Significance
Alopecia areata (AA) is a common autoimmune disorder, but progress in developing novel therapeutics has been hampered by an incomplete understanding of disease mechanisms. Using scRNAseq single-cell RNA sequencing (scRNAseq), we dissected the immune landscape of murine and human AA skin and depleted specific cell types in murine AA to interrogate their function. We demonstrated that CD8+ T cells are the predominant disease-driving cell type, whose heterogeneity is defined by an “effectorness gradient” that informs their trajectory of differentiation. Our results also uncovered Izumo1r/FR4 as a marker for Treg (regulatory T cells), whose selective depletion accelerated AA onset, demonstrating intact immunosuppressive Treg function in AA. Common mechanisms underlying murine and human AA highlight the translational impact of our work.
Keywords: alopecia areata, autoimmunity, hair follicle, single-cell RNA sequencing, T cells
Abstract
Alopecia areata (AA) is among the most prevalent autoimmune diseases, but the development of innovative therapeutic strategies has lagged due to an incomplete understanding of the immunological underpinnings of disease. Here, we performed single-cell RNA sequencing (scRNAseq) of skin-infiltrating immune cells from the graft-induced C3H/HeJ mouse model of AA, coupled with antibody-based depletion to interrogate the functional role of specific cell types in AA in vivo. Since AA is predominantly T cell-mediated, we focused on dissecting lymphocyte function in AA. Both our scRNAseq and functional studies established CD8+ T cells as the primary disease-driving cell type in AA. Only the depletion of CD8+ T cells, but not CD4+ T cells, NK, B, or γδ T cells, was sufficient to prevent and reverse AA. Selective depletion of regulatory T cells (Treg) showed that Treg are protective against AA in C3H/HeJ mice, suggesting that failure of Treg-mediated immunosuppression is not a major disease mechanism in AA. Focused analyses of CD8+ T cells revealed five subsets, whose heterogeneity is defined by an “effectorness gradient” of interrelated transcriptional states that culminate in increased effector function and tissue residency. scRNAseq of human AA skin showed that CD8+ T cells in human AA follow a similar trajectory, underscoring that shared mechanisms drive disease in both murine and human AA. Our study represents a comprehensive, systematic interrogation of lymphocyte heterogeneity in AA and uncovers a novel framework for AA-associated CD8+ T cells with implications for the design of future therapeutics.
Alopecia areata (AA) is one of the most common autoimmune disorders (1, 2), in which immune-mediated attack of the hair follicle (HF) results in nonscarring hair loss that can range from well-demarcated patches on the scalp to total scalp and/or body hair loss. Despite its wide prevalence, the therapeutic options for AA remain limited, largely due to an incomplete understanding of disease mechanisms. We previously demonstrated that autoreactive NKG2D+ CD8+ T cells are a hallmark of AA pathogenesis (3), an observation that led us to pioneer the use of Janus Kinase (JAK) inhibitors to treat AA in patients (4, 5). JAK inhibitors are the first FDA-approved agents for AA, representing a significant advancement in the therapeutic landscape of AA. However, AA often recurs upon cessation of treatment, underscoring the need to further investigate disease mechanisms to enable the development of novel therapeutics for AA.
The pathognomonic histologic feature of AA is a dense inflammatory infiltrate surrounding the lower part of the HF in a “swarm of bees” pattern (6). In healthy skin, the HF is a site of immune privilege, in which local immunoinhibitory signaling and the low expression of antigen presentation molecules allows the HF to evade immune recognition (7, 8). AA is thought to arise, at least in part, due to a collapse of immune privilege and subsequent immune-mediated attack (3, 7, 8). In addition to CD8+ T cells, the inflammatory infiltrate is also composed of CD4+ T helper (Th) cells, CD4+ regulatory T cells (Treg), and nonlymphocytic cell types such as monocytes, dendritic cells (DCs), and macrophages.
In light of the T cell-mediated nature of AA, numerous studies have been undertaken to dissect these immune cell subsets, with particular attention to T lymphocytes (9–13). However, aside from cytotoxic CD8+ T cells, the functional relevance of other T cell populations to AA and their potential as novel therapeutic targets remain poorly defined. For instance, although some studies have suggested a disease-promoting role for CD4+ Th cells in AA (12–15), it is unclear whether CD4+ Th cells contribute to disease onset, progression, neither, or both. There is also conflicting evidence on the role of CD4+ Treg in AA, with some studies suggesting that CD4+ Treg are lower in frequency and/or dysfunctional in AA (16–19), whereas others reported higher numbers of Treg or no difference in Treg frequency in AA relative to baseline (18, 20).
Understanding the complexities underlying autoimmune disorders such as AA has been impeded by the redundancy of the various cell surface markers used to isolate and profile specific subsets of interest. Such approaches often mask the contribution of rare cell types, which are difficult to isolate in sufficient numbers to analyze their transcriptional profiles. By providing transcriptomic information at the level of individual cells, single-cell RNA sequencing (scRNAseq) now affords an opportunity to deconvolute the diverse cell types within complex tissues and identify rare subpopulations involved in homeostasis or disease. Advances in single-cell technology have been transformative in uncovering novel insights into the immunopathology of various autoimmune disorders, including those that are genetically related to AA, such as rheumatoid arthritis and type 1 diabetes (21, 22).
Previous efforts to investigate lymphocyte function in AA largely relied on the adoptive transfer of exogenously derived cells, often stimulated and expanded ex vivo (3, 14, 15, 23, 24). Here, we utilized scRNAseq to dissect the immune landscape of AA skin, using both the graft-induced C3H/HeJ mouse model of AA (3, 23, 25–28), as well as human AA scalp biopsies. Furthermore, to interrogate the requirement of specific lymphocyte subsets and their potential contribution to disease, we coupled our scRNAseq studies with functional antibody-mediated cellular depletion studies in vivo using the graft-induced C3H/HeJ mouse model of AA.
Our findings demonstrated that CD8+ T cells are the predominant disease-driving cell type in AA, with an accessory role for CD4+ T cells and no obvious role for NK, B, or γδ T cells. We also identified Izumo1r/FR4 as a novel and highly specific marker of Treg in murine AA. Using anti-FR4-mediated depletion of Treg, we demonstrated that Treg-mediated immunosuppression is intact in C3H/HeJ mice. A focused analysis of CD8+ T cells uncovered an “effectorness gradient” that underlies CD8+ T cell heterogeneity in AA skin and informs their trajectory of differentiation, culminating in increased tissue residency and effector T cell function. The effectorness gradient is applicable to CD8+ T cells in both murine and human AA and provides a novel framework for understanding the diverse CD8+ T cell states that define AA, with implications for the future development of novel therapies.
Results
Single-Cell RNA Sequencing Reveals the Immune Landscape of Murine AA.
The C3H/HeJ mouse model of AA involves grafting full-thickness skin or transferring T cells from a diseased donor onto a naive C3H/HeJ recipient. This model demonstrates high concordance with human AA and has been widely used in translational studies by our group and others (3, 23, 25–28). In this model, grafted recipients develop patchy AA at approximately 5 to 6 wk post-engraftment and typically progress to total body hair loss by 24 wk. To comprehensively profile the immunological mechanisms underlying AA, we performed droplet-based scRNAseq (10× Genomics) on skin-infiltrating CD45+ immune cells isolated from grafted C3H/HeJ mice exhibiting total body hair loss (24 wk post-engraftment, as a model of alopecia universalis in AA patients, the most severe form of disease) versus ungrafted (UG) C3H/HeJ mice (without disease) as a control (Fig. 1A and SI Appendix, Fig. S1A), using three biological replicates per condition. Following measures to exclude doublets and low-quality cells (SI Appendix, Fig. S1B), cells from each reaction were merged and aligned using the Seurat-based canonical correlation analysis (CCA) workflow to correct batch effects (Materials and Methods). The resulting dataset encompassed 20,321 total cells, composed of 11,219 cells from the three AA mice and 9,102 cells from the three UG control mice.
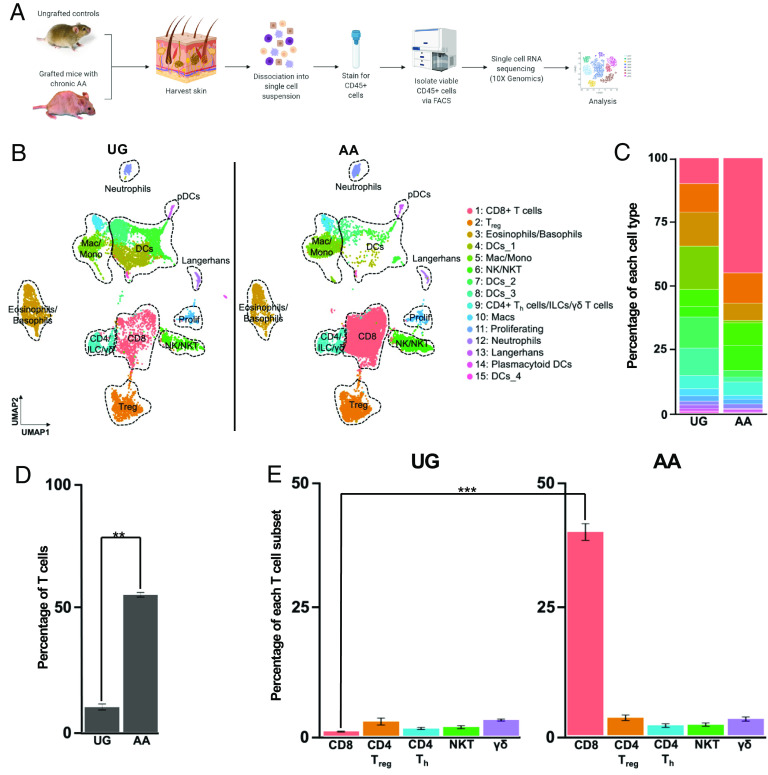
Fig. 1.
