Abstract
The identity of the physiologically important Cry1A receptor protein(s) in the lepidopteran Manduca sexta has been a matter of dispute due to the multiple proteins which bind the Cry1Ac toxin. Cry1Aa, Cry1Ab, and Cry1Ac exhibit essentially identical toxicities toward M. sexta larvae and show a high degree of sequence and presumed structural identities. These similarities make it likely that there is a common mechanism of toxicity in these lepidopteran-specific toxins in terms of both mode of action and the receptor proteins through which these toxins exert their lepidopteran-specific toxicity. Investigators in our laboratory previously demonstrated that the cloned 210-kDa glycoprotein BT-R1 binds all three Cry1A toxins (T. P. Keeton and L. A. Bulla, Jr., Appl. Environ. Microbiol. 63:3419–3425, 1997). This protein remains a common binding protein even after being subjected to various midgut membrane preparation and processing protocols. The method used to isolate proteins from the M. sexta larval midgut in no significant way affects the results of ligand binding and vacuum blotting experiments, and we have been unable to detect specific, high-affinity binding of any Cry1A toxin to Cry1Ac binding proteins other than BT-R1. Alterations in blot substrate and blocking, hybridization, and washing buffers support these conclusions. Collectively, these results indicate that in M. sexta the cadherin-like BT-R1 protein is a common high-affinity receptor protein for the Cry1A family of toxins.
Parasporal crystalline inclusions of Bacillus thuringiensis subspecies are among the most promising bacterial biopesticides available for use today. As a whole, these proteins (B. thuringiensis toxins) demonstrate great specificity toward certain orders of insects and, to date, have shown no known side effects for nontarget animals. Currently, one of the major drawbacks to the use of B. thuringiensis toxins as externally applied biopesticides is their lack of persistence in the field due to factors such as rain washout and degradation by UV irradiation. In part, these problems are being addressed by the production of transgenic food and textile crops expressing B. thuringiensis toxin genes in their own tissues. Agricultural biotechnology companies are pursuing these transgenic methodologies in the hope of producing food and textile crops which will be resistant to major insect pests without the need for externally applied pesticides.
To date, field trials of such plants have resulted in mixed success due, in part, to one of the most attractive advantages of the toxins as externally applied pesticides, i.e., a narrow spectrum of toxicity. This situation can be addressed by the engineering of transgenic plants expressing more than one toxin or by the use of novel B. thuringiensis subspecies producing toxins with multiple specificities. As the use of transgenic crops increases, however, insect resistance may become a problem. A few species of insects have already demonstrated increased tolerance for B. thuringiensis toxins, either in the field or in the laboratory. As suggested in these studies, decreased susceptibility to B. thuringiensis toxins may be due to a variety of factors, including alterations in insect gut physiology (14, 30, 37) or alterations of the ligand binding characteristics of the toxin receptor(s) involved (8, 32, 47).
To understand the development of receptor-mediated resistance, investigators first need to identify and characterize the physiologically important receptor molecules for each class of B. thuringiensis toxins from a background of low-affinity toxin binding proteins. That specific protein receptors are involved in Cry toxin killing of target insects has been known since the mid-1980s. Studies of the binding of radiolabeled Cry toxins in suspensions of insect midgut proteins isolated by various procedures have generated a rather extensive list of putative receptor molecules (4, 7, 15, 26, 41, 45, 46) without simultaneously identifying the binding protein(s) in question by use of midgut protein sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) blots. SDS-PAGE blots, when incubated with radiolabeled toxins, at least permit a visual estimation of both the number of proteins involved and their molecular masses. Ligand blots of Manduca sexta midgut proteins have been used successfully to identify and partially characterize for this particular insect binding proteins for the Cry1A lepidopteran-specific toxins (3, 10, 11, 19, 23, 24, 33, 42, 43).
Cry1Aa, Cry1Ab, and Cry1Ac demonstrate 82 to 90% amino acid identity to one another and, when compared directly, exhibit indistinguishable toxicities toward M. sexta larvae (16, 17, 45). Cry1Aa and Cry1Ab recognize a single midgut protein in M. sexta, the 210-kDa cadherin-like glycoprotein BT-R1 (42, 43). For Cry1Ac, however, at least two populations of receptor protein have been identified by SDS-PAGE ligand blot analyses of whole midgut protein preparations, including BT-R1 (10, 19, 28, 33) and other proteins with molecular masses ranging from 85 to 120 kDa (5, 11, 19, 21, 33, 44). BT-R1 and at least two aminopeptidases of approximately 120 kDa have now been subjected to partial purification, and their ligand binding characteristics have been described in some detail (10, 12, 13, 29, 34, 39). Both BT-R1 and a 120-kDa aminopeptidase from M. sexta have also been cloned, and their complementary DNA sequences have been reported (22, 43).
BT-R1 has been shown to specifically bind with high affinity the three tested Cry1A toxins (Cry1Aa, Cry1Ab, and Cry1Ac) both in M. sexta midgut protein preparations (19, 33) and in heterologous cell cultures expressing the BT-R1 cDNA (19). BT-R1 is also the only Cry1A binding protein that has been shown to have Cry1A-specific ligand binding characteristics when expressed in mammalian and insect cell cultures. This glycoprotein binds Cry1Aa, Cry1Ab, and Cry1Ac with extremely high and virtually equal affinities and specificities in both heterologous and homologous competition binding experiments with membrane proteins prepared from M. sexta larval midguts and transiently transfected Sf21 insect cells (19). The experiments in that report were the first to show a positive correlation between the binding affinities of M. sexta midgut protein suspensions and the identity of a single binding protein from whole midgut protein preparations immobilized on polyvinylidene difluoride (PVDF) filters. This correlation is of paramount importance for understanding the conflicting reports that appear when binding data are indirectly compared with the identification of a given binding protein on ligand blots, as pointed out recently by Lee and Dean (27).
To resolve the confusion surrounding the identification of the relevant M. sexta midgut protein receptor(s) which binds the Cry1A toxins of B. thuringiensis, including Cry1Ac, we have designed experiments with different ligand binding protocols to determine whether various procedures affect toxin binding by M. sexta midgut proteins. The results of our work clearly demonstrate that BT-R1 is a common high-affinity receptor for the Cry1A B. thuringiensis toxins and that the binding properties of BT-R1 are not affected by any procedures commonly accepted and used to study ligand-receptor properties of lepidopteran insects.
MATERIALS AND METHODS
Toxin purification.
Recombinant protoxins Cry1Aa, Cry1Ab, and Cry1Ac (Bacillus Genetic Stock Center, Ohio State University, Columbus) were prepared from Escherichia coli JM103 and trypsinized essentially as described by Lee et al. (26). In addition, the soluble trypsinized 60-kDa toxins were subjected to fast protein liquid chromatography (FPLC) NaCl gradient purification over an HR-5/5 Mono-Q anion-exchange column (Pharmacia) prior to quantitation, radioiodination, and use in bioassays. All toxin protein quantitations were performed by the bicinchoninic acid method (Pierce Chemical Co.) with bovine serum albumin (BSA; fraction V) as a standard.
Radioiodination.
