonstrated to be a driver of malignant behavior, enhancing its invasiveness and metastatic potential. Additionally, sialic acid is of particular importance for the interaction between cancer cells and immune system. Highly sialylated cancer cells are recognized as “self” and therefore escape immune surveillance and evade immune response, promoting tumor cell survival and retained malignancy.4,5 Recent studies showed that human glioma cells have a high level of sialylation.9 Increased sialylation was associated with invasiveness and dissemination and correlates with poor patient prognosis.10 In particular, GBM cells exhibit high levels of polysialylated neural adhesion molecule that appears to regulate the expression of oligodendrocyte lineage transcription factor 2 (Olig2),10 which is essential for the survival of glioblastoma stem cells and promotes GBM tumor formation, invasiveness and tumor relapse.11 Recent reports confirmed that OPC- and NPC-like cells are the driver of the invasive growth pattern of GBM.12
On the cell surface, the composition of sialoglycans is determined by a dynamic balance between sialylation by sialyltransferases and desialylation by sialidases. Both enzymes are known to exhibit aberrant expression in tumor cells.13 In humans, the biosynthesis of sialylated glycoproteins and glycolipids is mediated by 20 sialyltransferases,14 of which ST3Gal1 and ST3Gal3 have been demonstrated to be upregulated in GBM tumors and cell lines.15,16 The overexpression of these sialyltransferases, enhances the sialylation of GBM cells and is associated with poor prognosis in glioma patients.16 In particular, high expression of ST3Gal1 promotes self-renewal of GBM cells,15,16 while increased expression of ST3Gal3 increases GBM invasiveness.17
In contrast, sialylation is reduced by terminal sialic acid cleaving sialidases (also known as neuraminidases), 3 of which are found in the brain: NEU1, NEU3, and NEU4.18 The dysregulation of sialidases promotes the progression of both solid tumors and blood cancers by enhancing their malignant phenotype, leading to uncontrolled growth, increased invasiveness and metastasis formation.19 However, the role of sialidases in GBM is poorly described. A single study indicated that human glioblastoma tumors have reduced expression of NEU3.20In vitro, restored expression of NEU3 reduces GBM-cell migration and invasiveness, suggesting that sialidase activity may counteract tumorigenesis.20 Therefore, reduced sialidase activity in GBM may contribute to glioma tumor formation and growth.
Importantly, the tumorigenesis of glioma also depends on the interaction of GBM cells with each other and their crosstalk with non-glioma brain cells.21–23 GBM cells develop functional multicellular network structures that transmit long-range signals by intercellular calcium waves,24,25 which promote tumor growth and contribute to the resistance to the treatment.21 Functional connectivity within GBM tumor also promotes glioma cell proliferation that maintains tumorigenesis.22 Different types of cell-to-cell connections in GBM have been described, including gap junctions and tumor microtubes as well as heterotypic synapses between GBM cells and neurons.22,23 In neuronal networks, it has been shown that sialic acid is highly important for cell excitability and functional connectivity.26–28 Removal of sialic acid downregulates neuronal activity,26 suggesting that sialylation may regulate multicellular network properties. However, whether sialic acid defines the excitability and network connectivity of GBM cells remains to be investigated.
We hypothesized that sialic acid is an important player in GBM intra- and inter-tumor network formation, contributing to GBM pathology. Therefore, the aim of this study was to investigate the role of sialic acid turnover in GBM tumor growth and network connectivity.
Methods
Ethics
Detailed information’s of the permissions is provided in the Supplementary Methods.
Tissue Samples
Human neocortical access tissues were sampled from access cortex tissue from epilepsy/GBM surgeries obtained during resection. Detailed information regarding the donors is provided in Supplementary Table S1 (details Supplementary Methods).
Cell Culture
Patient-derived proneural glioblastoma stem cell line and mesenchymal glioblastoma cell line (#BTSC233), expressing ZsGreen cytosolic fluorescent protein, were previously established by Ravi et al.29 For Ca2+ imaging, cells were transduced with a LV-CAG-GCaMP6f lentivirus, following manufacturers recommendations. Further details are described in the Supplementary Method part.
Organotypic Brain Slice Preparation and Glioma Invasion
Organotypic slice cultures were prepared from cortical access tissue as previously published by Ravi et al.29 A detailed description is in the Supplementary Methods.
Metabolic Sialic Acid Labeling and Inhibitor Treatment
#BTSC168 or #BTSC233 cells were labeled by incubating them with Ac4ManNAz for 24 h, washed with growth medium and then incubated with DBCO-Cy5.5. A detailed description is in the Supplementary Methods.
Immunofluorescent Labeling
Fixed cells were permeabilized in Triton-X in PBS for 15 min, blocked in blocking buffer for 30 min, incubated with primary antibodies in blocking buffer overnight at 4 °C, visualized by secondary antibodies in blocking buffer for 2 h at room temperature, stained with DAPI, washed and mounted (details: Supplementary Methods).
Fluorescence Imaging and Quantification
De novo sialic acid labeling on GBM cells was imaged on TCS SP8 resonant scanner confocal microscope (Leica Microsystems). Images in 2D were analyzed on CellProfiler30, 31 software by measuring the signal of interest after identifying the mask of cell bodies. Further detailed description of the imaging analysis is described in the Supplementary Method part.
Microelectrode Array Recordings
Extracellular recordings were performed using the MEA 1060 UP (Multi-Channel Systems) device. Cells were directly cultured on the recording electrode. The cells were perfused with carbogen saturated recording medium under a constant flow rate of 3 ml/min, and activity was recorded for a total of 60 min.32 Further details on the analysis and methods are given in the Supplementary Methods.
Electron Microscopy
Glioblastoma cells (BTSC#168) cultured on coverslips were immersion fixed in 4% paraformaldehyde. After fixation, cultures were washed and incubated in 1% osmium tetroxide (Electon Microscopy Sciences). After embedding for electron microscopy (EM) (detailed methods are described in the Supplementary Method part), ultra-thin sectioning was performed using a Leica UC6 Ultracut. Sections were mounted on copper grids (Plano), and additional contrasting was performed using lead citrate (Merck, 0.13 mM for 3 min). Electron micrographs were take using a Philips CM100 microscope equipped with a Gatan Orius SC600 camera at a magnification of 2950 to 15 500×.
Calcium Imaging and Quantification
GMB cells and neocortical slices with GBM cells, expressing GCaMP6f reporter, were imaged on EVOSTM M7000 Imaging System (Invitrogen) at a sampling rate of 3–5 Hz for 10 min. More details are given in the Supplementary Methods.33
Network Analysis of Calcium Imaging
For network analysis we developed a R based software tool NeuroPhysiologyLab (https://github.com/heilandd/NeuroPhysiologyLab). A detailed description of the downstream analysis is in the Supplementary Methods.
Single-Cell RNA Sequencing
Single-cell RNA sequencing was carried out using the Chromium Next GEM Single Cell 3’v3.1 protocol (10× Genomics), which is a droplet-based scRNA-sequencing method as described by the manufactor’s protocol. A detailed description is in the Supplementary Methods. The libraries were sequenced on an Illumina NextSeq 550 Sequencing System using the NextSeq 500/550 High Output kit v2.5, with 28 cycles for read 1, 8 cycles for the i7 index and 56 cycles for read 2.
