Abstract
Humus samples were collected 12 growing seasons after the start of a simulated acid rain experiment situated in the subarctic environment. The acid rain was simulated with H2SO4, a combination of H2SO4 and HNO3, and HNO3 at two levels of moderate acidic loads close to the natural anthropogenic pollution levels of southern Scandinavia. The higher levels of acid applications resulted in acidification, as defined by humus chemistry. The concentrations of base cations decreased, while the concentrations of exchangeable H+, Al, and Fe increased. Humus pH decreased from 3.83 to 3.65. Basal respiration decreased with decreasing humus pH, and total microbial biomass, measured by substrate-induced respiration and total amount of phospholipid fatty acids (PLFA), decreased slightly. An altered PLFA pattern indicated a change in the microbial community structure at the higher levels of acid applications. In general, branched fatty acids, typical of gram-positive bacteria, increased in the acid plots. PLFA analysis performed on the bacterial community growing on agar plates also showed that the relative amount of PLFA specific for gram-positive bacteria increased due to the acidification. The changed bacterial community was adapted to the more acidic environment in the acid-treated plots, even though bacterial growth rates, estimated by thymidine and leucine incorporation, decreased with pH. Fungal activity (measured as acetate incorporation into ergosterol) was not affected. This result indicates that bacteria were more affected than fungi by the acidification. The capacity of the bacterial community to utilize 95 different carbon sources was variable and only showed weak correlations to pH. Differences in the toxicities of H2SO4 and HNO3 for the microbial community were not found.
During the past few decades, the impact of acid deposition on the environment has been under intensive study. Changes in the soil chemical status and the function of the decomposer community may lead to imbalances in nutrient cycling and productivity of the ecosystem. Both direct and indirect soil-mediated effects of an increased acid load on the size, composition, and activity of the soil microbes have been reported (e.g. 19, 29), as have microbe-mediated changes in soil processes, such as litter decomposition (34).
Strong reductions in microbial activity will easily be detected if unrealistically high doses of acids are applied. The soil ecosystem may also be influenced by a subtle but permanent increase in the acidity of rain. However, to our knowledge no such studies exist. This acidification experiment in northern Finland is an attempt to study such subtle effects, since the annual sulfur and nitrogen loads applied are within the range of deposition over large areas in central Europe (25). The 12-year duration of the experiment is also important, since the impact of artificial acidification on soil microbes is suggested to be dependent on the duration of exposure to acid (26, 39) as well as on the rate of acid application (36).
Our main aim was to characterize the impact of artificial acidification on the humus microbial community structure by applying total-community-level techniques available now in microbial ecology. We used different fungus- and bacterium-specific measurements to study if acidification causes shifts in these main decomposer groups. We also used methods to determine microbial community structure in more detail without determining the exact species composition. One of these methods, the analysis of phospholipid fatty acids (PLFA), was shown earlier to be efficient in detecting the effects of increasing pH on the microbial community (9, 10, 22). Since the cultivable part of the bacterial community has been reported several times to be affected by acidification (e.g., 8, 38), we also analyzed the community structure of the cultivable bacteria by plate counting and subsequent PLFA analysis. Biolog GN microtiter plate incubation of the total humus suspension was performed to detect changes in the substrate utilization potential of the microbial community (24). The possible adaptation of the bacteria to the decreased humus pH was determined by the thymidine incorporation technique (6). We also compared bacterial activities with fungal activities to elucidate which of these organism groups might be mainly responsible for the previously detected decrease in soil respiration rate due to acidification in the study plots (39).
MATERIALS AND METHODS
Study area and sampling.
The study area is situated near Kevo Subarctic Research Station in northern Finland, where the growing season is 110 to 125 days. The area is a dry, nutrient-poor, mixed pine-mountain birch woodland (Pinus sylvestris, Betula pubescens subsp. czerepanovii). The ground floor forms patches of dwarf shrub and lichen.
The study plots belong to a large-scale investigation in which the effects of acid rain on the subarctic ecosystem have been monitored since 1985 (30). Each plot (5 by 5 m) supported at least one pine and one mountain birch. The total number of plots used in the present study was 60, situated in three adjacent subareas. The treatments were dry control (D), irrigated control (W) treated with spring water (pH 6), and two levels of simulated acid rain, medium (m) and high (h). Originally (1985 to 1988), simulated acid rain was prepared by adding both H2SO4 and HNO3 (1.9:1 [wt/wt]) (SN subarea). In 1989, the treatments were modified as follows. In the SN subarea, the treatments continued unchanged (SN-m, pH 3.8; SN-h, pH 2.9), but in the S subarea only H2SO4 (S-m, pH 4.1; S-h, pH 3.1) was applied and in the N subarea only HNO3 (N-m, pH 4.7; N-h, pH 3.4) was applied. Plots were treated two or three times per week during June, July, and August. The cumulative S and N loads are shown in Table 1.
TABLE 1.
Cumulative acid loads of the Kevo study area
| Treatment | Cumulative acid load, 1985–1995 (g/m2)
|
|
|---|---|---|
| S | N | |
| W | 0.93 | 0.17 |
| SN-m | 4.40 | 1.38 |
| SN-h | 22.1 | 7.59 |
| S-m | 4.40 | 0.60 |
| S-h | 22.1 | 2.79 |
| N-m | 2.13 | 1.32 |
| N-h | 8.38 | 7.25 |
Humus samples were collected with a soil corer (2.8-cm diameter, 30 cores from each plot) in July 1995. Samples were sieved (2.8-mm mesh) and stored at 4°C for 2 weeks before basal respiration, substrate-induced respiration (SIR), Biolog GN microtiter plate (Biolog Inc., Hayward, Calif.), or PLFA analyses were performed. Subsamples for determination of the incorporation of [3H]thymidine, [14C]leucine, and [14C]acetate, ergosterol measurement, and plate counts were frozen (−18°C) for 5 months before the analyses. The samples were thawed and kept at room temperature for 3 to 7 days before the analyses.
Chemical analyses.