Cry toxins were iodinated as described by Keeton and Bulla (19) by the chloramine-T method with 125I-Na purchased from NEN-DuPont. Ten micrograms of toxin was mixed with 5 μl of 125I-Na (approximately 0.5 mCi) in 100 μl of sodium phosphate buffer (100 mM, pH 7.0). To this mixture 100 μg of chloramine-T was added and allowed to react for 20 s with constant mixing. Label incorporation was halted by the addition of 200 μg of sodium metabisulfite in 50 μl of distilled H2O, and nonincorporated 125I was removed by use of a 2-ml Excellulose desalting column (Pierce) blocked with BSA and equilibrated with phosphate-buffered saline (PBS).
Insect rearing and processing.
M. sexta eggs were purchased from Carolina Biologicals, and the larvae were reared on Carolina Biologicals artificial diet at 30°C under continuous lighting conditions. Midguts used in this and subsequent preparations were excised from fifth-instar larvae that had been chilled on ice for 10 min prior to excision of the midguts. The peritrophic membrane was removed from the lumen of the gut, and the remaining tissue was washed briefly in ice-cold homogenization buffer (see individual preparations below). Proteins were prepared according to the protocols outlined below.
Adamo membrane protein preparation.
Membrane proteins from both isolated insect midguts and cell cultures were prepared as described by Adamo et al. (1), and total protein was determined by the bicinchoninic acid method. Briefly, tissues or cells were homogenized on ice in a tightly fitting glass Dounce homogenizer containing 10 volumes of a hypotonic buffer composed of 5 mM Tris-HCl (final pH, 7.4) supplemented with 1 mM phenylmethylsulfonyl fluoride (PMSF) and 3 mM dithiothreitol (DTT). The mixture was then diluted with an equal volume of ice-cold 5 mM Tris-HCl (final pH, 7.4) containing 5 mM EDTA, 1 mM PMSF, 3 mM DTT, and a cocktail of protease inhibitors (aprotinin, 10 μg/ml; benzamidine, 1 mM; leupeptin, 1 μg/ml). Low-speed centrifugation (1,000 × g for 10 min in a Beckman JA-20 rotor) was used to pellet heavier cellular debris, and a second, lighter fraction was pelleted by ultracentrifugation (100,000 × g for 30 min in a Beckman SW60 Ti rotor). The resulting high-speed pellet, which contained essentially all detectable BT-R1 binding activity, was suspended in 10 mM HEPES (final pH, 7.4) containing 10% glycerol, 130 mM KCl, and 3 mM DTT prior to being flash frozen in liquid nitrogen and stored at −80°C.
English-Readdy membrane protein preparation.
Midguts were isolated and prepared as described by English and Readdy (6). Dissected midguts were suspended in ice-cold 50 mM sucrose–1 mM PMSF–2 mM Tris-HCl (final pH, 7.4) and homogenized on ice. CaCl2 was added to a final concentration of 10 mM, followed by a 15-min incubation on ice. The homogenate was then centrifuged at 4,300 × g in a Beckman JA-20 rotor for 10 min at 4°C, and the supernatant was transferred to a fresh tube and centrifuged at 27,000 × g for 10 min. The final pellet was suspended in 10 mM HEPES (final pH, 7.4) containing 130 mM KCl and 10% glycerol, flash frozen in liquid nitrogen, and stored at −80°C.
Wolfersberger membrane protein preparation.
Midguts were homogenized in 10 volumes of ice-cold buffer (48) consisting of 300 mM mannitol, 5 mM EGTA, and 17 mM Tris base (final pH, 7.5) and supplemented with the protease inhibitor cocktail described above in a glass Dounce homogenizer. An equal volume of 24 mM MgCl2 solution was added and mixed thoroughly with the homogenate. This mixture was allowed to stand for 15 min on ice. Following low-speed centrifugation (2,500 × g for 10 min in a Beckman JA-20 rotor), the supernatant was removed, transferred to a fresh tube, and centrifuged for 30 min at 30,000 × g in the same rotor. The final pellet was suspended in 100 mM HEPES (pH 7.4), flash frozen in liquid nitrogen, and stored at −80°C.
Transient expression of BT-R1 cDNA in insect cell cultures.
The BT-R1 cDNA was expressed in insect cell cultures essentially as described by Keeton and Bulla (19). Cell culture flasks (25 cm2; Corning) were seeded with 3 × 106 Spodoptera frugiperda Sf21 cells in TNM-FH medium (Grace’s insect medium with 3.3 g of lactalbumin hydrolysate and 3.3 g of Yeastolate per liter; all ingredients purchased from Invitrogen; supplemented with 10% heat-inactivated fetal bovine serum from JRH Biosciences) 1 h prior to the addition of DNA. Sterile DNA was added to 25 mM HEPES (final pH, 7.1) containing 140 mM NaCl and 125 mM CaCl2. A precipitate was formed when this cocktail was added to cells which had been transferred into Grace’s insect medium (without supplements but containing 10% heat-inactivated fetal bovine serum) immediately prior to the addition of DNA. Cells were incubated with the DNA precipitate at 27°C for 4 h, at which time the transfection medium was washed from the cells and the cells were returned to TNM-FH medium. Membranes were prepared by the membrane preparation procedure of Adamo et al. (1) 48 h following transfection.
SDS-PAGE ligand blotting.
For ligand blotting studies, proteins were denatured by being boiled in SDS loading buffer and separated by electrophoresis through discontinuous SDS–7.5 or 10.0% polyacrylamide gels as described by Keeton and Bulla (19). Separated proteins were blotted to PVDF membranes (Millipore) with a Panther semidry electroblotter (Owl Scientific). Transfers were conducted with sheets of Whatman 3MM paper soaked in a buffer composed of 10 mM 3-(cyclohexylamino)-1-propanesulfonic acid (CAPS; pH 10.0) and 10% methanol for 90 min at room temperature (approximately 23°C). Blots were blocked with a variety of buffers for 2 h at room temperature or overnight at 4°C, incubated with 125I-Cry1A toxins (approximately 1 nM) for 2 h at room temperature, washed three times (5 to 30 min each), and used to expose Kodak X-Omat AR autoradiography film at −80°C. Our standard blocking buffer consisted of 0.5% Tween 20 (polyoxyethelene sorbitan monolaurate), 10 mM Tris base, 150 mM NaCl, 5% glycerol, 5% nonfat milk powder, and 0.025% sodium azide (final pH, 8.0).
Vacuum blotting.
Nondenatured whole midgut membrane proteins were transferred to nitrocellulose filters by vacuum blotting with a Minifold II apparatus and protocol (Schleicher & Schuell). Briefly, 10 μg of either midgut proteins or Sf21 cell membrane proteins was diluted in ice-cold PBS, and the mixture was placed in the Minifold wells just as vacuum was applied. This suspension was drawn through a distilled H2O-wetted nitrocellulose membrane, and the membrane was air dried for 15 min prior to being dipped in our standard nonfat milk blocking buffer. Solubilized, FPLC-fractionated midgut proteins (10) were vacuum blotted to nitrocellulose in the same manner. All filters then were hybridized with 125I-labeled toxins as described above.
Quantitation of 125I-labeled toxin bound to ligand blots.
Following exposure to X-Omat AR film, ligand blots were cut into strips and placed in a Beckman Gamma 5500 gamma counter. Radioactivity corresponding to the identified bands was counted and tabulated relative to the amount of 125I-labeled toxin bound to BT-R1 in the control lane as specified below (see Table 1). After identical sampling, clean areas of each blot were subtracted as background.