Single-Cell Transcriptomic Analysis
Single-cell RNA sequencing was carried out with 10× Genomics Cell Ranger 7.1.0. The data was then postprocessed using the MILO-pipeline (available at https://github.com/theMILOlab-/scPipelines). More details information are given in the Supplementary Methods.
RNA Sequencing
RNA sequencing was performed on the MinION Sequencing Device, the SpotON Flow Cell (R9.4.1) and analyzed by Vis_Lab (https://github.com-/heilandd/Vis_Lab1.5). A detailed description is in the Supplementary Methods.
Cell Motility Analysis
The motility of #BTSC168 cells was evaluated in the images obtained by longitudinal imaging on Incucyte S3 Live-Cell Analysis System. Image files were analyzed by CellTracker34 software and output track files were imported into R. Further descriptions on the analysis are given in the Supplementary Methods.
Imaging of 3D Tumor Growth and Morphology
Migration of GBM cells into the surrounding brain tissue was observed on DPI5 using two-photon Olympus FV1000 microscope with a Mai Tai DeepSee laser (Spectra Physics) with 20×/NA 1.0 water-immersion objective. Further descriptions on the methods and analysis are given in the Supplementary Methods.
Statistical Analysis
RStudio (2021.09.1 + 372) software was used for statistical analysis. The data are presented as mean ± SEM for parametric analysis and as median with quartiles for non-parametric analysis, as defined in figure legends. The levels of significance were defined as *P < .05, **P < .01, ***P < .001, ****P < .0001. Detailed descriptions of the statistics are presented in the Supplementary Methods.
Results
Sialylation and Desialylation Enzymes are Heterogeneously Expressed Across Cell States
To investigate the potential for the sialylation and desialylation in glioblastoma cells, we first evaluated the expression of sialyltransferases (ST3GAL1 and ST3GAL3), cytidine monophosphate N-acetylneuraminic acid synthetase (CMAS) and the desialylation enzymes neuraminidases (NEU1-4). Using the GBMap,35 a large reference dataset containing over 1M cells gathered from 240 patients, we identified glioblastoma-specific expression of ST3GAL3, confirming published results,15 and CMAS, suggesting enhanced sialylation in GBM cells. Exploring other genes involved in the sialic acid metabolism, we found increased expression in glioblastoma cells of other sialyltransferases ST8SIA1, ST8SIA2, the N-acetylmannosamine kinase (GNE) and the transporter SLC35A1, Supplementary Figure 1a. The analysis of known glioblastoma transcriptional subgroups revealed a significant increase in the expression of sialylation enzymes in the AC- and MES-like subgroups (ST3GAL1, ST3GAL3, and CMAS), as shown in Supplementary Figure 1b. Furthermore, in OPC- and NPC-like cells we observed elevated levels of NEU3 and NEU4 expression, which are enzymes that remove sialic acid residues from glycoproteins and glycolipids. These findings suggest that sialic acid metabolism is heterogeneous and plays a crucial role in glioblastoma, as illustrated in Figure 1a and Supplementary Figure 1b. To further explore the spatial distribution of enzymes that promote or reduce sialylation, we employed spatially resolved transcriptomics on 16 primary untreated IDH wild-type (WT) glioblastomas.3 This approach revealed a distinct enrichment of enzymes that promote sialylation in recently described reactive (immune and hypoxia) and radial glia niches, as depicted in Figure 1b. Conversely, desialylation enzymes were primarily expressed in OPC-like and neural niches. Based on these findings from single cell and spatially resolved transcriptomic studies, we hypothesize that high levels of sialylation contribute to specific cell-cell interactions in AC- and MES-like cells, as illustrated in Figure 1c.
Figure 1.
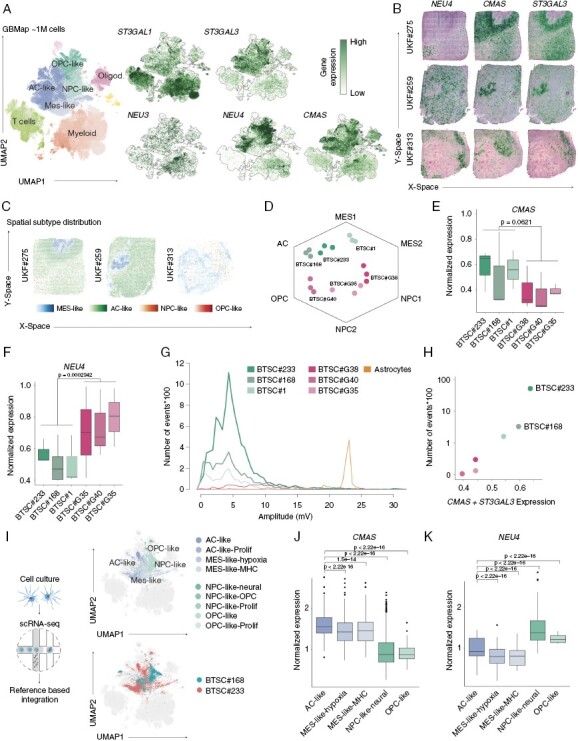
(a) UMAP representation of the GBMap scRNA-seq reference dataset. On the left side, colors represent the different cell type within glioblastoma and its microenvironment. In the right side, gene expression maps show the expression of genes involved in sialylation. (b) Surface plots of 3 representative spatially resolved transcriptomic samples demonstrating the expression of NEU4, CMAS, and ST3GAL3. (c) Cell type distribution of transcriptional states using single-cell deconvolution (robust decomposition of cell type mixtures, RCTD). Cell state enrichment scores are demonstrated by the color intensity indicated below. (d) Hexagon plot of 6 patient-derived cell lines (Neftel states). (e–f) Boxplots demonstrate the gene expression of CMAS (e) and NEU4 (f). Statistical evaluation was performed by Kruskal–Wallis test. (g) Evaluation of electrical activity of all patient-derived cell lines and astrocytes (Ctrl) using microelectrode array. The lineplot demonstrate the number of events (spikes*100) on the y-axis and the amplitude on the x-axis. (h) Scatter plot of the gene expression of sialylation enzymes (CMAS and ST3GAL3) and the logarithmic number of events. (i) Single cell RNA-seq of BTSC#233 and BTSC#168 cell lines annotated to the GBMap reference using azimuth. The UMAPs demonstrate the cell heterogeneity of both cell lines. (j–k) Boxplots demonstrate the gene expression of CMAS (e) and NEU4 (f) across cellular states from the single-cell RNA-seq. Statistical evaluation was performed by Kruskal–Wallis test.
Sialylation Enzyme Expression Correlates With Higher Electrical Activity in Cell Lines
Next, we quantified the gene expression levels of ST3GAL3, CMAS, and NEU4 in 6 patient-derived cell lines by RNA sequencing. We annotated the predominant cell state across all cell lines and confirmed the increased expression of ST3GAL3 and CMAS in AC- and MES-like cells and high expression of NEU4 in OPC/NPC-like cell lines, respectively (Figure 1d–f). Electrophysiological profiling using microelectrode array (MEA) of all cell lines showed a positive correlation of sialylation enzymes expression and number of spikes (R2 = 0.642, P = .0032) (Figure 1g, h). Based on these results, we used the 2 top candidates (BTSC#233 and BTSC#168) which revealed high levels of sialylation for further experiments. In order to investigate the cellular heterogeneity of both cell lines, we conducted single-cell RNA sequencing. The results revealed the preservation of transcriptomic heterogeneity, as evidenced by the presence of all cellular states within both cell lines, confirming recent reports3 (Figure 1i). Further examination of the expression of enzymes involved in sialylation and desialylation yielded consistent results, with higher expression of ST3GAL3 and CMAS observed in AC- and MES-like cells, and high expression of NEU4 found in OPC/NPC-like cell lines, as illustrated in Figure 1j, k.