CD8+ T cells are the predominant expanded cell type in AA skin. (A) Schematic of scRNAseq experiment. We harvested full-thickness skin from three skin-grafted C3H/HeJ mice with AA and three UG controls and isolated live CD45+ immune cells for scRNAseq (10× Genomics). Individual cDNA libraries were prepared for each sample prior to sequencing. (B) UMAP of scRNAseq data from (A) split across disease condition. Unsupervised clustering of 20,321 cells (9,102 UG; 11,219 AA) after quality control and CCA-based alignment uncovered 15 distinct populations. Note the marked increase in T cell infiltration in AA skin, especially that of CD8+ T cells. (C) Stacked bar plot showing distribution of each cluster relative to the total number of cells per disease condition. (D) Percentage of T cells relative to the total number of sequenced cells, averaged across the three replicates for each disease condition, showing a statistically significant (P = 0.002) increase in T cells in AA skin. Error bars represent SD. (E) Percentage of specific T cell subsets relative to the total number of sequenced cells, averaged across the three replicates for each disease condition, showing a statistically significant (P = 0.00089) increase in CD8+ T cells in AA skin. Error bars represent SD. DCs, dendritic cells; ILC, innate lymphoid cells; Mac, macrophages; Mono, Monocytes; NK, natural killer cells; NKT, natural killer T cells; Treg, regulatory T cells. Significance is indicated as follows: *P < 0.05; **P < 0.01; ***P < 0.001.
Uniform manifold approximation and projection (UMAP) dimensionality reduction and unsupervised graph-based clustering based on the first 30 principal components resulted in the identification of distinct clusters representing diverse immune cell types including CD8+ T cells, Treg, natural killer (NK) and NK T cells, non-Treg CD4+ Th cells, γδ T cells, innate lymphoid cells (ILCs), granulocytes, DCs, monocytes, and macrophages (Fig. 1B). Represented cell types and their distributions were broadly consistent across the three replicates for each disease condition (SI Appendix, Fig. S1C). Cell types were annotated using a combinatorial approach involving SingleR, an automatic annotation method for scRNAseq data that compares transcriptomic signatures to reference datasets such as the mouse Immunological Genome Project dataset (29, 30), the most highly expressed transcripts in each cluster, and the expression of canonical markers established in the literature (SI Appendix, Fig. S1 D–F). For instance, CD8+ T cells were characterized by high expression of Cd3e and Cd8a; Treg were defined by Cd3e, Cd4, and Foxp3 expression; and NK cells were defined by Nkg7, Xcl1, and Ncr1 expression in the absence of Cd3e expression.
CD8+ T Cells Are the Predominant Expanded Lymphocytic Cell Type in AA Skin.
Comparison across disease conditions revealed marked differences in cell-type distributions between AA and UG skin (Fig. 1 B and C), including a striking, statistically significant expansion of T lymphocytes, defined as all Cd3d, Cd3e, and/or Cd3g-positive cells, in AA skin (Fig. 1 D and E). On average, T cells represented 56.22% of the total skin-infiltrating immune cells sequenced from each AA replicate, whereas they represented only 10.83% of cells sequenced from each UG replicate, consistent with the hypothesis that AA is predominantly a T cell-mediated disease (3, 7). Closer inspection of the distribution of specific T lymphocyte subsets, including CD8+ T cells, CD4+ Th cells, Treg, NK T cells, and γδ T cells, showed that the expansion of T lymphocytes in AA skin was largely driven by an increase in the number of infiltrating CD8+ T cells (Fig. 1 D and E). We found a statistically significant enrichment of CD8+ T cells in AA skin, which represented 44.53% of sequenced cells, compared to 0.89% of cells sequenced from UG skin. In contrast, there was no significant change in the percentage of Treg (11.7% of AA cells, 10.8% of UG cells), CD4+ Th cells (2.2% of AA cells, 1.6% of UG cells), NK T cells (2.3% of AA cells, 1.8% of UG cells), or γδ T cells (3.51% of AA cells, 3.41% of UG cells) across disease conditions. Taken together, these results indicated that CD8+ T cells are the predominant expanded lymphocytic cell type infiltrating AA skin.
CD8+ T Cells Are Required for the Development of AA, and Their Depletion Is Sufficient for Disease Reversal.
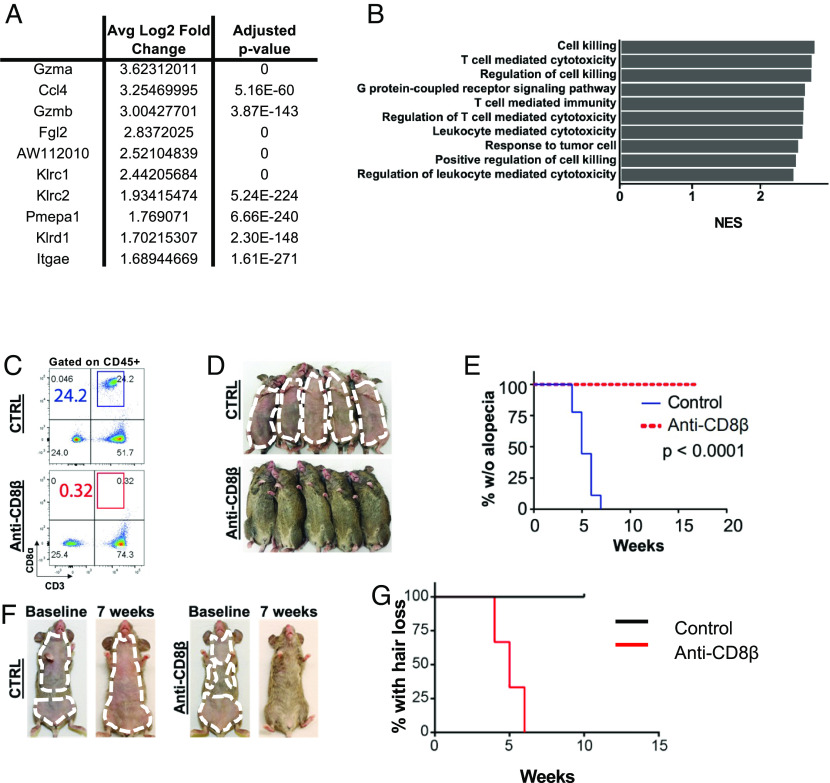
In addition to the dramatic expansion of CD8+ T cells in AA skin, differential gene expression analysis between CD8+ T cells in AA and UG skin showed upregulation of genes associated with T cell cytotoxicity and activation in AA CD8+ T cells, such as Gzma, Gzmb, and Ccl4 (Fig. 2A). Gene set enrichment analysis (GSEA) of statistically significant differentially expressed genes also showed enrichment of gene ontology (GO) terms involving T cell immunity and cytotoxicity (Fig. 2B and SI Appendix, Fig. S2B). These data support the pathogenic role of CD8+ T cells in AA that we and others previously established by utilizing the transfer of exogenously derived CD8+ T cells from disease donors that were stimulated and expanded in ex vivo culture (3, 14, 15, 25). To date, no studies have investigated whether endogenous CD8 + T cells are required for disease development and/or progression.
Fig. 2.
CD8+ T cells are required for disease development and progression in murine AA. (A) Top 10 genes up-regulated in AA-associated CD8+ T cells compared to UG-associated CD8+ T cells in differential gene expression analysis, showing upregulation of genes associated with T cell cytotoxicity and activation such as Ccl5Gzma, and Gzmb. “0” for adjusted P value indicates a near-zero value that was rounded to 0 by the R software. (B) Top 10 GO terms enriched in AA CD8+ T cells as identified by GSEA of differentially expressed genes between AA and UG CD8+ T cells, arranged via descending Normalized Enrichment Score (NES). Shown pathways were enriched in AA CD8+ T cells in a statistically significant manner, with P < 0.05. (C) Treatment of grafted C3H/HeJ mice with anti-CD8β efficiently depleted CD8+ T cells compared to isotype control. (D) Anti-CD8β administration in grafted C3H/HeJ mice prior to hair loss onset prevented AA compared to isotype control. (E) Kaplan–Meier curve for experiment shown in (D). (F) Anti-CD8β administration in grafted C3H/HeJ mice with established disease reversed AA and induced hair regrowth. Those treated with isotype control progressed to total body hair loss. (G) Kaplan–Meier curve for experiment shown in (F). P = 0.0246.
To interrogate the requirement of endogenous CD8+ T cells in the development of AA, we performed antibody-mediated depletion of CD8+ T cells in vivo and specifically tested whether the depletion of CD8+ T cells was sufficient for disease prevention. Treatment of C3H/HeJ mice with an anti-CD8β antibody at the time of skin engraftment (but prior to disease onset), twice weekly for 8 wk, completely prevented hair loss in all treated mice compared to those administered a vehicle control (Fig. 2 C–E). To ask whether depletion of CD8+ T cells was sufficient for disease reversal once the disease had been established, we treated grafted C3H/HeJ mice with the established disease at 5 to 7 wk with anti-CD8β, twice weekly for 10 wk. Anti-CD8β reversed AA and induced hair regrowth, whereas mice treated with the vehicle control progressed to total body hair loss (Fig. 2 F and G). Our findings demonstrated that CD8+ T cells were required for AA development and that their depletion was sufficient for reversal. Importantly, these data showed that the elimination of CD8+ T cells can eradicate AA in the setting of established disease.
Depletion of CD4+ T Cells Is Not Sufficient for AA Prevention or Reversal.