Total organic carbon and nitrogen were determined by dry combustion (Leco CHN-600). For nutrient analyses, air-dried humus was extracted with 0.1 M BaCl2 and the suspension was analyzed with an inductively coupled plasma emission spectrometer (ICP-AES, ARL 3580). The same suspension was also titrated with NaOH to measure exchangeable acidity. Effective cation-exchange capacity (CEC) was expressed as centimoles kilogram−1, and base saturation (BS) was expressed as a percentage of Ca, Mg, K, and Na of CEC. Humus pH was measured in a water (1:1.7 [vol/vol]) suspension. Dry weight was determined by drying duplicate subsamples at 105°C overnight, and organic matter content was obtained by heating at 550°C for 4 h. Detailed descriptions of the chemical extractions are given by Tamminen and Starr (37).
Microbiological analyses.
The water content of fresh humus was adjusted to 60% of the water-holding capacity before respiration determinations. Basal respiration was then measured as CO2 evolved in 39 h. SIR (2), which measures the biomass C of active microbes (Cmic) in soil, was determined as described by Priha and Smolander (35). Fresh humus, equaling 2 g (dry weight), with two replicates, was used in both analyses.
Phospholipid extraction and analysis of PLFA were done as previously described by Frostegård et al. (23). The total amount of PLFA was used to indicate the total microbial biomass, and the sum of PLFA considered to be predominantly of bacterial origin (i15:0, a15:0, 15:0, i16:0, 16:1ω9, 16:1ω7t, i17:0, a17:0, 17:0, cy17:0, 18:1ω7, and cy19:0) was chosen as an index of the bacterial biomass (21). The quantity of 18:2ω6,9 was used as an indicator of fungal biomass since 18:2ω6,9 has been suggested to be mainly of fungal origin in soil (18) and is known to correlate with the amount of ergosterol (21), a sterol found only in fungi.
Biolog GN microtiter plates containing 95 different carbon sources and a redox dye were inoculated with the homogenized and 10−4-diluted (0.9% NaCl) humus suspension. The plates were incubated at 20°C and read after 20, 46, 72, 92, 122, 140, and 164 h.
Bacterial growth rates were estimated by thymidine and leucine incorporation with the bacterial community extracted by homogenization-centrifugation as described by Bååth (4, 5). Two replicates were used for each humus sample.
The pH response of the bacterial community was measured, with some modifications, as described by Bååth (6). The pH of the extracted bacterial suspension was adjusted to approximately 3.8 with citrate-potassium phosphate buffer (final concentrations, 0.33 mM citric acid and 0.66 mM phosphate) or to 7.2 with potassium phosphate buffer (final concentration, 6.6 mM phosphate). The buffer concentrations were chosen in order to produce minimal inhibition of incorporation. The thymidine incorporation procedure was then performed as described above. The logarithm of the ratio of incorporation at pH 3.8 to that at pH 7.2 was used as an index of the bacterial community adaptation to pH, where a higher value indicates an increased tolerance for a lower pH (see reference 6 for further explanations).
Acetate incorporation into ergosterol (31) was used to estimate fungal growth rates. Humus suspensions (0.25 g [fresh weight] of humus and 1.5 ml of distilled water) were incubated for 18 h with labeled acetate (50 μl of [14C]acetate [2.11 GBq mmol−1] and 450 μl of 1 mM nonradioactive acetate) at 20°C. Incubation was stopped with 2 ml of 5% formalin. The humus slurry was centrifuged, and the supernatant was discarded. Ergosterol was then extracted and hydrolyzed according to Ek et al. (16), except that cyclohexane was used and the hydrolysis time at 70°C was 2 h. Ergosterol was analyzed by high-performance liquid chromatography. Ergosterol peaks were collected into scintillation vials. Relative {14C]acetate incorporation into ergosterol was expressed as disintegrations per minute per microgram of ergosterol.
The same homogenized bacterial suspension that was used for thymidine incorporation was used for plate counts. Suspensions were diluted with 0.1% peptone and spread on tryptone soy agar plates (0.2%). Plates were incubated at room temperature for 4 weeks, and the number of CFU, both total and yellow colonies, was calculated from plates with less than 80 CFU. Agar plates on which the number of CFU was high (about 100 to 300) were then flooded with citrate buffer, and a 1.5-ml portion of the bacterial suspension was taken for further analysis of PLFA as described above (CFU-PLFA).
Statistical analyses.
The results are expressed per organic matter content, since Vanhala et al. (39) found the organic C content of humus to be an important source of variation in the study plots. Subarea N had a lower cumulative proton load than subareas SN and S (Table 1), and the density of the natural vegetation was higher in subarea N than in the other two subareas. Therefore, the different treatments (D, W, m, and h) were not quite equivalent in the different subareas, and a two-factor analysis of variance (ANOVA) (including treatment and subarea) was not used. Instead, all of the measured variables, including the scores in the multivariate analysis (PLFA, CFU-PLFA, and Biolog measurements), were subjected to an analysis of covariance (ANCOVA). The analysis was performed with treatment and subarea as classifying variables and humus pH as the covariant. The interaction between subarea and treatment was also tested. The Newman-Keuls comparison of means was then performed if those sources of variation were found to be significant. A significant covariant revealed if the variable was related to pH and thus to acidification. A significant treatment effect indicated impacts which were due to something other than pH, e.g., the effect of watering. A significant subarea effect indicated a different response due to the composition of the acid. This kind of model thus enabled the testing of all 60 plots with the same analysis and allowed a comparison of the S and N subarea acids as well. Most of the results are presented by plotting all 60 study plots against humus pH. Variables which showed significant treatment effects are presented as an average for the five replicates of the treatments with standard error bars.
The moles percent of the individual PLFA were standardized by dividing them with their standard deviations before being subjected to principal-component analyses (PCA). Partial least-squares (PLS) regression analysis (Unscrambler II program) was used to compare the total humus PLFA and plated-community PLFA patterns. Calculation and standardization of the Biolog data on the basis of the reading time of the plates were performed following Garland (24). Reading times at which average well color development was between 0.7 and 1.0 were then selected for the statistical analysis. Absorbances of the separate wells were divided by the mean absorbance of the plate, and the results were subjected to PCA.
