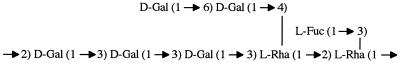Abstract
Recent work by our group has shown that an exopolysaccharide (EPS)-producing starter pair, Streptococcus thermophilus MR-1C and Lactobacillus delbrueckii subsp. bulgaricus MR-1R, can significantly increase moisture retention in low-fat mozzarella (D. B. Perry, D. J. McMahon, and C. J. Oberg, J. Dairy Sci. 80:799–805, 1997). The objectives of this study were to determine whether MR-1C, MR-1R, or both of these strains are required for enhanced moisture retention and to establish the role of EPS in this phenomenon. Analysis of low-fat mozzarella made with different combinations of MR-1C, MR-1R, and the non-EPS-producing starter culture strains S. thermophilus TA061 and Lactobacillus helveticus LH100 showed that S. thermophilus MR-1C was responsible for the increased cheese moisture level. To investigate the role of the S. thermophilus MR-1C EPS in cheese moisture retention, the epsE gene in this bacterium was inactivated by gene replacement. Low-fat mozzarella made with L. helveticus LH100 plus the non-EPS-producing mutant S. thermophilus DM10 had a significantly lower moisture content than did cheese made with strains LH100 and MR-1C, which confirmed that the MR-1C capsular EPS was responsible for the water-binding properties of this bacterium in cheese. Chemical analysis of the S. thermophilus MR-1C EPS indicated that the polymer has a novel basic repeating unit composed of d-galactose, l-rhamnose, and l-fucose in a ratio of 5:2:1.
Lactic acid bacteria (LAB) are a diverse group of industrially important, gram-positive, non-spore-forming microbes that produce lactic acid as a major product of carbohydrate fermentation. Many strains of LAB produce extracellular polysaccharides which may be tightly associated with the bacterial cell wall as capsules or liberated into the growth medium as a loose slime (5). The term exopolysaccharide (EPS) has been used to refer to either type of external polysaccharide. EPSs may be homopolysaccharides, composed of a single type of sugar monomer, or heteropolysaccharides, containing several types of sugar monomers (25). Extracellular homopolysaccharides are made by such LAB as Leuconostoc mesenteroides and Streptococcus mutans, while extracellular heteropolysaccharides are produced by several other species of LAB, including Streptococcus thermophilus and Lactobacillus delbrueckii subsp. bulgaricus (6).
The ability to produce EPS is unstable in LAB and may be lost following numerous transfers, prolonged periods of storage, or incubation at temperatures above that optimal for growth (6, 24). This instability of EPS production in mesophilic LAB has been attributed to the fact that the genes involved in polymer production are plasmid encoded. In contrast, genes for EPS production in thermophilic LAB, such as S. thermophilus and L. delbrueckii subsp. bulgaricus, are believed to be chromosomally encoded (6). Consequently, the unstable nature of the EPS phenotype in thermophilic strains is not understood, but it may be related to mobile genetic elements or genomic instability (24).
Because of the ability of EPSs to act as viscosifying, stabilizing, or water-binding agents in various foods, these polymers can act as effective natural alternatives to commercial stabilizers (6). For example, EPS-producing (EPS+) LAB are commonly used as starter cultures for yogurt manufacture because EPS improves the viscosity and texture of yogurt and decreases its susceptibility to syneresis (loss of whey from the curd) (14, 28).
Analysis of cheese microstructure has shown that in full-fat or part-skim mozzarella, the fat and a large portion of the water are located within channels that are formed by fat globules when the cheese curd is heated and stretched (18, 20). In low-fat mozzarella, however, there are very few fat globules to break up the protein matrix, resulting in less space for water retention (20). As a consequence, the cheese has a tough and rubbery texture and requires more heat for melting (19). Merrill et al. showed that procedures which increased moisture levels in reduced- and low-fat mozzarella improved the body, texture, and functional properties of the cheese (19). In addition to enhanced functionality, the ability to increase cheese moisture level (even by as little as 1%) gives processors an important economic advantage in the highly competitive mozzarella industry (27).
