Significance
Integrating immunostimulating activity into mRNA (Messenger RNA) packaging materials shows promise in designing nanoformulations for mRNA vaccines, enabling codelivery of antigen-encoding mRNA and adjuvants into the same antigen-presenting cells in a simple formulation. However, this orthodox approach requires elaborative optimization of the packaging materials to obtain mRNA delivery efficiency and adjuvant activity simultaneously. Alternatively, we developed a versatile adjuvant scheme to integrate immunostimulating functionalities into mRNA strands using RNA engineering. This methodology improved the adjuvant activity of three representative mRNA vaccine nanoformulations, i.e., anionic lipoplex, ionizable lipid–based lipid nanoparticles, and polyplex micelles, without compromising their delivery functionalities. Consequently, this approach potentiated the vaccination effect of these three nanoformulations and the anticancer effect of anionic lipoplex used for clinical trials.
Keywords: mRNA vaccine, RNA engineering, vaccine adjuvant, cancer vaccine, mRNA delivery
Abstract
Integrating antigen-encoding mRNA (Messenger RNA) and immunostimulatory adjuvant into a single formulation is a promising approach to potentiating the efficacy of mRNA vaccines. Here, we developed a scheme based on RNA engineering to integrate adjuvancy directly into antigen-encoding mRNA strands without hampering the ability to express antigen proteins. Short double-stranded RNA (dsRNA) was designed to target retinoic acid-inducible gene-I (RIG-I), an innate immune receptor, for effective cancer vaccination and then tethered onto the mRNA strand via hybridization. Tuning the dsRNA structure and microenvironment by changing its length and sequence enabled the determination of the structure of dsRNA-tethered mRNA efficiently stimulating RIG-I. Eventually, the formulation loaded with dsRNA-tethered mRNA of the optimal structure effectively activated mouse and human dendritic cells and drove them to secrete a broad spectrum of proinflammatory cytokines without increasing the secretion of anti-inflammatory cytokines. Notably, the immunostimulating intensity was tunable by modulating the number of dsRNA along the mRNA strand, which prevents excessive immunostimulation. Versatility in the applicable formulation is a practical advantage of the dsRNA-tethered mRNA. Its formulation with three existing systems, i.e., anionic lipoplex, ionizable lipid–based lipid nanoparticles, and polyplex micelles, induced appreciable cellular immunity in the mice model. Of particular interest, dsRNA-tethered mRNA encoding ovalbumin (OVA) formulated in anionic lipoplex used in clinical trials exerted a significant therapeutic effect in the mouse lymphoma (E.G7-OVA) model. In conclusion, the system developed here provides a simple and robust platform to supply the desired intensity of immunostimulation in various formulations of mRNA cancer vaccines.
Messenger RNA (mRNA) vaccines have promising potential in cancer immunotherapy, with numerous clinical trials in progress (1–3). Advantages of mRNA in cancer vaccination include efficient induction of cytotoxic T lymphocyte (CTL) immune responses via the major histocompatibility complex (MHC) class I pathway and flexible designing for targeting neoantigens just by changing mRNA sequences (4, 5). Consequently, various formulations for mRNA vaccine delivery have been developed to maximize antigen expression efficiency. In parallel, vigorous efforts have been devoted to designing safe and effective immunostimulatory adjuvants for robust immunization in cancer vaccines over the past years (2, 6, 7).
Numerous studies have demonstrated the benefit of coloading the antigen and immunostimulatory adjuvant into a single formulation, which ensures codelivery of these two essential components to the same antigen-presenting cells (8–10). The conventional approach is to coload adjuvant molecules with antigen mRNA into the same formulation using proper packaging materials (11, 12). An alternative approach that has emerged recently is to directly integrate adjuvancy into the packaging materials used for formulating mRNA (13–15). Ionizable lipids used for formulating lipid nanoparticles (LNPs) are typical examples of such materials with intrinsic immunostimulatory properties (16–18). Adjuvancy integration into packaging materials instead of using separate adjuvant molecules is beneficial for formulating clinically translatable vaccines because of the simplicity in the production process (19). Nevertheless, balancing the delivery efficiency and adjuvant activity is still challenging in this strategy, requiring elaborative optimization processes to prepare the packaging materials with desired functionalities. This issue motivated us to develop a simple methodology to maximize the adjuvancy of mRNA formulation just by using existing packaging materials already optimized in terms of delivery efficiency.