The study plots, although situated in a dry forest site, could be divided according to their original ground vegetation and moisture into three classes: dry, medium, and moist plots. The effect of vegetation was not included in the ANCOVA because of the uneven distribution of the vegetation within the treatments and also because it was already partly included in the model since it affected the soil pH. A separate ANOVA with vegetation, treatment, and subarea as sources of variation was, however, used to determine the reasons for the formation of principal components of the PLFA data.
RESULTS
Chemical analyses.
The results of the chemical analyses are presented in Table 2. At both the SN-h and the S-h plots, the pH of the humus layer decreased from 3.83 (mean for W plots) to 3.65 (mean for h plots). The N-h treatment, which had a considerably smaller proton load than the SN-h or S-h treatments and therefore a higher pH of the irrigation water, did not affect the humus pH. The concentrations of Ca, K, and Mg decreased with decreasing pH, leading to decreased BS and slightly lower CEC. The exchangeable acidity and the concentrations of Al and Fe increased due to acid treatments, but the trend was not significant in an ANCOVA model when pH was used as a covariant.
TABLE 2.
Chemical variables reported as means for five replicate plotsa
| Subarea | Treatment | pH | Organic matter | Corg | Ntot | C/N | H+ | Ca | K | Mg | Mn | Zn | Fe | Al | Na | CEC | BS |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SN | D | 3.79 | 67.1 | 36.2 | 0.99 | 36.5 | 122 | 3.47 | 752 | 643 | 301 | 126 | 61.9 | 329 | 94.6 | 42.2 | 58.7 |
| W | 3.85 | 64.7 | 33.1 | 0.95 | 34.5 | 106 | 3.83 | 794 | 738 | 339 | 181 | 48.3 | 218 | 92.1 | 42.1 | 65.2 | |
| m | 3.77 | 62.6 | 33.8 | 0.97 | 35.0 | 128 | 3.48 | 830 | 649 | 302 | 161 | 71.5 | 326 | 105.5 | 43.2 | 58.6 | |
| h | 3.66 | 61.2 | 30.9 | 0.90 | 34.4 | 146 | 2.45 | 757 | 423 | 207 | 146 | 100.1 | 464 | 75.0 | 39.0 | 45.9 | |
| S | D | 3.79 | 67.8 | 35.8 | 1.04 | 34.6 | 120 | 3.48 | 783 | 646 | 391 | 132 | 54.1 | 278 | 73.2 | 41.9 | 59.3 |
| W | 3.81 | 67.0 | 35.3 | 1.00 | 35.4 | 115 | 3.84 | 837 | 739 | 342 | 176 | 46.2 | 213 | 107.2 | 43.2 | 63.9 | |
| m | 3.78 | 58.6 | 32.9 | 0.92 | 35.8 | 119 | 3.46 | 752 | 633 | 337 | 145 | 80.7 | 394 | 93.1 | 42.8 | 57.9 | |
| h | 3.64 | 66.0 | 33.4 | 0.95 | 35.3 | 144 | 2.82 | 810 | 480 | 349 | 175 | 79.2 | 356 | 88.8 | 40.5 | 50.8 | |
| N | D | 3.87 | 61.9 | 33.3 | 0.93 | 35.6 | 114 | 3.84 | 952 | 651 | 413 | 140 | 69.0 | 286 | 63.4 | 43.7 | 62.1 |
| W | 3.85 | 62.8 | 33.4 | 0.92 | 36.1 | 129 | 3.85 | 925 | 674 | 374 | 201 | 55.4 | 256 | 113.7 | 45.1 | 61.4 | |
| m | 3.78 | 69.6 | 36.5 | 1.03 | 35.7 | 115 | 3.31 | 862 | 636 | 297 | 170 | 61.7 | 279 | 93.5 | 40.3 | 60.6 | |
| h | 3.81 | 72.6 | 37.8 | 1.08 | 35.1 | 117 | 3.57 | 937 | 690 | 409 | 181 | 62.2 | 211 | 91.8 | 42.2 | 62.1 | |
| SE of the error term | 0.56 | 0.017 | 0.28 | 2.87 | 0.06 | 18.7 | 12.8 | 18.1 | 5.36 | 3.37 | 19.6 | 2.57 | 0.51 | 0.80 | |||
| P for covariant soil pH | 0.00 | 0.05 | 0.00 | 0.08 | 0.00 | 0.01 | 0.00 | 0.00 | 0.51 | 0.20 | 0.70 | 0.40 | 0.00 | 0.00 | |||
| P for treatment effect | 0.12 | 0.77 | 0.05 | 0.36 | 0.48 | 0.63 | 0.08 | 0.20 | 0.01 | 0.00 | 0.22 | 0.00 | 0.87 | 0.04 |
Organic matter, organic C (Corg), and total N (Ntot) are given as percentages of dry matter. Most of the nutrients are expressed as micrograms gram of organic matter−1, except for Ca, which is expressed as milligrams gram of organic matter−1. Exchangeable acidity (H+) is given in microequivalents gram of organic matter−1, CEC is given in centimoles kilogram of organic matter−1, and BS is given as a percentage of Ca, Mg, K, and Na of CEC. The standard error (SE) of the error term was calculated by dividing the square root of the residual mean square by the square root of the number of study plots.
Microbial biomass and activity.