TABLE 1.
125I-labeled Cry1A toxin bound to 210- and 120-kDa proteins under different ligand blot conditions
| Membrane | Detergent | Blocking agent | % of the following toxin bound to the indicated protein:
|
|||
|---|---|---|---|---|---|---|
| Cry1Aca
|
Cry1Abb
|
|||||
| 210 kDa | 120 kDa | 210 kDa | 120 kDa | |||
| PVDF | None | 5% Milk | <1 | 20 | 5 | <1 |
| PVDF | 0.05% Tween 20 | 5% Milk | 95 | 80 | ||
| PVDF | 0.1% Tween 20 | 5% Milk | 100c | 85 | 100c | 2 |
| PVDF | 0.5% Tween 20 | 5% Milk | 120 | 75 | ||
| PVDF | 0.1% Deoxycholate | 5% Milk | 20 | 15 | 50 | <1 |
| PVDF | 0.1% Triton X-100 | 5% Milk | 95 | 40 | 55 | <1 |
| PVDF | 0.1% Nonidet P-40 | 5% Milk | 125 | 65 | 80 | <1 |
| PVDF | 0.1% SDS | 5% Milk | 90 | 55 | 90 | <1 |
| PVDF | 0.1% CHAPS | 5% Milk | 20 | 15 | 15 | <1 |
| PVDF | 0.1% Tween 20 | 3% BSA | 255 | 225 | ||
| PVDF | 0.1% Tween 20 | 2% Gelatin | 55 | 65 | ||
| Nitrocellulose | 0.1% Tween 20 | 5% Milk | 50 | 90 | ||
Data are compiled from ligand blots described in the text.
Radioactivity values were calculated relative to the amount of 125I-labeled toxin bound to the 210-kDa band (taken as 100%) when PVDF membranes and blocking and washing buffers containing 0.1% Tween 20 and 5% nonfat milk were used.
RESULTS
Binding of Cry1A toxins to M. sexta midgut membrane proteins prepared by three different procedures.
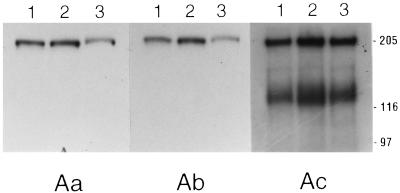
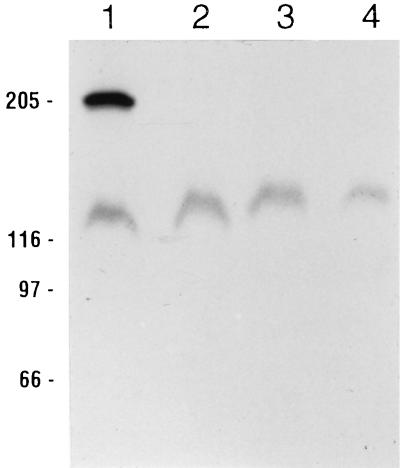
M. sexta midgut proteins were prepared by three different protocols prior to SDS-PAGE ligand blotting. 125I-labeled Cry1Aa, Cry1Ab, and Cry1Ac were then used as probes on identical M. sexta ligand blots of the prepared midgut proteins. As demonstrated in Fig. 1, there were no appreciable differences among the three preparations with regard to binding protein(s) detected by each toxin; Cry1Aa and Cry1Ab bound BT-R1 (210 kDa), whereas Cry1Ac bound BT-R1 as well as several proteins of lower molecular masses. We therefore did not perform the remaining experiments with all three buffer systems. To investigate the relative affinities of each of the midgut membrane proteins for the radiolabeled ligands, we carried out homologous and heterologous competition ligand blot experiments with 125I-labeled Cry1Ac in the presence of unlabeled Cry1Aa, Cry1Ab, and Cry1Ac as competing ligands (Fig. 2). The unlabeled competing ligands were present at 1,000 nM, or an approximately 1,000-fold excess relative to radiolabeled Cry1Ac. Although the number and relative intensities of bands that bound 125I-labeled Cry1Ac differed in this case from a previous report from our laboratory (19), the results in Fig. 2 do agree with our previous report in that only the 210-kDa band, corresponding to BT-R1, competed for binding in the presence of an approximately 1,000-fold excess of unlabeled competitor. Lanes 2 through 4 of Fig. 2 show that whereas the binding of 125I-labeled Cry1Ac to 210-kDa BT-R1 was undetectable in the presence of a 1,000-fold excess of unlabeled Cry1Aa, Cry1Ab, and Cry1Ac, binding to the ∼120-kDa protein in the presence of these unlabeled toxins was relatively unaffected. Previously, we demonstrated that the binding of 125I-labeled Cry1Ac toxin to BT-R1 was virtually eliminated in the presence of a 10-fold excess of unlabeled Cry1Ac (19). It is obvious from the results compiled in Fig. 1 and 2 that for a given Cry1A toxin, the three common midgut protein preparations produced essentially identical ligand binding results.
FIG. 1.
SDS-PAGE ligand blots of M. sexta midgut proteins. Membrane proteins were prepared by the Adamo (lane 1), English-Readdy (lane 2), or Wolfersberger (lane 3) method. Midgut proteins (50 μg) were solubilized in SDS loading buffer, separated by SDS-PAGE, and blotted semidry to PVDF filters. Identical blots were blocked, hybridized, and washed (see Materials and Methods) with Tween 20-TBS-5% nonfat milk buffer (pH 8.0). Panels Aa, Ab, and Ac show results obtained from hybridization with 125I-labeled Cry1Aa, Cry1Ab, and Cry1Ac, respectively. Positions of molecular size markers (in kilodaltons) are indicated on the right.
FIG. 2.
Competition ligand blot of M. sexta midgut proteins and Cry1Ac. M. sexta midgut proteins (100 μg) prepared by the Wolfersberger method were separated by SDS-PAGE and blotted to PVDF filters. Lanes were then cut into strips and hybridized with 125I-labeled Cry1Ac (lane 1) or a combination of 125I-labeled Cry1Ac and an approximately 1,000-fold excess of unlabeled competitor. Competing ligands in lanes 2 through 4 were Cry1Aa, Cry1Ab, and Cry1Ac, respectively. Positions of molecular size markers (in kilodaltons) are indicated on the left.
Results from various laboratories, including our own (10, 19), regarding the binding of Cry1Ac to whole midgut membrane protein preparations differ considerably. Having demonstrated that membrane preparation procedures alone are not responsible for such differences, we examined other parameters in experimental design that may be the cause of variation. In particular, we were interested in determining whether there were any conditions under which Cry1Ac bound preferentially to the lower-molecular-mass proteins relative to BT-R1. The variables included the extent to which blots were washed following hybridization, the kind of blocking proteins used, the composition of the blocking buffer detergent, the blotting procedure, and the substrate used for blotting.
Effect of blot hybridization and washing upon binding of Cry1Ac toxin to M. sexta midgut proteins.