Since EPS has the capacity to bind significant amounts of water, it was the hypothesis of our group that EPS+ LAB may be useful for the production of reduced- and low-fat mozzarella. Work by Perry et al. (21) recently showed that an EPS+ starter pair, S. thermophilus MR-1C and L. delbrueckii subsp. bulgaricus MR-1R, could be used to significantly increase moisture levels in low-fat mozzarella. The objectives of this study were to determine whether MR-1C, MR-1R, or both of these strains are required for enhanced moisture retention and to establish the role of EPS in this phenomenon. The results showed that S. thermophilus MR-1C was responsible for the increased cheese moisture level and demonstrated that this effect required the bacterium’s capsular EPS.
(Part of this research was presented at the 92nd Annual Meeting of the American Dairy Science Association, Guelph, Ontario, Canada, 22 to 25 June 1997.)
MATERIALS AND METHODS
Bacterial cultures.
The bacteria and plasmids used in this study are listed in Table 1. All bacterial cultures were incubated at 37°C, stored at 4°C, and maintained by biweekly transfer. S. thermophilus TA061, MR-1C, and DM10 were grown in M17 broth (26) containing 0.5% lactose, while Lactobacillus helveticus LH100 and L. delbrueckii subsp. bulgaricus MR-1R were cultured in MRS broth (8). Escherichia coli strains were grown in Luria-Bertani broth (16) with shaking.
TABLE 1.
Bacteria and plasmids used in this study
| Bacterium or plasmid | Relevant characteristic (source or reference)a |
|---|---|
| Strains | |
| S. thermophilus MR-1C | EPS+ (21) |
| S. thermophilus TA061 | EPS− mozzarella starter (21) |
| S. thermophilus DM10 | EPS− mutant of strain MR-1C (this study) |
| L. delbrueckii subsp. bulgaricus MR-1R | EPS+ (21) |
| L. helveticus LH100 | EPS− mozzarella starter (21) |
| E. coli INVαF′ | Cloning host (Invitrogen); F′ endA1 recA1 hsdR17 (rK− mK+) supE44 thi-1 gyrA96 relA1 φ80lacZΔM15Δ(lacZYA-argF)U169λ− |
| Plasmids | |
| pSA3 | Emr shuttle vector (3, 7) |
| pCR2.1 | Kanr cloning vector (Invitrogen) |
Kanr, kanamycin resistance.
Influence of individual EPS+ starter cultures on cheese moisture level.
To determine whether MR-1R, MR-1C, or both of these strains are required for increased cheese moisture retention, 10-kg vats of low-fat (6% fat) mozzarella were made by the method of Perry et al. (21) with different combinations of EPS+ and non-EPS-producing (EPS−) starters. Each starter pair consisted of a Lactobacillus species (MR-1R or LH100) and an S. thermophilus strain (MR-1C or TA061). Duplicate vats of mozzarella were manufactured on three separate occasions by using the four different starter combinations MR-1C plus MR-1R, MR-1C plus LH100, TA061 plus LH100, and TA061 plus MR-1R. Since the rate of culture acidification can influence cheese moisture composition (21), levels of starter inoculum (between 0.5 and 1.0%) were adjusted to obtain a uniform rate of acid production in all vats. The cheese moisture content was analyzed with a vacuum oven (15) after 1 day of storage at 4°C, and differences in moisture content were statistically evaluated by analysis of variance (ANOVA).
DNA isolation and manipulations.
Plasmid DNA was isolated from S. thermophilus MR-1C by the method of Anderson and McKay (1). Isolation of plasmid DNA from E. coli, restriction enzyme digestions, DNA ligation, and agarose gel electrophoresis were performed as described by Maniatis et al. (16).
Template DNA for PCR experiments was isolated by a method developed by Batt (2). Cells (250 μl) from an overnight culture were collected by centrifugation, washed once in 100 μl of phosphate-buffered saline (140 mM NaCl, 3 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4 [pH 7.4]), and suspended in a solution containing 10 μl of 10× PCR Buffer II (The Perkin-Elmer Corp., Norwalk, Conn.), 85 μl of sterile distilled H2O, and 4 μl of freshly prepared chicken egg white lysozyme (10 mg/ml) (Sigma Chemical Co., St. Louis, Mo.). After a 15-min incubation at 37°C, 1 μl of proteinase K (50 mg/ml) (Sigma Chemical Co.) was added, and the suspension was incubated at 50 to 55°C for 1 h and then boiled for 10 min to inactivate cellular enzymes. The template DNA was stored at −20°C until needed.