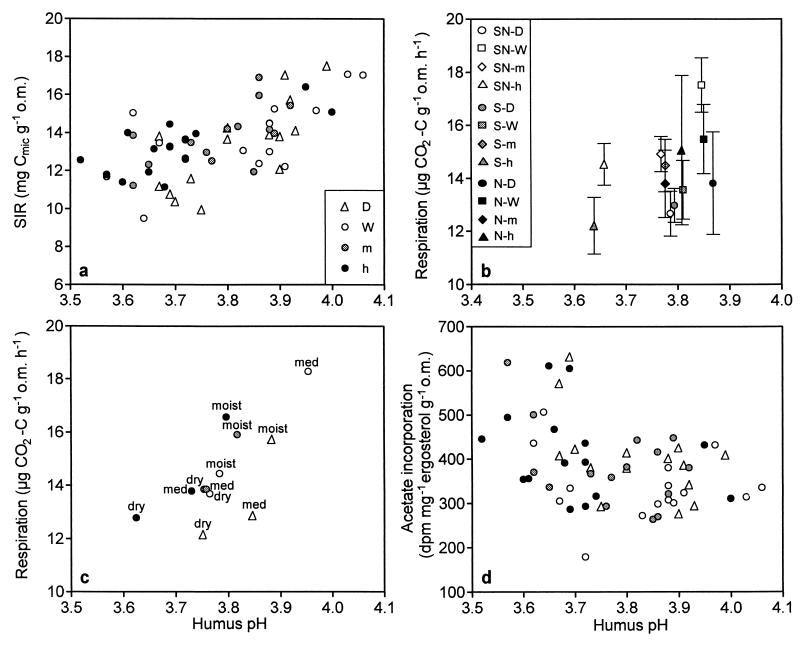
The total biomass of active microorganisms measured by SIR (Fig. 1a) and the total amount of microbe-derived PLFA (data not shown) were found to be dependent on the pH (P levels of the covariant pH were <0.001 and <0.01, respectively). SIR and total PLFA were positively correlated (r, 0.621). Actual reduction in the biomass due to the irrigation treatments was very small; e.g., the mean SIR biomass was 14.1 mg of Cmic g of organic matter−1 and the total PLFA biomass was 2.6 μmol g of organic matter−1 at SN-W plots, while the same values at SN-h plots were 13.1 mg of Cmic g of organic matter−1 and 2.4 μmol g of organic matter−1, respectively. Basal respiration rate was affected by both the humus pH and watering treatment (Fig. 1b). Respiration rate decreased (P < 0.001) with decreasing pH but increased (P < 0.05 for the treatment effect) with watering treatment. The changes in respiration rate were larger than the changes in biomass values, being 17.5, 12.7, and 14.5 μg of CO2-C g of organic matter−1 at SN-W, SN-D, and SN-h plots, respectively. The lower respiration rate of D plots (situated in dry and medium vegetation types) is also shown in Fig. 1c, where the respiration results with different treatments are presented according to the vegetation type of the study plot. In addition, the plots representing the driest vegetation type had relatively low humus pH and low respiration rate levels.
FIG. 1.
Effect of humus pH on total microbial biomass measured by SIR (a); basal respiration, where symbols indicate the mean for five replicate plots of the different treatments and bars indicate the standard error for the replicates (b); basal respiration, where symbols indicate the mean respiration level of the treatments situated within the same vegetation type, dry (dry), medium (med), or moist (moist) (c); and growth rate of fungi, estimated by [14C]acetate incorporation into ergosterol (d). Symbols for panels c and d are as described for panel a. o.m., organic matter.
Microbial community structure measured by PLFA.
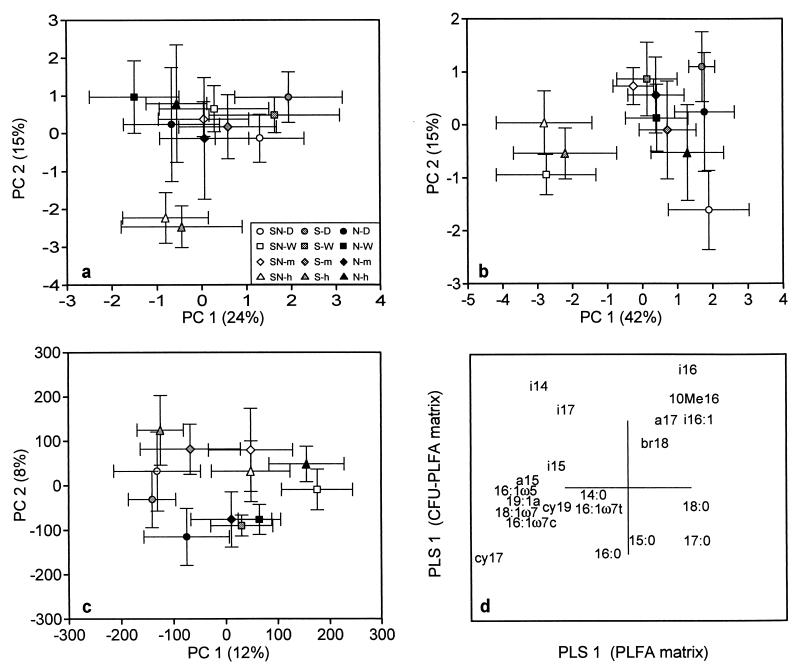
PCA of PLFA showed separation of high-acidity-level irrigated samples from the SN and S plots along the second principal component (PC 2) (Fig. 2a). When the scores of that component were subjected to ANCOVA, they were significantly affected by humus pH (P < 0.001). The individual PLFA responsible for the separation were 10Me16, i16:1, i16:0, and 18:1ω9, which showed a tendency for an increase in relative moles percent, whereas cy17, cy19, 19:1a, 18:1ω7, and 16:1ω7c decreased in the plots having a lower pH (data not shown). The actual differences in moles percent were small, however. The first principal component (PC 1) was not related to the humus pH. Scores of PC 1 were tested separately by ANOVA with treatment and vegetation as sources of variation. PC 1 had a significant vegetation effect. Samples from the driest vegetation type were found to the right side of the PCA plot, while samples from the moist vegetation type were found to the left side (data not shown). Thus, PC 1 for the PLFA pattern was at least partly influenced by vegetation, and PC 2 was influenced by humus pH. The two principal components together explained 39% of the variation.
FIG. 2.
