Abstract
Most proteins secreted into the extracellular space are first recruited from the endoplasmic reticulum into coat protein complex II (COPII)-coated vesicles or tubules that facilitate their transport to the Golgi apparatus. Although several secreted proteins have been shown to be actively recruited into COPII vesicles and tubules by the cargo receptors LMAN1 and SURF4, the full cargo repertoire of these receptors is unknown. We now report mass spectrometry analysis of conditioned media and cell lysates from HuH7 cells CRISPR targeted to inactivate the LMAN1 or SURF4 gene. We found that LMAN1 has limited clients in HuH7 cells, whereas SURF4 traffics a broad range of cargoes. Analysis of putative SURF4 cargoes suggests that cargo recognition is governed by complex mechanisms rather than interaction with a universal binding motif..
Keywords: secretome, COPII trafficking, cargo receptor
Introduction
Approximately a third of the proteins encoded by the mammalian genome are cotranslationally inserted into the endoplasmic reticulum (ER), from where they are subsequently trafficked to the plasma membrane or various intracellular organelles or secreted into the extracellular space.1–3 Properly folded proteins destined for secretion are transported from the ER to the Golgi via coat protein complex II (COPII)-coated vesicles or tubular structures.4–6 Entry into COPII vesicles/tubules is thought to occur passively via bulk flow or through active recruitment and concentration.7 Transmembrane cargoes can interact directly with the cargo-selective COPII component SEC24 on the cytoplasmic face of the ER, whereas soluble proteins restricted to the ER lumen require interaction with a membrane-spanning intermediary, referred to as a cargo receptor, to bridge this interaction. However, relatively few such cargo receptors have been discovered in mammalian cells to date, and the cargo receptor-dependence for most secreted proteins remains unclear.
LMAN1, also known as ERGIC-53, is a 53 kDa protein that localizes to the ER and ER-Golgi intermediate compartments (ERGIC).8 LMAN1 has been shown to function as a cargo receptor for coagulation factors V (F5) and VIII (F8),9,10 α1-antitrypsin (SERPINA1),11 Mac-2 binding protein (Mac-2BP),12 matrix metalloproteinase-9 (MMP9),13 cathepsin C (CTSC),14 cathepsin Z (CTSZ),15 and membrane protein γ-aminobutyric acid type A receptors (GABAARs).16In vivo studies in mice have confirmed the dependence of F5, F8, and SERPINA1, but not CTSC and CTSZ, on LMAN1 for secretion.17 No common LMAN1-binding motif has been identified. It is unclear whether there are other cargoes beyond those listed above that require LMAN1 for efficient secretion from the ER.
Another recently identified mammalian cargo receptor, SURF4, is a 29 kDa protein with multiple transmembrane domains that also localizes to the ER and ERGIC.18 SURF4 is highly conserved, with homologues in yeast (Erv29p), C. elegans (SFT-4), and Drosophila.19 Erv29p facilitates the secretion of yeast pro-α-factor, carboxypeptidase Y, and proteinase A.20–22 Multiple SURF4 cargoes in mammals have been identified to date, including PCSK9,23–25 apolipoprotein B (APOB)25–28 growth hormone,29 dentin sialophosphoprotein (DSPP),29 amelogenin,29 erythropoietin,30 pathogenic SERPINA1 polymers,31 sonic hedgehog,32 proinsulin,33 and the lysosomal proteins progranulin and prosaposin.34 Two SURF4-binding motifs on cargoes have been proposed, including an ER-ESCAPE tripeptide motif immediately downstream of the signal peptide sequence29 and a Cardin–Weintraub motif,32,35 though not all of the putative cargoes listed above carry one of these motifs, suggesting the presence of additional determinants for recognition by SURF4.24
Previous attempts to define a comprehensive cargo repertoire for LMAN1 and SURF4 have revealed additional putative cargoes. Using an in vitro vesicle formation assay and label-free mass spectrometry in cells depleted for LMAN1 or SURF4, Huang et al.(36) described 4 and 17 novel cargoes, respectively, for these receptors. Using SILAC labeling and mass spectrometry analysis of conditioned media following SURF4 knockdown, Gomez-Navarro and colleagues24 likewise identified 10 proteins in HEK293 cells and 18 proteins in HuH7 cells as potential SURF4 cargoes.
We and others have reported a mass spectrometry approach to identify secreted proteins by quantifying protein abundance in both conditioned media and cell lysates from cultured cells.37–39 We found that calculation of media to lysate (M/L) ratios (rather than using unadjusted protein abundance in the media alone) greatly improved the sensitivity and specificity for detecting secreted proteins.39 We now report the application of this approach to determine the cargo repertoire of LMAN1 and SURF4 in HuH7 cells.
Experimental Procedures
Cell Culture
HuH7 cells40 were cultured in DMEM supplemented with GlutaMAX (Thermo Fisher Scientific, Waltham, MA, 10569-044), 10% fetal bovine serum (MilliporeSigma, Burlington, MA, F8067), and penicillin/streptomycin (Thermo Fisher Scientific, Waltham, MA, 15140-122). Cells were passaged every 3–4 days and maintained between 20 and 80% confluence.
CRISPR-Mediated Inactivation of LMAN1 and SURF4
On day 0, cells were seeded at 20% confluence and infected with lentivirus generated from pLentiCRISPRv2 (Addgene plasmid #52961; http://n2t.net/addgene:52961; RRID:Addgene_52961; a gift from Feng Zhang)41 engineered to deliver Cas9 and a gRNA targeting LMAN1 (CCCCTTACACTATAGTGACG), SURF4 (TCCGAGCTGCATGTACTGTT), or a nontargeting gRNA (GTTCATTTCCAAGTCCGCTG), as previously described.23 On day 1, the medium was exchanged, and 1 μg/mL puromycin (MilliporeSigma, Burlington, MA, P8833) was added for 48 h. Following selection, surviving cells were passaged every 3–4 days until day 14 to allow for gene editing and protein turnover. On day 14, cells were washed three times with phosphate-buffered saline (PBS, Thermo Fisher Scientific, Waltham, MA, 10010-023) and switched to serum-free, phenol red-free DMEM prewarmed to 37 °C (Thermo Fisher Scientific, Waltham, MA, 31053-036) for 12 h.