GBM Cell Lines Demonstrate Highly Dynamic Sialic Acid Turnover
Building upon the previously observed high expression of genes involved in sialic acid turnover, we employed bioorthogonal CLICK chemistry to label de-novo synthesized sialic acid.36 Specifically, we utilized an artificial sugar containing an azide moiety (Ac4ManNAz), which serves as a substitute for acetylated mannose in sialic acid synthesis within cells. Once incorporated, Ac4ManNAz can be visualized by covalently binding its azide moiety to a fluorescently labeled alkyne (DBCO-Cy5), thus allowing for selective observation of sialylation within a restricted time frame, as illustrated in Figure 2a. Our cell line (BTSC#168) displayed highly efficient synthesis of sialic acid, which was dependent on the time and azido sugar concentration, as shown in Figure 2b, c. The concentration of azido sugar appeared to be a limiting factor for sialylation-specific signal, indicating high rate of de novo sialylation in GBM cells. In the absence of Ac4ManNAz, the fluorescent signal was week, indicating the high specificity of metabolic CLICK labeling (Figure 2c). To evaluate the rate of sialic acid turnover in GBM cell line, we then targeted sialic acid synthesis by the sialyltranferase inhibitor P-3FAX-Neu5Ac (FAX)37,38 and sialic acid cleavage by using sialidase inhibitor N-acetyl-2,3-dehydro-2-deoxyneuraminic acid (DANA)39 (Figure 2d). The application of FAX has been observed to result in a decrease in sialylation on the cell surface. However, the administration of DANA has been found to elicit an increase in the levels of sialic acid on the cell surface, owing to the suppression of cleavage processes. Quantitative analysis of longitude imaging of metabolically labeled de-novo synthesized sialic acid revealed that the inhibition of sialyltransferases by FAX significantly reduced (P = 0.0006) the amount of new sialic acid (Figure 2e, f). In contrast, inhibition of sialidases by DANA significantly increased (P < 0.0001) cell sialylation (Figure 2e, f). Inhibitory effect accumulated for 25 h and then remained stable for up to 45 h (Figure 2f). High-resolution imaging of GBM cells revealed that the inhibition of sialyltransferases led to the accumulation of fluorescent signal in the perinuclear compartment, suggesting that Ac4ManNAz was taken up but was not incorporated and distributed throughout the cell (Supplementary Figure 1d, e). Overall, our findings with the inhibitors of sialic acid turnover revealed that the sialylation of GBM cells is highly dynamic and can be modulated by chemical inhibitors.
Figure 2.
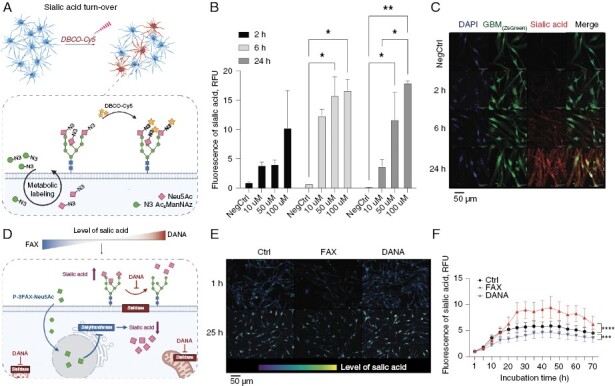
(a) Schematic illustration of metabolic sialic acid labeling by biorthogonal CLICK chemistry. (b) Quantification of metabolically labeled de novo sialic acid after incubation with 10 µM, 50 µM, or 100 µM of Ac4ManNAc for 2 h, 6 h, and 24 h. Two-way ANOVA, n = 2, mean ± SEM. (c) Metabolically labeled de novo sialic acid after treatment with 100 µM of Ac4ManNAc for 2 h, 6 h, and 24 h. (d) Schematic illustration of chemical inhibitors used to inhibit sialic acid synthesis (FAX, blue) or desialylation (DANA, red). (e–f) Longitudinal imaging of de novo sialylation in BTSC#168 cells after treatment with FAX or DANA. Two-way ANOVA, n = 3, mean ± SEM. *P < .05, **P < .01, ***P < .001, ****P < .0001
Sialylation Affect Cell Motility and Accumulate in the Cell Periphery
To further explore the temporal impact of decreased or enhanced sialylation, we performed live-cell imaging and analyzed the single cell tracks of BTSC#168 cells using the Cypro algorithm. We identified 3 phases of cell motility (Figure 3a). In the first phase, cells increased their velocity until a certain level of cell density is reached. In the second phase, cells exhibited a steady velocity due to the maintenance of an adequate spatial distance. In the final phase, the velocity of the cells decreased in response to high cell density. In the first phase, neither the inhibition nor activation of sialylation had any significant effect on cell velocity. However, during the second phase, the inhibition of sialylation (via FAX treatment) resulted in a significant increase in cell velocity (P = 0.0053), while hyper-sialylation (via DANA treatment) led to a decrease in cell velocity (P < 2.2 × 10−16). In the final phase, the reduced velocity after DANA treatment was maintained but the velocity increase induced by FAX treatment was lost (Figure 3a, b).
Figure 3.
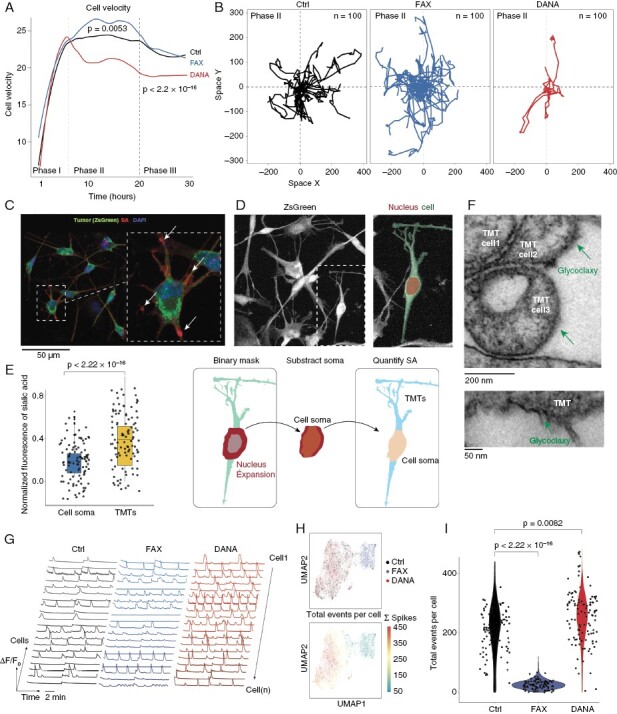
(a) Line plot of the cell velocity computed by Cypro comparing the control versus DANA and FAX treatment (n = 4). Dashed lines indicate the different phases of cell motility. (b) Spatial traces of representative cells (n = 100) in phase II after treatment with DANA and FAX. (c) Illustration of the imaging process pipeline to quantify the sialic acid accumulation in cell soma and branches. (d) Boxplot showing the significant accumulation of sialic acid in the cell periphery (Branches and TMT’s). (e) Electron microscopy representative images showing the thickening of glycocalyx in TMT’s (lower) and at cell-cell contacts (upper). (g) Representative Ca2+ traces extracted by the NeuroPhysiologyLab toolbox. (h) Scatter plots of low-dimensional embedding of Ca2+ traces using feature extraction and Uniform Manifold Approximation and Projection (UMAP). Upper UMAP, colors indicate the treatment groups. Bottom UMAP, colors indicate the total number of Ca2+ spikes per cell. (i) Violin plot of the total number of Ca2+ spikes per cell across treatment groups. Statistical evaluation was performed by Kruskal–Wallis test.