The oligonucleotide primers used to amplify fragments of the eps genes were designed with the S. thermophilus Sfi6 eps gene cluster sequence described by Stingele et al. (24) and are shown in Table 2. Thirty PCR cycles were carried out in a model 480 DNA thermal cycler (Perkin-Elmer, Foster City, Calif.) by using the following parameters: denaturation at 96°C for 15 s, annealing for 30 s, and extension at 72°C for 1.5 min. The primer annealing temperature ranged from 39 to 43°C (Table 2). The PCR amplicons were purified with a Bio-Rad (Hercules, Calif.) Prep-a-Gene kit and then sequenced by fluorescent dideoxy chain termination on a Perkin-Elmer Applied Biosystems model 373A automated DNA sequencer. Homology comparisons between S. thermophilus MR-1C and Sfi6 epsA to -F genes and deduced gene products were performed with GeneWorks version 2.3 software (IntelliGenetics, Inc., Mountain View, Calif.).
TABLE 2.
Oligonucleotide primers used for PCRs
| Primer | Sequence (5′ → 3′) | Annealing temp (°C) | Source or reference |
|---|---|---|---|
| epsB-f | TTATTATGGAGGTGAAC | 39 | 24 |
| epsE-r | ATATGCCACCGATTTTT | 39 | 24 |
| epsA-f | TAGTGACAACGGTTGTACTG | 40 | This study |
| epsA-r | GATCATTATGGACTGTCAC | 40 | This study |
| epsCD-f | ATCCAACCAACATATACATCA | 43 | This study |
| epsCD-r | AGGTGGAACAGGACCAGATG | 43 | This study |
| epsF-f | ACCAGATATTGTACATTGTC | 40 | This study |
| epsF-r | TGTCATAGGCTGTCACAAC | 40 | This study |
Suitability of pSA3 as an integration vector for S. thermophilus.
The streptococcus-E. coli shuttle vector pSA3 has been successfully used as a temperature-sensitive integration vector for gene replacement in L. helveticus (3). To determine whether pSA3 could also be used as an integration vector for S. thermophilus, its stability in S. thermophilus MR-1C at 37 and 45°C was investigated. S. thermophilus MR-1C competent cells were prepared as described by Marciset and Mollet (17), and electroporation with pSA3 was done with a Bio-Rad Gene Pulser apparatus set at the following parametric values: field strength, 10.25 kV/cm; resistance, 400 Ω, and capacitance, 25 μF. After electroporation, cells were plated on M17-lactose agar that contained 5 μg of erythromycin/ml and incubated at 37°C for 48 h. Erythromycin-resistant (Emr) colonies were isolated, and the presence of pSA3 in transformants was confirmed by agarose gel electrophoresis. S. thermophilus MR-1C(pSA3) was propagated in M17-lactose broth without erythromycin at 37 and 45°C, and after each transfer, cells were plated on M17-lactose agar with and without erythromycin to determine the fraction of the S. thermophilus MR-1C population that still contained pSA3.
Gene replacement.
To inactivate the MR-1C epsE gene, a 2.4-kbp fragment containing a portion of the epsE gene was obtained by PCR with S. thermophilus MR-1C template DNA and the primers epsB-f and epsE-r. This fragment was ligated into the vector pCR2.1 (Invitrogen, Carlsbad, Calif.) and cloned into E. coli INVαF′. An internal deletion in the cloned epsE gene was generated by digestion with HpaI (New England Biolabs, Beverly, Mass.) and treatment with exonuclease III by the method of Henikoff (13), except that mung bean nuclease (New England Biolabs) was used instead of S1 nuclease to generate blunt ends before ligation. The recombinant plasmid pST100 contained a 0.6-kb deletion in the cloned epsE gene fragment. An integration vector, pST101, was constructed by insertion of the 1.8-kb SphI-BamHI fragment of pST100 into SphI-BamHI-digested pSA3. The recombinant plasmid was introduced into S. thermophilus MR-1C by electroporation as described above. Integration of pST101 into the S. thermophilus MR-1C chromosome was induced by propagation of an Emr transformant, S. thermophilus MR-1C(pST101), in M17-lactose broth that contained 5 μg of erythromycin/ml at the nonpermissive temperature of 45°C. After five transfers under those conditions, the cells were incubated at 37°C to induce a second DNA recombination event that would result in gene replacement or reversion to the wild type (12). The ruthenium red plate assay of Stingele et al. (24) was used to screen colonies for an EPS− phenotype, and the Duguid staining method (11) was used to confirm capsule loss.