Conditioned Media and Cell Lysate Collection
Conditioned media and cell lysates were harvested as previously described.39 Briefly, conditioned media were collected from the cell culture dish and centrifuged at 2500g at 4 °C for 15 min to remove cell debris. The supernatant was then ultracentrifuged at 120,000g at 4 °C for 90 min to remove exosomes42 and concentrated using a 3 kDa molecular weight cutoff concentrator (MilliporeSigma, Burlington, MA, UFC900324). Cell lysates were collected in 2 mL of RIPA buffer (Thermo Scientific, Waltham, MA, 89900) containing a protease inhibitor cocktail (cOmplete Mini Protease Inhibitor Cocktail, Roche, Basel, Switzerland, 11836153001). Cell suspensions were sonicated, rotated end-over-end for 1 h, and centrifuged at 21,000g at 4 °C for 45 min. Supernatants were then transferred to new Eppendorf tubes. Protein concentrations in the conditioned media and lysates were determined by DC protein assay (Bio-Rad, Hercules, CA, 500-011).
Immunoblotting
Cell lysates (10 μg per sample) collected as above were resolved in a 4–20% Tris-glycine gel as previously described.25 Proteins were detected with antibodies against LMAN1 (Abcam, Cambridge, U.K., ab125006, 1:1000) or GAPDH (Abcam, Cambridge, U.K., ab181602, 1:10000).
Mass Spectrometry, Protein Identification, and Protein Quantification
Mass spectrometry, protein identification, and protein quantification were performed as previously described.39 Briefly, 75 μg of each lysate and 75 μg of each medium sample were proteolyzed, labeled with tandem mass tags (TMT) 10-plex (Thermo Fisher Scientific, Waltham, MA, 90110) according to the manufacturer’s protocol, and subjected to liquid chromatography–mass spectrometry analysis. Raw mass spectrometry files were converted into open mzML format and were analyzed using the FragPipe (https://fragpipe.nesvilab.org/) computational platform43–45 with the default TMT10-MS3 workflow. For proteins that were identified and quantified in both the media and lysate fractions, an M/L ratio was calculated using absolute intensity values. Signal peptide and transmembrane domain annotations were obtained from the UniProt database.46
Data Availability
Raw and processed mass spectrometry data have been deposited to the MassIVE database with the data set identifier: MSV000092642.
Statistical Analyses
The limma statistical package was used for comparison of protein abundance or M/L ratios between cells treated with a nontargeting, LMAN1-targeting, or SURF4-targeting gRNA, using a log2-transformed protein abundance or M/L ratios as input.47P-values were adjusted for multiple hypothesis testing using the Benjamini and Hochberg method. An adjusted p-value (q-value) of 0.05 or less was considered statistically significant.
Analysis of ER-ESCAPE Motifs
The human proteome reference database was downloaded from UniProt.46 Proteins were filtered for the presence of a signal peptide and the absence of a transmembrane domain(s). The tripeptide motif as proposed by Yin et al.(29) was extracted from the protein sequence. For proteins with a conventional signal peptide, the tripeptide motif is defined as the first three amino acid residues downstream of the annotated signal peptide sequence. For proteins with a propeptide domain, the tripeptide motif is defined as the first three amino acid residues downstream from the propeptide cleavage site. For rare cases of proteins with an uncleaved signal peptide (based on the UniProt annotation), the tripeptide motif is assigned as the first three amino acid residues of the protein. Each residue within the tripeptide motif was classified based on the classification system proposed by Yin et al.(29) For this analysis, SURF4 cargoes include putative SURF4 cargoes identified in this study as well as all previously reported cargoes.
Results
Identification of Bona Fide Secreted Proteins by Analysis of Cell Lysates and Conditioned Media
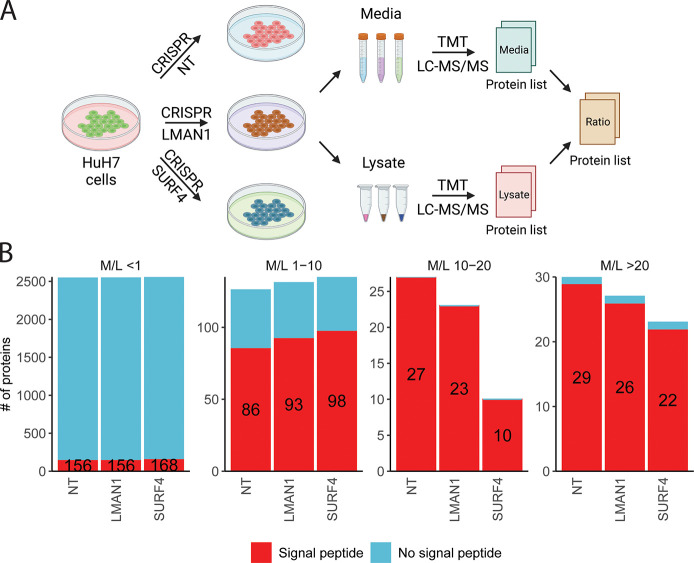
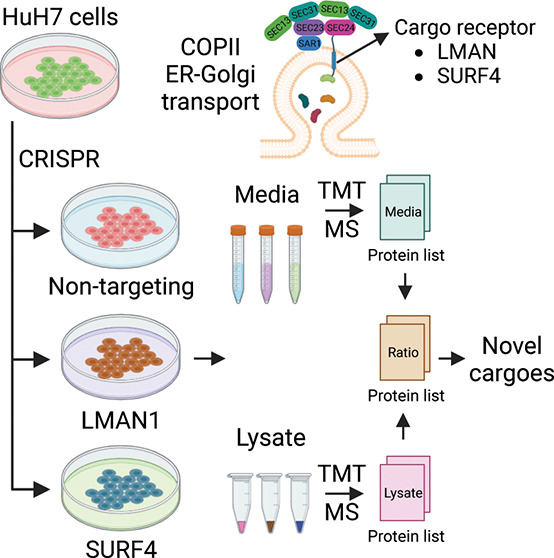
To identify secreted proteins that depend on either LMAN1 or SURF4 for secretion, we generated LMAN1- and SURF4-deficient HuH7 cells by CRISPR-mediated gene editing. We then collected conditioned media and cell lysates from cells receiving LMAN1-targeting, SURF4-targeting, or nontargeting (NT) gRNA for analysis by TMT mass spectrometry (n = 3 per group) (Figure 1A).
