Significance
Although vaccines against SARS-CoV-2 have been developed in record time, their protective effect against emerging variants of concern is not optimal. We previously developed an mRNA vaccine against HIV-1 that coexpresses the viral envelope with a retroviral Gag to induce the formation of virus-like particles (VLPs). Here, we applied a similar platform to SARS-CoV-2 to evaluate potential improvements. To facilitate the assembly of VLPs, we replaced the native cytoplasmic tail of the SARS-CoV-2 Spike protein with that of simian immunodeficiency virus (SIV), producing a hybrid Spike protein. In mice, an mRNA vaccine coexpressing Spike with SIV Gag was more effective than Spike alone. These results demonstrate that our VLP-forming mRNA vaccine platform can be extended to other pathogens besides HIV-1.
Keywords: COVID-19, vaccine, mRNA, neutralizing antibodies
Abstract
Vaccines have played a fundamental role in the control of infectious diseases. We previously developed a messenger RNA (mRNA) vaccine against HIV-1 that forms virus-like particles (VLPs) through coexpression of the viral envelope with Gag. Here, we applied the same principle to the design of a VLP-forming mRNA vaccine against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). To promote cognate interaction with simian immunodeficiency virus (SIV) Gag, we engineered different chimeric proteins encompassing the ectodomain and the transmembrane region of the SARS-CoV-2 Spike protein from the Wuhan-Hu-1 strain fused to the gp41 cytoplasmic tail of either HIV-1 (strain WITO) or SIV (strain mac239) with or without a partial truncation at amino acid 745 to enhance membrane expression. Upon cotransfection with SIV gag mRNA, the Spike-SIVCT.745 (SSt) chimera yielded the highest level of cell-surface expression and extracellular VLP release. Immunization of BALB/c mice with SSt+gag mRNA at 0, 4, and 16 wk induced higher titers of Spike-binding and autologous neutralizing antibodies at all time points compared to SSt mRNA alone. Furthermore, mice immunized with SSt+gag mRNA developed neutralizing antibodies effective against different variants of concern. These data demonstrate that the Gag/VLP mRNA platform can be successfully applied to vaccines against different agents for the prevention of infectious diseases of global relevance.
Although effective vaccines against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) have been developed with extraordinary speed (1–4), morbidity and mortality from coronavirus disease 2019 (COVID-19) remain at alarming levels. This is due to a number of factors, including the limited access or resistance to vaccination in some areas of the world, as well as the continuous viral evolution, which leads to the emergence of new variants of concern (VOC) that escape from the control of current vaccines and therapeutic neutralizing antibodies (5–7). In fact, the less-than-optimal breadth of neutralization and limited durability of protection against VOCs conferred by approved vaccines, despite a retained efficacy against severe disease (3, 8), make further improvements important for the global control and, ultimately, termination of the pandemic. All effective SARS-CoV-2 vaccines are based on the viral Spike (S) protein, which gives coronaviruses their characteristic crown-shaped structure and is the major target of neutralizing antibodies (1–4). An important advantage of mRNA technology is that it directs the endogenous production of immunogenic glycoproteins, such as the S protein of SARS-CoV-2, thereby ensuring their native glycosylation (9). Moreover, endogenous protein synthesis induces class I antigen presentation for the activation of CD8+ T cells (10, 11).
Major advances have been made over the past 30 y in the application of virus-like particles (VLPs) to the development of vaccines (12–14). Indeed, VLPs may offer important advantages over conventional subunit vaccines because of their larger size and repetitive antigen display, which effectively stimulate both the innate and the adaptive arms of the immune system. Approved VLP-forming vaccines against hepatitis B virus and human papilloma virus are highly effective and widely utilized on a global scale (12–14). We previously reported the design and preclinical evaluation of a VLP-forming HIV-1 mRNA vaccine obtained by simultaneous coexpression of the HIV-1 envelope (Env) and the simian immunodeficiency virus (SIV) Gag (15). The mRNA was nucleoside modified to reduce activation of innate immune responses and formulated in lipid nanoparticles (LNP) (9). In vitro, coexpression of Env with Gag within the same cells led to the efficient assembly and extracellular release of VLPs with abundant Env spikes on their surface. In vivo, a VLP-forming env-gag mRNA vaccine was more efficient than env mRNA alone in eliciting neutralizing antibodies in mice (15). Based on these observations, we sought to extend the same VLP-forming mRNA vaccine design to SARS-CoV-2 with the aim of obtaining an mRNA vaccine with increased efficacy, particularly in terms of neutralization breadth and durability. We report the engineering of chimeric SARS-CoV-2 S proteins bearing the cytoplasmic tail (CT) of either HIV-1 or SIV in order to facilitate the formation of Gag-based VLPs. Mice were immunized with an mRNA vaccine encoding a chimeric SARS-CoV-2 S protein bearing the truncated CT of SIV with or without a second mRNA encoding SIV Gag to promote VLP production. The results revealed a superior potency and breadth of a VLP-forming mRNA vaccine over an mRNA vaccine expressing the S protein alone. These results corroborate the use of particulate immunogens displaying multiplexed antigens for an optimized stimulation of antibody responses. Furthermore, they provide evidence that a Gag-based VLP-forming mRNA design is feasible for vaccines against diverse viral agents for the prevention of infectious diseases of global relevance.
Results
Engineering of Chimeric SARS-CoV-2 Spike Proteins with a Retroviral CT.