EPS composition.
EPS was isolated from S. thermophilus MR-1C by the method of Stingele et al. (24). For the identification of native sugar components, 7 mg of purified EPS was hydrolyzed with 2 ml of 2 M trifluoroacetic acid (TFA) for 1 h at 120°C in a sealed tube. The TFA was removed, and the sugars were converted to their peracetylated aldononitrile (PAAN) derivatives (23) and then analyzed by gas chromatography-mass spectroscopy (GC-MS). The GC-MS was done on a cross-linked methylsilicone capillary column (25-m length, 0.022-cm inside diameter, and 0.1-μm thickness) with a Perkin-Elmer Sigma 3B gas chromatograph coupled to a model 5970 mass selective detector (Hewlett-Packard, Wilmington, Del.) with an electron impact ion source of 70 eV. Helium was used as a carrier gas for all GC analyses. The column temperature was held at 160°C for 3 min and then raised 5°C/min to 185°C. The identity of each compound was determined by matching the mass spectrum to known standards.
By methylation of the intact native polysaccharide prior to TFA hydrolysis, the locations of linkage sites within the polysaccharide can be determined. Thirteen milligrams of the purified polymer was permethylated with sodium methylsulfinyl methanide and methyl iodide in dimethyl sulfoxide (23). The methylated EPS was hydrolyzed with TFA as described above, and the sugars were converted to their corresponding PAAN derivatives. For the separation of these compounds, the GC column temperature was held at 130°C for 3 min, raised 5°C/min to 165°C, and then held at 165°C for 10 min.
Nucleotide sequence accession number.
The MR-1C sequences are available under GenBank accession no. AF053346 to AF053351.
RESULTS
S. thermophilus MR-1C is responsible for increased cheese moisture levels.
Moisture analysis of replicate samples from three independent cheese-making trials revealed that cheeses made with MR-1C plus MR-1R, MR-1C plus LH100, TA061 plus MR-1R, and TA061 plus LH100 contained averages of 62.8, 62.7, 61.5, and 61.4% water, respectively. ANOVA showed that the coccus was the only factor which significantly influenced cheese moisture level (P < 0.01), and cheeses made with S. thermophilus MR-1C had an average of 1.3% more water than those made with S. thermophilus TA061 (Table 3). These data show that S. thermophilus MR-1C alone was responsible for the increased moisture levels noted previously in cheese made with the MR-1C and MR-1R starter pair (21).
TABLE 3.
Results of ANOVA for moisture of low-fat mozzarella as a function of starter culture
| Source | df | Sum of squares | Mean square | F | P |
|---|---|---|---|---|---|
| Model | 5 | 17.674 | 3.535 | 24.41 | 0.0006 |
| Error | 6 | 0.8688 | 0.145 | ||
| Corrected total | 11 | 18.542 | |||
| Rod | 1 | 0.017 | 0.017 | 0.12 | 0.7444 |
| Coccus | 1 | 4.877 | 4.877 | 33.68 | 0.0011 |
| Rod-coccus interaction | 1 | 0.002 | 0.002 | 0.01 | 0.9131 |
Partial characterization of the MR-1C eps gene cluster.
To determine whether the S. thermophilus MR-1C capsular EPS was responsible for the water-binding property of this bacterium, EPS production in MR-1C was inactivated by gene replacement. Stingele et al. recently characterized the eps gene cluster of S. thermophilus Sfi6 and showed that EPS biosynthesis involved at least 13 genes, designated epsA to -M, that are sequentially arranged on a 14-kbp fragment of the Sfi6 chromosome (24). In order to determine whether the MR-1C eps genes were identical to those of Sfi6, PCR was used to isolate fragments from individual S. thermophilus MR-1C eps genes, and then sequence alignments were performed to compare S. thermophilus MR-1C and Sfi6 eps genes and deduced gene products. To determine the orientation of MR-1C eps genes in the chromosome, PCR was performed with primers for the regions from epsA to -D, epsB to -E, epsC to -D, and epsD to -F. Those data showed that the MR-1C eps gene cluster was similar in organization to that of Sfi6 from epsA to epsF. Deduced amino acid sequences for the region from epsA to epsF of S. thermophilus MR-1C indicated that these proteins are between 95 and 99% identical to those in Sfi6 (Table 4). For reasons which are unclear at this time, efforts to characterize the region from epsG to epsM in MR-1C by a similar approach were unsuccessful, but experiments to characterize MR-1C genes downstream of epsF are under way.