Figure 1.
Identifying proteins dependent on LMAN1 or SURF4 for efficient secretion. (A) Experimental design to identify LMAN1 and SURF4 cargoes in the human hepatoma cell line (HuH7). HuH7 cells were infected with lentiviruses delivering CRISPR/Cas9 and guide RNAs targeting LMAN1, SURF4, or a nontargeting (NT) control. Following selection, cells were cultured for 2 weeks before being switched to serum-free media for 12 h. Conditioned media and cell lysates were collected for protein identification and quantification by liquid chromatography (LC) followed by tandem mass tag (TMT) mass spectrometry (MS). A protein abundance ratio was calculated for each protein that was detected in both the media and lysate fractions as described in Methods. (B) Number of proteins with (red) and without (blue) a signal peptide identified in different M/L fractions. The number of signal peptide-containing proteins in each bar is indicated.
Across all three sample groups, we identified and quantified 5858 and 2947 proteins in the lysate and media fractions, respectively, of which 2726 proteins were identified in both fractions (Figure S1A and Tables S1, S2). Consistent with our previous observations,39 the majority of the identified proteins lack a signal peptide, with increasing M/L ratio correlating with an increased proportion of proteins carrying a signal peptide, consistent with the expected enrichment for secretory proteins in the media relative to the cell lysates (Figure S1B). Comparisons between NT, LMAN1, and SURF4 samples revealed that there were fewer proteins with an M/L ratio of >10 in LMAN1- and SURF4-deficient samples (Figure 1B).
Few Proteins in HuH7 Cells Depend on LMAN1 for Secretion
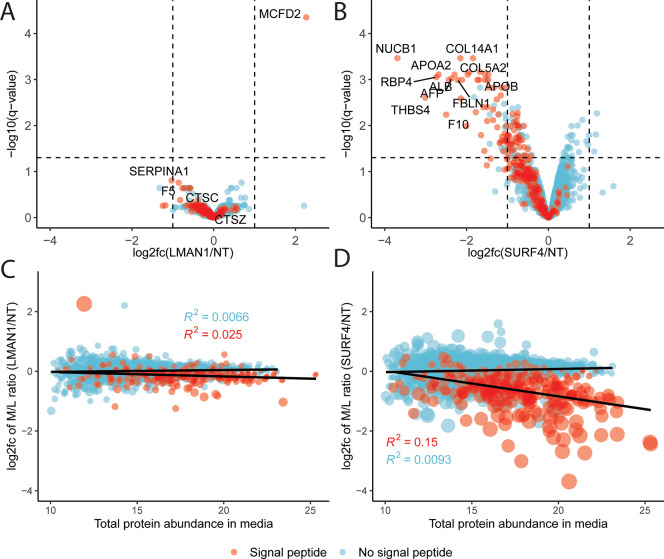
To identify secretory proteins that require LMAN1 for their secretion, we compared the M/L ratios of proteins collected from cells treated with LMAN1-targeting or NT gRNA. As shown in Figure 2A, the only protein demonstrating a significantly different M/L ratio following LMAN1 deletion was MCFD2, a 16 kDa soluble ER luminal protein that forms a complex with LMAN1 and acts as a cofactor for the secretion of factors V and VIII. MCFD2 lacks an ER retrieval motif, and its retention in the ER and ERGIC depends entirely on its interaction with LMAN1.48 The increased M/L ratio for MCFD2 in LMAN1-deficient cells (Figure 2A and Table S3), resulting from a decreased intracellular (Figure S2A and Table S4) and increased extracellular abundance (Figure S2B and Table S5), is therefore consistent with its release from the ER in the absence of LMAN1.36,48
Figure 2.
Differential effects on protein secretion in HuH7 cells following LMAN1 or SURF4 deletion. (A,B) Volcano plots comparing protein M/L ratios in LMAN1- (A) or SURF4- (B) deficient cells with those in controls. The log2 fold change (log2 fc) and statistical significance are plotted on the x- and y-axes, respectively. Proteins with a signal peptide are colored in red, and proteins without a signal peptide are colored in blue. Dashed vertical lines represent the log2 fc of 1 and −1. Dashed horizontal lines represent the −log10(q-value of 0.05). (C,D) Comparison of total abundance (estimation from FragPipe; see Methods) in the media with LMAN1 (C) or SURF4 (D) dependency. Trend lines represent linear regression. Dot sizes are proportional to −log10(q-value). R2 values in blue and red are for proteins without and with an annotated signal peptide, respectively.
Consistent with previously reported in vivo data in mice, we also found that genetic deletion of LMAN1 in HuH7 cells resulted in intracellular accumulation of SERPINA117 (Figure S2A) without a statistically significant reduction in media abundance (Figure S2B) or M/L ratio (Figure 2A). Though a trend was observed consistent with a decreased secretion of F5, a known LMAN1 cargo, these changes did not reach statistical significance. Taken together, these data suggest that LMAN1 traffics a small number of secretory cargoes in HuH7 cells. Though we cannot exclude technical limitations including incomplete LMAN1 deletion, the marked LMAN1 depletion observed in LMAN1-deficient cell lysates by mass spectrometry (Figure S2A) and immunoblotting (Figure S3) suggests that the latter explanation is unlikely.