We hypothesized that an elevation in the levels of sialic acid on the cell surface may augment the connections between glioblastoma cells, thereby diminishing their motility, consistent with recent findings indicating that cells that form functional networks are less invasive.12 This hypothesis is supported by literature suggesting that surface sialic acid not only enhances the migration and invasiveness of cancer cells5,8 but also actively influences cell-to-cell connectivity and neuronal activity.26–28 Therefore, we sought to investigate the role of sialylation in glioma cell communication. To examine the distribution of sialic acid abundance in relation to the morphological characteristics of the cell, specifically the cell body and its extensions, we employed high-resolution microscopy and quantified the enrichment of sialic acid puncta in the cell soma (nucleus extension) and cell branches and tumor microtubes (TMTs) (Figure 3c–e). Our analysis revealed a substantial enrichment of sialic acid in the cell branches and TMTs (Figure 2e). Given the observed enrichment of sialic acid in TMTs and at cell-cell contacts, we employed EM of BTSC#168 to visualize the glycocalyx, which confirmed a thick layer of glycocalyx in TMTs (Figure 2f).
Inhibited Salic Acid Turnover Perturb Cellular Network Formation and Communication
To investigate the impact of modified sialylation on glioblastoma networks, we examined intracellular calcium signaling, which exhibits robust activity at baseline and in response to l-glutamate stimulation.40 Short interval time-lapse imaging was utilized to capture and analyze individual intracellular Ca2+ spikes within #BTSC233_GCamp6F cells, and to quantitatively evaluate network properties. Cell sialylation and desialylation was decreased by the treatment with FAX and DANA, respectively. Results revealed a significant enhancement of cellular Ca2+ spikes when surface sialic acid levels were elevated by DANA treatment (Figure 3g). Subsequently, feature extraction and low-dimensional embedding of the calcium traces of each individual cell revealed 2 distinct clusters of cells highly correlated with treatment conditions (Figure 3h). Notably, control and DANA treatment displayed similarities in their signaling behavior due to the relatively high baseline levels of sialylation. Furthermore, DANA treatment led to a 3.8-fold increase in the frequency of Ca2+ spikes compared to control (P = 0.0043), while FAX treatment resulted in a reduction of spontaneous activity by 0.32-fold compared to control (P = 0.0082). This comparison of control to DANA treatment demonstrated a significant increase of Ca2+ events (P = 0.0082), as illustrated in Figure 3i.
Inhibited Sialic Acid Turnover Leads to Unspecific Gene Expression Changes
To evaluate the impact of decreased or enhanced sialylation on the cell surface, we performed RNA sequencing of BTSC#233 cells after FAX and DANA treatment (n = 4 RNA-seq per subgroup) and performed a weighted correlation network analysis (WGCNA), Supplementary Figure 2a. We correlated the eigen decompositions of each expression module (designated by colors) with the treatment condition and identified 2 modules that were significantly associated with desialylation (FAX: royal blue and light cyan) and 2 modules that were correlated with hyper-sialylation (DANA: purple and brown), as shown in Supplementary Figure 2b–d. Functional annotation of these expression modules revealed mainly unspecific enrichment of oligosaccharyl transferase activity (GO:0004576) after DANA treatment. After FAX treatment cells showed an enrichment of amino acid transporter activity (GO:0061459 and GO:0015190) (Supplementary Figure 2e). The transcriptomic data suggested that the perturbation of sialylation pathway has only minor effect on cell gene expression. The proliferation index was also not affected by the treatment (Supplementary Figure 2f).
Sialylation Affect Tumor Formation in Neocortical Ex Vivo Model of Glioblastoma
The structural and functional interactions of glioblastoma cells are greatly influenced by the regulatory signals from the surrounding tumor microenvironment.29 The regulatory signals from the tumor microenvironment that modulate the structural and functional interactions of glioblastoma cells cannot be fully captured in conventional in vitro cell culture systems. Therefore, the use of 3-dimensional brain culture models is necessary for a more comprehensive understanding of these signals.29 To investigate how sialylation modulates tumor growth in the tissue, we employed a well-established ex vivo model of glioblastoma, in which human glioblastoma cells are injected into neocortical sections of surgically resected access human brain tissue.29 #BTSC233_GCamp6F cells were injected into organotypic cortical slices and the sialylation of tumor cells as well as surrounding tissue in the natural tumor environment was investigated after the treatment with the inhibitors targeting the turnover of sialic acid (Figure 4a, b). We observed increased sialylation of GBM cells compared to the peritumoral region (Figure 4c). Higher sialylation of the tumor cells was retained even after the treatment with either DANA or FAX inhibitors, indicating enhanced and more robust de novo sialic acid synthesis in GBM cells compared to surrounding brain cells (P = 0.0034), Figure 4c, d. Next, we studied how inhibition of sialyltransferases or sialidases affects the formation of GBM tumor in the neocortical slice model. The growth and the migration of GBM cells injected into organotypic slice were imaged for up to 5 days post injection (Supplementary Figure 3a). Longitudinal imaging revealed that inhibition of sialic acid synthesis with FAX strongly limited the formation of GBM tumor (Supplementary Figure 3b.
Figure 4.
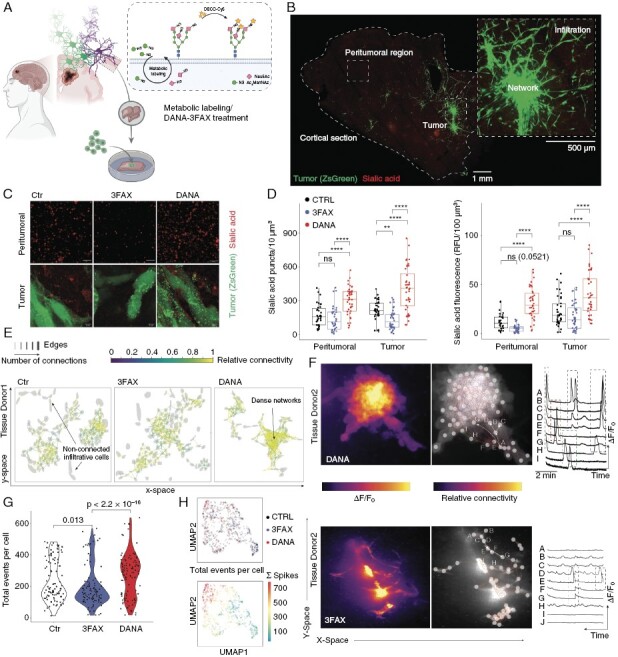
(a) Illustration of the workflow using a human neocortical slice model with metabolic labeling. (b) Representative overview of the slice model and spatially distinct regions. On the right side, a representative zoom-in to demonstrate the tumor architecture. (c) High-resolution images of GBM tumor sialylation after treatment with sialylation (FAX) and desialylation (DANA) inhibitors and quantification (d). (e) Tumor network plot indicating the Ca2+ signalling. The edges between tumor cells represent an existing functional connection, the colour indicate the frequency of co-appearing signals. (f) Representative Ca2+ traces after DANA and FAX treatment. (g) Quantification of the total Ca2+ events per cell across all the treatments. (h) Scatter plots of low-dimensional embedding of Ca2+ traces using feature extraction and Uniform Manifold Approximation and Projection (UMAP). Upper UMAP, colours indicate the treatment groups. Bottom UMAP, colours indicate the total number of Ca2+ spikes per cell.