TABLE 4.
Homologies between genes involved in EPS biosynthesis in S. thermophilus MR-1C and Sfi6
| Gene | Region sequenceda | % Homology
|
|
|---|---|---|---|
| DNA | Protein | ||
| epsA | 1227–2011 | 98.1 | 97.9 |
| epsB | 2679–3040 | 95.6 | 97.5 |
| epsC | 3330–3579 | 97.2 | 95.2 |
| epsD | 3840–4268 | 98.1 | 96.5 |
| epsE | 4627–5077 | 98.4 | 99.3 |
| epsF | 5656–6227 | 97.9 | 96.0 |
Nucleotide positions are based on S. thermophilus Sfi6 nucleotide sequences (24).
pSA3 stability in S. thermophilus MR-1C.
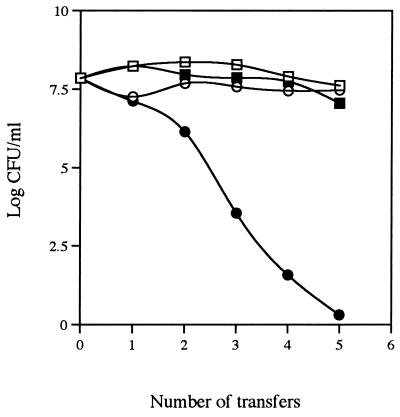
Growth studies demonstrated that pSA3 was stably maintained when S. thermophilus MR-1C(pSA3) was grown at 37°C, but the plasmid was rapidly lost when the bacterium was incubated at 45°C (Fig. 1). These data indicated that pSA3 could be used as a temperature-sensitive integration vector for gene replacement in S. thermophilus MR-1C.
FIG. 1.
Stability of pSA3 in S. thermophilus MR-1C grown at 37°C (▪ and □) and 45°C (• and ○). Closed and open symbols represent cell counts obtained from M17-lactose agar with and without 5 μg of erythromycin per ml, respectively.
Inactivation of the S. thermophilus MR-1C epsE gene.
At the conclusion of the epsE gene replacement experiments, an EPS− mutant of S. thermophilus MR-1C was isolated from milk agar which contained ruthenium red, and the absence of a capsule was confirmed with the Duguid capsule stain. DNA sequence analysis of the EPS− mutant, designated DM10, confirmed that the bacterium contained a frameshift mutation in the epsE gene. Interestingly, API 50 carbohydrate tests (bioMérieux Vitek, Inc., Hazelwood, Mo.) revealed that in addition to its native (wild-type) ability to ferment lactose, glucose, and sucrose, DM10 had acquired the ability to ferment galactose.
MR-1C capsular EPS is required for increased cheese moisture levels.
To establish whether the S. thermophilus MR-1C capsular EPS was responsible for increased moisture retention in low-fat mozzarella, cheese was made with L. helveticus LH100 and either S. thermophilus MR-1C or the MR-1C EPS− mutant, DM10. Moisture analysis showed that cheese made with DM10 and LH100 contained significantly less water (P < 0.01) than did cheese made with MR-1C and LH100 (Table 5).
TABLE 5.
Influence of the S. thermophilus MR-1C capsular EPS on the moisture content of low-fat mozzarella
| Starter combination | Cheese moisture level (%)a
|
||
|---|---|---|---|
| Trial 1 | Trial 2 | Trial 3 | |
| S. thermophilus DM10 + L. helveticus LH100 | 57.7 ± 0.2 | 58.8 ± 0.5 | 58.8 ± 0.1 |
| S. thermophilus MR-1C + L. helveticus LH100 | 61.2 ± 0.0 | 62.8 ± 0.5 | 61.8 ± 0.4 |
Means from duplicate samples ± standard deviations.
Composition of the MR-1C heteropolysaccharide.