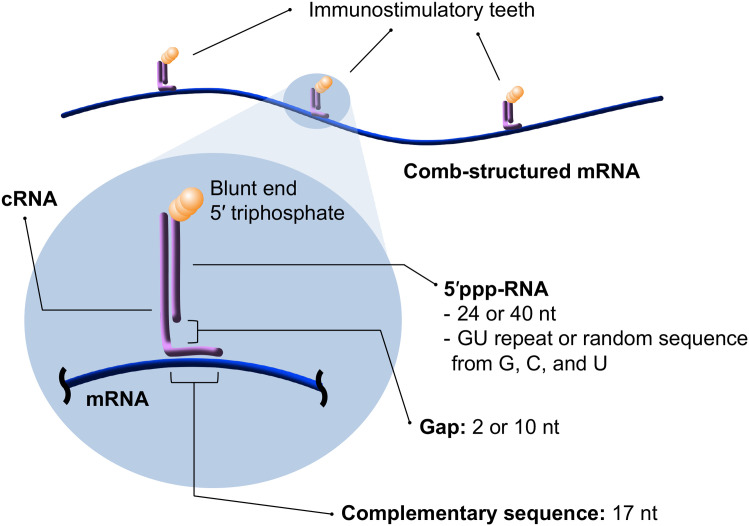
A key concept in this methodology is incorporating adjuvancy into mRNA itself rather than packaging materials, thereby maximizing immunostimulating and delivery efficiencies of mRNA formulations. For this purpose, comb-structured mRNA was designed by hybridizing antigen-encoding mRNA with immunostimulatory double-stranded RNA (dsRNA) teeth (Fig. 1). The dsRNA teeth activate innate immune receptors, functioning as adjuvants. This comb-structured mRNA can be encapsulated into various existing mRNA vaccine carriers without changing their original properties and functionalities. Furthermore, changing the tooth number enables the immunostimulation intensity of comb-structured mRNA to the optimal extent for each formulation prepared from different packaging materials. Notably, this methodology requires adding only a small amount of dsRNA, a nature-derived safe material, for potentiating existing mRNA vaccines. This feature allows for smooth clinical translation from a safety viewpoint.
Fig. 1.
Comb-structured mRNA with immunostimulatory dsRNA teeth. This formulation is prepared from in vitro transcribed RNA with 5′ triphosphate (5′ppp-RNA), mRNA, and a chemically synthesized counter-RNA strand complementary to the 5′ppp-RNA and particular sequences in mRNA (cRNA). dsRNA teeth from 5′ppp-RNA and cRNA possess blunt-ended 5′ triphosphate, a substrate of RIG-I. The length of the cRNA sequence complementary to mRNA is fixed to 17 nt to maintain mRNA translational activity. In addition, design parameters of comb-structured mRNA, including the lengths and sequences of dsRNA teeth, and lengths of gap sequence in cRNA between two regions complementary to 5′ppp-RNA and mRNA, were optimized.
Toward effective cancer immunotherapy, dsRNA teeth were designed to activate retinoic acid-inducible gene-I (RIG-I), an innate immune receptor inducing strong CTL responses (20, 21). RIG-I requires only short dsRNA with 20 base pairs (bp) for activation (22), minimally influencing the total RNA dose. Notably, this dsRNA-toothed approach has the versatility in fine-tuning the dsRNA structure and microenvironment by simply changing its length and sequence and a gap sequence length between dsRNA and mRNA (Fig. 1). A thorough examination of these variants specified the optimized comb-structure formulation for efficient and specific RIG-I stimulation. Ultimately, the comb-structured mRNA potentiated the following three existing mRNA vaccine systems, possessing completely different immunogenic properties, to successfully induce CTL immune responses: anionic lipoplex comparable with that used in clinical trials (23, 24), ionizable lipid–based LNP (iLNP), a prevalent platform of current mRNA vaccines (25), and polyplex micelle, a representative polymer-based delivery systems (26).