CD4+ T cells play central roles in autoimmunity and inflammation, via both the proinflammatory functions of Th cells as well as the immunomodulatory functions of Treg (31–33). Previous studies reported increased CD4+ T cell infiltration in murine AA lesional skin, and transfer of CD4+ T cells from diseased to naïve C3H/HeJ mice was shown to induce disease (15, 23). However, the rates of disease induction have been inconsistent, and their pathogenicity relative to CD8+ T cells has not been well established. Furthermore, whether endogenous CD4+ T cells are required for the development of disease remains an open question.
To examine the role of CD4+ T cells in AA, we first turned to our scRNAseq data and performed a differential gene expression analysis between AA-associated CD4+ T cells and their counterparts in UG skin. The top 10 up-regulated transcripts in AA-associated CD4+ T cells were composed of both genes associated with inflammation and T cell activation such as Ctsw, Ifng, Ccl5, and Kcnn4, as well as those classically associated with effector Treg function and immunosuppression, such as Fgl2, Pdcd1, and Ctla4 (SI Appendix, Fig. S2A). Normalized enrichment scores (NES) of GO terms associated with leukocyte-mediated cytotoxicity, T cell–mediated immunity, and T cell receptor signaling did not reach statistical significance and were lower than those derived from differentially expressed genes between AA and UG CD8+ T cells (P > 0.05, SI Appendix, Fig. S2B).
We next performed antibody-mediated depletion studies of CD4+ T cells in the graft-induced C3H/HeJ AA mouse model to investigate the efficacy of depleting CD4+ T cells in achieving AA prevention and/or reversal. To assess the role of CD4+ T cells in disease prevention, we administered anti-CD4 to skin-grafted C3H/HeJ mice prior to disease onset. While anti-CD4 delayed the timing of AA onset relative to isotype control, anti-CD4 did not prevent disease (SI Appendix, Fig. S2 C–E). To test the role of CD4+ T cells in disease reversal, we administered anti-CD4 to grafted C3H/HeJ mice with established AA, which also showed no effect on hair regrowth (SI Appendix, Fig. S2F). Our results are consistent with a potential secondary role for CD4+ T cells in disease initiation, as suggested by previous cell transfer studies (14, 15, 23), however, our studies showed that CD4+ T cells were not required for AA prevention or reversal.
CD4+ Treg Have Functional Immunosuppressive Capacity in AA.
Since anti-CD4 targets both CD4+ Th and Treg cells, we next aimed to parse the individual functions of these two CD4+ cell types in AA. Differential gene expression analysis between Treg of AA and UG mice demonstrated up-regulated expression of interferon (IFN) response genes such as Iigp1, Stat1, Igtp, and Irf1 in AA skin (SI Appendix, Fig. S3A). IFN-γ is not only one of the main disease-driving cytokines in AA (3) but also induces CD4+ Treg development, proliferation, and immunosuppressive function (34–37). AA Treg also showed up-regulated expression of genes associated with enhanced Treg function and migration such as Ly6a, Ccr2, Fgl2, Gzmb, and Satb1 (38–44). We observed no significant differences in the expression of Il-10 and Tgfb1, two anti-inflammatory cytokines that are critical to Treg-mediated immunosuppression. Taken together, these gene expression profiles suggested that AA-associated Treg have intact immunosuppressive capacity. To test this functionally, we performed a Treg suppression assay and found no significant difference in the ability of AA versus UG Treg to suppress CD4+ T cell proliferation in vitro (SI Appendix, Fig. S3B).
We next sought to selectively deplete Treg in C3H/HeJ mice to probe their function in vivo. Previous studies utilized anti-CD25 (encoded by Il2ra) to deplete Treg in the context of both autoimmunity and cancer (45–47). However, in addition to CD4+ Treg, our scRNAseq showed high Il2ra expression in other cell types, including CD4+ Th cells, NK/NK T cells, and CD8+ T cells (SI Appendix, Fig. S3C). Indeed, we found that administration of anti-CD25 in grafted C3H/HeJ mice significantly delayed and reversed disease in a manner indistinguishable from anti-CD8 treatment (SI Appendix, Fig. S3 D–H), suggesting that anti-CD25 depleted multiple cell types and therefore does not specifically target Treg in AA.
Interestingly, our differential gene expression analysis between AA and UG Treg revealed striking upregulation of Izumo1r in AA Treg (SI Appendix, Fig. S4A). Izumo1r encodes Folate Receptor 4 (FR4), a cell surface receptor that was previously used to target Treg in other contexts such as melanoma (48) but has not been previously studied in AA. Unlike other cell surface markers upregulated in Treg, our data showed that Izumo1r was expressed specifically in AA-associated Treg and was not observed in other non-Treg immune cell types. As predicted by our functional assays and consistent with their known immunosuppressive function, treatment of skin-grafted C3H/HeJ mice prior to disease onset with anti-FR4 efficiently depleted Treg in the skin and accelerated disease onset by approximately 2 wk compared to isotype control (SI Appendix, Fig. S4 B–D). Further, the delayed disease phenotype we observed upon anti-CD4 treatment in grafted C3H/HeJ mice (SI Appendix, Fig. S2 C–E) was likely due to the absence of CD4+ Th, as opposed to the depletion of dysfunctional Treg. These results demonstrated that AA is not driven by a failure in Treg-mediated immunosuppression but rather that Treg are protective in AA, consistent with the immunosuppressive role classically ascribed to Treg.
To systematically interrogate the function of the remaining skin-infiltrating lymphocyte subsets in AA, we performed additional antibody-based depletion of NK cells, B cells, and γδ T cells. We observed no effect of NK, B, or γδ T cell depletion on AA disease onset (SI Appendix, Fig. S5).
scRNAseq Uncovers CD8+ T Cell Heterogeneity in AA.
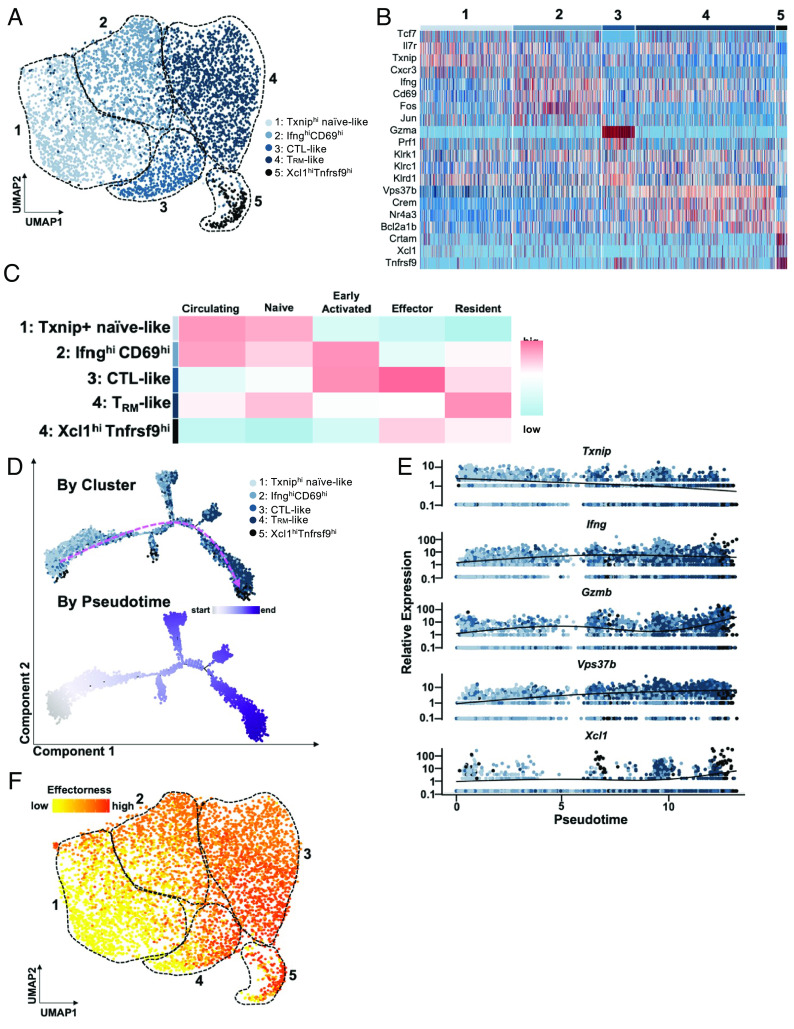
While the critical role of CD8+ T cells in AA has been corroborated by previous studies (3, 15, 23, 49), few studies to date have dissected the heterogeneity of CD8+ T cells in AA, especially as it relates to their potential function(s) in disease. To further define the diverse CD8+ T cell states in AA and their contribution to disease pathogenesis, we performed unsupervised reclustering of AA-associated CD8+ T cells, which identified five distinct subpopulations (Fig. 3A).
Fig. 3.
An effectorness gradient underlies CD8+ T cell heterogeneity in AA skin and identifies a potential trajectory for their differentiation. (A) UMAP of AA CD8+ T cell subsets. Unsupervised re-clustering of AA CD8+ T cells uncovered five subpopulations. (B) Heatmap of top highly expressed genes in each cluster. Cluster 1 showed higher expression of Tcf7, Il7r, Txnip, and Cxcr3; cluster 2 showed upregulation of Ifng, Cd69, Fos, and Jun; cluster 3 was enriched for Gzma, Prf1, Klrc1, and Klrd1; cluster 4 was up-regulated for Vps37b, Crem, Nr4a3, Bcl2a1b; cluster 5 showed higher expression of Crtam, Xcl1, and Tnfrsf9. Interestingly, expression of Klrk1, which encodes NKG2D, was not restricted to a single cluster. (C) Heatmap of scores for signatures of CD8+ T cell subpopulations described in previously published studies. Note that expression of gene signatures varies across clusters on a gradual continuum, as opposed to being exclusive to a particular cluster. (D) Pseudotime trajectory analysis of AA CD8+ T cells, colored by cluster (Top) and by position along pseudotime (Bottom). Pink dashed arrow denotes general direction of inferred differentiation across pseudotime. (E) Expression of select marker genes as identified in Fig. 3B along pseudotime, colored by cluster. (F) Pseudotime values were used to compute effectorness scores for each cell. Effectorness scores overlaid onto the UMAP of AA-associated CD8+ T cells demonstrated a high degree of correlation with the separation of the five CD8+ T cell subsets via unsupervised clustering of their transcriptional profiles, as shown in (A).