The GC-MS analysis showed that the most abundant monosaccharides in the MR-1C capsule were galactose (70.8 mol%) and rhamnose (18.3 mol%). An additional deoxysugar which was eluted at 4.0 min was identified as fucose (6.6 mol%). Traces of glucose and mannose were also found (3.5 and 1.0 mol%, respectively), but these sugars were believed to be contaminants from medium components that were not removed during the EPS purification procedure. This possibility was supported by the absence of any methylation (PAAN) derivatives for these sugars. The identities of the galactose, rhamnose, and fucose PAAN derivatives that were isolated from the S. thermophilus MR-1C EPS are listed in Table 6.
TABLE 6.
Identification of permethylated PAAN derivatives from the S. thermophilus MR-1C polysaccharide
| PAAN methyl sugar | Linkage site(s) | Mol% |
|---|---|---|
| 2,3,4-Tri-O-methyl-l-fucose | 1 | 4.8 |
| 4-Mono-O-methyl-l-rhamnose | 1, 2, 3 | 4.6 |
| 2,3,4,6-Tetra-O-methyl-d-galactose | 1 | 15.8 |
| 2-Mono-O-methyl-l-rhamnose | 1, 3, 4 | 8.64 |
| 2,4,6-Tri-O-methyl-d-galactose | 1, 3 | 33.7 |
| 3,4,6-Tri-O-methyl-d-galactose | 1, 2 | 14.1 |
| 2,3,4-Tri-O-methyl-d-galactose | 1, 6 | 11.7 |
DISCUSSION
This study investigated the individual contributions of the EPS+ bacteria S. thermophilus MR-1C and L. delbrueckii subsp. bulgaricus MR-1R to cheese moisture retention. Analysis of low-fat mozzarella made with EPS+ and EPS− starter combinations showed that S. thermophilus MR-1C, a strain known to produce a large capsular EPS, was exclusively responsible for the increased moisture level previously observed in cheese made with an MR-1C and MR-1R starter pair (21). To determine whether the S. thermophilus MR-1C capsule is essential for the water-binding property of this bacterium, the epsE gene of MR-1C was inactivated by gene replacement. Mozzarella made with L. helveticus LH100 and the EPS− mutant, S. thermophilus DM10, had significantly lower moisture level than did cheese made with LH100 and S. thermophilus MR-1C. The difference in the moisture contents of the cheeses made with MR-1C and with DM10 (Table 5) was greater than that noted for the cheeses made with TA061 and with MR-1C (Table 3). Because TA061 is not an isogenic derivative of MR-1C, and the rates of starter acidification in all cheeses were similar, the basis for this difference remains unknown. One possibility is that even though TA061 is phenotypically EPS−, this strain may still produce some type of cell surface carbohydrate that binds water. Nonetheless, the data in Table 3 demonstrate that the highest cheese moisture levels were attained by the addition of strain MR-1C rather than strain TA061, and the data in Table 5 show that this effect was due to the MR-1C capsular EPS.
Compositional analysis of the MR-1C capsule indicated that it had an octameric repeating unit composed of d-galactose, l-rhamnose, and l-fucose in a ratio of 5:2:1. Fucose has not been previously reported to occur in EPSs from LAB, but a polysaccharide from S. thermophilus OR 901 recently described by Bubb et al. (4) was found to contain galactose and rhamnose in a ratio of 5:2. Examination of EPS methylation data for the MR-1C capsule suggested that this EPS may be structurally similar to the OR 901 EPS. The OR 901 and MR-1C EPS each contained a nonreducing terminal galactose. A 2-methyl-l-rhamnose PAAN was detected in MR-1C, and this sugar is also found in the OR 901 EPS at the junction of the backbone and the digalactose side chain. The 2,4,6-trimethyl-d-galactose PAAN found in the MR-1C sample represents a galactose linked at its C-3 position, and the OR 901 heptasaccharide repeat unit has two backbone galactose moieties with linkages at this position. In addition, the MR-1C EPS yielded 2,3,4-trimethyl-d-galactose PAAN and 3,4,6-trimethyl-d-galactose PAAN, which are indicative of galactose moieties substituted at the C-6 position and linked at the C-2 position, respectively. The OR 901 EPS also includes galactose units with these two types of linkages in their structures. The fucose moiety from the MR-1C sample was a 2,3,4-trimethyl-l-fucose PAAN, indicating that it is located at the end of a side chain rather than at an internal linkage position. The presence of a 4-methyl-l-rhamnose PAAN in MR-1C indicated that the fucose in MR-1C may be attached to the rhamnose residue which is unsubstituted in the OR 901 polymer (4). This theory is supported by the finding that the 4-methyl-l-rhamnose PAAN and 2,3,4-trimethyl-l-fucose PAAN in MR-1C occurred in essentially equal molar ratios (Table 6).