Score plot of PCA showing the separation of the study plots along PC 1 and PC 2 determined with humus PLFA data (a), plated-community PLFA data (CFU-PLFA) (b), Biolog GN analysis (c), and comparison of humus PLFA and plated-community PLFA analyses with PLS regression (d). Bars indicate the standard error for five replicate plots.
Bacterial biomass and growth rate.
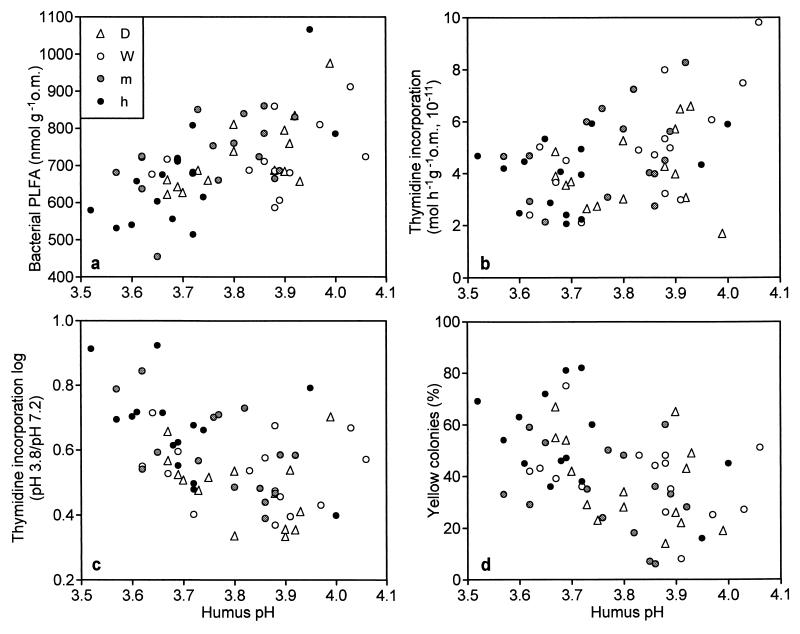
The amount of bacterial PLFA decreased (P < 0.001) in the study plots with decreasing pH (Fig. 3a). Also, the thymidine (Fig. 3b) and leucine (data not shown) incorporation rates decreased significantly (P < 0.001 for both) at the same plots, indicating a lower growth rate of bacteria at a lower pH. The decrease was also seen when the incorporation rates were calculated per microbial biomass C instead of per organic matter content (data not shown).
FIG. 3.
Effect of humus pH on PLFA of bacterial origin (see Materials and Methods) (a), thymidine incorporation rate for bacteria (b), bacterial community adaptation to pH (the growth rate index is the logarithm of the ratio of thymidine incorporation at pH 3.8 to that at pH 7.2) (c), and the proportion of yellow colonies among CFU (d). o.m., organic matter.
Adaptation of the bacterial community to pH.
Adaptation of the bacterial community to pH was expressed as the ratio of the thymidine incorporation rates in a humus suspension at low and high pHs. Higher values were found for the plots having a lower pH (Fig. 3c), indicating an increased tolerance of the bacterial community for a more acidic environment (P < 0.05).
Number and community structure of the colony-forming bacteria.
A tendency for a decrease, even though not significant, in viable counts of bacteria at a lower humus pH was seen (data not shown). An indication of an altered bacterial community was obtained by the increased percentage of yellow colonies (Fig. 3d) in the plots having a lower pH (P < 0.05). Also, the PLFA analysis of the bacteria growing on the agar plates revealed some changes in the bacterial community (Fig. 2b). D samples were found on the right side of the PCA plot, separating along PC 1 (explaining 42% of the variation). PC 1 also separated the SN-h and S-h irrigated samples on the left side of the plot, and both PC 1 and PC 2 showed a slight dependence on soil pH (P < 0.05 for both), even though many of the W plots (SN-W) also had a similar PLFA composition. The PCA plot of the individual PLFA from the plates indicated that the amount of the branched fatty acids increased in the samples with a lower pH (data not shown). The mean moles percent of i16:0, 10Me16, and i17:0 were 8.8, 5.8, and 3.6% at SN-W and S-W plots and 14.1, 7.5, and 4.9% at SN-h and S-h plots, respectively.
Comparison of the total humus community and the plated community.
In order to compare the changes in individual PLFA from direct humus analysis and the plated bacterial community, a PLS regression for the PLFA was performed. In the PLS regression, total humus PLFA data were used as the x matrix and plated-community PLFA data (CFU-PLFA) were used as the y matrix. As a result of the PLS regression, the score plot separated the acidified plots along the first PLS regression component (data not shown). PLS regression also created loading values for individual PLFA. The loading values for the first PLS regression component of the humus PLFA matrix are shown in the x axis, and the loading values for the first PLS regression component of the CFU-PLFA matrix are shown in the y axis (Fig. 2d). Especially i16:0, i16:1, and 10Me16 increased in both analyses, while cy17:0, cy19:0, 16:1ω7c, and 18:1ω7 decreased in both analyses. The changes in 15:0, 16:0, 17:0, 18:0, i14:0, and i17:0 did not follow this pattern.
Substrate utilization potential.
Study plots treated with high-level acid load were slightly separated along PC 2 (Fig. 2c). Scores of PC 1 and PC 2 were tested with ANCOVA, and the results showed them both to be slightly affected by soil pH (P < 0.05), indicating that there were some changes in substrate utilization due to humus pH. However, PC 1 mainly separated the D plots (P < 0.01 for the treatment effect). This finding, together with the fact that the two principal components explained only 20% of the variation in the Biolog GN data, makes the interpretation of the results difficult.
Fungal biomass and growth rate.
Fungal biomass, measured both with fungal PLFA 18:2ω6,9 and with ergosterol (data not shown), remained relatively unchanged during the experiment. The fungal growth rate was estimated by [14C]acetate incorporation into ergosterol (Fig. 1d). The changing pH did not have a significant impact on the incorporation rate, although the highest values were usually found at lower pHs.