Relative to other clusters, cluster 1 showed increased expression of Tcf7 and Il7r, which are upregulated on naive T cells (Fig. 3B) (50–52). Cluster 1 cells also showed high expression of Txnip, whose expression has been negatively associated with T cell activation (53–55), and Cxcr3, which we previously showed is expressed on CD8+ T cells migrating into the skin in AA (26). Cluster 2 showed high expression of Ifng, one of the main T cell-derived cytokines in AA (3), and Cd69, a marker of T cell activation and tissue retention (56). The AP-1 family transcription factors Fos and Jun, which mediate effector T cell differentiation, were also upregulated in cluster 2 (57, 58). Cluster 3 was enriched for the expression of genes classically associated with cytotoxic T lymphocyte (CTL) function such as Gzma and Prf1.
We previously established that CD8+ T cells expressing the activating NKG2 family receptor NKG2D are both necessary and sufficient for disease (3), and therefore, we anticipated that the CTL-like cells in cluster 3 would be enriched for Klrk1, which encodes NKG2D. Surprisingly, in addition to Klrk1, the CTL-like cells in cluster 3 also showed enriched expression of Klrc1 and Klrd1, which encode the NKG2A/CD94 complex, respectively, an inhibitory NKG2 family receptor that is known to dampen T cell activation and serve as an immune checkpoint (59, 60). Cluster 4 showed high expression of Vps37b, Crem, and Nr4a3, which have been associated with tissue-resident memory (TRM) cells (61–63), as well Bcl2a1b, an anti-apoptotic member of the Bcl2 family. Finally, cluster 5 was up-regulated for Crtam, Tnfrsf9, and Xcl1, which have also been reported to be upregulated in activated CD8+ T cells (22, 64, 65). A recent scRNAseq study identified Xcl1hiTnfrsf9 hi CD8+ T cells in a mouse model of type 1 diabetes, an autoimmune disease that is genetically related to AA (22, 66).
To characterize these subsets in terms of known CD8+ T cell functions, we scored the cells in each cluster based on previously published transcriptomic signatures of CD8+ T cell subsets (Fig. 3C and SI Appendix, Table S1) (67–69). Our results were consistent with the characteristics ascribed to the marker genes associated with each cluster. For instance, cluster 1 scored highly for the “Circulating” (nontissue resident) and “Naive” signatures, while cluster 3 scored highly for the “Effector” signature, and cluster 4 for the “Resident” signature. Recently, Jaiswal et al. (70) performed a kinetic scoring analysis to query the transcriptomic programming of melanoma tumor-infiltrating lymphocytes against datasets of CD8+ T cell differentiation from naive to memory states in murine and human viral infections (70–74). An analysis of our CD8+ T cell data from AA skin using this approach produced results consistent with our annotation of each of the five subsets (SI Appendix, Fig. S6) (70). For example, cluster 1 genes were significantly down-regulated in activated T cells during murine lymphocytic choriomeningitis virus infection, as well as progressively suppressed as T cells differentiated from naive status after murine vaccinia virus infection and human yellow fever (YF) vaccination. Meanwhile, cluster 4 programs were strongly enforced as CD8+ T cells differentiated toward TRM signature and long-term memory after vaccinia virus skin scarification and YF vaccination, respectively.
An Effectorness Gradient Underlies CD8+ T Cell Heterogeneity in AA.
In addition to identifying the likely functions of each CD8+ T cell subset in AA, our analyses showed that expression of these gene signatures was not exclusive to a single cluster but instead was distributed between clusters across a gradual continuum (Fig. 3C), which we termed an effectorness gradient. This concept stems from a previous scRNAseq study in CD4+ T cells from healthy individuals demonstrated an effectorness gradient underlying CD4+ T cell heterogeneity, characterized by a transcriptional continuum from naive to effector and memory states alongside the increased expression of cytokines and chemokines (75). A recent study extended this concept to CD8+ T cells in human inflammatory bowel disease (76). Our results suggested that this framework could also apply to CD8+ T cells in AA skin, in which CD8+ T cells comprise interrelated transcriptional states of increasing “effectorness” as opposed to forming discrete, mutually exclusive populations.
To examine the relationship among the CD8+ T cell subsets, we performed a pseudotime analysis, which inferred the progression of naive-like cells in cluster 1 toward IfnghiCD69hi cluster 2 cells that scored highly on the “Early Activated” signature and CTL-like cluster 3 cells, ending with TRM-like cluster 4 cells and Tnfrsf9hiXclhi cluster 5 cells (Fig. 3D). Additionally, cells located at the beginning of the trajectory expressed higher levels of the naive gene Txnip, followed by increased expression of Ifng, which was enriched in cluster 2 cells that scored highly for the Early Activated signature (Fig. 3E). Cells located toward the end of the trajectory were enriched for the effector and TRM genes Gzmb, Vps37b, and Xcl1.
Given these results, we computed and scored the effectorness of each cell based on the pseudotime values, as described in previous studies (75, 76). We observed a strong correlation between effectorness scores and the separation of the five subsets in the UMAP (Fig. 3F). This analysis therefore demonstrated that the relationship between heterogeneous CD8+ T cell populations infiltrating AA skin can also be distributed along an effectorness gradient and revealed a potential trajectory of CD8+ T cell differentiation in AA.
Shared Immunological Mechanisms Underlie Murine and Human AA.
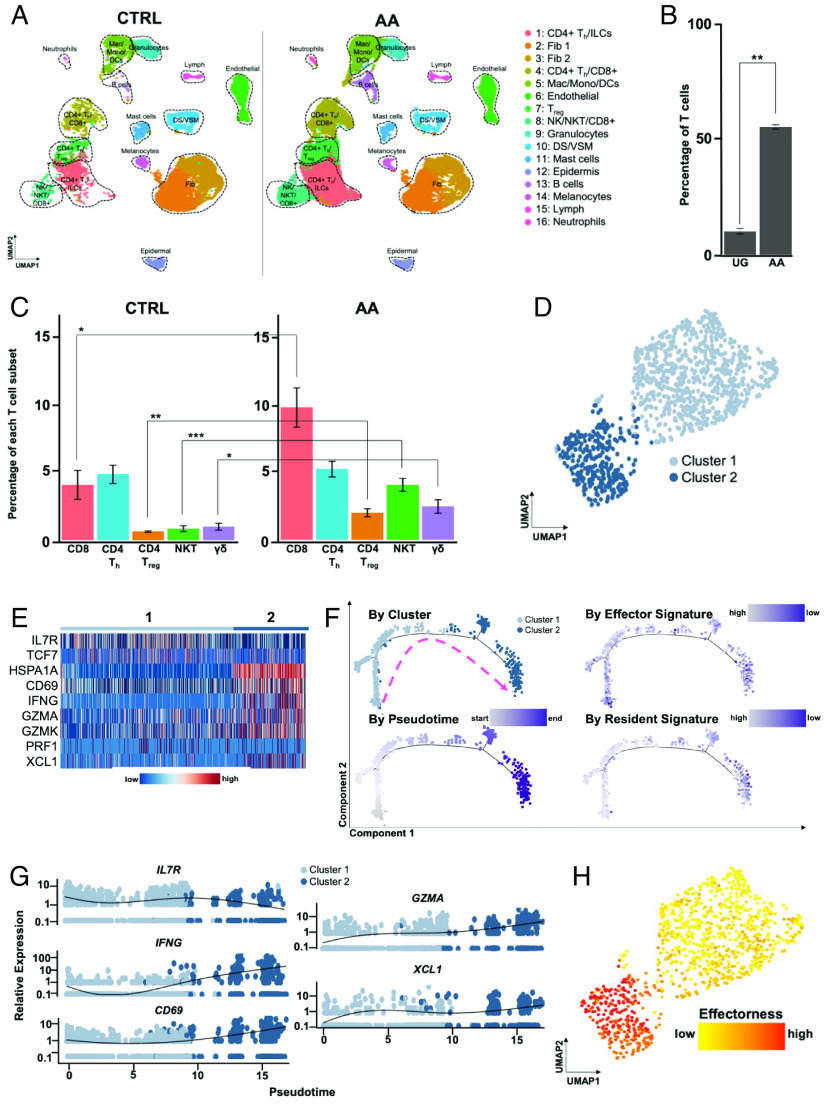
To investigate the translational potential of our scRNAseq findings in the graft-induced C3H/HeJ mouse model to AA patients, we performed scRNAseq (10× Genomics) of scalp biopsies from five AA patients with active disease (from lesional scalp) and two healthy individuals without AA (CTRL). cDNA libraries were prepared independently for each of the seven samples. Following quality control methods and batch effect correction, UMAP projection and unsupervised clustering identified 16 clusters representing diverse skin cell types including various lymphocyte populations, myeloid lineages, fibroblasts, epidermal cells, endothelial cells, and melanocytes (Fig. 4A and SI Appendix, Fig. S7A). Each cluster was annotated based on SingleR predictions (29, 30), the expression of canonical marker genes established in the literature, and the top most highly expressed genes in each cluster, similar to the approach we undertook for scRNAseq of murine AA skin immune infiltrates (SI Appendix, Fig. S7 B–D).
Fig. 4.