Results
In Vitro Screening Identifies Highly Immunostimulating Comb-Structured mRNA.
RIG-I prefers dsRNA with 5′ triphosphate (5′ppp) at the blunt end as a substrate (22, 27). For the preparation of comb-structured mRNA stimulating RIG-I, we designed a short RNA with 5′ triphosphate (5′ppp-RNA) and a counter-RNA strand complementary both to this 5′ppp-RNA and the particular sequences in mRNA (cRNA) (Fig. 1). While in vitro transcription (IVT) without a 5′ cap analog is a cost-effective method to prepare 5′ppp-RNA, there is an issue of producing contaminant RNA strand complementary to the intended RNA strand mainly via RNA-templated RNA transcription (28–30). Contaminant dsRNA formed from this complementary RNA may negatively influence the hybridization process and immunostimulatory property of the RNA. Here, we circumvented this issue by designing the sequence of 5′ppp-RNA to contain no adenine (A) residue and several uracil (U) residues. IVT of 5′ppp-RNA without A residue circumvents the formation of complementary contaminant RNA, which should have several A residues in its sequence. The risk of complementary RNA formation may be further reduced by avoiding self-complementary sequences in 5′ppp-RNA, as RNA hybridization to self-RNA in cis or trans through self-complementary sequences plays a significant role in complementary RNA synthesis (31). Accordingly, as for 5′ppp-RNA, we prepared 24 nucleotides (nt) or 40 nt GU-repeat RNA, lacking a self-complementary sequence. We also prepared RNA with a random sequence from guanine (G), cytosine (C), and U residues containing self-complementary sequences as control (SI Appendix, Table S1). cRNA was chemically synthesized to avoid the formation of IVT by-products (SI Appendix, Table S2). The length of the complementary sequence in cRNA hybridized to mRNA was fixed to 17 nt because both translational activity and immunogenicity of mRNA were still preserved even after hybridization of 17-nt complementary RNA (32). We placed 2-nt or 10-nt gap sequences between complementary sequences to 5′ppp-RNA and mRNA in the cRNA strand, hypothesizing that such a difference in the gap sequence length might influence the microenvironment of dsRNA teeth for recognition by innate immune receptors. Gaussia luciferase (gLuc) mRNA was used as a standard to quantify the protein translational activity of comb-structured mRNA. Successful hybridization was confirmed by ultrafiltration, which separates free-formed dsRNA passing through the filter from those hybridized with mRNA using Cy5-labeled dsRNA. Eventually, in this experiment, Cy5-fluorescence was undetected in the flow-through, showing that almost all Cy5-labeled dsRNA was successfully hybridized to mRNA (SI Appendix, Fig. S1).
There are several design parameters in immunostimulatory dsRNA teeth, including the lengths and sequences of dsRNA and the lengths of gap sequence in cRNA between two regions complementary to 5′ppp-RNA and mRNA (Fig. 1). These parameters were optimized based on the immunostimulating property of comb-structured mRNA. For high throughput screening of comb-structured mRNA design requiring a considerable number of cells, we selected an immortalized murine dendritic cell line, DC2.4 cells. Lipofectamine LTX, a commonly used lipid-based transfection reagent, was used for mRNA introduction, as its efficient mRNA introduction capability in vitro allows for high-throughput evaluation of immunostimulatory and translational properties of mRNA. Notably, the formulation materials used in Lipofectamine LTX exert negligible immunostimulatory properties. Actually, as shown in Figs. 2 and 3, Lipofectamine LTX loading mRNA without immunostimulatory dsRNA tooth showed a negligible immunostimulating effect in vitro. Thus, Lipofectamine LTX formulation is suitable for examining the inherent immunostimulatory and translational properties of loaded mRNA with varying structures of dsRNA teeth without bias. Transcripts of proinflammatory genes, i.e., interferon β (IFN-β) and interleukin 6 (IL-6), were quantified 4 h after transfection to evaluate the immunostimulatory property of mRNA with dsRNA teeth.