Wide Range of Proteins Depend on SURF4 for Secretion in HuH7 Cells
In contrast to LMAN1, SURF4 disruption in HuH7 cells caused a significant decrease in the M/L ratio for numerous signal peptide-containing proteins [log2(fc) < −1 and adjust p-value < 0.05]. For most proteins, this change was associated with both an increase in lysate abundance and a decrease in media abundance (Figure S4 and Tables S6 and S7). Affected proteins include known SURF4 cargoes such as APOB, APOA1, and APOA2,27,28 several putative cargoes identified in other mass spectrometry based studies,24,36 as well as a number of potentially novel SURF4 cargoes (Figure 2B and Table S8). We conducted Gene Ontology (GO) enrichment analysis49,50 using PANTHER51 to assess the biological processes affected by potential SURF4 cargoes. Analyzing secreted proteins sensitive to SURF4 deletion (i.e., showing a significant increase in lysate abundance and a decrease in media abundance and M/L ratios) revealed significant enrichment of proteins involved in multiple lipoprotein and cholesterol metabolism pathways, consistent with the hepatocyte origin of the HuH7 cell line (Figure S5A–C) and the previously reported role of SURF4 in the secretion of lipid-related proteins.23–28
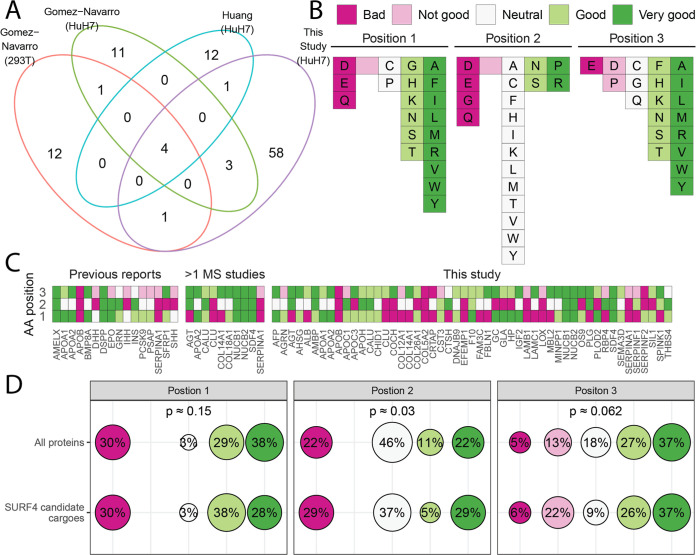
Though PCSK9 has been demonstrated to be dependent on SURF4 for efficient secretion in vivo(25) and in HEK293T cells in vitro,23 we did not identify PCSK9 as a SURF4 cargo in HuH7 cells, consistent with a previous report.52 7 out of 2726 proteins in our data set were among the other known SURF4 cargoes from previously published studies, 4 of which showed a significantly decreased M/L ratio. Comparison of protein abundance in the media with the effect of cargo receptor deletion suggested a greater dependency of highly abundant proteins on SURF4, but not LMAN1, for their secretion (Figure 2C,D). A comparison of the putative SURF4 cargoes identified here with those in two other recent reports24,36 identified 4 secreted proteins shared between all data sets (Figure 3A and Table S9) and 10 between two or more data sets (Figure 3A,C).
Figure 3.
Most SURF4 cargoes do not contain an ER-ESCAPE motif.29 (A) Venn diagram of putative SURF4 cargoes that were identified in the current study, or previously reported by Gomez-Navarro et al.(24) in HEK293T and HuH7 cells following analyses of conditioned media, or by Huang et al.(36) using an in vitro COPII vesicle formation assay. (B) Classification system for the three amino acid residues immediate downstream of the signal peptide cleavage site (ER-ESCAPE tripeptide motif) proposed by Yin et al.(29) (C) Tripeptide motifs in previously reported SURF4 cargoes,23–34 in cargoes that were identified in more than one MS-based data sets, and in this study color-coded according to (B). (D) Percentage of each amino acid class [as described in (B)] at each position in the tripeptide motif for all secreted proteins with an annotated signal peptide in the proteome (n = 2110, see Methods) and for SURF4 candidate cargoes (n = 86). P-values were obtained from Fisher’s exact test.
Most SURF4-Dependent Secreted Proteins Lack an ER-ESCAPE Motif
Previously, Yin et al.(29) proposed a tripeptide motif (ER-ESCAPE) as being responsible for the recognition and recruitment of SURF4 cargoes. This motif is located immediately downstream of the signal peptide and is characterized by a proline residue flanked on either side by a hydrophobic amino acid (Figure 3B). Qualitative analysis of candidate SURF4 cargoes identified in our study and in previous studies revealed a wide range of sequences at the predicted site of the ER-ESCAPE motif, including some that fit the motif well (NUCB1, NUCB2, and SDF4) and others with no apparent overlap (COL5A2 and APOB)25–28 (Figure 3C and Table S10).
To determine whether the ER-ESCAPE motif was significantly enriched among SURF4 cargoes, we compared the tripeptide sequences from the full set of candidate SURF4 cargoes in relation to all proteins in the human proteome with an annotated signal peptide (Tables S11, S12). As shown in Figure 3D, there are no significant differences in the distribution of each amino acid class within the tripeptide motif between SURF4 candidate cargoes that have been reported to date and all secreted proteins with an annotated signal peptide in the proteome. In addition, we also found that among the proteins detected in our data sets, those that are not SURF4 cargoes generally carry “stronger” ER-ESCAPE motifs relative to those of SURF4 cargoes (Figure S6). These analyses suggest that the ER-ESCAPE motif may be relevant only for a limited subset of SURF4 cargoes.
Recent studies have also suggested a key role for the Cardin–Weintraub (CW) motif [K/R][K/R][K/R]XX[K/R][K/R] in recognition by SURF4 for cargoes such as the hedgehog family.32,35 Analysis of the human proteome identified 267 proteins carrying a CW motif among 7327 proteins that carry a signal peptide or a transmembrane domain. Among the 115 SURF4 candidate cargoes identified by our and previous studies (Figure 3C), seven carry a CW motif, though only one of these seven proteins was identified in the current study (Table S13).
Discussion
In this study, we applied a broad, whole proteome approach39 to profile the cargo repertoires for two well-characterized cargo receptors, LMAN1 and SURF4, in the human hepatocellular carcinoma cell line, HuH7. Though only a limited set of LMAN1-dependent cargoes was identified in HuH7 cells, SURF4 facilitated secretion for a broad range of proteins. A subset of putative SURF4 cargoes carry the previously described ER-ESCAPE29 or CW motifs32,35 potentially mediating interaction with SURF4. However, most do not, suggesting a more complex mechanism governing the SURF4 cargo recognition.