DISCUSSION
According to the soil acidification hypothesis, which includes the loss of base cations and increases in exchangeable H+, Al, and Fe, possibly resulting in a lowered pH (37), the humus was acidified as a result of the acid treatments (Table 2). A small decrease in the humus pH at the higher acid load was also seen even though the natural variation in pH was large compared to the effect of the treatments.
Anderson and Domsch (3) found the total microbial biomass and respiration rate to be lower in naturally acidic soils than in soils with a neutral pH. In our study, the basal respiration rate (Fig. 1b) and to some extent the total biomass (Fig. 1a) were also dependent on pH and therefore on acidification, even though the pH range studied by us was much smaller. Respiration rate was also affected by irrigation (see results from the ANCOVA) and vegetation type (Fig. 1c). It is likely that the lack of watering was the reason for the lower respiration rate in the D plots, since increased moisture has been reported several times to raise microbial activity (40, 42). The lowest respiration rates were found at the plots situated in the dry vegetation type, irrespective of the treatment (Fig. 1c). Humus pH at the dry vegetation type tended to be lower than that at the other vegetation types, indicating that the lower respiration rate at the dry vegetation type might have been due to an originally lower pH at those areas. This lower pH might also be the reason why the dry vegetation type was more sensitive to the effects of acidification (30).
The analysis of humus PLFA revealed changes in microbial community structure due to pH (Fig. 2a). The individual PLFA more typical of the acid-treated plots separated along PC 2 and were considered to be of bacterial origin. Most of them, e.g., branched PLFA i16:0, i16:1, 10Me16, and 10Me17, are common in gram-positive bacteria (32). The differences in moles percent were, however, small. PLFA of eucaryotic origin, e.g., 18:2ω6,9 and 20:4 (1, 21), did not respond to pH but separated to the left along PC 1, the vegetation axis, indicating the connection of the fungal biomass to different vegetation types.
An altered humus bacterial community composition due to pH was also supported by the PLFA pattern of the cultivable bacterial population (Fig. 2b), since the S or SN plots treated with higher acid levels (in addition to some W plots) separated in the PCA. The increase in relative moles percent of branched PLFA in low-pH humus was larger but was much more variable in this analysis than in the humus PLFA analysis. PLS regression of the plated and direct humus PLFA analyses showed similarities between PLFA affected by pH (Fig. 2d). Most of the PLFA increased (i16:0, i16:1, and 10Me16) or decreased (cy17:0, cy19:0, 16:1ω7c, and 18:1ω7) with decreasing humus pH in both analyses. Some differences were also seen; for example, PLFA i14:0 and i17:0 increased in acidified plots when the cultivable community was analyzed and decreased in the same plots in the humus PLFA analysis. This result might indicate that some parts of the bacterial population are excluded or enriched on the agar plates. The differences in saturated straight-chain PLFA were probably due to these PLFA being present in eucaryotes in soil. As a whole, it appears that plate counts were not solely a result of selection by the plating technique, since the bacteria responsible for the greatest changes in bacterial PLFA due to acidification were probably able to grow on agar media.
In addition to the changes in humus and plated-community PLFA patterns, the proportion of yellow colonies among total CFU increased with decreasing humus pH (Fig. 3d), indicating an altered bacterial community composition. The percentage of yellow colonies has been reported to decrease with lime and ash treatments of coniferous forest soil resulting in a higher pH (7). An increase in coniferous forest humus pH has been reported to change the microbial community measured by PLFA analysis toward more gram-negative and fewer gram-positive bacteria (9, 10, 22). The abundance of PLFA common in gram-negative bacteria (cy17:0, 18:1ω7, 16:1ω5, and 16:1ω7c) (41) increased with increasing pH (22), while in our study these PLFA were more typical in the control areas with a higher pH. On the other hand, the levels of most of the fatty acids more abundant at the control areas of the liming treatments (i15:0, i16:0, and 10Me16) were found to be increased due to acidification. Consequently, coniferous forest humus seems to contain a bacterial group, consisting mainly of gram-positive bacteria, which easily adapts to an acid environment, and a group of bacteria, mainly gram-negative ones, which more easily adapts to humus with a more neutral pH. However, whether this situation represents a direct pH effect or an effect of pH altering carbon availability and thus selecting for more active bacteria, as suggested earlier (10), cannot be elucidated from the present results.
The potential of the bacterial community to degrade different carbon sources appeared to be only slightly affected by pH (Fig. 2c). However, the suitability or sensitivity of the Biolog method for bulk humus samples might be questioned, since reductions in the number of bacteria utilizing starch, protein, pectin, xylan, and cellulose after 4 years of irrigation in these same study plots were found (27).
When the bacterial community incorporated thymidine in a solution in which pH was decreased or increased with buffers, the community of the plots having a lower humus pH showed greater adaptation to the more acidic environment than did the community of the plots having a higher humus pH (Fig. 3c). However, the bacterial growth rate in general, as indicated by the thymidine incorporation rate (Fig. 3b), decreased due to acidification in spite of the fact that at least part of the bacterial community was adapted to the altered environment. Since changes in the PLFA pattern and percentage of yellow colonies indicated a change in bacterial community structure, it is likely that the increased acid tolerance of the bacterial community resulted at least partly from a shift in species composition.
Fungal biomass appeared not to suffer from acidification to the same extent as bacterial biomass, since the amounts of both fungal PLFA 18:2ω6,9 and ergosterol were unaffected by the treatments. Thus, it is likely that the decrease in total activity (basal respiration) was due to pH affecting mainly the bacterial community. These results are therefore in accordance with several previous reports about unchanged (11) or even increased (28) fungal biomass or a higher fungal/bacterial biomass ratio (13) caused by acidification. On the contrary, results showing decreased fungal biomass (17, 20) or decreased fluorescein diacetate-active fungi (8, 33) also have been reported, but these results are mainly from experiments with high acid loads. The fungal growth rate tested with [14C]acetate incorporation indicated a very slight increase with decreasing humus pH (Fig. 1d), but such a small difference in this new method must be considered tentative. At least it can be stated that the fungal activity was not negatively affected by the acidity.