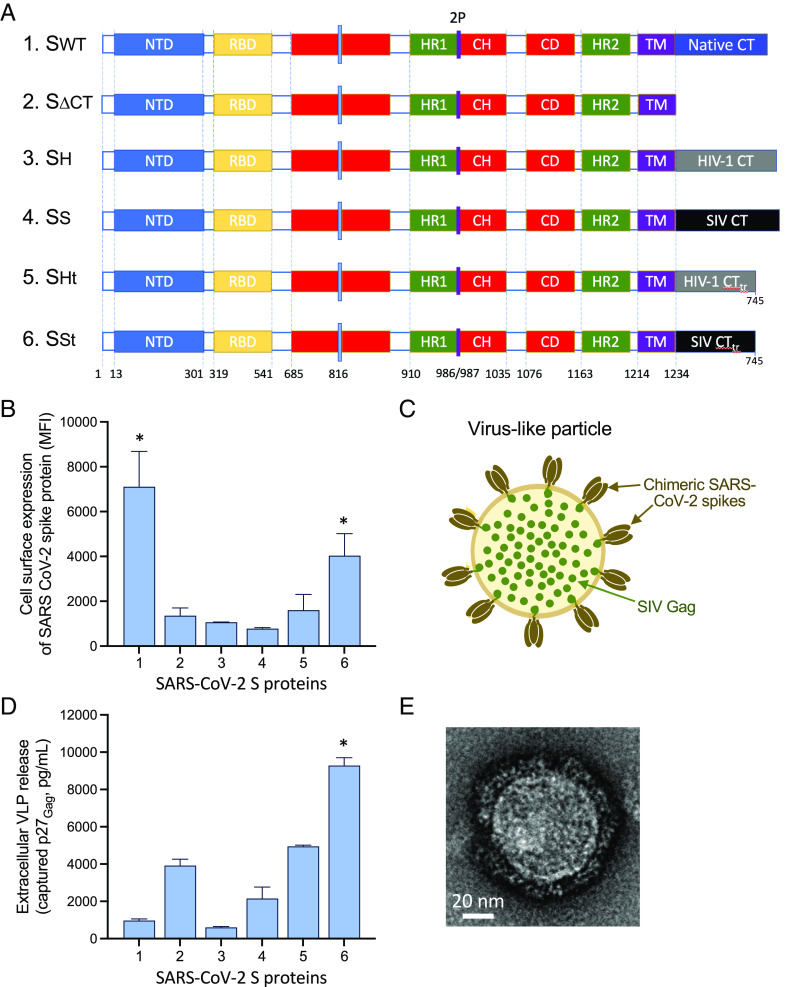
The SARS-CoV-2 S protein is a type-I transmembrane protein comprising an extracellular domain, a transmembrane domain, and a CT. Since homologous recognition between HIV-1 or SIV Gag and their respective gp41 CTs facilitates virion assembly and extracellular release (16, 17), we replaced the entire native CT of the SARS-CoV-2 S protein from the Wuhan-Hu-1 strain, modified by proline substitutions of K986 and V987 (2P) and deletion of the furin cleavage site, with the CT of the gp41 transmembrane Env protein of either HIV-1 (strain WITO) or SIV (strain mac239). The retroviral CTs were fused to the last residue of the S-protein transmembrane domain either in their full-length or truncated form (at aa. 745 of the HXB2 reference sequence) in order to increase cell-surface expression (Fig. 1A).
Fig. 1.
(A) Engineering of chimeric Spike constructs encompassing the ectodomain and the transmembrane portion of the Wuhan-Hu-1 SARS-CoV-2 Spike with cytoplasmic tails (CT) from the following: 1. Native SARS-CoV-2 Spike; 2. no CT; 3. HIV-1 gp41, full length; 4. SIV gp41, full length; 5. HIV-1 gp41, truncated at aa. 745 of the HXB2 reference sequence; and 6. SIV gp41, truncated at aa. 745. (B) Cell-surface expression of different Spike constructs after mRNA transfection into 293-T cells. The asterisks denote statistical significance (P < 0.05) for the comparison by the two-tailed t test with each of the other conditions. (C) Schematic representation of hybrid VLPs with chimeric Spike and SIV Gag. (D) Formation of extracellular VLPs by different Spike constructs after cotransfection into 293-T cells together with SIV gag mRNA. The asterisk denotes statistical significance (P < 0.05) for the comparison by the two-tailed t test with each of the other conditions. (E) Negative-stain EM of a hybrid VLP produced with the SSt Spike.
Codon-optimized mRNA was produced for the four chimeric S proteins as well as for the full-length S protein bearing the native CT and a CT-truncated form as controls. The mRNAs were transfected into 293-T cells, and the cell-surface expression of the S proteins was evaluated by flow cytometry using monoclonal antibodies against the S ectodomain. The CT-truncated S protein (SΔCT; no. 2) showed a dramatic reduction of cell-surface expression, compared to the full-length S protein (Swt; no. 1), pointing to the importance of the CT domain for export to the cellular membrane (Fig. 1B). Among the chimeric S proteins, the highest expression was seen with the SIV CT-truncated chimera (SSt; no. 6), which reached about half the level seen with Swt, while the full-length CT SIV chimera (SS; no. 5) and both the full-length CT (SH; no. 3) and truncated CT (SHt; no. 4) HIV-1 chimeras showed a significantly reduced expression (Fig. 1B).
Production of Hybrid VLPs Formed by a Retroviral Gag and Engineered SARS-CoV-2 Spike Proteins.
Previously, HIV-1 Gag-based chimeric VLPs have successfully been produced using the SARS-CoV (18, 19) or SARS-CoV-2 (20) S proteins without altering the native CT domain. However, the efficiency of VLP production was not directly addressed in those studies. To evaluate the ability to assemble hybrid VLPs and the efficiency of their extra-cellular release, we cotransfected mRNA expressing the different SARS-CoV-2 S proteins along with SIV gag mRNA at a 1:2 wt:wt ratio (Fig. 1C). This ratio was determined to be optimal for VLP formation based on previous in vitro experiments with HIV-1 Env and SIV Gag. In spite of its high expression on the cellular surface, very low levels of secreted VLPs were detected with the S protein bearing the native CT (Swt), whereas the CT-truncated S protein, despite its lower expression on the cell membrane, yielded higher levels of extracellular VLPs (Fig. 1D). Among the chimeric S proteins, the two CT-truncated chimeras (SSt and SHt) yielded higher levels of VLP production than their counterparts with full-length CTs (SS and SH), with SSt showing a greater efficiency than SHt (Fig. 1D). These results confirmed that cognate interaction between SIV Gag and the homologous SIV CT leads to higher VLP production. Negative-staining electron microscopy was used to morphologically characterize the hybrid SSt-Gag VLPs. The ultrastructural images of a VLP produced with the SSt engineered Spike revealed the presence of round-shaped particles with multiple spike-like projections on their surface (Fig. 1E).