In contrast, there was no difference in the tumor area after the treatment with DANA, Supplementary Figure 3b. However, inhibition of sialidases led to a different GBM migration pattern, in which GBM cells are more spread, Supplementary Figure 3a. These results demonstrated that levels of sialylation regulate GBM tumor formation, and sialylation pathway can be chemically targeted to limit GBM tumor growth. To assess the role of sialic acid synthesis and elimination in GBM tumor formation in more detail, we performed 2-photon imaging and 3D analysis of GBM tumors in neocortical slices (Supplementary Figure 3c). We found that the core of the tumor was denser after the treatment with FAX, suggesting that reduced sialylation limits GBM-cell migration (Supplementary Figure 3c, d). Despite the lack of alterations in the 3-dimensional volume of the tumor subsequent to the inhibition of sialic acid synthesis or cleavage, our findings indicated that the morphological characteristics of the tumor were contingent in response to the changes of sialylation (Supplementary Figure 3e–h). Specifically, the application of DANA resulted in the formation of tumors that were more elongated and flattened in comparison to the control group. Conversely, the treatment with FAX did not affect the 3-dimensional form of the tumor, suggesting that the observed disparities are specifically attributed to the excessive sialylation of glioblastoma cells, as depicted in Supplementary Figure 3e–h. In conclusion, our analysis of glioblastoma tumor formation in the neocortical slice model indicates that the growth and development of the tumor is closely regulated by the sialylation levels of glioblastoma cells and/or the cells in the surrounding microenvironment.
Our findings from cell culture experiments, in which altered surface levels of sialic acid impacted the calcium signaling network of glioblastoma cells (Figure 3g–i), prompted us to investigate the extent to which sialylation dictates intratumoral cell activity in the human neocortical slice model. Calcium imaging in organotypic slices that were injected with GCamp6F-expressing glioblastoma cells confirmed the results obtained from the cell culture experiments, as depicted in Figure 4e, f. The inhibition of sialic acid synthesis with FAX significantly diminished the spontaneous activity of glioblastoma cells within the tumor and hindered the formation of a glioblastoma cell network. Although a subset of glioblastoma cells remained active, the number of actively signaling cells was significantly reduced (P = 0.013), Figure 4g. Conversely, the inhibition of sialidases with DANA resulted in an enhancement of calcium spiking and the formation of a highly connected glioblastoma cell tumor network (Figure 4e, f). The inhibition of desialylation significantly increased the number of signaling cells (P = 2.2 × 10−16) (Figure 4g). Feature extraction followed by low-dimensional embedding revealed an amplified signal frequency in tumor cells treated with DANA (Figure 4h). Collectively, these findings demonstrate a crucial role of sialylation in the establishment of functional connectivity within the glioblastoma cell network in both cell culture and tumor models.
Knocking-Out Sialidases Increases GBM Cell Sialylation and Enhances Cellular Activity and Network Connectivity In Vitro
Our investigation revealed an increase in sialidase expression within glioblastoma tumors and associated cell lines, as depicted in Figure 1. Additionally, DANA inhibition of sialidase activity resulted in an enhancement of functional network connectivity. In order to further explore the specific role of sialidases NEU1 and NEU4 in this context, knock-out (KO) lines of #BTSC233_GCamp6F cells were generated through the use of CRISPR/Cas9 gene editing techniques. Perturbation of either NEU1 or NEU4 led to significantly higher de-novo cell sialylation as demonstrated by metabolic labeling (Figure 5a, b). The loss of NEU1 or NEU4 neither increased nor compromised the proliferation of GBM cells, as Ki67 expression did not differ between WT and KO cells (Supplementary Figure 4a). We then used sialidase KO cells in 3D neocortical glioma model to distinguish, whether our observed tumor growth reliance on sialylation is mediated by GBM cell-autonomous or tumor microenvironment-dependent mechanisms. Either NEU1 KO or NEU4 KO cells were injected into organotypic slices and tumor formation was monitored for 6 days (Figure 5c). We found that loss of either NEU1 or NEU4 promoted tumor growth (Figure 5c, d), indicating that over-sialylation of GBM cells enhances tumorigenesis. In concordance with prior discoveries, the deletion of either NEU1 or NEU4 resulted in an elevated level of intracellular Ca2+ activity when compared to control samples, as illustrated in Figure 5e. Additionally, a higher frequency of Ca2+spikes per cell was observed in both NEU1 and NEU4 knockouts (Figure 5f), thereby confirming that cell surface sialylation plays a crucial role in modulating functional glioblastoma multiforme connectivity and tumorigenesis in a cell-autonomous manner.
Figure 5.
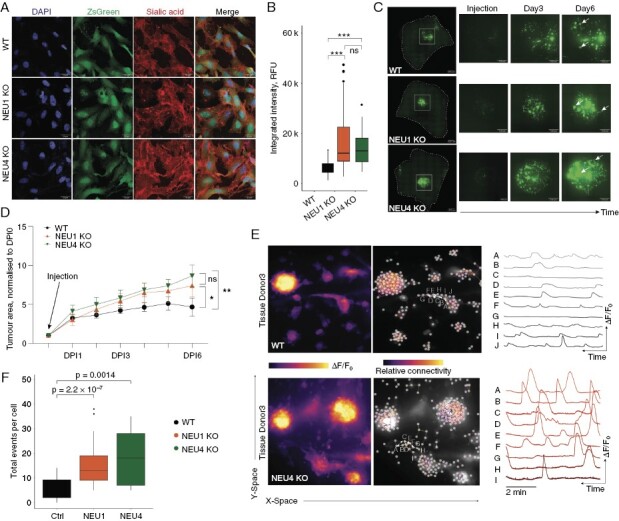
a–b) Representative images and quantification of metabolically labeled sialic acid in WT, NEU1 KO, and NEU4 KO GBM cells. c–d) Representative images and quantification of tumor growth of WT, NEU1 KO, and NEU4 KO cells in the human neocortical slice model. e) Representative Ca2+ traces in WT and NEU4 KO cells. f) Quantification of the total Ca2+ events per cell in WT, NEU1 KO, and NEU4 KO cells.