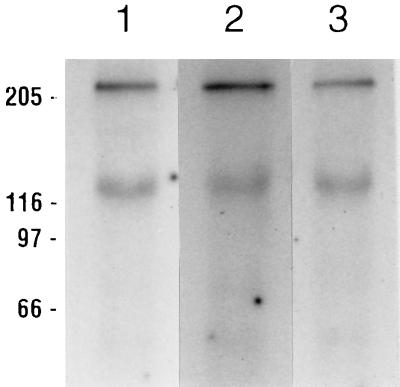
Figure 3 shows the results obtained from an experiment in which the effects of various washing techniques on identical blots of Adamo-prepared M. sexta midgut proteins were compared. We could not detect any differences among experiments performed with or without nonfat milk proteins or Tween 20 in the hybridization and washing procedures. In this experiment, three lanes of an SDS-polyacrylamide gel of Adamo-prepared M. sexta midgut proteins were transferred to a PVDF filter, which was then cut into strips. All three strips were blocked in our standard Tween 20–Tris-buffered saline (TBS)–nonfat milk buffer (10, 19, 42, 43). The ligand blots in lane 1 of Fig. 3 were hybridized with 125I-labeled Cry1Ac and washed with this same buffer. Lane 2 of Fig. 3 is a strip hybridized and washed with Tween 20-TBS without the nonfat milk proteins, and the strip in lane 3 was hybridized and washed with TBS alone. It is clear that there were no significant differences in the results obtained, despite the different hybridization and washing procedures used.
FIG. 3.
Comparison of various hybridization and washing conditions for M. sexta midgut ligand blots with 125I-labeled Cry1Ac. Midgut proteins were prepared by the Adamo method, and 50 μg per lane was separated by SDS-PAGE, blotted to PVDF filters, and blocked overnight with Tween 20-TBS-5% nonfat milk buffer (pH 8.0). Hybridizations were then performed for 2 hours with either the same buffer, a buffer composed of TBS without milk proteins but supplemented with 0.05% Tween 20, or TBS alone. Three washes with a buffer of the same composition followed, each approximately 5 min at room temperature. Hybridization and washing conditions were Tween 20-TBS-5% nonfat milk buffer (lane 1), Tween 20-TBS (lane 2), and TBS alone (lane 3). Positions of molecular size markers (in kilodaltons) are indicated on the left.
Comparison of blocking proteins.
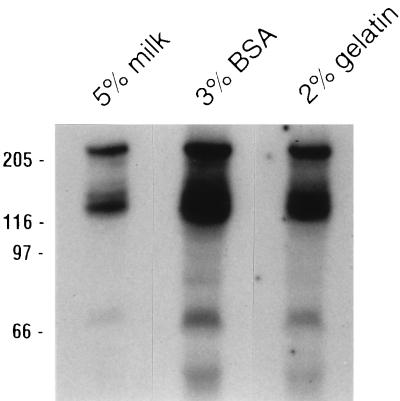
Next, we investigated different blocking agents which may alter results based on their various blocking capacities with PVDF. The most commonly used blocking proteins are those found in nonfat dried milk, often used at a 5% (wt/vol) concentration, although some laboratories use BSA or gelatin as filter blocking agents. An SDS-PAGE ligand blot of three identical lanes was cut into strips and blocked, hybridized, and washed with Tween 20-TBS containing one of the aforementioned blocking agents (Fig. 4). In Fig. 4, lane 1 was blocked, hybridized, and washed with 5% nonfat dry milk, lane 2 was processed with 3% BSA, and lane 3 was processed with 2% gelatin. As was true for the various hybridization and washing conditions, blocking agents had no influence on the relative abilities of 210-kDa BT-R1 and the other Cry1Ac binding proteins to bind Cry1Ac, although large differences in the overall amount of labeled toxin which bound to the blots were detected. Presumably this finding was due to the various overall abilities of the agents to block binding to proteins as well as to the PVDF filter itself.
FIG. 4.
Comparison of Cry1Ac binding to M. sexta midgut proteins in the presence of different blocking reagents. Proteins (80 μg) prepared by the Wolfersberger method were separated by SDS-PAGE following solubilization in SDS loading buffer. Following semidry blotting to PVDF filters, lanes were cut into strips and blocked overnight with Tween 20-TBS buffers containing various blocking agents (listed above the lanes). Following hybridization with 125I-labeled Cry1Ac for 2 h, the strips were washed three times, 5 min each, with the same buffers. Positions of molecular size markers (in kilodaltons) are indicated on the left.
Comparison of various blocking buffer detergents.
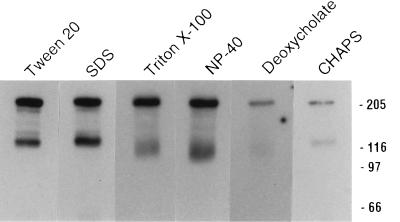
An important component of most blocking and hybridization buffers used for Cry toxin ligand blot studies is some form of detergent, which can be an ionic or a nonionic agent, including zwitterionic molecules. In most published work involving the Cry1A toxins and M. sexta, the nonionic detergent Tween 20 is used in concentrations ranging from 0.01 to 0.5%. To investigate the effect of various detergents on Cry1Ac toxin ligand blotting, multiple identical M. sexta midgut protein blots were blocked, hybridized, and washed with buffers consisting of Tris-buffered saline (final pH, 7.4), 5% glycerol, 5% nonfat milk powder, and a 0.1% concentration of one of the following detergents: (i) Tween 20, (ii) SDS, (iii) Triton X-100, (iv) Nonidet P-40, (v) deoxycholate, and (vi) 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS). The results of these experiments are shown in Fig. 5; lane 1 is the result of an experiment carried out in the presence of our standard detergent, Tween 20, and the remaining lanes (lanes 2 through 6) represent identical PVDF ligand blot strips treated with the various detergents listed above. As with the blocking agent experiments, some differences in the overall amount of labeled Cry1Ac toxin which bound to the blots were apparent, although there was no difference in the relative abilities of BT-R1 and the other proteins to bind this ligand. This finding appeared true even with the cholesterol-derived detergents deoxycholate and CHAPS, which allowed much lower overall levels of 125I-labeled Cry1Ac binding (Fig. 5, lanes 5 and 6).
FIG. 5.
Comparison of Cry1Ac binding to midgut proteins in the presence of different detergents. Proteins (80 μg) prepared by the Wolfersberger method were separated by SDS-PAGE following solubilization in SDS loading buffer. Following semidry blotting to PVDF filters, lanes were cut into strips and blocked overnight with TBS-5% nonfat milk buffers containing 0.1% the detergent listed above each lane. Following hybridization with 125I-labeled Cry1Ac for 2 h, the strips were washed three times, 5 min each, with the same buffers. Positions of molecular size markers (in kilodaltons) are indicated on the right. NP-40, Noninet P-40.