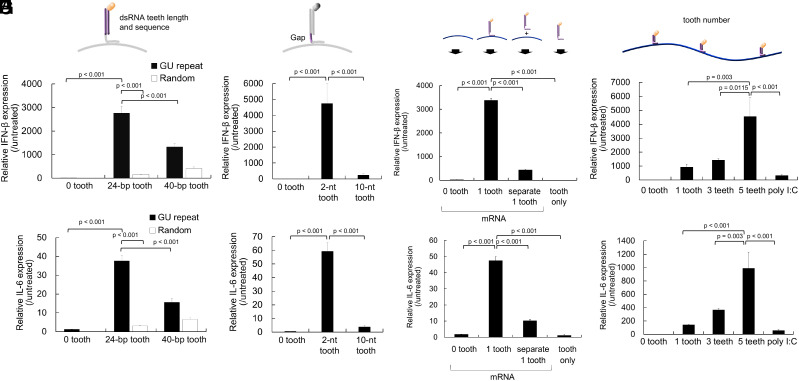
Fig. 2.
Optimization of tooth design for efficient immunostimulation. (A and B) Effect of dsRNA lengths and sequences. (C and D) Effect of gap RNA lengths in cRNA between two regions complementary to 5′ppp-RNA and mRNA. (E and F) Effect of the dsRNA tooth and mRNA codelivery method. Introduction of a 24-nt GU-repeat tooth alone and separate introduction of mRNA and a 24-nt GU-repeat tooth were performed. (G and H) Effect of tooth numbers. The teeth used possessed dsRNA with 24-bp GU-repeat and a 2-nt gap (dsRNA24-GUrepeat/2-gap). Transcript levels of interferon (IFN)-β (A, C, E, and G) and interleukin (IL)-6 (B, D, F, and H) were measured using quantitative PCR 4 h after mRNA introduction to DC2.4 cells. n = 5 in (A–D) and n = 6 in (E–H).
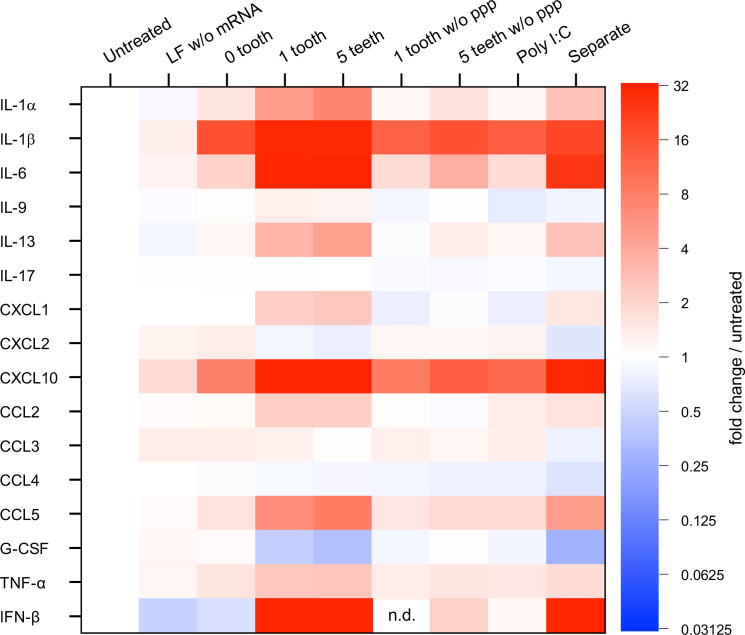
Fig. 3.
Immunological profiling of BMDCs after mRNA treatment. Protein expression levels of 25 types of cytokines, interferons, and chemokines were quantified 24 h after mRNA introduction to mouse BMDCs. Colors in the heatmap represent protein levels relative to untreated control. Among 25 types of molecules, nine types (IFN-γ, IL-2, 4, 5, 7, 10, 12p40, 12p70, and 15) were below detection limits, and data from the other 16 are shown. n = 6. LF, Lipofectamine; PPP, 5′ triphosphate; G-CSF, granulocyte colony-stimulating factor. In Separate, mRNA and tooth with tooth amount equal to that in 1 tooth were separately formulated with LF for addition to BMDCs.