Instead of direct toxicity of protons or anions, reduction in the availability of carbon for microbes has been suggested to be the main reason for the adverse effects of acidification (15, 33). Reduced substrate availability of heterotrophs may thus determine microbial activity; this situation may be evidenced as lowered respiration rate or bacterial activity, as shown in the present study. Another reason for the changes in the microbial community may be the effects of acid on the plants. The same factors that influence plant growth, e.g., environmental stress, may affect plant root exudation (14). Bäck et al. (12) reported reduced branch and needle growth of the pines of these Kevo study plots as a consequence of a decline in photosynthetic activity caused by irrigation of S and SN plots. However, since we did not find any differences among S, SN, and N plots that could not be attributed to an altered pH, we suggest that the acidification effect on the microorganisms was not mediated through the plants.
The present study showed that even though the northern European woodlands are naturally very acidic, the present low-level but prolonged acid rain resulted in clear signs of acidification, as defined by soil chemical variables. Litter decomposition rate, the ultimate sign of disturbances in the decomposition process, had earlier been shown to be decreased in these study plots (30), yet only marginal effects on total microbial biomass and basal respiration rate were found. The community-level techniques, however, were sensitive enough to reveal disturbances in the structure and general activity of the bacterial community, even though at least some of the bacteria were found to be adapted to an altered humus pH. Fungal biomass and activity were not affected or were affected only to a minor extent. This result could, of course, be due to the more specific character of bacterial methods (PLFA, thymidine incorporation, and Biolog analyses), while fungal community structure was not analyzed to the same extent. However, at least the total biomass and the growth rate of the fungi were unchanged, while those of the bacteria were reduced.
ACKNOWLEDGMENTS
We thank Outi Priha and Janna Pietikäinen for valuable comments during the preparation of the manuscript, Claire Gower and Suzanna Meekins for revising the English, and Anne Siika for helping with the figures.
This study was financially supported by the Academy of Finland.
REFERENCES
- 1.Amano N, Shinmen Y, Akimoto K, Kawashima H, Amichi T. Chemotaxonomic significance of fatty acid composition in the genus Mortierella (Zygomycetes, Mortierellaceae) Mycotaxon. 1992;44:257–265. [Google Scholar]
- 2.Anderson J P E, Domsch K H. A physiological method for the quantitative measurement of microbial biomass in soils. Soil Biol Biochem. 1978;10:214–221. [Google Scholar]
- 3.Anderson T-H, Domsch K H. The metabolic quotient for CO2 (qCO2) as a specific activity parameter to assess the effects of environmental conditions, such as pH, on the soil microbial biomass of forest soils. Soil Biol Biochem. 1993;25:393–395. [Google Scholar]
- 4.Bååth E. Thymidine incorporation into macromolecules of bacteria extracted from soil by homogenization-centrifugation. Soil Biol Biochem. 1992;24:1157–1165. [Google Scholar]
- 5.Bååth E. Measurement of protein synthesis by soil bacterial assemblages with the leucine incorporation technique. Biol Fertil Soils. 1994;17:147–153. [Google Scholar]
- 6.Bååth E. Adaptation of soil bacterial communities to prevailing pH in different soils. FEMS Microbiol Ecol. 1996;19:227–237. [Google Scholar]
- 7.Bååth E, Arnebrandt K. Growth rate and response of bacterial communities to pH in limed and ash treated forest soils. Soil Biol Biochem. 1994;26:995–1001. [Google Scholar]
- 8.Bååth E, Berg B, Lohm U, Lundgren B, Lundkvist H, Rosswall T, Söderström B, Wirén A. Effects of experimental acidification and liming on soil organisms and decomposition in a Scots pine forest. Pedobiologia. 1980;20:85–100. [Google Scholar]
- 9.Bååth E, Frostegård Å, Fritze H. Soil bacterial biomass, activity, phospholipid fatty acid pattern, and pH tolerance in an area polluted with alkaline dust deposition. Appl Environ Microbiol. 1992;58:4026–4031. doi: 10.1128/aem.58.12.4026-4031.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bååth E, Frostegård Å, Pennanen T, Fritze H. Microbial community structure and pH response in relation to soil organic matter quality in wood-ash fertilized, clear-cut or burned coniferous forest soil. Soil Biol Biochem. 1995;27:229–240. [Google Scholar]
- 11.Bååth E, Lundgren B, Söderström B. Effects of artificial acid rain on microbial activity and biomass. Bull Environ Contam Toxicol. 1979;23:737–740. doi: 10.1007/BF01770034. [DOI] [PubMed] [Google Scholar]
- 12.Bäck J, Neuvonen S, Huttunen S. Pine needle growth and fine structure after prolonged acid rain treatment in the subarctic. Plant Cell Environ. 1994;17:1009–1021. [Google Scholar]
- 13.Bewley R J F, Parkinson D. Bacterial and fungal activity in sulphur dioxide polluted soils. Can J Microbiol. 1985;31:13–15. [Google Scholar]
- 14.Bolton H J, Fredrickson J K, Elliott L F. Microbial ecology of the rhizosphere. In: Metting F B Jr, editor. Soil microbial ecology. New York, N.Y: Marcel Dekker, Inc.; 1993. pp. 27–63. [Google Scholar]
- 15.Chang F-H, Alexander M. Effects of simulated acid precipitation on decomposition and leaching of organic carbon in forest soils. Soil Sci. 1984;138:226–234. [Google Scholar]