Discussion
The hallmark of brain tumors is their exclusive presence within the CNS. Metastasis to extracranial sites occurs only in rare occasions. This exclusivity has biological roots that are poorly understood. For instance, glioblastoma and pediatric brain tumors have recently been described to establish synaptic connections to neurons and thus maintain a direct functional connection to the CNS.22,23 Therefore, over the past few years, there has been a particular focus on the interaction between malignant brain tumors and the interfaces with their neuronal environment. It has been demonstrated that tumor cells form functional syncytia within so-called tumor networks22,23; however, the underlying reasons are still largely controversial. Recently, Venkataramani et al. have uncovered the functional context of transcriptional diversity and demonstrated the difference of tumor cells that actively participate in tumor networks and those that escape the network formation.12 Strikingly, the unconnected cells, transcriptionally defined as NPC- and OPC-like cells, abandon the functional network to invade the brain by hijacking mechanisms from early progenitor cells.12 At present, it is still ambiguous for what reasons tumor cells are arranged in functional networks and, more importantly, why they abandon these networks at a given time to drive invasive growth. The importance and role of surface molecules in this complex interaction, in particular the glycocalyx, is controversially discussed in many cancers and potentially represents a more important contributor than previously appreciated.4,5 Sialic acid is a part of the glycocalyx and were shown to contribute to the immune escape mechanisms,4,5 but their role in glioblastoma network formation and invasion remains unexplored.
In the healthy brain, sialic acid is of paramount importance for brain homeostasis, cell-cell interactions, neuronal growth, alteration of synaptic connectivity, memory formation and regulation of the brain immunity.41,42 From the field of cancer neuroscience, we have learned that brain tumors largely mimic mechanisms of healthy brain to drive invasion, growth, and cell survival.2,3 In light of the previous findings, we hypothesized that sialic acid metabolism and turnover is potentially altered to promote malignancy in glioblastoma. To this end, we explored the gene expression pattern of malignant and microenvironmental cells at single-cell resolution and quantified the sialic acid turnover in primary glioblastoma cell lines, demonstrating an enhanced accumulation of sialic acids along with tumor-specific gene expression, dysregulating the enzymes important for the sialic acid metabolism. In line with previous reports,15 we confirmed a tumor-cell specific up-regulation of sialyltransferase ST3GAL3. However, we indicated that the increase of ST3GAL1 expression can be attributed to immune cells (myeloid and T cells) rather than GBM cells themselves (Figure 1a), further supporting the role of sialic acid in the interaction of tumor and its microenvironment. In summary, our gene expression analysis showed that sialic acid synthesis is highly upregulated in GBM tumor cells. In particular, sialidases were upregulated in a subpopulation of AC-like and OPC-NPC-like cells demonstrating the heterogeneity in sialic acid metabolism across transcriptional subgroups. The functional experiments using bioothogonal CLICK chemistry to metabolically label de novo synthesized sialic acid confirmed a high sialic acid turnover. To further explore the biological consequences for increased or decreased sialylation, we performed a large number of validations revealing the importance of sialic acid for GBM network structure and function. Confirming the importance of sialic acid in cell-cell interactions, we demonstrated an accumulation of sialic acid at the intercellular GBM cell contacts, which have also been reported in other cancers.43 Further functional imaging displayed a major impact of inhibition and enhancing of sialylation on the functional signaling within the tumor network. Strikingly, inhibition of sialylation leads to almost complete loss of intercellular Ca2+ signaling. Since the cell culture conditions do not resemble the human brain microenvironment, we applied the experimental pipeline of a human neocortical slice model, which we have described and extensively studied previously.3,29,40,44–47 We found that the level of GBM cell sialylation defines tumor growth, as inhibition of sialylation reduced the size of tumor in neocortical slices. However, loss of sialidases had an opposite effect, suggesting that sialylation may drive propensity to migrate in GBM. Our observation that the intratumoral network activity is as well driven by sialylation, contradicts the hypothesis that only unconnected cells migrate12 and demonstrates that the detailed relationships between signaling and tumor migration are not definitively understood. Finally, we sought to distinguish whether the effect of chemical inhibition of sialic acid metabolism is facilitated by the tumor or passively by the microenvironment. For this purpose, we performed a KO of sialidases (NEU1 and NEU4), which confirmed our previous experiments at all levels. This demonstrated that observed effects of changed sialic acid metabolism on GBM tumor growth and network formation are GBM-cell autonomous rather than depending on tumor microenvironment, suggesting sialylation as a potentially druggable target for GBM treatment. Our results reveal a new pathway required for tumor network formation and suggest an important role for glycocalyx in neurological cancers.
Supplementary Material
Contributor Information
Ugne Kuliesiute, Microenvironment and Immunology Research Laboratory, Medical Center, University of Freiburg, Freiburg, Germany; Department of Neurosurgery, Medical Center, University of Freiburg, Freiburg, Germany; Faculty of Medicine, Freiburg University, Freiburg, Germany; Institute of Biosciences, Life Sciences Center, Vilnius University, Vilnius, Lithuania; VU LSC-EMBL Partnership for Genome Editing Technologies, Life Sciences Center, Vilnius University, Vilnius, Lithuania.
Kevin Joseph, Microenvironment and Immunology Research Laboratory, Medical Center, University of Freiburg, Freiburg, Germany; Department of Neurosurgery, Medical Center, University of Freiburg, Freiburg, Germany; Faculty of Medicine, Freiburg University, Freiburg, Germany; Department of Neuroanatomy, Institute of Anatomy and Cell Biology, Faculty of Medicine, University of Freiburg, Freiburg, Germany; Center Brain Links Brain Tools, University of Freiburg, Freiburg, Germany.
Jakob Straehle, Department of Neurosurgery, Medical Center, University of Freiburg, Freiburg, Germany; Faculty of Medicine, Freiburg University, Freiburg, Germany; Department of Neuroanatomy, Institute of Anatomy and Cell Biology, Faculty of Medicine, University of Freiburg, Freiburg, Germany.
Vidhya Madapusi Ravi, Microenvironment and Immunology Research Laboratory, Medical Center, University of Freiburg, Freiburg, Germany; Department of Neurosurgery, Medical Center, University of Freiburg, Freiburg, Germany; Faculty of Medicine, Freiburg University, Freiburg, Germany; Department of Neuroanatomy, Institute of Anatomy and Cell Biology, Faculty of Medicine, University of Freiburg, Freiburg, Germany; Center Brain Links Brain Tools, University of Freiburg, Freiburg, Germany.
Jan Kueckelhaus, Microenvironment and Immunology Research Laboratory, Medical Center, University of Freiburg, Freiburg, Germany; Department of Neurosurgery, Medical Center, University of Freiburg, Freiburg, Germany; Faculty of Medicine, Freiburg University, Freiburg, Germany; Department of Neuroanatomy, Institute of Anatomy and Cell Biology, Faculty of Medicine, University of Freiburg, Freiburg, Germany; Center Brain Links Brain Tools, University of Freiburg, Freiburg, Germany.
Jasim Kada Benotmane, Microenvironment and Immunology Research Laboratory, Medical Center, University of Freiburg, Freiburg, Germany; Department of Neurosurgery, Medical Center, University of Freiburg, Freiburg, Germany; Faculty of Medicine, Freiburg University, Freiburg, Germany; Department of Neuroanatomy, Institute of Anatomy and Cell Biology, Faculty of Medicine, University of Freiburg, Freiburg, Germany; Center Brain Links Brain Tools, University of Freiburg, Freiburg, Germany.