First, we optimized sequences and lengths in the dsRNA tooth using comb-structured mRNA with one tooth. GU-repeat tooth induced enhanced expression of proinflammatory transcripts compared to random sequence (Fig. 2 A and B), possibly because GU-repeat sequence might avoid the formation of complementary RNA strand by-products as aforementioned. Elongation of GU-repeat tooth from 24 bp to 40 bp resulted in a slight reduction in proinflammatory transcripts. Although the mechanism underlying this result is unclear, longer complementary RNA strands might have more freedom to hybridize with each other at unintended positions, hampering the formation of intended dsRNA possessing 5′-triphosphate at the blunt end, a preferred ligand of RIG-I. Then, gap RNA length in cRNA between two regions complementary to 5′ppp-RNA and mRNA was optimized for the dsRNA tooth with 24-bp GU-repeat. As seen in Fig. 2 C and D, the 2-nt gap revealed higher proinflammatory transcript levels than the 10-nt gap. Presumably, gap length might alter the local environment of dsRNA and, as a result, affect the process of dsRNA recognition by innate immune receptors. Interestingly, the introduction of only a free-formed 24-bp GU-repeat tooth and the separate introduction of mRNA and 24-bp GU-repeat tooth failed to induce strong immunostimulation (Fig. 2 E and F). From a mechanistic viewpoint, cellular uptake efficiency of the dsRNA tooth could cause a difference in the immunostimulation intensity between formulations in this experiment. To check this possibility, we evaluated the cellular uptake efficiency of fluorescently labeled dsRNA tooth. Notably, compared to the nonannealed formulation, the annealed formulation showed only a 1.5-fold increase in uptake in flow cytometry (SI Appendix, Fig. S2) while inducing fourfold-to-eightfold higher levels of the proinflammatory transcripts (IFN-β and IL-6). This large discrepancy between the uptake efficiency and immunostimulation capability strongly suggests that the annealing approach is key to increasing immunostimulatory activity. mRNA introduction using other transfection reagents, including anionic lipoplex used for in vivo vaccination study (Fig. 4) and linear poly(ethyleneimine) also showed the benefit of attaching a tooth to mRNA compared to codelivering mRNA and a tooth without annealing each other (SI Appendix, Fig. S3). These results demonstrated the benefit of hybridizing dsRNA to mRNA strands for stimulating innate immunity. Accordingly, we selected to use dsRNA with 24-bp GU-repeat and a 2-nt gap (dsRNA24-GUrepeat/2-gap) in the following experiments. As seen in Fig. 2 G and H, expression levels of proinflammatory transcripts increased with the number of dsRNA24-GUrepeat/2-gap teeth from 1 to 5. Notably, comb-structured mRNA induced more intense immunostimulation than poly I:C, a widespread RNA adjuvant. Dose–response curves also showed enhanced adjuvanting efficiency of comb-structured mRNA over poly I:C (SI Appendix, Fig. S4).
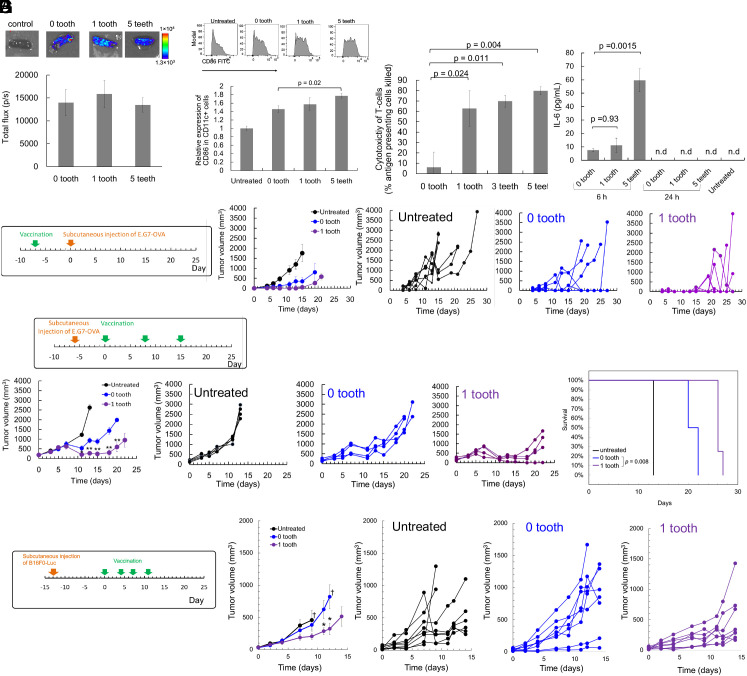
Fig. 4.