While several established LMAN1 cargoes, including F5 and SERPINA1, were detected in our data set, with a trend toward a reduced M/L ratio in LMAN1-deleted cells, these changes did not reach statistical significance. These findings suggest limited statistical power, potentially due to only partial secretion blockade in LMAN1-deficient HuH7 cells and/or overlap with other cargo receptors. Indeed, humans and mice with complete LMAN1 deficiency exhibit incomplete reduction in plasma levels for two key LMAN1 cargoes, F5 and F8, to only ∼10% (humans) and 50% (mice) of those in wild-type controls.17,53 Similarly, levels for the LMAN1 cargo SERPINA1 are unchanged in the plasma of Lman1–/– mice, though modest accumulation is observed in hepatocytes.17 Although the lysosomal proteins CTSC and CTSZ have also been proposed as cargoes for LMAN1,14,15 our experiment approach would not be expected to distinguish protein accumulation in different cellular compartments (i.e., the ER vs the lysosome). Taken together, these data suggest that LMAN1 traffics only a small subset of proteins in HuH7 cells.
In contrast to LMAN1, the depletion of SURF4 significantly impeded the secretion of 52 proteins, suggesting a much broader cargo repertoire for SURF4 than for LMAN1. SERPINA1, a previously reported LMAN1 cargo,11 also exhibited a reduced M/L ratio following SURF4 deletion, suggesting dependence on SURF4 as well as LMAN1 for efficient secretion. These findings are consistent with a recent report demonstrating a role for SURF4 in the secretion of pathogenic SERPINA1 polymers as well as SERPINA1 monomers, albeit to a lesser extent than LMAN1.24,31 The large difference in the number of potential clients for LMAN1 and SURF4 could also help explain the normal development and only mild bleeding defect observed in LMAN1-deficient mice and human17,53 in contrast to the early embryonic lethality of Surf4–/– mice54 and the lack of human disorders associated with mutations in SURF4.
Though the role of the ER-ESCAPE motif proposed by Yin et al.(29) in the efficient SURF4-mediated trafficking of PCSK9 and NUCB1 has been confirmed by Gomez-Navarro and colleagues,24 our analysis suggests a more complex process for SURF4 cargo selection. We failed to confirm a general enrichment for the ER-ESCAPE motif among the broader repertoire of potential SURF4 cargoes. However, we cannot exclude the possibility that the tripeptide motif is shifted from the expected starting position in the majority of cargoes.24 It is also possible that SURF4 indirectly interacts with cargoes via a cofactor, with each cofactor having a different recognition motif. Lastly, the broad range of proteins that rely on SURF4 for secretion also suggests the possibility that SURF4 could function as a general mediator of ER-Golgi transport, promoting secretion in a way other than directly binding to its cargoes. For example, a recent study reported that SURF4 regulates the entrance of secretory proteins into a tubular network lacking LMAN1 and extending from the ER, which expedites protein delivery to the Golgi, suggesting a distinct SURF4 trafficking route.55
Our data demonstrated that genetic inactivation of LMAN1 and SURF4 using the lentiCRISPR system is efficient and specific.41 The only proteins that significantly decrease in cell lysates from LMAN1-null cells are LMAN1 and MCFD2, with SURF4 being the only such protein in SURF4-deleted cells. We also previously reported that the secretion defect seen in SURF4-deficient HEK293T cells generated with the same SURF4 gRNA used in this study was efficiently rescued by expression of a CRISPR-resistant SURF4 variant.23 Taken together, these data suggest minimal off-target effects with this CRISPR-mediated gene targeting approach. Finally, our data confirm a previously proposed model that LMAN1 and MCFD2 form a cargo receptor complex and that MCFD2 is retained in the ER solely due to its interaction with LMAN1.36,48 We found no evidence for a similar SURF4 cofactor with SURF4 being the only protein demonstrating a significant decrease in protein abundance in the cell lysates of SURF4 null cells.
In this report, we demonstrated the analysis of protein abundance in both cell lysates and condition media to identify secreted proteins that depend on LMAN1 and SURF4 for secretion. This approach could also be applied to other cargo receptors and cell types. In addition, a similar strategy could also be extended to study changes in cell secretomes resulting from disruptions in the conventional and unconventional secretory pathways as well as in response to external stimuli.
Acknowledgments
This work was supported by grants from the National Institute of Health (R35HL135793 to D.G., R01GM094231 to A.I.N., K08HL148552 to B.T.E., R01HL157062 and R01HL148333 to R.K., and K99GM141268 to P.S.A.). V.T.T. was supported by fellowships from the American Heart Association (20PRE35210706) and the University of Michigan Horace H. Rackham School of Graduate Studies. D.G. is a Howard Hughes Medical Institute Investigator.
Supporting Information Available
The following Supporting Information is available free of charge at ACS Web site. The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jproteome.3c00259.
Comparison of protein abundance in the cell media and lysates improves the identification of secreted proteins, comparison of protein abundance in the lysates and media between LMAN1-deficient and control cells, efficient depletion of LMAN1 in cells treated with LMAN1-targeting gRNA, comparison of protein abundance in the lysates and media between SURF4-deficient and control cells, gene ontology (GO) enrichment analysis for biological process terms, and proteins that are not SURF4 cargoes on average carrying a better ER-ESCAPE motif than putative SURF4 cargoes (PDF)
Protein abundance in cell lysate and media fractions (FragPipe output), M/L ratio for proteins that were identified in both fractions, comparison of the M/L ratio between LMAN1-deficient and NT cells (limma output), comparison of the protein abundance in cell lysates between LMAN1-deficient and NT cells (limma output), comparison of the protein abundance in conditioned media between LMAN1-deficient and NT cells (limma output), comparison of the protein abundance in cell lysates between SURF4-deficient and NT cells (limma output), comparison of the protein abundance in conditioned media between SURF4-deficient and NT cells (limma output), comparison of the M/L ratio between SURF4-deficient and NT cells (limma output), putative SURF4 cargoes that were identified in Gomez-Navarro et al., Huang et al., and this study, SURF4 cargoes that are identified in previous studies, >2MS studies, this study, and their tripeptide motif, putative SURF4 cargoes and their tripeptide motif, proteins with a signal peptide in the proteome and their tripeptide motif, and proteins with a signal peptide or a transmembrane domain in the proteome and their CW motif (XLSX)
Author Present Address
†† Room 5028 Life Sciences Institute 210 Washtenaw Avenue Ann Arbor, MI
The authors declare no competing financial interest.