An effectorness gradient underlies CD8+ T cell heterogeneity in human AA skin. (A) UMAP of scRNAseq data obtained from scalp biopsies of 5 AA patients and 2 healthy controls, split across disease condition. Unsupervised clustering of 32,134 cells (21,766 AA; 32,134 CTRL) after quality control and CCA- based alignment uncovered 16 distinct populations. Note the marked increase in T cell infiltration in AA skin. (B) Percentage of T cells relative to the total number of sequenced immune cells, averaged across the replicates for each disease condition, showing a statistically significant (P = 0.002) increase in T cells in AA skin. (C) Percentage of specific T cell subsets relative to the total number of sequenced immune cells, averaged across the replicates for each disease condition. We observed statistically significant increases in the frequency of CD8+ T cells (P = 0.017), CD4+ Treg (P = 0.0051), NK T cells (P = 0.0016) and γδ T cells (P = 0.047) in AA skin. Significance is indicated as follows: *P < 0.05; **P < 0.01; ***P < 0.001. (D) UMAP of AA CD8+ T cell subsets. Unsupervised reclustering of AA CD8+ T cells uncovered two subpopulations. (E) Heatmap showing enriched expression of IL7R and TCF7 in cluster 1, while cluster 2 cells showed enriched expression of HSPA1ACD69IFNGGZMAGZMKPRF1, and XCL1. (F) Pseudotime trajectory analysis of AA CD8+ T cells, colored by cluster (Top Left), position along pseudotime (Bottom Left), Effector signature scores (Top Right), and Resident signature scores (Bottom Right). Pink dashed arrow denotes general direction of inferred differentiation across pseudotime. (G) Expression of select genes as identified in (B) along pseudotime, colored by cluster. (H) Pseudotime values were used to compute effectorness scores for each cell. Effectorness scores overlaid onto the UMAP of AA-associated CD8+ T cells demonstrated a high degree of correlation with the separation of the two CD8+ T cell subsets via unsupervised clustering of their transcriptional profiles, as shown in (D). Error bars represent SD. DS, dermal sheath; Fib, fibroblasts; Lymph, lymphatic vessel; VSM, vascular smooth muscle.
As observed in our murine scRNAseq data, AA skin overall was enriched for T lymphocytes compared to healthy skin, with T cells representing 23.74% of all sequenced immune cells, compared to 11.49% of all immune cells in CTRL skin (Fig. 4 B and C). CD8+ T cells were significantly enriched in AA skin (9.85% AA vs. 4.11% CTRL), while there was no statistically significant difference in the frequency of CD4+ Th cells between AA and CTRL. In contrast to our murine dataset, we also observed a small but statistically significant increase in the frequency of Treg (2.02% AA vs. 0.64% CTRL), as well as NK T cells (4.1% AA vs. 0.85% CTRL) and γδ T cells (2.49% AA vs. 1.00% CTRL).
Differential gene expression analyses of CD8+ T and CD4+ Th cells between AA and CTRL skin, followed by GSEA, showed upregulation of cytotoxicity and NKG2 family genes such as GZMK and KLRK1 in AA-associated CD8+ T cells. AA-associated CD8+ T cells also showed statistically significant enrichment in GO terms for “regulation of leukocyte-mediated cytotoxicity,” “NK cell-mediated immunity,” and “adaptive immune response” (SI Appendix, Fig. S8 A and B). Meanwhile, although AA CD4+ Th showed upregulation of genes associated with T cell activation and migration such as IL32, CXCR4, and CD69 (56, 77, 78), they were not enriched for cytotoxicity or NKG2 family-related genes, and the top enriched GO terms implicated pathways that were associated with general cellular processes such as protein translation and translocation (SI Appendix, Fig. S8 C and D). We were unable to perform differential gene expression analysis between AA and CTRL-associated Treg due to the low number of Treg captured in our dataset. Taken together, these results suggested that CD8+ T cells were the predominant disease-driving cell type in human AA, consistent with our findings in murine AA.
We next performed a focused analysis of CD8+ T cells, which both our mouse scRNAseq data and accompanying in vivo studies established as a predominant pathogenic cell type in AA. Unsupervised clustering of human AA-associated CD8+ T cells yielded two clusters, which largely diverged based on the expression of marker genes we identified in our previous mouse sequencing data (Fig. 4 D and E). For instance, TXNIP and TCF7, which were associated with the Naive-like cluster in murine AA CD8+ T cells (Fig. 3), were more highly expressed in cluster 1, while genes we previously associated with effector and activated T cell clusters such as CD69, IFNG, GZMA, and XCL1 were enriched in cluster 2. Similar to our approach for murine CD8+ T cells in AA, we further corroborated this annotation by analyzing our human CD8+ T cells in AA against signatures derived from various T cell states in murine and human viral infections (70–74), which showed that cluster 1 overall scored more highly for naive T cell states relative to activation and memory states, whereas the opposite was true for cluster 2 (SI Appendix, Fig. S9).
Our mouse scRNAseq studies suggested that CD8+ T cells in AA are composed of interrelated cellular states that form a transcriptional continuum of effectorness, as opposed to distinct subsets with mutually exclusive gene signatures. To examine whether this phenomenon held true in human AA-associated CD8+ T cells, we performed a pseudotime trajectory analysis, which inferred the gradual progression of cells in cluster 1 to those in cluster 2 (Fig. 4F). Plotting gene expression along pseudotime showed that Naive-like genes such as TXNIP were expressed highly at the beginning of pseudotime, whereas CD69, IFNG, GZMA, and XCL1 expression increased with the progression of pseudotime (Fig. 4G). Based on the pseudotime values, we then scored each cell’s effectorness, showed a strong correlation between effectorness scores, and the separation of the two human CD8+ T cell clusters in AA (Fig. 4H). These results underscore the presence of shared immunological mechanisms underlying murine and human AA.
Discussion
The immune landscape in AA is highly complex, characterized by the infiltration of diverse immune cell populations in the skin microenvironment that already exhibits exquisite immunological diversity at baseline. Here, we leveraged recent advances in single-cell sequencing technology to dissect the immune composition of both murine and human AA. Our single-cell studies guided in vivo functional experiments in the graft-induced C3H/HeJ mouse model, where we employed antibody-based cell depletion to systematically interrogate the requirement of specific immune cell populations. In contrast to previous functional studies, which relied on adoptive cell transfer and ex vivo expansion of specific populations of interest (3, 14, 15, 23, 24), our antibody-mediated depletion studies tested the requirement of endogenous lymphocyte populations in disease onset and reversal in the in vivo setting. Our results also expand upon previously published depletion studies in other rodent models of AA, such as the DEBR rat model and C3H/HeJ mice that spontaneously develop AA with age, albeit at an unpredictable rate (79–81).
Both our scRNAseq and depletion experiments established the predominant role of CD8+ T cells in AA, which were the only cell type whose depletion resulted in both AA prevention and disease reversal. Although anti-CD4 delayed AA onset, its failure to prevent or reverse AA suggested an accessory role for CD4+ T cells in disease initiation relative to CD8+ T cells, whereas depletion of NK, B, and γδ T cells had no effect on disease. While previous studies using the spontaneous C3H/HeJ model of AA did not observe complete disease reversal upon CD8+ T cell depletion, the authors reported markedly higher rates of AA reversal with CD8+ T cell depletion compared to CD4+ T cell depletion, consistent with our results and supporting the predominant role of CD8+ T cells in AA pathogenesis (81). These results invite future drug discovery efforts focused on the role of CD8+ T cells and, importantly, provide proof-of-concept for the design of therapeutic approaches that eliminate pathogenic CD8+ T cells.
To date, no studies have clearly resolved the heterogeneity of AA-associated CD8+ T cells in a manner that informs their function and potential trajectory of differentiation. Our focused analysis of AA-associated murine CD8+ T cells identified five subsets, whose gene expression profiles showed high concordance with established signatures for naive, early activated, effector, and resident CD8+ T cells in other contexts (67–69). The dissection of CD8+ T cell heterogeneity as it relates to the specific function of each subset affords an opportunity to target these populations therapeutically. For instance, clusters 2 and 3, which show relative upregulation of Ifng, Prf1, and Gzmb compared to other clusters, may be readily targeted by currently available immunosuppressive agents such as steroids and JAK inhibitors. However, while JAK inhibitors represent a significant advancement in the therapeutic landscape of AA, the majority of AA patients exhibit disease relapse upon discontinuation of their prescribed treatment regimen. This may be mediated in part by the persistence of TRM in the skin (82–85), which have not been previously described or identified in AA. Our scRNAseq data demonstrate that TRM (cluster 4) are numerous in AA skin and are characterized by the relative upregulation of genes previously associated with TRM in other contexts such as Crem, Vps37b, and Nr4a3. Additional studies are warranted to define their role in AA, particularly with regard to the potential for these markers as targets for future therapeutic modalities aimed at eliminating TRM to achieve durable treatment responses.
Further analysis revealed that while CD8+ T cell subsets in both murine and human AA have distinct transcriptional profiles, these profiles are not mutually exclusive but rather exist on a continuum that infers a differentiation trajectory for these subsets from naive to effector and resident T cell populations. A recent study using human CD4+ T cells demonstrated that CD4+ T cells undergoing polarization form a continuous transcriptional gradient of interrelated states and referred to this phenomenon as an effectorness gradient, in which “high effectorness” reflects high levels of cytokine secretion and an increased potential to initiate a robust effector response upon stimulation (75). This concept has also been applied to CD4+ T cells in colorectal cancer and both CD4+ and CD8+ T cells in inflammatory bowel disease (75, 86). These findings present a framework for understanding CD8+ T cell heterogeneity in AA that has implications for the development of future therapeutic interventions.