Cancer vaccines using lipoplex in mice. (A) Expression of fLuc in the spleen 24 h after i.v. injection of lipoplex. n = 4. (B) Activation of DC in the spleen 24 h after i.v. injection of lipoplex. n = 4. The expression level of CD86 in CD11c positive splenocytes. (C) CTL immunity against OVA 7 d after mRNA vaccination, evaluated by the in vivo CTL assay. n = 4. (D) Serum levels of IL-6 were measured using ELISA 6 h and 24 h after i.v. injection of lipoplexes. n = 4. n.d.: not detected. (E) Prophylactic model of subcutaneously inoculated lymphoma expressing OVA, treated using OVA mRNA. n = 6 for untreated, n = 4 for the other two groups. (F) Therapeutic model of subcutaneously inoculated lymphoma expressing OVA, treated using OVA mRNA. n = 4. (G) Therapeutic model of subcutaneously inoculated melanoma, treated using Trp2 mRNA. †Average tumor volume was not shown at later time points because of the death of one or more mice. n = 8. *P < 0.05 versus 0 tooth. (E−G) The first chart from the left shows the average tumor volumes in each group. The second to fourth charts from the left shows the tumor volume of individual mice (Right). (F) The right figure shows the mouse survival. (B−F) OVA mRNA dose for each injection was 5 μg, and tooth dose was 0.23 μg for 1 tooth, 0.70 μg for 3 teeth, and 1.2 μg for 5 teeth, respectively. (G) The dose was 10 μg for Trp2 mRNA and 0.34 μg for a tooth.
Although 0 tooth, introduction of unmodified mRNA, apparently induced low expression of proinflammatory transcripts, the level of INF-β was significantly higher than the untreated cells (SI Appendix, Fig. S5). This result is consistent with a previous report showing type I interferon induction after unmodified mRNA treatment (33).
Comb-Structured mRNA Activates RIG-I for Immunostimulation.
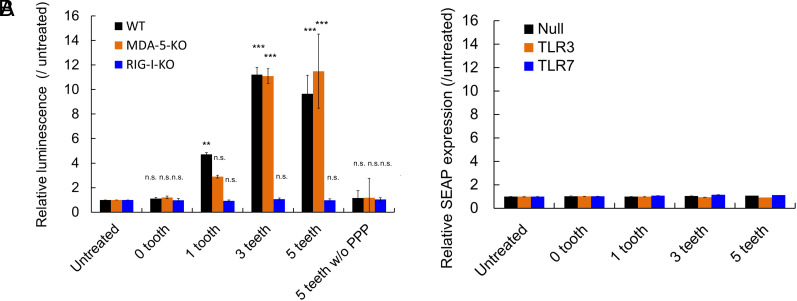
Several innate immune receptors potentially recognize comb-structured mRNA. They include dsRNA receptors [RIG-I (22), melanoma differentiation-associated gene (MDA)-5 (34), Toll-like receptor (TLR) 3 (35)], and a single-strand RNA receptor [TLR7 (36)]. RIG-I involvement in inflammatory response due to mRNA transfection was evaluated using the RIG-I knockout RAW-Lucia cell. RAW-Lucia cell is a macrophage-derived cell line genetically modified to express Lucia luciferase (lLuc) responding to proinflammatory stimuli under the promoter responsive to interferon regulatory factors. As seen in Fig. 5A, without RIG-I knockout (WT), comb-structured mRNA with 1, 3, and 5 teeth showed enhanced lLuc expression in RAW-Lucia cells compared to untreated control and mRNA without a tooth. In contrast, comb-structured mRNA with 1, 3, and 5 teeth did not increase lLuc expression levels in RIG-I knockout cells. Further notably, without 5′ppp, a critical motif in dsRNA for RIG-I recognition (22), lLuc expression levels were comparable between comb-structured mRNA and untreated control in RAW-Lucia cells without RIG-I knockout. These results highlight a pivotal role of RIG-I in the immunostimulation induced by comb-structured mRNA. On the contrary, knockout of MDA-5 in RAW Lucia cells showed almost no influence on lLuc expression after treatment with comb-structured mRNA. This result suggests that comb-structured mRNA possessing dsRNA24-GUrepeat/2-gap teeth does not stimulate MDA-5, which requires dsRNA longer than 2 kb for its stimulation (34).