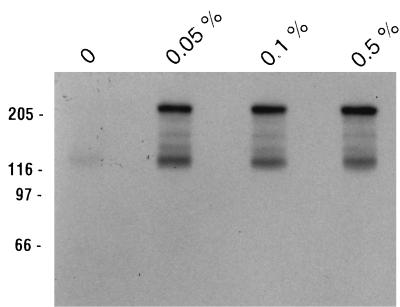
Having determined that the only difference in the ligand binding results obtained with the battery of detergents used was in the overall intensity of the signals and not in the specific binding of Cry1Ac to either 210-kDa BT-R1 or the other Cry1Ac binding proteins, we wanted to know whether we could optimize binding by altering the concentration of our standard detergent, Tween 20. As shown in lane 1 of Fig. 6, in the absence of detergent no binding to 210-kDa BT-R1 was detected and greatly reduced amounts of 125I-labeled Cry1Ac were bound to the ∼120-kDa protein relative to the results in previous experiments. Lanes 2 through 4 of Fig. 6 show binding results obtained with increasing concentrations of Tween 20, 0.05 to 0.5%, and clearly show that increasing the concentration of Tween 20 above 0.05% in no way affected binding. Following exposure to X-Omat AR autoradiography film, the blot strips shown in Fig. 5 and 6 were cut into sections, and the exact amount of bound toxin was determined with a scintillation counter. A tabulation of the data is presented in Table 1 as an aid to understanding the ligand blot autoradiogram results discussed to this point, along with data compiled from several such experiments with 125I-labeled Cry1Ac (data not shown). Four main facts stand out in Table 1: (i) Cry1Ab toxin binds essentially only to 210-kDa BT-R1 in M. sexta, (ii) at least a 0.05% concentration of almost any non-cholesterol-based detergent is required for optimal ligand binding to M. sexta midgut proteins on PVDF blots, (iii) Cry1A binding differences that do appear with various detergents and blocking buffers represent effects only on the overall intensity of binding to all proteins and do not preferentially affect the 210- or 120-kDa binding proteins individually, and (iv) the cholesterol-based detergents deoxycholate and CHAPS allow for less than 35% maximal Cry1A binding, i.e., that which is detected in the presence of Tween 20. Neither the absolute requirement for detergent nor the significance of the lack of effectiveness of cholesterol-based detergents in facilitating maximal binding can be explained at this time.
FIG. 6.
Comparison of Cry1Ac binding to midgut proteins in the presence of different concentrations of Tween 20. M. sexta midgut proteins were prepared by the Wolfersberger method, and 80 μg was separated by SDS-PAGE following solubilization in SDS loading buffer. After semidry blotting to PVDF filters, lanes were cut into strips and blocked with TBS-5% nonfat milk buffers containing various Tween 20 concentrations (listed above the lanes) overnight. Following hybridization with 125I-labeled Cry1Ac for 2 h, the strips were washed three times, 5 min each, with the same buffers. Positions of molecular size markers (in kilodaltons) are indicated on the left.
BT-R1 ligand binding following denaturing and nondenaturing treatments.
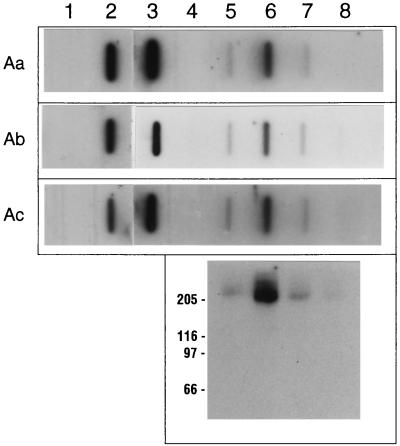
The last variable that we investigated involved vacuum blotting of nondenatured M. sexta midgut proteins and Sf21-expressed BT-R1. To date, 125I-labeled ligand blot experiments conducted in our laboratory with either material have involved the separation of midgut proteins by denaturing SDS-PAGE and subsequent transfer to PVDF filters (10, 19, 42, 43). While M. sexta midgut proteins separated by SDS-PAGE and semidry transferred to nitrocellulose filters have produced results identical to those obtained with PVDF filters under similar conditions (33), we believed it important to demonstrate that the same results can be obtained with a nondenaturing blot in conjunction with a different blotting substrate, such as nitrocellulose. Figure 7 shows the results of such experiments with nondenatured proteins for vacuum blot ligand binding. Sf21 cell culture-expressed BT-R1 was compared directly to both whole M. sexta midgut membrane preparations and solubilized, size-fractionated samples originally described by Francis and Bulla (10). Three horizontal strips represented identical vacuum blots hybridized with 125I-labeled Cry1Aa, Cry1Ab, and Cry1Ac. Vacuum blots of mock-transfected Sf21 cells (Fig. 7, lane 1) were used as controls to show that the Cry1A toxins do not bind any proteins in these cells unless they are transfected with the cDNA encoding BT-R1 (lanes 1 and 2). Lane 3 of Fig. 7 contained 10 μg of nondenatured M. sexta midgut proteins; as can be seen by comparing this lane to lane 2, both the natural and the cell-culture-expressed glycoproteins were capable of binding the three Cry1A ligands under these conditions. Previously we demonstrated the partial purification of BT-R1 by Superdex-200 FPLC size fractionation (10). Lanes 4 through 8 of Fig. 7 show that the peak of Cry1A binding activity in the present work eluted in fraction 8 (lane 6), and an SDS-PAGE ligand blot of these same materials (Fig. 7, bottom panel) clearly identified the protein responsible for this binding as 210-kDa BT-R1. In our previous work, it was demonstrated that the only Cry1A binding protein present in these particular fractions was BT-R1 and that it bound not only Cry1Ab but also Cry1Aa and Cry1Ac (10). While the experiment reported here was not intended to be a quantitative study of Cry1A toxin binding, it is the first report of BT-R1 Cry1A binding activity with nondenatured materials on nitrocellulose vacuum blots. However, it was previously shown that BT-R1 binds Cry1A toxins (i) under initially denaturing SDS-PAGE ligand blot conditions (10, 19, 33, 42, 43), (ii) in Western blotting (31, 33), (iii) in affinity column chromatography (35), and (iv) in immunoprecipitation (10, 42), the latter two procedures with nondenatured materials.
FIG. 7.
Nitrocellulose vacuum blots of nondenatured BT-R1 ligand binding activity. (Top panel) Horizontal strips are autoradiograms of nitrocellulose vacuum blots incubated with 125I-labeled Cry1Aa, Cry1Ab, and Cry1Ac, as indicated on the left. Lanes: 1, 10 μg of cell membrane proteins prepared by the Adamo method from mock-transfected Sf21 cells; 2, 10 μg of proteins from Sf21 cells transfected with the BT-R1 insect cell expression construct; 3, 10 μg of midgut membrane proteins from M. sexta; 4 through 8, M. sexta midgut proteins solubilized with 0.5% CHAPS and then FPLC fractionated with a Superdex-200 FPLC column (10). (Bottom panel) Autoradiogram of an SDS-PAGE ligand blot of material from the Superdex-200 fractions hybridized with 125I-labeled Cry1Ab. Fractions 7 through 10 are shown in alignment with those on the vacuum blots. Positions of molecular size markers (in kilodaltons) are indicated on the left.
DISCUSSION
Understanding the molecular biology of insect receptors for Cry toxins will be a crucial part of gaining better control over the use of these biopesticides by allowing the engineering of more effective toxins in terms of longer persistence in the field, higher toxicity, customized spectrum of toxicity, and ultimately control of resistance development in a given crop pest. For these reasons, it is imperative to gain a complete understanding of toxin mode of action and the role that receptors play in this mechanism. In this study, we showed that BT-R1 is a common receptor for the lepidopteran-specific Cry1Aa, Cry1Ab, and Cry1Ac toxins of B. thuringiensis. The specific high-affinity binding of these toxins to BT-R1 is unaffected by either the method used to isolate midgut proteins, the kind of blocking agents used, the type of blocking buffer detergents used, blotting procedure, or blotting substrate. While we cannot explain the absolute requirement for detergents in the SDS-PAGE ligand blots described by our laboratory and others, we have reported elsewhere competition binding assays with suspensions of native and cell culture-expressed binding proteins in PBS without detergents (19). These experiments provided the same degree of competition as that reported in SDS-PAGE ligand blot competition experiments both in this study and in the earlier study (19).