Junyi Zhang, Microenvironment and Immunology Research Laboratory, Medical Center, University of Freiburg, Freiburg, Germany; Department of Neurosurgery, Medical Center, University of Freiburg, Freiburg, Germany; Faculty of Medicine, Freiburg University, Freiburg, Germany; Department of Neuroanatomy, Institute of Anatomy and Cell Biology, Faculty of Medicine, University of Freiburg, Freiburg, Germany; Center Brain Links Brain Tools, University of Freiburg, Freiburg, Germany.
Andreas Vlachos, Department of Neuroanatomy, Institute of Anatomy and Cell Biology, Faculty of Medicine, University of Freiburg, Freiburg, Germany; Center Brain Links Brain Tools, University of Freiburg, Freiburg, Germany; Center for Basics in Neuromodulation, Faculty of Medicine, University of Freiburg, Freiburg, Germany.
Juergen Beck, Department of Neurosurgery, Medical Center, University of Freiburg, Freiburg, Germany; Faculty of Medicine, Freiburg University, Freiburg, Germany; Department of Neuroanatomy, Institute of Anatomy and Cell Biology, Faculty of Medicine, University of Freiburg, Freiburg, Germany.
Oliver Schnell, Microenvironment and Immunology Research Laboratory, Medical Center, University of Freiburg, Freiburg, Germany; Department of Neurosurgery, Medical Center, University of Freiburg, Freiburg, Germany; Faculty of Medicine, Freiburg University, Freiburg, Germany.
Urte Neniskyte, Institute of Biosciences, Life Sciences Center, Vilnius University, Vilnius, Lithuania; VU LSC-EMBL Partnership for Genome Editing Technologies, Life Sciences Center, Vilnius University, Vilnius, Lithuania.
Dieter Henrik Heiland, Microenvironment and Immunology Research Laboratory, Medical Center, University of Freiburg, Freiburg, Germany; Department of Neurosurgery, Medical Center, University of Freiburg, Freiburg, Germany; Faculty of Medicine, Freiburg University, Freiburg, Germany; Department of Neurological Surgery, Lou and Jean Malnati Brain Tumor Institute, Robert H. Lurie Comprehensive Cancer Center, Feinberg School of Medicine, Northwestern University, Chicago, Illinois; Comprehensive Cancer Center Freiburg (CCCF), Faculty of Medicine and Medical Center—University of Freiburg, Freiburg, Germany; German Cancer Consortium (DKTK), partner site, Freiburg.
Funding
This project was funded by the German Cancer Consortium (DKTK), Else Kröner-Fresenius Foundation (DHH). The work is part of the MEPHISTO project (DHH), funded by BMBF (iGerman Ministry of Education and Research) (project number: 031L0260B). UK was funded by International Brain Research Organization Pan-European Regional Committee InEurope Short Stay Grant, Baltic-German University Liaison Office supported by the German Academic Exchange Service (DAAD) with funds from the Foreign Office of the Federal Republic Germany, EU’s Erasmus+ programme, German Academic Exchange Service (DAAD) Research Grant – Short - Term Grant, 2021, No 57552336. UK and UN were funded by the European Regional Development Fund under grant agreement No 01.2.2-CPVA-V-716-01-0001 with the Central Project Management Agency (CPVA), Lithuania.
Data Availability
The single-cell sequencing and spatially resolved transcriptomic data underlying this article are available in Dryad Digital Repository, at https://zenodo.org/record/6962901 and https://datadryad.org/stash/dataset/doi:10.5061/dryad.h70rxwdmj. Used code and software is available at https://github.com/theMILOlab/SPATA2. Imaging and bulk RNA-sequencing data available on request.
Conflict of interest statement
No potential conflicts of interest were disclosed by the authors.
Authorship statement
U.K., D.H.H., U.N. conceived the idea, designed the experiments, and composed the paper. J.K., J.K.B. conducted the computational analysis of RNA-seq. K.J. and U.K. single cell transcripomics, J.S. and A. V. performed and analyzed electronmicroscopy, D.H.H. conducted the development of the NeuroPhysiology software pipeline. U.K. conducted cell and slice culture experiments. U.K., K.J. conducted calcium imaging. A.U.A., O.S., J.B., J.Z., V.M.R., K.J. contributed to the interpretation of the results. D.H.H., U.N. supervised the project.
References
- 1. D’Alessio A, Proietti G, Sica G, Scicchitano BM.. Pathological and molecular features of glioblastoma and its peritumoral tissue. Cancers (Basel). 2019;11(4). doi: 10.3390/cancers11040469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Neftel C, Laffy J, Filbin MG, et al. An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell. 2019;178(4):835–849.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ravi VM, Will P, Kueckelhaus J, et al. Spatially resolved multi-omics deciphers bidirectional tumor-host interdependence in glioblastoma. Cancer Cell. 2022;40(6):639–655.e13. [DOI] [PubMed] [Google Scholar]
- 4. Büll C, Stoel MA, den Brok MH, Adema GJ.. Sialic acids sweeten a tumor’s life. Cancer Res. 2014;74(12):3199–3204. [DOI] [PubMed] [Google Scholar]
- 5. Zhou X, Yang G, Guan F.. Biological functions and analytical strategies of sialic acids in tumor. Cells. 2020;9(2):273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pinho SS, Reis CA.. Glycosylation in cancer: mechanisms and clinical implications. Nat Rev Cancer. 2015;15(9):540–555. [DOI] [PubMed] [Google Scholar]
- 7. Rodrigues JG, Balmaña M, Macedo JA, et al. Glycosylation in cancer: Selected roles in tumour progression, immune modulation and metastasis. Cell Immunol. 2018;333:46–57. doi: 10.1016/j.cellimm.2018.03.007. [DOI] [PubMed] [Google Scholar]
- 8. Kang H, Wu Q, Sun A, et al. Cancer cell glycocalyx and its significance in cancer progression. Int J Mol Sci. 2018;19(9). doi: 10.3390/ijms19092484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wielgat P, Trofimiuk E, Czarnomysy R, Braszko JJ, Car H.. Sialic acids as cellular markers of immunomodulatory action of dexamethasone on glioma cells of different immunogenicity. Mol Cell Biochem. 2019;455(1-2):147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Amoureux M-C, Coulibaly B, Chinot O, et al. Polysialic acid neural cell adhesion molecule (PSA-NCAM) is an adverse prognosis factor in glioblastoma, and regulates olig2 expression in glioma cell lines. BMC Cancer. 2010;10:91. doi: 10.1186/1471-2407-10-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Suzuki M, Suzuki M, Nakayama J, et al. Polysialic acid facilitates tumor invasion by glioma cells. Glycobiology. 2005;15(9):887–894. [DOI] [PubMed] [Google Scholar]
- 12. Venkataramani V, Yang Y, Schubert MC, et al. Glioblastoma hijacks neuronal mechanisms for brain invasion. Cell. 2022;185(16):2899–2917.e31. [DOI] [PubMed] [Google Scholar]
- 13. Li F, Ding J.. Sialylation is involved in cell fate decision during development, reprogramming and cancer progression. Protein Cell. 2019;10(8):550–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Harduin-Lepers A, Vallejo-Ruiz V, Krzewinski-Recchi MA, et al. The human sialyltransferase family. Biochimie. 2001;83(8):727–737. [DOI] [PubMed] [Google Scholar]
- 15. Yamamoto H, Saito T, Kaneko Y, et al. α2,3-Sialyltransferase mRNA and α2,3-linked glycoprotein sialylation are increased in malignant gliomas. Brain Res. 1997;755(1):175–179. [DOI] [PubMed] [Google Scholar]