Enhanced Immunogenicity of a VLP-Forming SARS-CoV-2 Spike mRNA Vaccine in Mice.
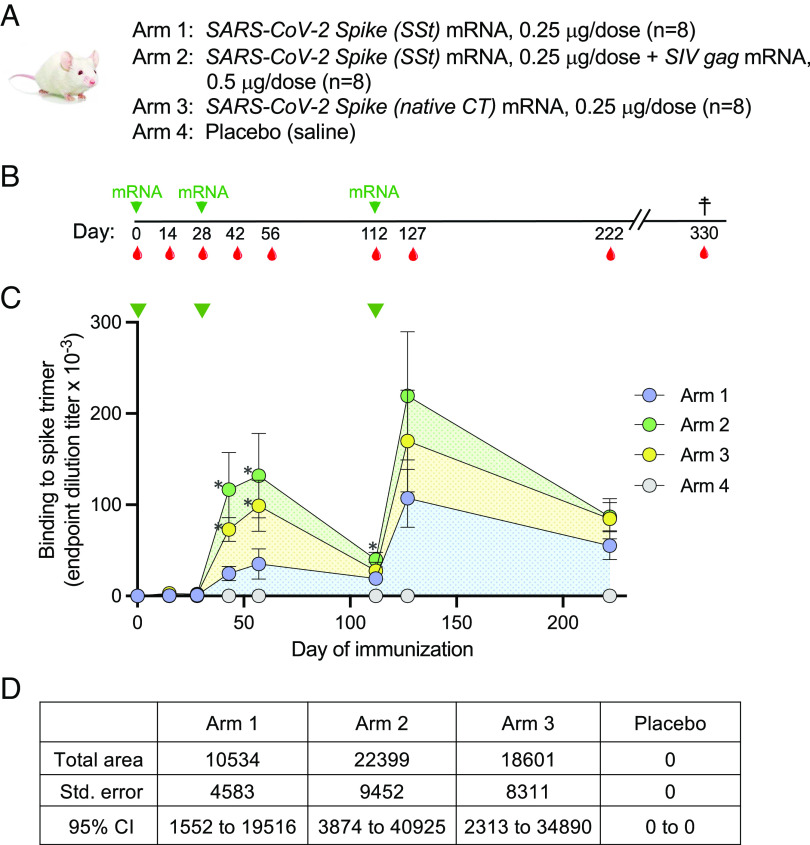
Based on the efficiency of VLP production observed in vitro, the SSt chimeric protein was tested in immunization studies in mice. Thus, wild-type BALB/c mice (n = 8 per group) were sequentially immunized with SSt mRNA alone (arm 1; non-VLP forming) or with mixed SSt+gag mRNA (arm 2; VLP forming); as controls, we immunized two additional groups: one with mRNA encoding the SARS-CoV-2 S protein with its native CT (Swt; arm 3) and one with saline solution as placebo (arm 4) (Fig. 2A). The two mRNAs used in arm 2 were formulated in LNP individually. All mice were immunized three times: at weeks 0, 4, and 16 (Fig. 2B). To better highlight the potential difference in efficiency among the various vaccine formulations, we used a low amount of SARS-CoV-2 S mRNA in each study arm (0.25 µg/inoculation), while gag mRNA was used at 0.5 µg/inoculation for arm 2 and premixed with S mRNA before inoculation. Antibody titers against the homologous (Wuhan-Hu-1) trimeric S protein were measured in serum by limiting-dilution enzyme-linked immunosorbent assay (ELISA) at weeks 0, 2, 4, 6, 8, 16, 18, and 32. No specific antibodies were detected after a single inoculation (day 28), while they started to appear in all mRNA-immunized mice, with the exception of one mouse in arm 1 and one in arm 2, after the second inoculation (day 42) (Fig. 2C). The mean reciprocal titer of S-trimer-binding antibodies was significantly higher in mice immunized with SSt+gag mRNA (arm 2) than in mice immunized with SSt mRNA alone (arm 1); the titers were also higher in the control group immunized with Swt mRNA (arm 3) than in the SSt mRNA group, reflecting the higher in vitro expression levels of Swt vs. SSt (Fig. 1B), while no specific antibodies were detected in mice inoculated with placebo (arm 4). The antibody titers slightly increased in all three groups 2 wk later (day 56) and remained significantly higher in arms 2 and 3 compared to arm 1. At the time of the third immunization (day 112), the antibody titers had markedly declined but remained significantly higher in arm 2 than in arm 1. A marked increase in titers was observed 2 wk after the third immunization (day 127), reaching higher mean levels in arm 2 than in arms 1 and 3; however, the difference was not statistically significant, presumably due to the wide titer range. Likewise, there were higher mean titers in arms 2 and 3 than in arm 1 16 wk after the second boost (day 222), although the difference did not reach statistical significance. Analysis of the area under the curve (AUC) showed that the VLP-forming arm 2 had more than twice the AUC of arm 1 (22,399, 95% CI 3,874-40,925 vs. 10,534, 95% CI 1,552-19,516), while the control arm 3 had an intermediate value (18,601, 95% CI 2,313-34,890) (Fig. 2D). These results provide evidence that the VLP-forming SSt+gag mRNA vaccine had a higher immunogenicity than the SSt mRNA-only vaccine.
Fig. 2.
(A) Design of the mouse immunization study. Each study arm included eight wild-type BALB/c mice. (B) Sequence of mRNA immunizations (green arrows) and bleedings (red drops). (C) Induction of SARS-CoV-2 Spike-binding antibodies in the four study arms over time, as assessed by ELISA on plastic-immobilized Wuhan-Hu-1 Spike trimers. The time of immunization is indicated by the green arrows at the top. The data represent mean values from each study arm (±SEM). The asterisks indicate a P-value below 0.05 as tested by the unpaired nonparametric two-tailed t test (Mann–Whitney). (D) Calculated area under the curve for the level of Spike-binding antibodies shown in (C).
Enhanced Autologous Neutralization in Mice Immunized with a VLP-Forming SARS-CoV-2 mRNA Vaccine.