Our laboratory was the first to show a direct correlation between ligand blot competition data and competition binding assays performed in suspension (19). Demonstrating a positive correlation between these experiments is essential and is the only way to verify the significance of the identification of a novel toxin binding protein short of its cloning and functional expression. While the Cry1Ac ligand blots in the present work correlate well with some of those reported elsewhere (3, 10, 26), they differ somewhat from those reported earlier by our laboratory (19) in the total number of proteins detected. While all of the competition ligand blot experiments demonstrated here were done with materials initially denatured by SDS-PAGE separations, we previously demonstrated corroborating competition data with nondenatured suspensions of the same binding proteins (19, 42, 43). Also, these nondenaturing competition assays generated plots which provided no indication of multiple high-affinity binding sites for the three Cry1A toxins examined (19). When considered together, the data indicate that among the binding proteins which appear on Cry1Ac SDS-PAGE ligand blots, BT-R1 demonstrates the highest degree of specificity for this ligand among all of the Cry1Ac binding proteins, as determined by competition in both heterologous and homologous competition ligand blot experiments.
For the lepidopteran Bombyx mori, a 180-kDa cadherin-like protein demonstrating amino acid sequence similarity to BT-R1 has been identified, purified, and partially sequenced (18). Although 125I-labeled Cry1Aa binding to a 180-kDa cadherin-like protein was eliminated by the presence of unlabeled competing Cry1Aa, an additional band(s) of approximately 110 kDa which was also identified by 125I-labeled Cry1Aa on midgut protein ligand blots failed to demonstrate a detectable degree of competition under identical circumstances. Thus, it now appears that B. mori, like M. sexta, contains both high-affinity and low-affinity binding proteins for at least one Cry1A toxin. In both insects, the high-affinity receptor appears to be a cadherin-like protein with a large molecular mass. At this time, we can only speculate on the role of these cadherin-like proteins in the mode of action of B. thuringiensis toxins, which are generally thought to disrupt ionic balance in the midgut epithelium (25). However, at least one type of cadherin has been shown to be the crucial receptor for the binding of the gram-positive intracellular pathogen Listeria monocytogenes to the plasma membrane of nonphagocytotic epithelial cells (9, 36). It is conceivable that in acting as a receptor for the Cry1A toxins, BT-R1 is responsible, either directly or indirectly, for mediating the intercalation of the lepidopteran-specific toxins into the brush border membranes of intoxicated larvae.
Of the Cry1Ac binding proteins identified to date, none have been shown to exhibit the specific, high-affinity binding demonstrated by BT-R1. The Cry1Ac binding 120-kDa aminopeptidase from M. sexta has been partially purified and used in competition binding assays (38), liposome reconstitution experiments (39), and surface plasmon resonance experiments (34, 39). These experiments have served to confuse matters in that the affinity of the aminopeptidase for Cry1Ac was determined to be relatively low and the number of binding sites for Cry1Aa and Cry1Ac, as determined by surface plasmon resonance experiments, differed between reports (34, 39). These inconsistencies may have arisen, in part, from the different degrees of enrichment to which the binding proteins were subjected in each study or from differences in the degree of saturation encountered by the Cry1Ac binding proteins in these experiments. However, studies with Cry1Ac have been shown elsewhere to be complicated by Cry1Ac binding to a biotinylated 120-kDa protein which was shown not to be an aminopeptidase as well as to the biotinylated forms of albumin, ovalbumin, and pyruvate carboxylase (5). We do not believe, however, that the method of labeling the Cry1A toxins has been a factor in this and other studies. Our laboratory previously demonstrated that chloramine-T iodination of Cry1Ab did not affect the toxicity of this toxin for M. sexta larvae (42), nor would one expect to see the high specificity and affinity of competition binding with a nonlabeled competitor reported here and elsewhere if the iodination reaction altered either the affinity or the specificity of the toxins for the binding proteins discussed in this work.
Considering the similarities in toxicity and amino acid sequence among the three Cry1A toxins discussed in this report, we propose that until shown otherwise, it is reasonable to conclude that these toxins act through a common receptor and a common mode of action in M. sexta. Recently, various phylogenetic relationships among Cry toxins were discussed in some detail by Bravo (2). This elegant review detailed one obvious difference among the Cry1A toxins which is especially relevant to the topic of binding protein specificity. In an analysis of the phylogeny of the three domains of the Cry1A toxins, the origins of only domains I and II appear to be shared: domain III of Cry1Ac resides on a unique branch in the phylogenetic trees described by Bravo (2), indicating it has an origin unlike that of any other Cry toxin. It is interesting to note that in receptor binding studies of M. sexta, Cry1Ab-Cry1C hybrids showed that domain II of Cry1Ab was responsible for binding to 210-kDa BT-R1 (3). In the same study, it was shown that Cry1Ac binding to BT-R1 was dependent upon Cry1Ac-specific sequences in domain I and/or domain II, while binding to the 120-kDa aminopeptidase was dependent upon Cry1Ac-specific sequences in domain III. Similar studies of BT-R1 expressed in insect cell cultures also highlighted the importance of domain II in specific binding to the 210-kDa Cry1A binding protein (20). While it has generally been accepted that domain I is involved in toxin insertion into the brush border membrane and domain II is believed to be the major determinant in receptor recognition and hence toxin specificity, the role of domain III is less agreed upon. Generally, it is believed to be involved in the maintenance of toxin structure and stability, although recent reports have implied an involvement in the formation of membrane ion channels (40). We find it unlikely that the unique domain III of Cry1Ac could be ultimately responsible for determining the M. sexta-specific toxicity of this single toxin and not that of Cry1Aa and Cry1Ab as well.
There remains considerable debate over both the identity and the function of the physiologically relevant Cry1A receptor(s) in M. sexta. Until it can be demonstrated that such a toxin binding protein can impart sensitivity to a cell which is normally insensitive, absolute declarations regarding the mode of action of the Cry toxins will remain difficult, if not impossible, to make. At this point in the understanding of Cry1A receptors and toxin mode of action, it is difficult to understand the significance of multiple low-affinity Cry1Ac binding proteins. Nonetheless, on the basis of the plethora of ligand binding, toxicity, and sequence data currently available, it is reasonable to conclude that the highly similar Cry1A toxins most likely exhibit their pesticidal activity, at least in M. sexta, through a common cadherin-like high-affinity receptor such as BT-R1.
ACKNOWLEDGMENTS
This work was supported by a grant from Pioneer Hi-Bred International, Inc., to L.A.B. and a fellowship from the National Institutes of Health (F32 AI09582-02) to T.P.K.
We extend our gratitude to Hideshi Ihara and colleagues (Osaka Prefecture University, Osaka, Japan) for sharing results with us prior to publication. We thank Ruud De Maagd and Dirk Bosch (Center for Plant Breeding and Reproduction Research, Wageningen, The Netherlands) for performing Cry1 chimera binding studies using the insect cell culture-expressed BT-R1 Cry1A receptor and Terry Meyer (Pioneer Hi-Bred International, Inc., Johnston, Iowa) for critical discussions of the manuscript.