Supplementary Material
References
- Uhlen M.; Fagerberg L.; Hallstrom B. M.; Lindskog C.; Oksvold P.; Mardinoglu A.; Sivertsson A.; Kampf C.; Sjostedt E.; Asplund A.; et al. Tissue-based map of the human proteome. Science 2015, 347 (6220), 1260419. 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- Uhlen M.; Karlsson M. J.; Hober A.; Svensson A. S.; Scheffel J.; Kotol D.; Zhong W.; Tebani A.; Strandberg L.; Edfors F.; et al. The human secretome. Sci. Signal. 2019, 12 (609), eaaz0274 10.1126/scisignal.aaz0274. [DOI] [PubMed] [Google Scholar]
- Tang V. T.; Ginsburg D. Cargo selection in endoplasmic reticulum-to-Golgi transport and relevant diseases. J. Clin. Invest. 2023, 133 (1), e163838 10.1172/JCI163838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanetti G.; Pahuja K. B.; Studer S.; Shim S.; Schekman R. COPII and the regulation of protein sorting in mammals. Nat. Cell Biol. 2012, 14 (1), 20–28. 10.1038/ncb2434. [DOI] [PubMed] [Google Scholar]
- Markova E. A.; Zanetti G. Visualizing membrane trafficking through the electron microscope: Cryo-tomography of coat complexes. Acta Crystallogr. Sect. D: Struct. Biol. 2019, 75 (5), 467–474. 10.1107/S2059798319005011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel A. V.; Chang C. L.; Shtengel G.; Xu C. S.; Hoffman D. P.; Freeman M.; Iyer N.; Aaron J.; Khuon S.; Bogovic J.; et al. ER-to-Golgi protein delivery through an interwoven, tubular network extending from ER. Cell 2021, 184 (9), 2412–2429.e16. 10.1016/j.cell.2021.03.035. [DOI] [PubMed] [Google Scholar]
- Barlowe C.; Helenius A. Cargo Capture and Bulk Flow in the Early Secretory Pathway. Annu. Rev. Cell Dev. Biol. 2016, 32 (1), 197–222. 10.1146/annurev-cellbio-111315-125016. [DOI] [PubMed] [Google Scholar]
- Schindler R.; Itin C.; Zerial M.; Lottspeich F.; Hauri H. P. ERGIC-53, a membrane protein of the ER-Golgi intermediate compartment, carries an ER retention motif. Eur. J. Cell Biol. 1993, 61 (1), 1–9. [PubMed] [Google Scholar]
- Nichols W. C.; Seligsohn U.; Zivelin A.; Terry V. H.; Hertel C. E.; Wheatley M. A.; Moussalli M. J.; Hauri H. P.; Ciavarella N.; Kaufman R. J.; et al. Mutations in the ER-Golgi intermediate compartment protein ERGIC-53 cause combined deficiency of coagulation factors V and VIII. Cell 1998, 93 (1), 61–70. 10.1016/S0092-8674(00)81146-0. [DOI] [PubMed] [Google Scholar]
- Neerman-Arbez M.; Johnson K. M.; Morris M. A.; McVey J. H.; Peyvandi F.; Nichols W. C.; Ginsburg D.; Rossier C.; Antonarakis S. E.; Tuddenham E. G. Molecular analysis of the ERGIC-53 gene in 35 families with combined factor V-factor VIII deficiency. Blood 1999, 93 (7), 2253–2260. 10.1182/blood.V93.7.2253. [DOI] [PubMed] [Google Scholar]
- Nyfeler B.; Reiterer V.; Wendeler M. W.; Stefan E.; Zhang B.; Michnick S. W.; Hauri H. P. Identification of ERGIC-53 as an intracellular transport receptor of α1-antitrypsin. J. Cell Biol. 2008, 180 (4), 705–712. 10.1083/jcb.200709100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.; Hojo S.; Matsumoto N.; Yamamoto K. Regulation of Mac-2BP secretion is mediated by its N-glycan binding to ERGIC-53. Glycobiology 2013, 23 (7), 904–916. 10.1093/glycob/cwt027. [DOI] [PubMed] [Google Scholar]
- Duellman T.; Burnett J.; Shin A.; Yang J. LMAN1 (ERGIC-53) is a potential carrier protein for matrix metalloproteinase-9 glycoprotein secretion. Biochem. Biophys. Res. Commun. 2015, 464 (3), 685–691. 10.1016/j.bbrc.2015.06.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollenweider F.; Kappeler F.; Itin C.; Hauri H. P. Mistargeting of the lectin ERGIC-53 to the endoplasmic reticulum of HeLa cells impairs the secretion of a lysosomal enzyme. J. Cell Biol. 1998, 142 (2), 377–389. 10.1083/jcb.142.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyfeler B.; Michnick S. W.; Hauri H. P. Capturing protein interactions in the secretory pathway of living cells. Proc. Natl. Acad. Sci. U.S.A. 2005, 102 (18), 6350–6355. 10.1073/pnas.0501976102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y. L.; Zhang B.; Mu T. W. LMAN1 (ERGIC-53) promotes trafficking of neuroreceptors. Biochem. Biophys. Res. Commun. 2019, 511 (2), 356–362. 10.1016/j.bbrc.2019.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B.; Zheng C.; Zhu M.; Tao J.; Vasievich M. P.; Baines A.; Kim J.; Schekman R.; Kaufman R. J.; Ginsburg D. Mice deficient in LMAN1 exhibit FV and FVIII deficiencies and liver accumulation of α1-antitrypsin. Blood 2011, 118 (12), 3384–3391. 10.1182/blood-2011-05-352815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrovic S.; Ben-Tekaya H.; Koegler E.; Gruenberg J.; Hauri H. P. The Cargo Receptors Surf4, Endoplasmic Reticulum-Golgi Intermediate Compartment (ERGIC)-53, and p25 Are Required to Maintain the Architecture of ERGIC and Golgi. Mol. Biol. Cell 2008, 19 (5), 1976–1990. 10.1091/mbc.e07-10-0989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armes N.; Fried M. Surfeit locus gene homologs are widely distributed in invertebrate genomes. Mol. Cell. Biol. 1996, 16 (10), 5591–5596. 10.1128/MCB.16.10.5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belden W. J.; Barlowe C. Role of Erv29p in Collecting Soluble Secretory Proteins into ER-Derived Transport Vesicles. Science 2001, 294 (5546), 1528–1531. 10.1126/science.1065224. [DOI] [PubMed] [Google Scholar]