Based on the similarities between the sugars in the S. thermophilus OR 901 and MR-1C EPSs, we propose that the S. thermophilus MR-1C EPS has a basic repeating unit very similar to that described by Bubb et al. (4), except that l-fucose is substituted on the terminal l-rhamnose residue (Fig. 2).
FIG. 2.
Proposed structure of the S. thermophilus MR-1C capsular heteropolysaccharide. Abbreviations: Gal, galactose; Rha, rhamnose; Fuc, fucose.
An interesting and unexpected attribute of S. thermophilus DM10 was the expression of galactose fermention (Gal+). Strains of S. thermophilus typically are unable to ferment galactose, yet genes encoding the key Leloir enzymes galactokinase (GalK), galactose-1-phosphate uridyltransferase (GalT), uridine diphospho-galactose-4-epimerase (GalE), and mutarotase (GalM) have been isolated from this species (9, 10). This finding has led to suggestions that in S. thermophilus, the Leloir enzymes may function for the production of uridinediphosphosugar precursors for EPS biosynthesis (22). Nevertheless, De Vos (9) has reported that Gal+ S. thermophilus can be obtained by mutations in the gal promoter region which increase transcriptional activity and suggested that the Gal+ phenotype of this bacterium was due to a very low level of expression of Leloir enzymes. The S. thermophilus epsE gene is thought to encode the galactosyltransferase that catalyzes the first step in the assembly of the EPS basic repeating unit, i.e., transfer of galactose-1-phosphate from UDP-galactose to the undecaprenyl-phosphate lipid carrier (24). For this reason, inactivation of epsE may be expected to result in an accumulation of UDP-galactose within the cell. Since overproduction of phosphorylated sugars can be toxic, the survival and growth of S. thermophilus DM10 might require a Gal+ phenotype. Experiments are now under way to determine whether the acquisition of this phenotype was a primary (e.g., UDP-galactose was an inducer of the gal operon) or secondary (e.g., coincidental mutation in the gal promoter region) effect of epsE inactivation in S. thermophilus DM10.
ACKNOWLEDGMENTS
This work was supported in part by Dairy Management, Inc. (through the Western Dairy Center), a Gandhi graduate fellowship to D. Low, the Utah Agricultural Experiment Station, and Weber State University College of Science sabbatical assistance to D. Horne.
We thank the Utah State University Biotechnology Center for automated DNA sequence analysis.
Footnotes
Journal paper 6079 of the Utah Agricultural Experiment Station, Utah State University.
REFERENCES
- 1.Anderson D G, McKay L L. Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl Environ Microbiol. 1983;46:549–552. doi: 10.1128/aem.46.3.549-552.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Batt, C. A. 1995. Personal communication.
- 3.Bhowmik T, Fernández L, Steele J L. Gene replacement in Lactobacillus helveticus. J Bacteriol. 1993;175:6341–6344. doi: 10.1128/jb.175.19.6341-6344.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bubb W A, Urashima T, Fujiwara R, Shinnai T, Ariga H. Structural characterisation of the exocellular polysaccharide produced by Streptococcus thermophilus OR 901. Carbohydr Res. 1997;301:41–50. doi: 10.1016/s0008-6215(97)00083-9. [DOI] [PubMed] [Google Scholar]
- 5.Cerning J. Production of exopolysaccharides by lactic acid bacteria and dairy propionibacteria. Lait. 1995;75:463–472. [Google Scholar]
- 6.Cerning J. Exocellular polysaccharides produced by lactic acid bacteria. FEMS Microbiol Rev. 1990;87:113–130. doi: 10.1111/j.1574-6968.1990.tb04883.x. [DOI] [PubMed] [Google Scholar]