REFERENCES
- 1.Adamo H, Caride A J, Penniston J T. Use of expression mutants and monoclonal antibodies to map the erythrocyte Ca2+ pump. J Biol Chem. 1992;267:14244–14249. [PubMed] [Google Scholar]
- 2.Bravo A. Phylogenetic relationships of Bacillus thuringiensis δ-endotoxin family proteins and their functional domains. J Bacteriol. 1997;179:2793–2801. doi: 10.1128/jb.179.9.2793-2801.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Maagd R A, Van der Klei H, Bakker P L, Stiekema W J, Bosch D. Different domains of Bacillus thuringiensis δ-endotoxins can bind to insect midgut membrane proteins on ligand blots. Appl Environ Microbiol. 1996;62:2753–2757. doi: 10.1128/aem.62.8.2753-2757.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Denolf P, Jansens S, Peferoen M, Degheele D, Van Rie J. Two different Bacillus thuringiensis delta-endotoxin receptors in the midgut brush border membrane of the European corn borer, Ostrinia nubilalis (Hübner) (Lepidoptera: Pyralidae) Appl Environ Microbiol. 1993;59:1828–1837. doi: 10.1128/aem.59.6.1828-1837.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Du C, Nickerson K W. The Bacillus thuringiensis insecticidal toxin binds biotin-containing proteins. Appl Environ Microbiol. 1995;62:2932–2939. doi: 10.1128/aem.62.8.2932-2939.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.English L H, Readdy T L. Delta endotoxin inhibits a phosphatase in midgut epithelial membranes of Heliothis virescens. Insect Biochem. 1989;19:145–152. [Google Scholar]
- 7.Escriche B, Ferré J, Silva F J. Occurrence of a common binding site in Mamestra brassicae, Phthorimaea operculella, and Spodoptera exigua for the insecticidal crystal proteins CryIA from Bacillus thuringiensis. Insect Biochem Mol Biol. 1997;27:651–656. doi: 10.1016/s0965-1748(97)00039-8. [DOI] [PubMed] [Google Scholar]
- 8.Ferré J, Real M D, Van Rie J, Jansens S, Peferoen M. Resistance to the Bacillus thuringiensis bioinsecticide in a field population of Plutella xylostella is due to a change in a midgut membrane receptor. Proc Natl Acad Sci USA. 1991;88:5119–5123. doi: 10.1073/pnas.88.12.5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finlay B B, Cossart P. Exploitation of mammalian host cell functions by bacterial pathogens. Science. 1997;276:718–725. doi: 10.1126/science.276.5313.718. [DOI] [PubMed] [Google Scholar]
- 10.Francis B R, Bulla L A., Jr Further characterization of BT-R1, the cadherin-like receptor for Cry1Ab toxin in tobacco hornworm (Manduca sexta) midguts. Insect Biochem Mol Biol. 1997;27:541–550. doi: 10.1016/s0965-1748(97)00029-5. [DOI] [PubMed] [Google Scholar]
- 11.Garczynski S F, Crim J W, Adang M J. Identification of putative insect brush border membrane-binding molecules specific to Bacillus thuringiensis δ-endotoxin by protein blot analysis. Appl Environ Microbiol. 1991;57:2816–2820. doi: 10.1128/aem.57.10.2816-2820.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garczynski S F, Adang M J. Bacillus thuringiensis CryIA(c) δ-endotoxin binding aminopeptidase in the Manduca sexta midgut has a glycosyl-phosphatidylinositol anchor. Insect Biochem Mol Biol. 1995;25:409–415. [Google Scholar]
- 13.Gill S S, Cowles E A, Francis V. Identification, isolation, and cloning of a Bacillus thuringiensis CryIAc toxin-binding protein from the midgut of the lepidopteran insect Heliothis virescens. J Biol Chem. 1995;270:27277–27282. doi: 10.1074/jbc.270.45.27277. [DOI] [PubMed] [Google Scholar]
- 14.Gould F, Martínez-Ramírez A, Anderson A, Ferré J, Silva F J, Moar W J. Broad-spectrum resistance to Bacillus thuringiensis toxins in Heliothis virescens. Proc Natl Acad Sci USA. 1992;89:7986–7990. doi: 10.1073/pnas.89.17.7986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hofmann C, Vanderbruggen H, Höfte H, Van Rie J, Jansens S, Van Mellaert H. Specificity of Bacillus thuringiensis δ-endotoxins is correlated with the presence of high-affinity binding sites in the brush border membrane of target insect midguts. Proc Natl Acad Sci USA. 1988;85:7844–7848. doi: 10.1073/pnas.85.21.7844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Höfte H, Van Rie J, Jansens S, Van Houtven A, Vanderbruggen H, Vaeck M. Monoclonal antibody analysis and insecticidal spectrum of three types of lepidopteran-specific insecticidal crystal proteins of Bacillus thuringiensis. Appl Environ Microbiol. 1988;54:2010–2017. doi: 10.1128/aem.54.8.2010-2017.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Höfte H, Whiteley H R. Insecticidal crystal proteins of Bacillus thuringiensis. Microbiol Rev. 1989;53:242–255. doi: 10.1128/mr.53.2.242-255.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ihara, H., T. Uemura, M. Masuhara, S. Ikawa, K. Sugimoto, A. Wadano, and M. Himeno. Purification and partial amino acid sequences of the binding protein from Bombyx mori for Cry1Aa δ-endotoxin of Bacillus thuringiensis. Submitted for publication. [DOI] [PubMed]
- 19.Keeton T P, Bulla L A., Jr Ligand specificity and affinity of BT-R1, the Bacillus thuringiensis Cry1A toxin receptor from Manduca sexta, expressed in mammalian and insect cell cultures. Appl Environ Microbiol. 1997;63:3419–3425. doi: 10.1128/aem.63.9.3419-3425.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keeton, T. P., R. A. De Maagd, and D. Bosch. Unpublished observations.