Redefining CD8+ T cell heterogeneity through the lens of an effectorness gradient represents a major conceptual shift from current efforts to disrupt CD8+ T cell function in AA, which largely rely on the identification and targeting of a single or small group of genes whose expression is thought to be perturbed in a mutually exclusive, binary manner. Instead, this framework provides an opportunity to discover cytokines and other inflammatory signaling pathways that shape CD8+ T cell differentiation trajectories and enable the development of approaches to potentially block the emergence of pathogenic CD8+ T cell subsets from their naive precursors. Furthermore, this framework enables a more nuanced interpretation of the different outcomes of cell depletion among our studies here and previously published studies (79–81): It implies that the effectorness gradient of CD8+ T cells targeted by depleting antibodies also varies as a function of time, such that results may differ depending on when and at what stage CD8+ T cells were targeted for depletion. Of note, the CD8+ and CD4+ T cell depletion studies in the spontaneous C3H/HeJ model of AA were performed in 5 to 10-mo-old mice, since spontaneous AA occurs in an unpredictable fashion over a wide age range (81).
A longstanding question in the field has been whether dysfunctional Treg contribute to loss of tolerance or immune privilege underlying disease pathogenesis. Our previous genome-wide association studies identified AA risk variants within Treg-associated genes such as CTLA4, ICOS, and LRRC32 (66, 87–89), and previous studies suggested Treg dysfunction and/or deficiency in human AA (16–19). The role of Treg in AA has remained elusive in part due to the lack of reliable methods to interrogate Treg function in C3H/HeJ mice since genetic mouse models are limited in the C3H/HeJ background, and anti-CD25, which has previously been used to target Treg in other contexts (45–47), also targets CD8+ T cells in AA (SI Appendix, Fig. S3 D–H). Interestingly, our single-cell data uncovered Izumo1r/FR4 as a highly specific marker of Treg in murine AA, providing a powerful tool for selective interrogation of Treg in AA mice for the first time. Anti-FR4-mediated Treg depletion in murine AA significantly accelerated disease onset, demonstrating that Treg play a protective role in AA in vivo, as expected. This finding was consistent with the results of our in vitro Treg suppression assay and suggests that the loss of immune tolerance in AA is not driven by Treg dysfunction and may instead be mediated by properties intrinsic to the HF itself or other cell types within the complex HF microenvironment (8). This also supports the results of previous cell-transfer studies, in which the transfer of CD4+ CD25+ Treg from AA mice could prevent disease induction by CD8+ T cell transfer in recipient C3H/HeJ mice (12). While we did not capture IZUMO1R expression in human AA Treg, the detection of IZUMO1R expression in human Treg has proven difficult in numerous studies, perhaps due to the presence of a pseudogene or an alternatively spliced variant not captured by conventional sequencing and alignment methods (90). Additionally, our human data showed statistically significant increases in the proportion of CD4+ Treg, as well as NK T and γδ T cells, which was not observed in our murine model. Although we did not observe a significant increase in the proportion of CD4+ Th in lesional skin, the proportion of CD4+ Th in control skin was greater than what was observed in murine skin, which is consistent with studies that have demonstrated substantial CD4+ T cell infiltration in human skin, even at baseline in noninflamed states (91, 92). Thus, despite the shared mechanisms between murine and human AA demonstrated in this study, these observations point to differences that warrant further study in human samples. Overall, our findings represent a comprehensive analysis of the diverse lymphocyte subsets in mouse and human AA and illustrate the utility of single-cell RNA sequencing as a powerful approach to resolve cellular heterogeneity in complex tissues and inform functional studies. Our work addresses numerous longstanding questions within the field of AA research, including CD8+ T cell heterogeneity and differentiation in AA skin, the potential role for TRM in disease relapse with currently available modalities of treatment, as well as excluding a key role for CD4+ Th and Treg. The discovery of shared mechanisms between murine and human AA underscores the translational potential of our findings and provides a road map for future studies aimed at developing novel therapeutic strategies for targeting specific CD8+ T cell subsets in a disease with a high unmet need.
Materials and Methods
Mice.
Female C3H/HeJ mice were obtained from The Jackson Laboratory and were maintained under pathogen-free conditions and a 12-h light/dark cycle at an ambient temperature of 22 to 26 °C. Female mice were used because like numerous other autoimmune disorders (93), AA has a slight female predominance (94, 95). This female predominance is observed in the skin-graft C3H/HeJ mouse model of AA (96), in which the literature reported that 95 to 100% of grafted female C3H/HeJ mice developed AA, whereas the incidence was 30% in male C3H/HeJ mice (23, 96). To induce AA in C3H/HeJ mice, we grafted full-thickness skin (approximately 1 cm in diameter) from donor mice with established disease onto 8- to 10-wk-old naive syngeneic recipients, as previously described (3). Animals were housed and all experiments were performed in compliance with institutional guidelines as approved by the Institutional Animal Care and Use Committee at Columbia University.
Human Subjects.
All human studies were conducted in compliance with the Columbia University Institutional Review Board-approved protocol, AAAI0706. AA patients and healthy individuals were recruited from the Department of Dermatology at Columbia University Irving Medical Center. Informed written consent was obtained from each individual prior to participation in the study.
In Vivo Mouse Studies.
For antibody-based cell depletion studies in the context of AA prevention, skin-grafted C3H/HeJ mice were treated with the antibody of interest or an isotype control IgG antibody (Bio X Cell, #BP0089) via intraperitoneal (IP) injection, 250 μg twice weekly for 8 wk starting from the day of grafting. For antibody-based cell depletion studies in the context of AA reversal, skin-grafted C3H/HeJ mice with established disease (approximately 5 to 7 wk after grafting) were treated with the antibody of interest or an IgG isotype control via IP injection, 500 μg twice weekly for 10 wk. Experiments were designed to validate the efficacy of in vivo antibodies in depleting the target cell type, and then investigate whether depletion led to an effect on either AA prevention or acceleration. Based on preliminary experiments, a type I error (α) of 0.05, and a β of 0.2, we calculated that at least five mice were required in prevention experiments and two mice were required in reversal experiments to detect an effect in at least 50% of animals included in each experiment with statistical significance. The primary outcome variable between two groups was the presence of hair loss.
A list of antibodies can be found in SI Appendix, Table S2.
Single-Cell 3’ mRNA Sequencing.
All scRNAseq experiments were performed at the JP Sulzberger Columbia Genome Center Single Cell Analysis Core. Libraries were prepared using the 10× Genomics Chromium Single Cell 3′ Reagent Kits according to the manufacturer’s instructions, with a target of 5,000 cells and 350 million reads. Libraries were sequenced on an Illumina NextSeq 500/550.
Statistics.
Statistical analyses were performed using the GraphPad Prism 7.0 software, unless otherwise noted. Comparisons between the distributions of cell types were performed using a two-tailed unpaired student’s t test with Welch’s correction. Log-rank tests were used to analyze the hair loss or regrowth curves. Data in bar and dot graphs are means ± SD. Significance is indicated as follows: *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Materials and methods for the preparation of single-cell suspensions from mouse and human skin, fluorescence-activated cell sorting and flow cytometry, Treg suppression assay, and processing and analysis of scRNAseq data can be found in SI Appendix, Supplementary Materials and Methods.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank E. Chang, J. Huang, M. Zhang, and W. Zeng for their expert technical assistance in the laboratory. We thank A. Obradovic for guidance on our single-cell analysis workflow, I. Krupska and E. Bush from the JP Sulzberger Genome Center for assistance with single-cell sequencing, M. Kissner from the Columbia Stem Cell Initiative for assistance with fluorescence-activated cell sorting and flow cytometry, and S. Youssef and L. Bordone for help with acquisition of human samples. This study was supported by NIH/NIAMS P50AR070588 AA Center for Research Translation (AACORT) and the Locks of Love Foundation. E.Y.L was supported by NIH/NIAMS F31AR077409. Z.D. was supported by a mentored young investigator grant award from the National AA Foundation and NIH/NIAMS K01AR070291. NIH/National Institute of Arthritis and Musculoskeletal and Skin Diseases grant P50AR070588 AACORT (A.M.C.) Locks of Love Foundation grant (A.M.C.) NIH/National Institute of Arthritis and Musculoskeletal and Skin Diseases grant F31AR077409 (E.Y.L.) NIH/National Institute of Arthritis and Musculoskeletal and Skin Diseases grant K01AR070291 (Z.D.) National AA Foundation Young Investigator Grant Award (Z.D.)
Author contributions
E.Y.L., Z.D., E.H.C.W., and A.M.C. designed research; E.Y.L., Z.D., and E.H.C.W. performed research; A.J., N.A., and A.M.C. contributed new reagents/analytic tools; E.Y.L., Z.D., A.J., and E.H.C.W. analyzed data; A.J., E.H.C.W., and N.A. revised the manuscript; N.A. and A.M.C. supervised the study; and E.Y.L., Z.D., and A.M.C. wrote the paper.
Competing interests
N.A. is a scientific advisor and an equity holder in Shennon Biotechnologies and is a consultant for Janssen, Immunitas, 23 and me, and Cellino Pharmaceuticals. A.M.C. is a consultant/scientific advisor for Almirall S.A., Arcutis Biotherapeutics, Inc., Intrinsic Medicine, Inc., and Pfizer, Inc.; is a shareholder of Aclaris Therapeutics, Inc. and Intrinsic Medicine, Inc.; has received research grant support from Pfizer, Inc.; is a coinventor on several patents filed by Columbia University on the use of JAK inhibitors in treating hair loss disorders, which have been licensed to Aclaris Therapeutics, Inc. and sublicensed from Aclaris to Eli Lilly & Co., resulting in intellectual property payments to the coinventors and to Columbia University; serves on the scientific advisory boards for the Dystrophic EB Research Association of America and the National AA Foundation; and is currently President of the American Hair Research Society.
Footnotes
Reviewers: C.A.J., Durham University; and N.W., Vanderbilt University Medical Center.
Data, Materials, and Software Availability
Raw sequencing data have been deposited in GEO with the accession number GSE233906 (97). All other data needed to evaluate the conclusions in the paper are present in the paper and/or the SI Appendix. Additional data related to this paper may be requested from the authors.