Fig. 5.
Contribution of innate immune receptors for immunostimulation by mRNA. (A) Comb-structured mRNA was added to RAW cells without knockout of immune receptors (WT) and with MDA-5 or RIG-I knockout (MDA-5-KO, RIG-I-KO). The cells were genetically modified to express Lucia luciferase (lLuc) after proinflammatory stimulation for quantifying immunostimulation intensity based on lLuc expression. n = 6. (B) Comb-structured mRNA was added to HEK 293 cells, genetically modified to express TLR3 or TLR7, or without transformation to express TLRs (Null). The cells were transformed to express SEAP reporter after nuclear factor-κB (NF-κB) stimulation. n = 6. PPP, 5′ triphosphate. **P < 0.01; ***P < 0.001, n.s., nonsignificant versus untreated, respectively.
The involvement of TLRs was studied using a human embryonic kidney-derived cell line, HEK293 cells, genetically modified to express human TLR3 or TLR7, denoted as HEK-TLR3 or HEK-TLR7, respectively. Note that original HEK293 cells, denoted as HEK-null, exhibited a negligible level of TLR expression. These three cell lines were further transformed to express secreted embryonic alkaline phosphatase (SEAP) reporter after nuclear factor-κB (NF-κB) stimulation. In HEK-TLR3 and HEK-TLR7, mRNA with 1, 3, and 5 teeth and without teeth provided SEAP expression levels comparable with those in untreated control, indicating a minimal role of TLR3 and 7 in immunostimulation by comb-structured mRNA (Fig. 5B).
The immunological signaling assay described above (Fig. 5) did not reveal the innate immunity pathway induced by mRNA without a tooth, which provided significant immunostimulation (Fig. 3 and SI Appendix, Fig. S5). Therefore, we improved assay sensitivity by increasing incubation time in the experiments using Raw-Lucia cells and increasing mRNA doses in the experiments using HEK293 cells expressing SEAP. After treatment with mRNA without a tooth, RAW-Lucia WT cells showed enhanced Lucia expression than RAW-Lucia cells with RIG-I knockout, indicating the involvement of RIG-I (SI Appendix, Fig. S6A). Meanwhile, mRNA without tooth induced comparable SEAP expression in HEK293 cells with and without TLR3 and TLR7 overexpression (SI Appendix, Fig. S6B), indicating a nonprimary role of TLR3 and TLR7 in innate immune activation after unmodified mRNA introduction.
Comb-Structured mRNA Efficiently Activates DCs with Minimal Influence on Translational Activity.
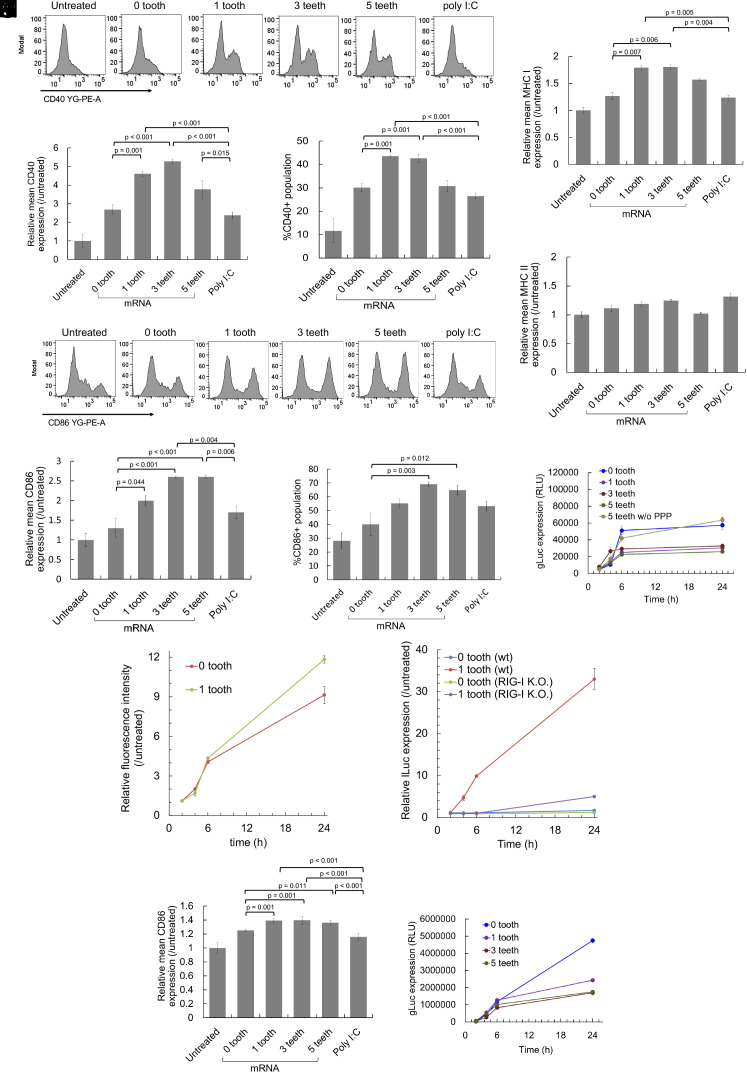