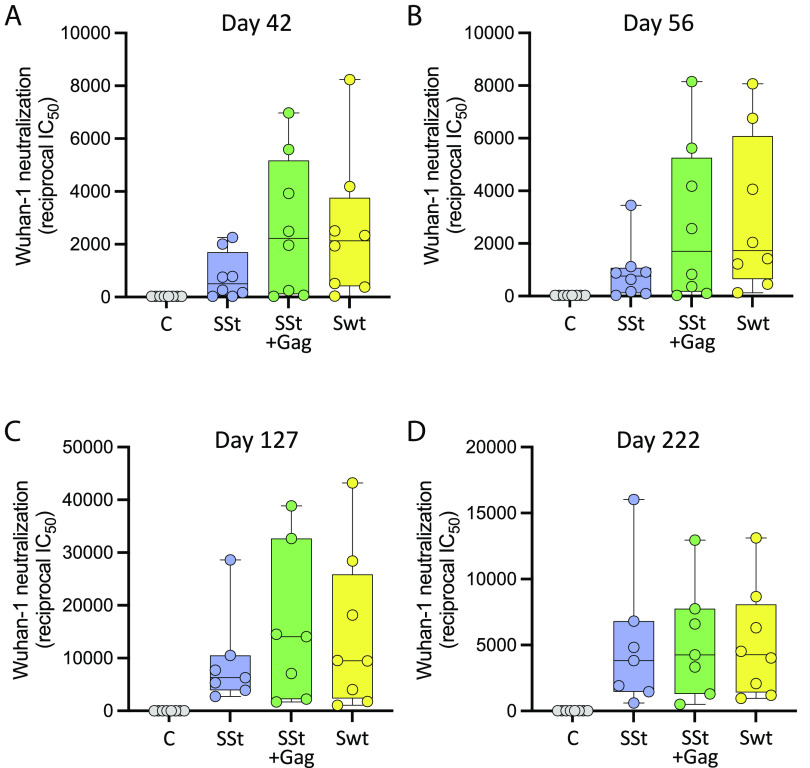
Neutralization of the autologous SARS-CoV-2 pseudovirus (Wuhan-Hu-1 strain) was sequentially tested in the serum of immunized mice. Autologous neutralizing antibodies were not detected after the first immunization (day 28) but started to appear 2 wk after the second immunization (day 42), although there were highly heterogenous responses within individual groups, with a few animals showing no serum neutralization (two in arm 1, one in arm 2, and one in arm 3), most likely due to the low dose of mRNA inoculated (0.5 µg) (Fig. 3A). The mean reciprocal titer of neutralization (IC50) was markedly higher in arm 2, immunized with SSt+gag mRNA (mean ± SEM: 2,662 ± 933), than in arm 1, immunized with SSt mRNA alone (mean IC50, 786 ± 312); however, the difference was not statistically significant presumably due to the wide data dispersion. Of note, the autologous neutralization titers elicited in arm 3 (Swt mRNA; mean IC50, 2,507 ± 950) were comparable to those in arm 2, again reflecting the higher in vitro expression of Swt vs. SSt and corroborating the efficiency of the original SARS-CoV-2 vaccine even in the absence of Gag. No neutralization was seen in the placebo group (arm 4). Similar levels of neutralization were detected in the various groups 2 wk later (day 56) (Fig. 3B), while a brisk rise in titers was seen after the second vaccine boost (day 127), again with higher mean IC50 values in arm 2 (15,874 ± 5,162) and arm 3 (14,458 ± 5,230) than in arm 1 (9,301 ± 3,141), although the differences did not reach statistical significance (Fig. 3C). The neutralization titers remained high in all immunized groups both at week 16 after the second boost (day 222) (Fig. 3D) and at study termination 16 wk later (day 330) (SI Appendix, Fig. S1A), indicating that the responses were durable. The mean IC50 values at study termination were 5,732 ± 1,754 (arm 1), 8,088±2,052 (arm 2), and 7,094 ± 1,675 (arm 3).
Fig. 3.
Neutralization of the autologous SARS-CoV-2 strain (Wuhan-Hu-1) in mice immunized with the vaccines indicated below the x axis at different time points. (A) Day 42 (14 d after the first vaccine boost). (B) Day 56 (28 d after the first vaccine boost). (C) Day 127 (14 d after the second vaccine boost). (D) Day 222 (16 wk after the second vaccine boost). The median titer is indicated by a horizontal line in each box. No statistical differences were seen by either parametric or nonparametric t tests presumably due to the wide data dispersion.
Enhanced Heterologous Neutralization in Mice Immunized with a VLP-Forming SARS-CoV-2 Spike mRNA Vaccine.