- Caldwell S. R.; Hill K. J.; Cooper A. A. Degradation of Endoplasmic Reticulum (ER) Quality Control Substrates Requires Transport between the ER and Golgi. J. Biol. Chem. 2001, 276 (26), 23296–23303. 10.1074/jbc.M102962200. [DOI] [PubMed] [Google Scholar]
- Otte S.; Barlowe C. Sorting signals can direct receptor-mediated export of soluble proteins into COPII vesicles. Nat. Cell Biol. 2004, 6 (12), 1189–1194. 10.1038/ncb1195. [DOI] [PubMed] [Google Scholar]
- Emmer B. T.; Hesketh G. G.; Kotnik E.; Tang V. T.; Lascuna P. J.; Xiang J.; Gingras A. C.; Chen X. W.; Ginsburg D. The cargo receptor SURF4 promotes the efficient cellular secretion of PCSK9. Elife 2018, 7, 1–18. 10.7554/eLife.38839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Navarro N.; Maldutyte J.; Poljak K.; Peak-Chew S. Y.; Orme J.; Bisnett B. J.; Lamb C. H.; Boyce M.; Gianni D.; Miller E. A. Selective inhibition of protein secretion by abrogating receptor-coat interactions during ER export. Proc. Natl. Acad. Sci. U.S.A. 2022, 119 (31), e2202080119 10.1073/pnas.2202080119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang V. T.; McCormick J.; Xu B.; Wang Y.; Fang H.; Wang X.; Siemieniak D.; Khoriaty R.; Emmer B. T.; Chen X. W.; et al. Hepatic inactivation of murine Surf4 results in marked reduction in plasma cholesterol. Elife 2022, 11, e82269 10.7554/eLife.82269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saegusa K.; Sato M.; Morooka N.; Hara T.; Sato K. SFT-4 Surf4 control ER export of soluble cargo proteins and participate in ER exit site organization. J. Cell Biol. 2018, 217, 2073–2085. 10.1083/jcb.201708115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.; Wang H.; Xu B.; Huang D.; Nie C.; Pu L.; Zajac G. J. M.; Yan H.; Zhao J.; Shi F.; et al. Receptor-Mediated ER Export of Lipoproteins Controls Lipid Homeostasis in Mice and Humans. Cell Metab. 2021, 33 (2), 350–366.e7. 10.1016/j.cmet.2020.10.020. [DOI] [PubMed] [Google Scholar]
- Wang B.; Shen Y.; Zhai L.; Xia X.; Gu H.-m.; Wang M.; Zhao Y.; Chang X.; Alabi A.; Xing S.; et al. Atherosclerosis-associated hepatic secretion of VLDL but not PCSK9 is dependent on cargo receptor protein Surf4. J. Lipid Res. 2021, 62, 100091. 10.1016/j.jlr.2021.100091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y.; Garcia M. R.; Novak A. J.; Saunders A. M.; Ank R. S.; Nam A. S.; Fisher L. W. Surf4 (Erv29p) binds amino-terminal tripeptide motifs of soluble cargo proteins with different affinities, enabling prioritization of their exit from the endoplasmic reticulum. PLoS Biol. 2018, 16 (8), e2005140 10.1371/journal.pbio.2005140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z.; King R.; Tang V.; Myers G.; Balbin-Cuesta G.; Friedman A.; McGee B.; Desch K.; Ozel A. B.; Siemieniak D.; et al. The Endoplasmic Reticulum Cargo Receptor SURF4 Facilitates Efficient Erythropoietin Secretion. Mol. Cell. Biol. 2020, 40 (23), 001800-20. 10.1128/MCB.00180-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordonez A.; Harding H. P.; Marciniak S. J.; Ron D. Cargo receptor-assisted endoplasmic reticulum export of pathogenic α1-antitrypsin polymers. Cell Rep. 2021, 35 (7), 109144. 10.1016/j.celrep.2021.109144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X.; Chen R.; Mesias V. S. D.; Wang T.; Wang Y.; Poljak K.; Fan X.; Miao H.; Hu J.; Zhang L.; et al. A SURF4-to-proteoglycan relay mechanism that mediates the sorting and secretion of a tagged variant of sonic hedgehog. Proc. Natl. Acad. Sci. U.S.A. 2022, 119 (11), e2113991119 10.1073/pnas.2113991119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saegusa K.; Matsunaga K.; Maeda M.; Saito K.; Izumi T.; Sato K. Cargo receptor Surf4 regulates endoplasmic reticulum export of proinsulin in pancreatic beta-cells. Commun. Biol. 2022, 5 (1), 458. 10.1038/s42003-022-03417-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devireddy S.; Ferguson S. M. Efficient progranulin exit from the ER requires its interaction with prosaposin, a Surf4 cargo. J. Cell Biol. 2022, 221 (2), e202104044 10.1083/jcb.202104044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X.; Wang T.; Guo Y. Export of polybasic motif-containing secretory proteins BMP8A and SFRP1 from the endoplasmic reticulum is regulated by surfeit locus protein 4 (SURF4): SURF4 regulates ER export of BMP8A and SFRP1. J. Biol. Chem. 2022, 298, 102687. 10.1016/j.jbc.2022.102687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.; Yin H.; Li B.; Wu Q.; Liu Y.; Poljak K.; Maldutyte J.; Tang X.; Wang M.; Wu Z.; et al. An in vitro vesicle formation assay reveals cargo clients and factors that mediate vesicular trafficking. Proc. Natl. Acad. Sci. U.S.A. 2021, 118 (35), e2101287118 10.1073/pnas.2101287118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grube L.; Dellen R.; Kruse F.; Schwender H.; Stühler K.; Poschmann G. Mining the Secretome of C2C12 Muscle Cells: Data Dependent Experimental Approach to Analyze Protein Secretion Using Label-Free Quantification and Peptide Based Analysis. J. Proteome Res. 2018, 17 (2), 879–890. 10.1021/acs.jproteome.7b00684. [DOI] [PubMed] [Google Scholar]