- 21.Knight P J K, Crickmore N, Ellar D J. The receptor for Bacillus thuringiensis CryIA(c) delta endotoxin in the brush border membrane of the lepidopteran Manduca sexta is aminopeptidase N. Mol Microbiol. 1994;11:429–436. doi: 10.1111/j.1365-2958.1994.tb00324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knight P J K, Knowles B H, Ellar D J. Molecular cloning of an insect aminopeptidase N that serves as a receptor for Bacillus thuringiensis CryIA(c) toxin. J Biol Chem. 1995;270:17765–17770. doi: 10.1074/jbc.270.30.17765. [DOI] [PubMed] [Google Scholar]
- 23.Knowles B H, Ellar D J. Characterization and partial purification of a plasma membrane receptor for Bacillus thuringiensis var. kurstaki lepidopteran-specific δ-endotoxin. J Cell Sci. 1986;83:89–101. doi: 10.1242/jcs.83.1.89. [DOI] [PubMed] [Google Scholar]
- 24.Knowles B H, Knight P J K, Ellar D J. N-acetyl galactosamine is part of the receptor in insect gut epithelia that recognizes an insecticidal protein from Bacillus thuringiensis. Proc R Soc London. 1991;245:31–35. doi: 10.1098/rspb.1991.0084. [DOI] [PubMed] [Google Scholar]
- 25.Knowles B H. Mechanism of action of Bacillus thuringiensis insecticidal δ-endotoxins. Adv Insect Physiol. 1994;24:275–308. [Google Scholar]
- 26.Lee M K, Milne R E, Ge A Z, Dean D H. Location of a Bombyx mori receptor binding region on a Bacillus thuringiensis δ-endotoxin. J Biol Chem. 1992;267:3115–3121. [PubMed] [Google Scholar]
- 27.Lee M K, Dean D H. Inconsistencies in determining Bacillus thuringiensis toxin binding sites relationship by comparing competition assays with ligand blotting. Biochem Biophys Res Commun. 1996;220:575–580. doi: 10.1006/bbrc.1996.0445. [DOI] [PubMed] [Google Scholar]
- 28.Lee M K, You T H, Young B A, Cotrill J A, Valaitis A P, Dean D H. Aminopeptidase N purified from gypsy moth brush border membrane vesicles is a specific receptor for Bacillus thuringiensis CryIAc toxin. Appl Environ Microbiol. 1996;62:2845–2849. doi: 10.1128/aem.62.8.2845-2849.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo K, Lu Y-J, Adang M J. A 106 kDa form of aminopeptidase is a receptor for Bacillus thuringiensis Cry1C δ-endotoxin in the brush border membrane of Manduca sexta. Insect Biochem Mol Biol. 1996;26:783–791. [Google Scholar]
- 30.Luo K, Tabashnik B E, Adang M J. Binding of Bacillus thuringiensis Cry1Ac toxin to aminopeptidase in susceptible and resistant diamondback moths (Plutella xylostella) Appl Environ Microbiol. 1997;63:1024–1027. doi: 10.1128/aem.63.3.1024-1027.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maaty, W. S. A., and L. A. Bulla, Jr. Unpublished observations.
- 32.MacIntosh S C, Stone T B, Jokerst R S, Fuchs R L. Binding of Bacillus thuringiensis proteins to a laboratory-selected line of Heliothis virescens. Proc Natl Acad Sci USA. 1991;88:8930–8933. doi: 10.1073/pnas.88.20.8930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martínez-Ramírez A C, González-Nebauer S, Escriche B, Real M D. Ligand blot identification of a Manduca sexta midgut binding protein specific to three Bacillus thuringiensis CryIA-type ICPs. Biochem Biophys Res Commun. 1994;201:782–787. doi: 10.1006/bbrc.1994.1769. [DOI] [PubMed] [Google Scholar]
- 34.Masson L, Lu Y J, Mazza A, Brousseau R, Adang M J. The CryIA(c) receptor purified from Manduca sexta displays multiple specificities. J Biol Chem. 1995;270:20309–20315. doi: 10.1074/jbc.270.35.20309. [DOI] [PubMed] [Google Scholar]
- 35.Meng, J., and L. A. Bulla, Jr. Unpublished observations.
- 36.Mengaud J, Ohayon H, Gounon P, Mège R-M, Cossart P. E-cadherin is the receptor required for internalin, a surface protein required for entry of L. monocytogenes into epithelial cells. Cell. 1996;84:923–932. doi: 10.1016/s0092-8674(00)81070-3. [DOI] [PubMed] [Google Scholar]
- 37.Oppert B, Kramer K J, Johnson D E, MacIntosh S C, McGaughey W H. Altered protoxin activation by midgut enzymes from a Bacillus thuringiensis resistant strain of Plodia interpunctella. Biochem Biophys Res Commun. 1994;198:940–947. doi: 10.1006/bbrc.1994.1134. [DOI] [PubMed] [Google Scholar]
- 38.Sangadala S, Walters F S, English L H, Adang M J. A mixture of Manduca sexta aminopeptidase and phosphatase enhances Bacillus thuringiensis insecticidal CryIA(c) toxin binding and 86Rb-K+ efflux in vitro. J Biol Chem. 1994;269:10088–10092. [PubMed] [Google Scholar]
- 39.Schwartz J-L, Lu Y-J, Söhnlein P, Brousseau R, Laprade R, Masson L, Adang M J. Ion channels formed in planar lipid bilayers by Bacillus thuringiensis toxins in the presence of Manduca sexta midgut receptors. FEBS Lett. 1997;412:270–276. doi: 10.1016/s0014-5793(97)00801-6. [DOI] [PubMed] [Google Scholar]
- 40.Schwartz J-L, Potvin L, Chen X J, Brousseau R, Laprade R, Dean D H. Single-site mutations in the conserved alternating-arginine region affect ionic channels formed by Cry1Aa, a Bacillus thuringiensis toxin. Appl Environ Microbiol. 1997;63:3978–3984. doi: 10.1128/aem.63.10.3978-3984.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simpson R M, Burgess E P J, Markwick N P. Bacillus thuringiensis δ-endotoxin binding sites in two lepidoptera, Wiseana spp. and Epiphyas postvittana. J Invertebr Pathol. 1997;70:136–142. doi: 10.1006/jipa.1997.4680. [DOI] [PubMed] [Google Scholar]
- 42.Vadlamudi R K, Ji T H, Bulla L A., Jr A specific binding protein from Manduca sexta for the insecticidal toxin of Bacillus thuringiensis subsp. berliner. J Biol Chem. 1993;268:12334–12340. [PubMed] [Google Scholar]
- 43.Vadlamudi R K, Weber E, Ji I, Ji T H, Bulla L A., Jr Cloning and expression of a receptor for an insecticidal toxin of Bacillus thuringiensis. J Biol Chem. 1995;270:5490–5494. doi: 10.1074/jbc.270.10.5490. [DOI] [PubMed] [Google Scholar]
- 44.Valaitis A P, Lee M K, Rajamohan F, Dean D H. Brush border membrane aminopeptidase-N in the midgut of the gypsy moth serves as the receptor for the CryIA(c) δ-endotoxin of Bacillus thuringiensis. Insect Biochem Mol Biol. 1995;25:1143–1151. doi: 10.1016/0965-1748(95)00050-x. [DOI] [PubMed] [Google Scholar]
- 45.Van Rie J, Jansens S, Höfte H, Degheele D, Van Mellaert H. Specificity of Bacillus thuringiensis δ-endotoxins. Eur J Biochem. 1989;186:239–247. doi: 10.1111/j.1432-1033.1989.tb15201.x. [DOI] [PubMed] [Google Scholar]
- 46.Van Rie J, Jansens S, Höfte H, Degheele D, Van Mellaert H. Receptors on the brush border membrane of the insect midgut as determinants of the specificity of Bacillus thuringiensis delta-endotoxins. Appl Environ Microbiol. 1990;56:1378–1385. doi: 10.1128/aem.56.5.1378-1385.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van Rie J, McGaughey W H, Johnson D E, Barnett B D, Van Mellaert H. Mechanism of insect resistance to the microbial insecticide Bacillus thuringiensis. Science. 1990;247:72–74. doi: 10.1126/science.2294593. [DOI] [PubMed] [Google Scholar]
- 48.Wolfersberger M, Luethy P, Maurer A, Parenti P, Sacchi F V, Giordana B, Hanozet G M. Preparation and partial characterization of amino acid transporting brush border membrane vesicles from the larval midgut of the cabbage butterfly (Pieris brassicae) Comp Biochem Physiol. 1987;86A:301–308. [Google Scholar]