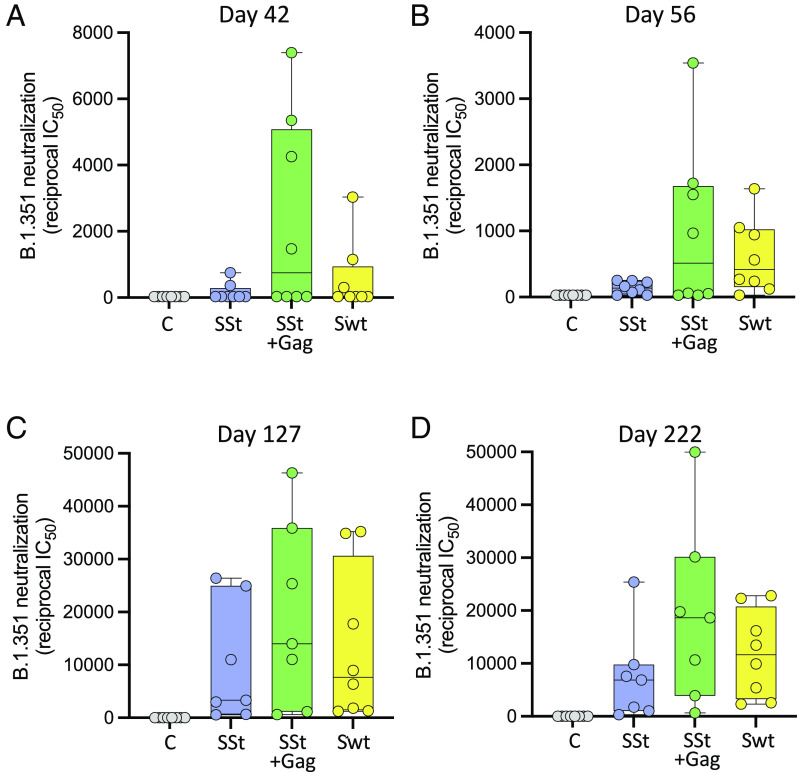
To evaluate the breadth of neutralization elicited by the mRNA vaccines, we tested the mouse sera against a heterologous SARS-CoV-2 strain (Beta variant or B.1.531) that is particularly difficult to neutralize by both vaccine sera and monoclonal antibodies (21). Heterologous neutralizing antibodies started to appear 2 wk after the second immunization (day 42), but only in four of eight mice (50%) from arm 2 (immunized with SSt+gag mRNA) and in 2 of eight mice (25%) from arm 3 (immunized with Swt mRNA) were neutralization titers greater than 1:1000 detected (Fig. 4A). The mean reciprocal titer was markedly higher in arm 2 (2,323 ± 1,038), than in arm 1, immunized with SSt mRNA alone (161 ± 94) and in arm 3 (330 ± 170). The neutralization titers slightly declined on day 56 (Fig. 4B) but were markedly enhanced by the second booster immunization (day 127), again with higher mean IC50 values in arm 2 (19,194 ± 6,150) than in arm 1 (9,981 ± 3,987) and arm 3 (13,444 ± 5,093) (Fig. 4C), although the differences did not reach statistical significance. Interestingly, at this time point, heterologous neutralizing antibodies were detected in all immunized mice, confirming the importance of the second boost for the induction of neutralization breadth (22). Moreover, heterologous neutralization titers were durable over time as they remained sustained 16 wk after the second boost (day 222), especially in arm 2 (Fig. 4D), confirming the increased efficacy of the VLP-forming mRNA vaccine formulation. To further assess the breadth of neutralization elicited by the mRNA vaccines, we also tested the Omicron variant (B.1.1.529) and two of its subvariants (BA.2.12.1 and BA.4) at the time of study termination (day 330). The results confirmed that mice immunized with SSt+gag mRNA consistently developed a broader neutralization response than mice immunized with SSt mRNA alone, although some immunized mice failed to neutralize the Omicron subvariants BA.2.12.1 and BA.4. Of note, BA.2.12.1 and especially BA.4 were neutralized most efficiently by mice immunized with Swt mRNA (arm 3; SI Appendix, Fig. S1 available with the full text of this article at PNAS.org).
Fig. 4.
Neutralization of a heterologous SARS-CoV-2 strain [B.1.351 (Beta) strain] in mice immunized with the vaccines indicated below the x axis at different time points. (A) Day 42 (14 d after the first vaccine boost). (B) Day 56 (28 d after the first vaccine boost). (C) Day 127 (14 d after the second vaccine boost). (D) Day 222 (16 wk after the second vaccine boost). The median titer is indicated by a horizontal line in each box. No statistical differences were seen by either parametric or nonparametric t tests presumably due to the wide data dispersion.
Discussion
We report herein the design and preclinical evaluation in a mouse model of an mRNA vaccine platform against SARS-CoV-2 that generates VLPs displaying on their surface the immunogenic S protein. To achieve the production of VLPs, we used the same strategy that we have previously employed for the design of an HIV-1 mRNA vaccine via coexpression of a retroviral Gag protein to drive the assembly and extracellular release of hybrid VLPs (15). The production of hybrid VLPs based on HIV-1 Gag has previously been reported for both SARS-CoV and SARS-CoV-2 S proteins (18, 19, 20), attesting to the versatility of retroviral Gag proteins for VLP production. However, no attempts were made to optimize VLP production by facilitating cognate Gag interaction, and no preclinical studies of immunogenicity and efficacy were performed. In our approach, we aimed at maximizing the efficiency of VLP production by modifying the SARS-CoV-2 S protein to facilitate homologous recognition by retroviral Gag. Thus, we replaced the native CT domain with the CT domain of either HIV-1 or SIV. In vitro cotransfection experiments clearly illustrated the advantage of using the Gag-homologous CT since the most effective combination to produce SIV Gag-based VLPs was the S protein bearing the CT domain of SIV. Moreover, we found that truncation of the SIV CT domain at aa. 745 in gp41 significantly augmented both the cell-surface expression of the S protein and the yield of extracellular VLPs. A potential drawback of the engineered S protein that we used in this study (SSt) was its reduced expression efficiency compared to the S protein bearing its native CT. This limitation might be addressed by structure-guided optimization of the chimeric S protein, for example, by elimination of known endocytosis motifs and/or by only partial replacement of the native CT with shorter segments of the SIV CT. However, this optimization was beyond the scope of the present study whose major aim was to investigate whether a VLP-forming Spike+Gag mRNA vaccine platform was superior to the standard mRNA vaccine expressing only the homologous S protein.