Supporting Information
References
- 1.Villasante Fricke A. C., Miteva M., Epidemiology and burden of alopecia areata: A systematic review. Clin. Cosmet. Investig. Dermatol. 8, 397–403 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jabbari A., Petukhova L., Cabral R. M., Clynes R., Christiano A. M., Genetic basis of alopecia areata: A roadmap for translational research. Dermatol. Clin. 31, 109–117 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xing L., et al. , Alopecia areata is driven by cytotoxic T lymphocytes and is reversed by JAK inhibition. Nat. Med. 20, 1043–1049 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mackay-Wiggan J., et al. , Oral ruxolitinib induces hair regrowth in patients with moderate-to-severe alopecia areata. JCI Insight 1, e89790 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jabbari A., et al. , An open-label pilot study to evaluate the efficacy of tofacitinib in moderate to severe patch-type alopecia areata, totalis, and universalis. J. Invest. Dermatol. 138, 1539–1545 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pratt C. H., King L. E., Messenger A. G., Christiano A. M., Sundberg J. P., Alopecia areata. Nat. Rev. Dis. Primers 3, 1–17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anzai A., Wang E. H. C., Lee E. Y., Aoki V., Christiano A. M., Pathomechanisms of immune-mediated alopecia. Int. Immunol. 31, 439–447 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bertolini M., McElwee K., Gilhar A., Bulfone-Paus S., Paus R., Hair follicle immune privilege and its collapse in alopecia areata. Exp. Dermatol. 29, 703–725 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Hashimoto K., et al. , Altered T cell subpopulations and serum anti-TYRP2 and tyrosinase antibodies in the acute and chronic phase of alopecia areata in the C3H/HeJ mouse model. J. Dermatol. Sci. 104, 21–29 (2021). [DOI] [PubMed] [Google Scholar]
- 10.Valsecchi R., et al. , Peripheral T-cell subsets in patients with alopecia areata in different clinical phases. Dermatologica 171, 170–174 (1985). [DOI] [PubMed] [Google Scholar]
- 11.Gu S. Q., Ros A. M., von Stedingk L. V., Thyresson N., Wasserman J., T cell subpopulations and their functions in vitro: A study in patients with alopecia areata and alopecia universalis. Int. Arch. Allergy Appl. Immunol. 66, 208–217 (1981). [DOI] [PubMed] [Google Scholar]
- 12.McElwee K. J., et al. , Transfer of CD8(+) cells induces localized hair loss whereas CD4(+)/CD25(-) cells promote systemic alopecia areata and CD4(+)/CD25(+) cells blockade disease onset in the C3H/HeJ mouse model. J. Invest. Dermatol. 124, 947–957 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Borcherding N., et al. , A transcriptomic map of murine and human alopecia areata. JCI Insight 5, 137424 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilhar A., et al. , Mediation of alopecia areata by cooperation between CD4+ and CD8+ T lymphocytes: Transfer to human scalp explants on Prkdcscid mice. Arch. Dermatol. 138, 916–922 (2002). [DOI] [PubMed] [Google Scholar]
- 15.Connell S. J., et al. , CD4 T cell-mediated induction of alopecia areata is dose dependent and requires CD8 T cells. J. Immunol. 206, 21.14 (2021). [Google Scholar]
- 16.Speiser J. J., et al. , Regulatory T-cells in alopecia areata. J. Cutan. Pathol. 46, 653–658 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Czarnowicki T., et al. , Alopecia areata is characterized by expansion of circulating Th2/Tc2/Th22, within the skin-homing and systemic T-cell populations. Allergy 73, 713–723 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Hamed F. N., et al. , Alopecia areata patients show deficiency of FOXP3+CD39+ T regulatory cells and clonotypic restriction of Treg TCRβ-chain, which highlights the immunopathological aspect of the disease. PLoS One 14, e0210308 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zöller M., McElwee K. J., Vitacolonna M., Hoffmann R., Apoptosis resistance in peripheral blood lymphocytes of alopecia areata patients. J. Autoimmun. 23, 241–256 (2004). [DOI] [PubMed] [Google Scholar]
- 20.Shin B. S., Furuhashi T., Nakamura M., Torii K., Morita A., Impaired inhibitory function of circulating CD4+CD25+ regulatory T cells in alopecia areata J. Dermatol. Sci. 70, 141–143 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Zhang F., et al. , Defining inflammatory cell states in rheumatoid arthritis joint synovial tissues by integrating single-cell transcriptomics and mass cytometry. Nat. Immunol. 20, 928–942 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zakharov P. N., Hu H., Wan X., Unanue E. R., Single-cell RNA sequencing of murine islets shows high cellular complexity at all stages of autoimmune diabetes. J. Exp. Med. 217, e20192362 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McElwee K. J., Boggess D., King L. E., Sundberg J. P., Experimental induction of alopecia areata-like hair loss in C3H/HeJ mice using full-thickness skin grafts. J. Invest. Dermatol. 111, 797–803 (1998). [DOI] [PubMed] [Google Scholar]
- 24.Wang E. H. C., McElwee K. J., Nonsurgical induction of alopecia areata in C3H/HeJ mice via adoptive transfer of cultured lymphoid cells. Methods Mol. Biol. 2154, 121–131 (2020). [DOI] [PubMed] [Google Scholar]
- 25.McElwee K. J., et al. , Alopecia areata in C3H/HeJ mice involves leukocyte-mediated root sheath disruption in advance of overt hair loss. Vet. Pathol. 40, 643–650 (2003). [DOI] [PubMed] [Google Scholar]
- 26.Dai Z., et al. , CXCR3 blockade inhibits T cell migration into the skin and prevents development of alopecia areata. J. Immunol. 197, 1089–1099 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dai Z., Chen J., Chang Y., Christiano A. M., Selective inhibition of JAK3 signaling is sufficient to reverse alopecia areata. JCI Insight 6, e142205 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dai Z., et al. , Blockade of IL-7 signaling suppresses inflammatory responses and reverses alopecia areata in C3H/HeJ mice. Sci. Adv. 7, eabd1866 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aran D., et al. , Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 20, 163–172 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heng T. S. P., Painter M. W.; Immunological Genome Project Consortium, The Immunological Genome Project: Networks of gene expression in immune cells. Nat. Immunol. 9, 1091–1094 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Dittel B. N., CD4 T cells: Balancing the coming and going of autoimmune-mediated inflammation in the CNS. Brain. Behav. Immun. 22, 421–430 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raphael I., Joern R. R., Forsthuber T. G., Memory CD4+ T cells in immunity and autoimmune diseases Cells 9, 531 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dominguez-Villar M., Hafler D. A., Regulatory T cells in autoimmune disease. Nat. Immunol. 19, 665–673 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Z., et al. , Role of IFN-gamma in induction of Foxp3 and conversion of CD4+ CD25- T cells to CD4+ Tregs. J. Clin. Invest. 116, 2434–2441 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen M., et al. , IFN-β induces the proliferation of CD4+CD25+Foxp3+ regulatory T cells through upregulation of GITRL on dendritic cells in the treatment of multiple sclerosis. J. Neuroimmunol. 242, 39–46 (2012). [DOI] [PubMed] [Google Scholar]
- 36.Metidji A., et al. , IFN-α/β receptor signaling promotes regulatory T cell development and function under stress conditions. J. Immunol. 194, 4265–4276 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Korporal M., et al. , Interferon beta-induced restoration of regulatory T-cell function in multiple sclerosis is prompted by an increase in newly generated naive regulatory T cells. Arch. Neurol. 65, 1434–1439 (2008). [DOI] [PubMed] [Google Scholar]
- 38.Zhang Z., et al. , Activation and functional specialization of regulatory T cells lead to the generation of Foxp3 instability. J. Immunol. 198, 2612–2625 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Grinberg-Bleyer Y., et al. , Pathogenic T cells have a paradoxical protective effect in murine autoimmune diabetes by boosting Tregs. J. Clin. Invest. 120, 4558–4568 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhan Y., et al. , CCR2 enhances CD25 expression by FoxP3+ regulatory T cells and regulates their abundance independently of chemotaxis and CCR2+ myeloid cells. Cell Mol. Immunol. 17, 123–132 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chruscinski A., et al. , Role of regulatory T cells (Treg) and the Treg effector molecule fibrinogen-like protein 2 in alloimmunity and autoimmunity. Rambam Maimonides Med. J. 6, e0024 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cao X., et al. , Granzyme B and perforin are important for regulatory T cell-mediated suppression of tumor clearance. Immunity 27, 635–646 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Loebbermann J., et al. , Regulatory T cells expressing granzyme B play a critical role in controlling lung inflammation during acute viral infection. Mucosal. Immunol. 5, 161–172 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kitagawa Y., et al. , Guidance of regulatory T cell development by Satb1-dependent super-enhancer establishment. Nat. Immunol. 18, 173–183 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Setiady Y. Y., Coccia J. A., Park P. U., In vivo depletion of CD4+FOXP3+ Treg cells by the PC61 anti-CD25 monoclonal antibody is mediated by FcgammaRIII+ phagocytes Eur. J. Immunol. 40, 780–786 (2010). [DOI] [PubMed] [Google Scholar]
- 46.Onda M., Kobayashi K., Pastan I., Depletion of regulatory T cells in tumors with an anti-CD25 immunotoxin induces CD8 T cell-mediated systemic antitumor immunity. Proc. Natl. Acad. Sci. U.S.A. 116, 4575–4582 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Solomon I., et al. , CD25-Treg-depleting antibodies preserving IL-2 signaling on effector T cells enhance effector activation and antitumor immunity. Nat. Cancer 1, 1153–1166 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamaguchi T., et al. , Control of immune responses by antigen-specific regulatory T cells expressing the folate receptor. Immunity 27, 145–159 (2007). [DOI] [PubMed] [Google Scholar]