- 16.Ek H, Sjögren M, Arnebrant K, Söderström B. Extramatrical myceliar growth, biomass allocation and nitrogen uptake in ectomycorrhizal systems in response to collembolan grazing. Appl Soil Ecol. 1994;1:155–169. [Google Scholar]
- 17.Esher R J, Marx D H, Ursic S J, Baker R L, Brown L R, Coleman D C. Simulated acid rain effects in fine roots, ectomycorrhizae, microorganisms, and invertebrates in pine forests of the southern United States. Water Air Soil Pollut. 1992;61:269–278. [Google Scholar]
- 18.Federle T W. Microbial distribution in soil—new techniques. In: Megusar F, Gantar M, editors. Perspectives in microbial ecology. Ljubljana, Yugoslavia: Slovene Society for Microbiology; 1986. pp. 493–498. [Google Scholar]
- 19.Fritze H. Effects of environmental pollution on forest soil microflora—review. Silva Fenn. 1992;26:37–48. [Google Scholar]
- 20.Fritze H, Kiikkilä O, Pasanen J, Pietikäinen J. Reaction of forest soil microflora to environmental stress along a moderate pollution gradient next to an oil refinery. Plant Soil. 1992;140:175–182. [Google Scholar]
- 21.Frostegård Å, Bååth E. The use of phospholipid fatty acid analysis to estimate bacterial and fungal biomass in soil. Biol Fertil Soils. 1996;22:59–65. [Google Scholar]
- 22.Frostegård Å, Bååth E, Tunlid A. Shifts in the structure of soil microbial communities in limed forests as revealed by phospholipid fatty acid analysis. Soil Biol Biochem. 1993;25:723–730. [Google Scholar]
- 23.Frostegård Å, Tunlid A, Bååth E. Phospholipid fatty acid composition, biomass, and activity of microbial communities from two soil types experimentally exposed to different heavy metals. Appl Environ Microbiol. 1993;59:3605–3617. doi: 10.1128/aem.59.11.3605-3617.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garland J L. Analytical approaches to the characterization of samples of microbial communities using patterns of potential C source utilization. Soil Biol Biochem. 1996;28:213–221. [Google Scholar]
- 25.Kauppi P E, Mielikäinen K, Kuusela K. Biomass and carbon budget of European forests, 1971 to 1990. Science. 1992;256:70–74. doi: 10.1126/science.256.5053.70. [DOI] [PubMed] [Google Scholar]
- 26.Kelly J M, Strickland R C. CO2 efflux from deciduous forest litter and soil in response to simulated acid rain treatment. Water Air Soil Pollut. 1984;23:431–440. [Google Scholar]
- 27.Kytöviita M-M, Fritze H, Neuvonen S. The effects of acid irrigation on soil microorganisms at Kevo, northern Finland. Environ Pollut. 1990;66:21–31. doi: 10.1016/0269-7491(90)90196-j. [DOI] [PubMed] [Google Scholar]
- 28.Mancinelli R L. Alpine tundra soil bacterial responses to increased soil loading rates of acid precipitation, nitrate, and sulfate, Front Range, Colorado, U.S.A. Arct Alp Res. 1986;18:269–275. [Google Scholar]
- 29.Myrold D D, Nason G E. Effect of acid rain on soil microbial processes. In: Mitchell R, editor. Environmental microbiology. New York, N.Y: Wiley-Liss, Inc.; 1992. pp. 59–82. [Google Scholar]
- 30.Neuvonen S, Suomela J. The effect of simulated acid rain on pine needle and birch leaf litter decomposition. J Appl Ecol. 1990;27:857–872. [Google Scholar]
- 31.Newell S Y, Fallon R D. Toward a method for measuring instantaneous fungal growth rates in field samples. Ecology. 1991;72:1547–1559. [Google Scholar]
- 32.O’Leary W M, Wilkinson S G. Gram-positive bacteria. In: Ratledge C, Wilkinson S G, editors. Microbial lipids. Vol. 1. London, England: Academic Press Ltd.; 1988. pp. 117–185. [Google Scholar]
- 33.Persson T, Lundkvist H, Wirén A, Hyvönen R, Wessén B. Effects of acidification and liming on carbon and nitrogen mineralization and soil organisms in mor humus. Water Air Soil Pollut. 1989;45:77–96. [Google Scholar]
- 34.Prescott C E, Parkinson D. Effects of sulphur pollution on rates of litter decomposition in a pine forest. Can J Bot. 1985;63:1436–1443. [Google Scholar]
- 35.Priha O, Smolander A. FE- and SIR-derived microbial biomass C, and respiration rate in limed soil of Scots pine sapling stands. Biol Fertil Soils. 1994;17:301–308. [Google Scholar]
- 36.Strayer R F, Alexander M. Effects of simulated acid rain on glucose mineralization and some physicochemical properties of forest soils. J Environ Qual. 1981;10:460–464. [Google Scholar]
- 37.Tamminen P, Starr M R. A survey of forest soil properties related to soil acidification in southern Finland. In: Kauppi P, Anttila P, Kenttämies K, editors. Acidification in Finland. Berlin, Germany: Springer-Verlag KG; 1990. pp. 234–251. [Google Scholar]
- 38.Thompson I P, Blackwood I L, Davies T D. Soil bacterial changes upon snowmelt: laboratory studies of the effects of early and late meltwater fractions. FEMS Microbiol Ecol. 1987;45:269–274. [Google Scholar]
- 39.Vanhala P, Fritze H, Neuvonen S. Prolonged simulated acid rain treatment in the subarctic: effect on the soil respiration rate and microbial biomass. Biol Fertil Soils. 1996;23:7–14. [Google Scholar]
- 40.von Lützow M, Zelles L, Scheunert I, Ottow J C G. Seasonal effects of liming, irrigation, and acid precipitation on microbial biomass N in a spruce (Picea abies L.) forest soil. Biol Fertil Soils. 1992;13:130–134. [Google Scholar]
- 41.Wilkinson S G. Gram-negative bacteria. In: Ratledge C, Wilkinson S G, editors. Microbial lipids. Vol. 1. London, England: Academic Press Ltd.; 1988. pp. 299–489. [Google Scholar]
- 42.Zelles L, Scheunert I, Kreutzer K. Effect of artificial irrigation, acid precipitation and liming on the microbial activity in soil of a spruce forest. Biol Fertil Soils. 1987;4:137–143. [Google Scholar]