To evaluate the immunogenicity and efficacy of the VLP-forming SSt+Gag mRNA vaccine platform in a pre-clinical model in vivo, we inoculated it into wild-type BALB/c mice and compared it side by side with SSt-only mRNA. The results documented an increased immunogenicity of the VLP-forming SSt+Gag platform, which elicited significantly higher S-trimer-binding antibodies than SSt alone. Furthermore, the SSt+Gag vaccine induced higher levels of neutralizing antibodies both against the autologous strain (Wuhan-1) and especially against a difficult-to-neutralize heterologous SARS-CoV-2 VOC. We note that there was a marked variability in neutralization titers within individual study arms, likely as a consequence of the low Spike mRNA dose used for immunization (0.25 µg). This variability complicates the interpretation of the neutralization results because none of the differences between groups reached statistical significance despite a clear difference in the mean neutralization titers. As a control, mice were immunized with mRNA expressing the S protein bearing its native CT (Swt), which had shown higher expression levels in vitro. Accordingly, Swt mRNA induced consistently higher levels of specific antibodies, both binding and neutralizing, compared to the chimeric SSt mRNA in the absence of gag. It should be emphasized that a comparison between Swt and SSt mRNA was not a specific objective of our study, where Swt mRNA was only included as a positive control. Nevertheless, it is noteworthy that in spite of the reduced cell-surface expression of the modified S protein, the VLP-forming vaccine performed as well or even better than Swt mRNA, as shown by higher titers of S-binding antibodies at all time points except at day 222 and by higher titers of heterologous neutralizing antibodies against the Beta and Omicron variants. Only against the two Omicron subvariants tested, BA.2.12.1 and BA.4, was Swt mRNA slightly more effective that SST+gag mRNA. Thus, one might expect that a VLP-forming vaccine platform using an engineered S protein optimized for cell-surface expression would perform even better than in the present study with regard to both potency and breadth of neutralization. Interestingly, in all vaccine arms, the greatest increase in neutralization titers was observed after the third immunization, with a brisk rise also seen in the SSt-only group (arm 1), confirming the clinical data on the importance of the second boost to increase neutralization titers and breadth (22).
The precise mechanisms underlying the greater efficacy of our VLP-forming S/Gag vaccine, compared to the conventional S-only vaccine, are not yet fully clarified. The prevalent view is that VLP-based vaccines are more efficacious because VLPs closely resemble native viral particles in terms of size, shape, and antigenic configuration (12–14). The large size and the rounded shape of VLPs appear to facilitate their recognition, uptake, and processing by antigen-presenting cells such as dendritic cells, thereby fostering optimal antigen presentation. However, the property that may have the greatest impact on vaccine efficacy is the presentation of membrane-anchored proteins in their native antigenic conformation and in repetitive arrays on the surface of VLPs, which is known to provide the most effective modality for triggering B-cell stimulation (23). Multiplex antigen presentation can also be achieved by cell-surface expression, although with in vivo–transfected cells, both the membrane clustering of antigens and the likelihood to reach naive B cells bearing the appropriate B-cell receptor may be lower than with the highly mobile VLPs. In addition, VLPs typically encompass more than one antigenic protein, such as the SARS-CoV-2 S protein and the SIV Gag protein in our platform, which increases the overall immunogenicity of the vaccine resulting in both antigen-specific (antiviral) and nonspecific (bystander immune activation) potentiating effects. A limitation of the present study is that we did not collect suitable samples to investigate the cellular immune responses elicited by our vaccine. Although the VLP platform is likely to preferentially affect antibody responses via a more effective B-cell stimulation, the enhanced antigen presentation may also affect T-cell responses, further contributing to the induction of protective immunity. Additional studies in larger animal models such as hamsters or nonhuman primates which are suitable for virus challenge experiments are needed to evaluate the protective efficacy of our VLP-forming SARS-CoV-2 vaccine platform.
In summary, we have developed a VLP-forming SARS-CoV-2 mRNA vaccine that, in a mouse model, elicited antibodies with greater potency and breadth compared to an mRNA vaccine encoding the S protein alone. This vaccine warrants further investigation in preclinical models and, potentially, in future clinical trials. The mRNA platform has once again proven its extraordinary versatility and rapidity of development for the implementation of vaccine designs that would have posed significant problems of industrial manufacturing for a protein-based vaccine. Furthermore, our study illustrates how our Gag-based VLP-forming vaccine platform, owing to its efficiency and versatility, can be extended to pathogens other than retroviruses and therefore might be used to generate effective vaccines against a wide array of infectious agents of global relevance.
Materials and Methods
mRNA Production and SARS-CoV-2 S Protein Engineering.
Sequence-optimized mRNAs encoding the immunogens were synthesized in vitro using an optimized T7 RNA polymerase-mediated transcription reaction with N1-methylpseudouridine-5′-triphosphate utilized in place of uridine-5′-triphosphate (24). The in vitro transcription DNA template contained the target open reading frame flanked by 5′ and 3′ untranslated regions and an encoded polyA tail. After transcription, a cap 1 structure was added enzymatically using vaccine capping enzyme and 2′O-methyltransferase (New England Biolabs, Ipswich, MA). Purified mRNAs were sterilized by filtration and either used for in vitro transfection formulated into LNPs using a method described previously (25). Briefly, mRNA was diluted in acetate, pH 5.0, and mixed with lipids (SM-102: DSPC: Cholesterol: PEG2000-DMG) dissolved in neat ethanol at a ratio of 3:1 (mRNA:lipids). The product was then dialyzed against phosphate buffered saline (PBS), pH 7.4, tested for encapsulation, particle size/polydispersity, mRNA purity, and endotoxin, and was deemed acceptable for in vivo use.
The sequence of the SARS-CoV-2 S protein from the Wuhan-Hu-1 strain [GenBank accession number MN908947; S-protein sequence identical to that of the of USA/WA-1/2020 (WA-1); GenBank accession number, MN985325], modified by proline substitutions of K986 and V987 (2P) and deletion of the furin cleavage site, was engineered by either truncating the entire CT domain after the last residue of the transmembrane domain (Leu1235) or by replacing it with a retroviral gp41 CT. For this purpose, the sequence of the gp41 CT from either HIV-1 (strain WITO 4160.27, GenBank: DQ821488) or SIV (strain mac239, GenBank: M33262) was fused to Leu1235. The retroviral CTs were inserted either full length or truncated after the codon for aa. 745 (HXB2 reference sequence numbering) in order to increase cell-surface expression, as previously reported (26).
In Vitro mRNA Transfection and Flow Cytometry Assays.