- 49.de Jong A., et al. , High-throughput T cell receptor sequencing identifies clonally expanded CD8+ T cell populations in alopecia areata. JCI Insight 3, 121949 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao X., Shan Q., Xue H.-H., TCF1 in T cell immunity: A broadened frontier. Nat. Rev. Immunol. 22, 147–157 (2021), 10.1038/s41577-021-00563-6. [DOI] [PubMed] [Google Scholar]
- 51.Schluns K. S., Kieper W. C., Jameson S. C., Lefrançois L., Interleukin-7 mediates the homeostasis of naïve and memory CD8 T cells in vivo. Nat. Immunol. 1, 426–432 (2000). [DOI] [PubMed] [Google Scholar]
- 52.Xue H.-H., et al. , IL-2 negatively regulates IL-7 receptor alpha chain expression in activated T lymphocytes. Proc. Natl. Acad. Sci. U.S.A. 99, 13759–13764 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Muri J., et al. , The thioredoxin-1 system is essential for fueling DNA synthesis during T-cell metabolic reprogramming and proliferation. Nat. Commun. 9, 1851 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Muri J., Thut H., Kopf M., The thioredoxin-1 inhibitor Txnip restrains effector T-cell and germinal center B-cell expansion. Eur. J. Immunol. 51, 115–124 (2021). [DOI] [PubMed] [Google Scholar]
- 55.Levring T. B., et al. , Tumor necrosis factor induces rapid down-regulation of TXNIP in human T cells. Sci. Rep. 9, 16725 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cibrián D., Sánchez-Madrid F., CD69: From activation marker to metabolic gatekeeper. Eur. J. Immunol. 47, 946–953 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Papavassiliou A. G., Musti A. M., The multifaceted output of c-Jun biological activity: Focus at the junction of CD8 T cell activation and exhaustion. Cells 9, 2470 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Waugh K. A., Leach S. M., Slansky J. E., Targeting transcriptional regulators of CD8+ T cell dysfunction to boost anti-tumor immunity Vaccines (Basel) 3, 771–802 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Creelan B. C., Antonia S. J., The NKG2A immune checkpoint—a new direction in cancer immunotherapy. Nat. Rev. Clin. Oncol. 16, 277–278 (2019). [DOI] [PubMed] [Google Scholar]
- 60.Kamiya T., Seow S. V., Wong D., Robinson M., Campana D., Blocking expression of inhibitory receptor NKG2A overcomes tumor resistance to NK cells. J. Clin. Invest. 129, 2094–2106 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Miragaia R. J., et al. , Single-cell transcriptomics of regulatory T cells reveals trajectories of tissue adaptation. Immunity 50, 493–504.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Beura L. K., et al. , T cells in nonlymphoid tissues give rise to lymph-node-resident memory T cells. Immunity 48, 327–338.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wakim L. M., et al. , The molecular signature of tissue resident memory CD8 T cells isolated from the brain. J. Immunol. 189, 3462–3471 (2012), 10.4049/jimmunol.1201305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li Y., et al. , Tumor-infiltrating TNFRSF9+ CD8+ T cells define different subsets of clear cell renal cell carcinoma with prognosis and immunotherapeutic response. Oncoimmunology 9, 1838141 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Takeuchi A., et al. , CRTAM confers late-stage activation of CD8+ T cells to regulate retention within lymph node. J. Immunol. 183, 4220–4228 (2009). [DOI] [PubMed] [Google Scholar]
- 66.Petukhova L., et al. , Genome-wide association study in alopecia areata implicates both innate and adaptive immunity. Nature 466, 113–117 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Milner J. J., et al. , Runx3 programs CD8+ T cell residency in non-lymphoid tissues and tumours. Nature 552, 253–257 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Milner J. J., et al. , Heterogenous populations of tissue-resident CD8+ T cells are generated in response to infection and malignancy. Immunity 52, 808–824.e7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Andreatta M., et al. , Interpretation of T cell states from single-cell transcriptomics data using reference atlases. Nat. Commun. 12, 2965 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jaiswal A., et al. , An activation to memory differentiation trajectory of tumor-infiltrating lymphocytes informs metastatic melanoma outcomes. Cancer Cell 40, 524–544.e5 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sarkar S., et al. , Functional and genomic profiling of effector CD8 T cell subsets with distinct memory fates. J. Exp. Med. 205, 625–640 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Doering T. A., et al. , Network analysis reveals centrally connected genes and pathways involved in CD8+ T cell exhaustion versus memory. Immunity 37, 1130–1144 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Akondy R. S., et al. , Origin and differentiation of human memory CD8 T cells after vaccination. Nature 552, 362–367 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pan Y., et al. , Survival of tissue-resident memory T cells requires exogenous lipid uptake and metabolism. Nature 543, 252–256 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cano-Gamez E., et al. , Single-cell transcriptomics identifies an effectorness gradient shaping the response of CD4+ T cells to cytokines. Nat. Commun. 11, 1801 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang M. J., et al. , Polygenic enrichment distinguishes disease associations of individual cells in single-cell RNA-seq data. Nat. Genet. 54, 1572–1580 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Snyder M. E., et al. , Generation and persistence of human tissue-resident memory T cells in lung transplantation. Sci. Immunol. 4, eaav5581 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Goedhart M., et al. , CXCR4, but not CXCR3, drives CD8+ T-cell entry into and migration through the murine bone marrow. Eur. J. Immunol. 49, 576–589 (2019). [DOI] [PubMed] [Google Scholar]
- 79.McElwee K. J., Spiers E. M., Oliver R. F., In vivo depletion of CD8+ T cells restores hair growth in the DEBR model for alopecia areata Br. J. Dermatol. 135, 211–217 (1996). [PubMed] [Google Scholar]
- 80.McElwee K. J., Spiers E. M., Oliver R. F., Partial restoration of hair growth in the DEBR model for alopecia areata after in vivo depletion of CD4+ T cells Br. J. Dermatol. 140, 432–437 (1999). [DOI] [PubMed] [Google Scholar]
- 81.Carroll J. M., McElwee K. J., King L. E., Byrne M. C., Sundberg J. P., Gene array profiling and immunomodulation studies define a cell-mediated immune response underlying the pathogenesis of alopecia areata in a mouse model and humans. J. Invest. Dermatol. 119, 392–402 (2002). [DOI] [PubMed] [Google Scholar]
- 82.Almutairi N., Nour T. M., Hussain N. H., Janus kinase inhibitors for the treatment of severe alopecia areata: An open-label comparative study. Dermatology 235, 130–136 (2019). [DOI] [PubMed] [Google Scholar]
- 83.Aşkın Ö., Özkoca D., Uzunçakmak T. K., Serdaroğlu S., Evaluation of the alopecia areata patients on tofacitinib treatment during the COVID-19 pandemic. Dermatol. Ther. 34, e14746 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen L., Shen Z., Tissue-resident memory T cells and their biological characteristics in the recurrence of inflammatory skin disorders. Cell Mol. Immunol. 17, 64–75 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ryan G. E., Harris J. E., Richmond J. M., Resident memory T cells in autoimmune skin diseases. Front. Immunol. 12, 652191 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang M. J., et al. , Polygenic enrichment distinguishes disease associations of individual cells in single-cell RNA-seq data. Nat. Genet. 54, 1572–1580 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wing K., et al. , CTLA-4 control over Foxp3+ regulatory T cell function. Science 322, 271–275 (2008). [DOI] [PubMed] [Google Scholar]
- 88.Vocanson M., et al. , Inducible costimulator (ICOS) is a marker for highly suppressive antigen-specific T cells sharing features of TH17/TH1 and regulatory T cells. J. Allergy Clin. Immunol. 126, 280–289, 289.e1-7 (2010). [DOI] [PubMed] [Google Scholar]
- 89.Nasrallah R., et al. , A distal enhancer at risk locus 11q13.5 promotes suppression of colitis by Treg cells. Nature 583, 447–452 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tian Y., et al. , A novel splice variant of folate receptor 4 predominantly expressed in regulatory T cells. BMC Immunol. 13, 30 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Clark R. A., et al. , The vast majority of CLA+ T cells are resident in normal skin. J. Immunol. 176, 4431–4439 (2006). [DOI] [PubMed] [Google Scholar]
- 92.Bos J. D., et al. , The skin immune system (SIS): Distribution and immunophenotype of lymphocyte subpopulations in normal human skin. J. Invest. Dermatol. 88, 569–573 (1987). [DOI] [PubMed] [Google Scholar]
- 93.Angum F., Khan T., Kaler J., Siddiqui L., Hussain A., The prevalence of autoimmune disorders in women: A narrative review. Cureus 12, e8094 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Caldwell C. C., Saikaly S. K., Dellavalle R. P., Solomon J. A., Prevalence of pediatric alopecia areata among 572,617 dermatology patients. J. Am. Acad. Dermatol. 77, 980–981 (2017). [DOI] [PubMed] [Google Scholar]
- 95.Lundin M., et al. , Gender differences in alopecia areata. J. Drugs Dermatol. 13, 409–413 (2014). [PubMed] [Google Scholar]
- 96.Silva K. A., Sundberg J. P., Surgical methods for full-thickness skin grafts to induce alopecia areata in C3H/HeJ mice. Comp. Med. 63, 392–397 (2013). [PMC free article] [PubMed] [Google Scholar]
- 97.Lee E. Y., Dai Z., Wang E. H. C., Christiano A. M., Functional interrogation of lymphocyte subsets in alopecia areata using single-cell RNA sequencing. GEO4. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE2339065. June 1 2023. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
Raw sequencing data have been deposited in GEO with the accession number GSE233906 (97). All other data needed to evaluate the conclusions in the paper are present in the paper and/or the SI Appendix. Additional data related to this paper may be requested from the authors.