- 16. Chong YK, Sandanaraj E, Koh LWH, et al. ST3GAL1-associated transcriptomic program in glioblastoma tumor growth, invasion, and prognosis. J Natl Cancer Inst. 2016;108(2). doi: 10.1093/jnci/djv326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yamamoto H, Oviedo A, Sweeley C, Saito T, Moskal JR.. Alpha2,6-sialylation of cell-surface N-glycans inhibits glioma formation in vivo. Cancer Res. 2001;61(18):6822–6829. [PubMed] [Google Scholar]
- 18. Pshezhetsky AV, Ashmarina M.. Keeping it trim: roles of neuraminidases in CNS function. Glycoconj J. 2018;35(4):375–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Miyagi T, Takahashi K, Hata K, Shiozaki K, Yamaguchi K.. Sialidase significance for cancer progression. Glycoconj J. 2012;29(8-9):567–577. [DOI] [PubMed] [Google Scholar]
- 20. Takahashi K, Proshin S, Yamaguchi K, et al. Sialidase NEU3 defines invasive potential of human glioblastoma cells by regulating calpain-mediated proteolysis of focal adhesion proteins. Biochim Biophys Acta Gen Subj. 2017;1861(11 Pt A):2778–2788. [DOI] [PubMed] [Google Scholar]
- 21. Osswald M, Jung E, Sahm F, et al. Brain tumour cells interconnect to a functional and resistant network. Nature. 2015;528(7580):93–98. [DOI] [PubMed] [Google Scholar]
- 22. Venkatesh HS, Morishita W, Geraghty AC, et al. Electrical and synaptic integration of glioma into neural circuits. Nature. 2019;573(7775):539–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Venkataramani V, Tanev DI, Strahle C, et al. Glutamatergic synaptic input to glioma cells drives brain tumour progression. Nature. 2019;573(7775):532–538. [DOI] [PubMed] [Google Scholar]
- 24. Gritsenko PG, Atlasy N, Dieteren CEJ, et al. p120-catenin-dependent collective brain infiltration by glioma cell networks. Nat Cell Biol. 2020;22(1):97–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Osswald M, Solecki G, Wick W, Winkler F.. A malignant cellular network in gliomas: potential clinical implications. Neuro Oncol. 2016;18(4):479–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Isaev D, Isaeva E, Shatskih T, et al. Role of extracellular sialic acid in regulation of neuronal and network excitability in the rat hippocampus. J Neurosci. 2007;27(43):11587–11594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Boll I, Jensen P, Schwämmle V, Larsen MR.. Depolarization-dependent induction of site-specific changes in sialylation on N-linked glycoproteins in rat nerve terminals. Mol Cell Proteomics. 2020;19(9):1418–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kulkarni RU, Wang CL, Bertozzi CR.. Subthreshold voltage analysis demonstrates neuronal cell-surface sialic acids modulate excitability and network integration. bioRxiv. 2020. [Google Scholar]
- 29. Ravi VM, Joseph K, Wurm J, et al. Human organotypic brain slice culture: a novel framework for environmental research in neuro-oncology. Life Sci Alliance. 2019;2(4):e201900305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stirling DR, Carpenter AE, Cimini BA.. CellProfiler Analyst 3.0: accessible data exploration and machine learning for image analysis. Bioinformatics. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schindelin J, Arganda-Carreras I, Frise E, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Joseph K, Mottaghi S, Christ O, Feuerstein TJ, Hofmann UG.. When the ostrich-algorithm fails: blanking method affects spike train statistics. Front Neurosci. 2018;12:293. doi: 10.3389/fnins.2018.00293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Patel TP, Man K, Firestein BL, Meaney DF.. Automated quantification of neuronal networks and single-cell calcium dynamics using calcium imaging. J Neurosci Methods. 2015;243:26–38. doi: 10.1016/j.jneumeth.2015.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Piccinini F, Kiss A, Horvath P.. CellTracker (not only) for dummies. Bioinformatics. 2016;32(6):955–957. [DOI] [PubMed] [Google Scholar]
- 35. Ruiz Moreno C, Stunnenberg HG, Nilsson M, et al. Harmonized single-cell landscape, intercellular crosstalk and tumor architecture of glioblastoma. bioRxiv. 2022. [Google Scholar]
- 36. Laughlin ST, Baskin JM, Amacher SL, Bertozzi CR.. In vivo imaging of membrane-associated glycans in developing zebrafish. Science. 2008;320(5876):664–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Heise T, Pijnenborg JFA, Büll C, et al. Potent metabolic sialylation inhibitors based on C-5-modified fluorinated sialic acids. J Med Chem. 2019;62(2):1014–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Büll C, Boltje TJ, Wassink M, et al. Targeting aberrant sialylation in cancer cells using a fluorinated sialic acid analog impairs adhesion, migration, and in vivo tumor growth. Mol Cancer Ther. 2013;12(10):1935–1946. [DOI] [PubMed] [Google Scholar]
- 39. Minami A, Fujita Y, Shimba S, et al. The sialidase inhibitor 2,3-dehydro-2-deoxy-N-acetylneuraminic acid is a glucose-dependent potentiator of insulin secretion. Sci Rep. 2020;10(1):5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schneider M, Vollmer L, Potthoff A-L, et al. Meclofenamate causes loss of cellular tethering and decoupling of functional networks in glioblastoma. Neuro Oncol. 2021;23(11):1885–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liao H, Klaus C, Neumann H.. Control of innate immunity by sialic acids in the nervous tissue. Int J Mol Sci. 2020;21(15). doi: 10.3390/ijms21155494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang B. Sialic acid is an essential nutrient for brain development and cognition. Annu Rev Nutr. 2009;29:177–222. doi: 10.1146/annurev.nutr.28.061807.155515. [DOI] [PubMed] [Google Scholar]
- 43. Li Q, Xie Y, Xu G, Lebrilla CB.. Identification of potential sialic acid binding proteins on cell membranes by proximity chemical labeling. Chem Sci. 2019;10(24):6199–6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Henrik Heiland D, Ravi VM, Behringer SP, et al. Tumor-associated reactive astrocytes aid the evolution of immunosuppressive environment in glioblastoma. Nat Commun. 2019;10(1):2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Potthoff A-L, Heiland DH, Evert BO, et al. Inhibition of gap junctions sensitizes primary glioblastoma cells for temozolomide. Cancers (Basel). 2019;11(6). doi: 10.3390/cancers11060858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Maier JP, Ravi VM, Kueckelhaus J, et al. Inhibition of metabotropic glutamate receptor III facilitates sensitization to alkylating chemotherapeutics in glioblastoma. Cell Death Dis. 2021;12(8):723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ravi VM, Neidert N, Will P, et al. T-cell dysfunction in the glioblastoma microenvironment is mediated by myeloid cells releasing interleukin-10. Nat Commun. 2022;13(1):925. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The single-cell sequencing and spatially resolved transcriptomic data underlying this article are available in Dryad Digital Repository, at https://zenodo.org/record/6962901 and https://datadryad.org/stash/dataset/doi:10.5061/dryad.h70rxwdmj. Used code and software is available at https://github.com/theMILOlab/SPATA2. Imaging and bulk RNA-sequencing data available on request.