HEK293T/17 cells [obtained from the American Type Culture Collection (ATCC)] were transiently transfected with mRNA encoding different SARS-CoV-2 S proteins using the TransIT-mRNA Transfection Kit (Mirus Cat# MIR2250). After 48 h, the cells were collected by mechanical shaking and pipetting, washed with PBS, and incubated with anti-SARS-Cov-2 S mouse monoclonal antibodies 1A9 (GeneTex Cat#GTX632604, 1 μg mL−1) or human antibody S-118 (gift from Vaccine Research Center, NIH) in flow cytometry buffer [1 × PBS with 2% fetal bovine serum (FBS)] at room temperature for 30 min. After washing with flow cytometry buffer twice, the cells were incubated with phycoerythrin-conjugated goat anti-mouse or anti-human IgG (Southern Biotech) at room temperature for 15 min. The cells were then washed once with PBS, fixed in 4% paraformaldehyde, and analyzed on a BD LSR Fortessa (BD Biosciences). Data analysis and geometric mean fluorescence intensity measurement were performed using FlowJo v10.7.1.
Virion-Capture Assays.
SARS-CoV-2 VLPs were produced by transiently transfected SARS-CoV-2 S and SIV gag mRNAs in HEK293/17 cells. Protein G–conjugated immunomagnetic beads (Dynabeads, Life Technologies) were armed with the appropriate antibody (10 µg/mL) for 20 min at room temperature (30 µL per reaction) and then washed three times with PBS containing 0.025% (w/v) casein (Vector laboratories Cat#SP-5020-250) to remove the unbound antibody. VLP capture was performed by incubating the armed beads with culture supernatants for 30 min under continuous low-speed rotation. After magnet concentration and washing, captured VLPs were lysed with RIPA buffer on the beads, and the released p27 Gag protein concentration was measured using an SIV p27 antigen in-house ELISA. Serial dilutions of reference p27 protein and lysed VLP p27 were coated onto 96-well ELISA plates (Corning Cat#9018) at 4 °C overnight. Then, the plates were blocked with 1× casein at room temperature for 1 h, followed by the addition of mouse anti-p27 monoclonal antibody (AIDS Reagent Program, Cat#3537) at 1 µg ml−1. After washing, horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G (IgG) (Invitrogen, Cat#A16072) was added for 1 h at room temperature. The optical density at 450 nm was measured. The VLP p27 concentrations were calculated using a dose–response curve fit with a five-parameter nonlinear function with Prism software (Graphpad Software, Inc., San Diego, CA).
Negative-Stain Electron Microscopy.
A 4.8-µl drop of the undiluted sample was applied to a glow-discharged carbon-coated copper grid and incubated for approximately 15 s. The drop was then removed using blotting paper, and the grid was washed consecutively with three drops of buffer containing 10 mM HEPES, pH 7.0, and 150 mM NaCl. Material adsorbed to the carbon was negatively stained with 0.75% uranyl acetate, and the grid was allowed to air-dry. Images were taken using a Thermo Scientific Talos F200C transmission electron microscope operated at 200 kV and equipped with a Thermo Scientific Ceta camera.
Mouse Immunization Protocol.
BALB/c mice were injected with mRNA formulations in a volume of 0.05 mL using a sterile syringe by intramuscular injection in the hind leg. The animals were immunized at weeks 0, 4 and 16 with 0.25 µg SARS-CoV-2 S mRNA alone (arms 1 and 3; n = 8) or mixed with 0.5 µg SIVmac239 gag mRNA (arm 2; n = 8). Control mice (n = 4) received PBS because previous studies have proven that the mRNA formulations being tested do not create substantial levels of nonspecific immunity. Blood was collected on days −1, 14, 28, 42, 56, 112, 127, and 330. Blood collections did not exceed 1% of the total body weight or 10 mL kg−1 over a 2-wk period, per Animal Care & Use Committee (ACUC) protocol. Regarding the sample size, a Wilcoxon sum rank test with a two-tailed level of significance of 0.05 would have >80% power to detect an effect size of 1.6 between the SARS-CoV-2 SSt and SARS-CoV-2 SSt–SIV Gag vaccine arms, each with a sample size of eight, and to detect an effect size of two between a vaccine arm and the control arm, with sample sizes of eight and four, respectively.
Immunoassays.
For conventional antigen-binding ELISA, 96-well plates were coated with 50 μL/well containing 1 μg/mL protein [SARS-Cov-2S(2P)] in 1 × PBS (pH 7.4) and incubated at 4 °C overnight. Serial dilutions of heat-inactivated sera were added to the wells, and plates were incubated for 2 h at room temperature. The plates were washed with ELISA wash buffer (R&D System, Cat # WA126), and the secondary conjugated antibody (goat anti-mouse Invitrogen Cat#A16072) was added and incubated for 1 h at room temperature. The plates were washed, and the color was developed by adding substrate reagent (R&D System Cat#DY999); the development was stopped with 2N Sulfuric Acid (R&D System Cat#DY994) after 15 min. The plates were read at OD450 in an ELISA plate reader. The data were plotted, and the dose–response curves were generated with Prism.
Pseudotype Neutralization Assay.
The SARS-Cov-2 pseudovirus neutralization assays were performed as previously reported (21). Briefly, single-round luciferase-expressing pseudoviruses were generated by cotransfection of plasmids encoding the full-length SARS-CoV-2 S protein (Wuhan-Hu-1, GenBank accession number, B.1.351/Beta; Omicron B.1.1.529, BA.2.12.1 and BA.4 sublineages; luciferase (pHR′ CMV Luc), a lentivirus backbone (pCMV ΔR8.2), and human transmembrane protease serine 2 (TMPRSS2) at a ratio of 1:20:20:0.3 into HEK293T/17 cells (ATCC) using the transfection reagent LiFect293™(LifeSct LLC). The pseudoviruses were harvested after 72 h. The supernatants were collected by centrifugation at 1,500 rpm for 10 min at room temperature, filtered through a 0.45-μm filter, aliquoted, and titrated before the neutralization assay. To test serum-mediated neutralization, a six-point, threefold dilution series was prepared for each serum in culture medium [Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum, 1% penicillin–streptomycin, and 3 μg/mL puromycin]. Each dilution (50 μL) was mixed with 50 μL of diluted pseudovirus in 96-well plates and incubated for 30 min at room temperature. The mixture was then incubated with 104 ACE-2-expressing 293T cells (293T-hACE2.MF) in a final volume of 200 μL. A total of 150 µL of supernatant was carefully removed after 72 h. The cells were lysed with 50 μL Bright-Glo™ Luciferase Assay substrate (Promega) for 5 min, and the luciferase activity (relative light units, RLU) was measured. Percent neutralization was normalized considering uninfected cells as 100% neutralization and cells infected in the absence of antibodies as 0% neutralization. Half-maximal (IC50) titers were determined using an asymmetrical (five-parameter) logistic dose–response curves function in Prism.