- 7.Dao M L, Ferretti J J. Streptococcus-Escherichia coli shuttle vector pSA3 and its use in the cloning of streptococcal genes. Appl Environ Microbiol. 1985;49:115–119. doi: 10.1128/aem.49.1.115-119.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeMan J, Rogosa M, Sharpe M E. A medium for the cultivation of lactobacilli. J Appl Bacteriol. 1960;23:130–135. [Google Scholar]
- 9.De Vos W M. Metabolic engineering of sugar catabolism in lactic acid bacteria. Antonie Leeuwenhoek. 1996;70:223–242. doi: 10.1007/BF00395934. [DOI] [PubMed] [Google Scholar]
- 10.De Vos W M, Vaughan E E. Genetics of lactose utilization in lactic acid bacteria. FEMS Microbiol Rev. 1994;15:217–237. doi: 10.1111/j.1574-6976.1994.tb00136.x. [DOI] [PubMed] [Google Scholar]
- 11.Duguid J P. The demonstration of bacterial capsules and slime. J Pathol Bacteriol. 1951;63:673. doi: 10.1002/path.1700630413. [DOI] [PubMed] [Google Scholar]
- 12.Hamilton C M, Aldea M, Washburn B K, Babitzke P, Kushner S R. New method for generating deletions and gene replacements in Escherichia coli. J Bacteriol. 1989;171:4617–4622. doi: 10.1128/jb.171.9.4617-4622.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henikoff S. Unidirectional digestion with exonuclease III in DNA sequence analysis. Methods Enzymol. 1987;155:156–165. doi: 10.1016/0076-6879(87)55014-5. [DOI] [PubMed] [Google Scholar]
- 14.Kang K S, Cottrell I W. Polysaccharides. In: Peppler H J, Perlman D, editors. Microbial technology: microbial processes. 2nd ed. Vol. 1. New York, N.Y: Academic Press, Inc.; 1979. pp. 417–481. [Google Scholar]
- 15.Kosikowski F V. Cheese and fermented milk foods. 2nd ed. Brooktondale, N.Y: Kosikowski and Associates; 1982. [Google Scholar]
- 16.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 17.Marciset O, Mollet B. Multifactorial experimental designs for optimizing transformation of Streptococcus thermophilus. Biotechnol Bioeng. 1994;43:490–496. doi: 10.1002/bit.260430609. [DOI] [PubMed] [Google Scholar]
- 18.McMahon D J, Oberg C J, McManus W. Functionality of mozzarella cheese. Aust J Dairy Technol. 1993;48:99–104. [Google Scholar]
- 19.Merrill R K, Oberg C J, McMahon D J. A method for manufacturing reduced fat mozzarella cheese. J Dairy Sci. 1994;77:1783–1789. [Google Scholar]
- 20.Oberg C J, Broadbent J R. Thermophilic starter cultures: another set of problems. J Dairy Sci. 1993;76:2392–2406. [Google Scholar]
- 21.Perry D B, McMahon D J, Oberg C J. Effect of exopolysaccharide producing cultures on moisture retention in low-fat mozzarella cheese. J Dairy Sci. 1997;80:799–805. [Google Scholar]
- 22.Poolman B. Energy transduction in lactic acid bacteria. FEMS Microbiol Rev. 1993;12:125–147. doi: 10.1111/j.1574-6976.1993.tb00015.x. [DOI] [PubMed] [Google Scholar]
- 23.Seymour F, Plattner R D, Slodki M E. Gas-liquid chromatography-mass spectrometry of methylated and deuteriomethylated per-O-acetyl-aldonitriles from d-mannose. Carbohydr Res. 1975;44:181–198. [Google Scholar]
- 24.Stingele F, Neeser J-R, Mollet B. Identification and characterization of the eps (exopolysaccharide) gene cluster from Streptococcus thermophilus Sfi6. J Bacteriol. 1996;178:1680–1690. doi: 10.1128/jb.178.6.1680-1690.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sutherland I W. Bacterial exopolysaccharides. Adv Microb Physiol. 1972;8:143–212. doi: 10.1016/s0065-2911(08)60190-3. [DOI] [PubMed] [Google Scholar]
- 26.Terzaghi B E, Sandine W E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975;29:807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thunell, R. K. 1998. Personal communication.
- 28.van den Berg D J C, Robijn G W, Janssen A C, Giuseppin M L F, Vreeker R, Kamerling J P, Vliegenthart J F G, Ledeboer A M, Verrips C T. Production of a novel extracellular polysaccharide by Lactobacillus sake 0-1 and characterization of the polysaccharide. Appl Environ Microbiol. 1995;61:2840–2844. doi: 10.1128/aem.61.8.2840-2844.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]