- Poschmann G.; Brenig K.; Lenz T.; Stühler K. Comparative Secretomics Gives Access to High Confident Secretome Data: Evaluation of Different Methods for the Determination of Bona Fide Secreted Proteins. Proteomics 2021, 21 (2), e2000178 10.1002/pmic.202000178. [DOI] [PubMed] [Google Scholar]
- Abbineni P. S.; Tang V. T.; da Veiga Leprevost F.; Basrur V.; Xiang J.; Nesvizhskii A. I.; Ginsburg D. Identification of secreted proteins by comparison of protein abundance in conditioned media and cell lysates. Anal. Biochem. 2022, 655, 114846. 10.1016/j.ab.2022.114846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabayashi H.; Miyano K.; Sato J.; Yamane T.; Taketa K. Growth of human hepatoma cell lines with differentiated functions in chemically defined medium. Cancer Res. 1982, 42 (9), 3858–3863. [PubMed] [Google Scholar]
- Sanjana N. E.; Shalem O.; Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 2014, 11 (8), 783–784. 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purushothaman A. Exosomes from Cell Culture-Conditioned Medium: Isolation by Ultracentrifugation and Characterization. Methods Mol. Biol. 2019, 1952, 233–244. 10.1007/978-1-4939-9133-4_19. [DOI] [PubMed] [Google Scholar]
- da Veiga Leprevost F.; Haynes S. E.; Avtonomov D. M.; Chang H. Y.; Shanmugam A. K.; Mellacheruvu D.; Kong A. T.; Nesvizhskii A. I. Philosopher: a versatile toolkit for shotgun proteomics data analysis. Nat. Methods 2020, 17 (9), 869–870. 10.1038/s41592-020-0912-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A. T.; Leprevost F. V.; Avtonomov D. M.; Mellacheruvu D.; Nesvizhskii A. I. MSFragger: ultrafast and comprehensive peptide identification in mass spectrometry-based proteomics. Nat. Methods 2017, 14 (5), 513–520. 10.1038/nmeth.4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djomehri S. I.; Gonzalez M. E.; da Veiga Leprevost F.; Tekula S. R.; Chang H. Y.; White M. J.; Cimino-Mathews A.; Burman B.; Basrur V.; Argani P.; et al. Quantitative proteomic landscape of metaplastic breast carcinoma pathological subtypes and their relationship to triple-negative tumors. Nat. Commun. 2020, 11 (1), 1723. 10.1038/s41467-020-15283-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A.; Martin M. J.; Orchard S.; Magrane M.; Agivetova R.; Ahmad S.; Alpi E.; Bowler-Barnett E. H.; Britto R.; Bursteinas B.; et al. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49 (D1), D480–D489. 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie M. E.; Phipson B.; Wu D.; Hu Y.; Law C. W.; Shi W.; Smyth G. K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43 (7), e47 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyfeler B.; Zhang B.; Ginsburg D.; Kaufman R. J.; Hauri H. P. Cargo selectivity of the ERGIC-53/MCFD2 transport receptor complex. Traffic 2006, 7 (11), 1473–1481. 10.1111/j.1600-0854.2006.00483.x. [DOI] [PubMed] [Google Scholar]
- Ashburner M.; Ball C. A.; Blake J. A.; Botstein D.; Butler H.; Cherry J. M.; Davis A. P.; Dolinski K.; Dwight S. S.; Eppig J. T.; et al. Gene Ontology: tool for the unification of biology. Nat. Genet. 2000, 25 (1), 25–29. 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleksander S. A.; Balhoff J.; Carbon S.; Cherry J. M.; Drabkin H. J.; Ebert D.; Feuermann M.; Gaudet P.; Harris N. L.; et al. The Gene Ontology knowledgebase in 2023. Genetics 2023, 224 (1), iyad031. 10.1093/genetics/iyad031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P. D.; Ebert D.; Muruganujan A.; Mushayahama T.; Albou L. P.; Mi H. PANTHER: Making genome-scale phylogenetics accessible to all. Protein Sci. 2022, 31 (1), 8–22. 10.1002/pro.4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y.; Wang B.; Deng S.; Zhai L.; Gu H. m.; Alabi A.; Xia X.; Zhao Y.; Chang X.; Qin S.; et al. Surf4 regulates expression of proprotein convertase subtilisin/kexin type 9 (PCSK9) but is not required for PCSK9 secretion in cultured human hepatocytes. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2020, 1865 (2), 158555. 10.1016/J.BBALIP.2019.158555. [DOI] [PubMed] [Google Scholar]
- Zhang B.; McGee B.; Yamaoka J. S.; Guglielmone H.; Downes K. A.; Minoldo S.; Jarchum G.; Peyvandi F.; de Bosch N. B.; Ruiz-Saez A.; et al. Combined deficiency of factor V and factor VIII is due to mutations in either LMAN1 or MCFD2. Blood 2006, 107 (5), 1903–1907. 10.1182/blood-2005-09-3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmer B. T.; Lascuna P. J.; Kotnik E. N.; Saunders T. L.; Khoriaty R.; Ginsburg D. Murine Surf4 is essential for early embryonic development. PLoS One 2020, 15, e0227450 10.1371/journal.pone.0227450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R.; Chen K.; Wang B.; Xu K. SURF4-induced tubular ERGIC selectively expedites ER-to-Golgi transport. Dev. Cell 2022, 57 (4), 512–525.e8. 10.1016/j.devcel.2021.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw and processed mass spectrometry data have been deposited to the MassIVE database with the data set identifier: MSV000092642.