Statistical Analysis.
Comparisons between different mRNA constructs (Fig. 1) and different groups of immunized animals (Figs. 2–4) were performed by either parametric or nonparametric (Mann–Whitney) t tests with all P values two-tailed. A P value of less than 0.05 was considered to indicate statistical significance.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank H. Mu and M. Farzan for the 293T-ACE2 stable cells; K. Corbett, W. Shi, L. Wang, and N. Doria-Rose for SARS-CoV-2 Spike-expressing plasmids; and B. Zhang and P. D. Kwong for human antibody S-118. This work was supported in part by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases.
Author contributions
P.Z., S.F., I.R., E.N., G.S.-J., S.H., A.C., A.S.F., and P.L. designed research; P.Z., Y.T., M.S., V.G., H.M., Y.S., D.R., and Y.-T.L. performed research; P.Z., A.S.F., and P.L. analyzed data; and P.Z. and P.L. wrote the paper.
Competing interests
Several authors are employees of Moderna, Inc. and hold stocks from the company. Some authors have filed patents on mRNA vaccines.
Footnotes
Reviewers: N.P., University of Pennsylvania; and M.P., Uniformed Services University of the Health Sciences.
Contributor Information
Anthony S. Fauci, Email: afauci@niaid.nih.gov.
Paolo Lusso, Email: plusso@niaid.nih.gov.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Folegatti P. M., et al. , Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: A preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet 396, 467–478 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jackson L. A., et al. , An mRNA vaccine against SARS-CoV-2 - preliminary report. N. Engl. J. Med. 383, 1920–1931 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Polack F. P., et al. , Safety and efficacy of the BNT162b2 mRNA covid-19 vaccine. N. Engl. J. Med. 383, 2603–2615 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sadoff J., et al. , Interim results of a phase 1–2a trial of Ad26.COV2.S covid-19 vaccine. N. Engl. J. Med. 384, 1824–1835 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corti D., Purcell L. A., Snell G., Veesler D., Tackling COVID-19 with neutralizing monoclonal antibodies. Cell 184, 4593–4595 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karim S. S. A., Karim Q. A., Omicron SARS-CoV-2 variant: A new chapter in the COVID-19 pandemic. Lancet 398, 2126–2128 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mascola J. R., Graham B. S., Fauci A. S., SARS-CoV-2 viral variants-tackling a moving target. JAMA 325, 1261–1262 (2021). [DOI] [PubMed] [Google Scholar]
- 8.Baden L. R., et al. , Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 384, 403–416 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pardi N., Hogan M. J., Porter F. W., Weissman D., mRNA vaccines —A new era in vaccinology. Nat. Rev. Drug. Discov. 17, 261–279 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corbett K. S., et al. , Evaluation of the mRNA-1273 vaccine against SARS-CoV-2 in nonhuman primates. N. Engl. J. Med. 383, 1544–1555 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sahin U., et al. , COVID-19 vaccine BNT162b1 elicits human antibody and T(H)1 T cell responses. Nature 586, 594–599 (2020). [DOI] [PubMed] [Google Scholar]
- 12.Ludwig C., Wagner R., Virus-like particles-universal molecular toolboxes. Curr. Opin. Biotechnol. 18, 537–545 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roldao A., Mellado M. C., Castilho L. R., Carrondo M. J., Alves P. M., Virus-like particles in vaccine development. Expert Rev. Vaccines 9, 1149–1176 (2010). [DOI] [PubMed] [Google Scholar]
- 14.Qian C., et al. , Recent progress on the versatility of virus-like particles. Vaccines (Basel) 8, 139 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang P., et al. , A multiclade env-gag VLP mRNA vaccine elicits tier-2 HIV-1-neutralizing antibodies and reduces the risk of heterologous SHIV infection in macaques. Nat. Med. 27, 2234–2245 (2021). [DOI] [PubMed] [Google Scholar]
- 16.Freed E. O., Martin M. A., Domains of the human immunodeficiency virus type 1 matrix and gp41 cytoplasmic tail required for envelope incorporation into virions. J. Virol. 70, 341–351 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wyma D. J., Kotov A., Aiken C., Evidence for a stable interaction of gp41 with Pr55(Gag) in immature human immunodeficiency virus type 1 particles. J. Virol. 74, 9381–9387 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simmons G., et al. , Characterization of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike glycoprotein-mediated viral entry. Proc. Natl. Acad. Sci. U.S.A. 101, 4240–4245 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glowacka I., et al. , Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J. Virol. 85, 4122–4134 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boix-Besora A., Lorenzo E., Lavado-Garcia J., Godia F., Cervera L., Optimization, production, purification and characterization of HIV-1 GAG-based virus-like particles functionalized with SARS-CoV-2. Vaccines (Basel) 10, 250 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Z., et al. , Potent monoclonal antibodies neutralize omicron sublineages and other SARS-CoV-2 variants. Cell Rep. 41, 111528 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muecksch F., et al. , Increased memory B cell potency and breadth after a SARS-CoV-2 mRNA boost. Nature 607, 128–134 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bachmann M. F., et al. , The influence of antigen organization on B cell responsiveness. Science 262, 1448–1451 (1993). [DOI] [PubMed] [Google Scholar]
- 24.Nelson J., et al. , Impact of mRNA chemistry and manufacturing process on innate immune activation. Sci. Adv. 6, eaaz6893 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hassett K. J., et al. , Optimization of lipid nanoparticles for intramuscular administration of mRNA vaccines. Mol. Ther. Nucleic Acids 15, 1–11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zingler K., Littman D. R., Truncation of the cytoplasmic domain of the simian immunodeficiency virus envelope glycoprotein increases env incorporation into particles and fusogenicity and infectivity. J. Virol. 67, 2824–2831 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All study data are included in the article and/or SI Appendix.