Detailed functional analyses of comb-structured mRNA were performed using mouse primary bone marrow-derived dendritic cells (BMDCs), which retain intrinsic in vivo characteristics of dendritic cells. First, the capability of comb-structured mRNA for activating BMDCs was evaluated by measuring expression levels of surface marker proteins, including CD86, CD40, and MHC class I and II (MHC I and MHC II). After introducing into BMDCs, mRNA with a tooth of dsRNA24-GUrepeat/2-gap significantly increased CD86, CD40, and MHC I expression levels compared to mRNA without tooth (Fig. 6 A–G), demonstrating efficient activation of BMDCs by comb-structured mRNA. In most cases, these markers' expression level tends to become maximal at the three teeth. Meanwhile, the teeth introduction into mRNA has minimal effect on MHC II expression (Fig. 6H). Notably, compared to polyinosinic-polycytidylic acid (poly I:C), mRNA possessing 1 to 5 teeth showed enhanced CD86, CD40, and MHC I expression in BMDCs, demonstrating intense activity of comb structure as an immunostimulatory adjuvant.
Fig. 6.
Activation of dendritic cells by comb-structured mRNA. mRNA was added to mouse BMDCs (A–K) and human BMDCs (L and M). (A–H and J) Expression of surface markers, CD40 (A–C), CD86 (D–F and L), MHC I (G), and MHC II (H) was quantified using immunocytochemistry 24 h after mRNA addition. n = 4 (I and M) Expression of gLuc was measured using the cultured medium. n = 6. PPP, 5′ triphosphate. (J and K) The kinetics of innate immune activation and antigen presentation. (J) For evaluating the kinetics of antigen presentation, OVA epitope (SIINFEKL)/MHC class I H-2Kb complexes on the surface of DC2.4 cells were quantified using flow cytometry after the introduction of OVA mRNA with 0 and 1 tooth. n = 4. (K) For evaluating the kinetics of innate immune activation, OVA mRNA with 0 and 1 tooth was added to reporter RAW-Lucia cell lines with and without RIG-I knockout. Lucia luciferase (lLuc) expression was measured as a marker of innate immune activation. n = 6.
Then, the efficiency of antigen expression was studied using gLuc as a reporter. gLuc expression efficiency of comb-structured mRNA with 1, 3, and 5 teeth was preserved to approximately 50% of the efficiency of mRNA without a tooth in BMDCs (Fig. 6I). According to previous studies, activation of innate immune responses harms translational activity of mRNA (37, 38), which may reduce gLuc expression efficiency after teeth attachment in the present experiment. To study this issue, we introduced gLuc mRNA with 5 teeth without 5′ppp, which lacks the immunostimulating ability (Fig. 5). Notably, mRNA having 5 teeth without 5′ triphosphate exerted similar gLuc expression to mRNA without a tooth (Fig. 6I). These results indicate that the reduction of translational activity after the teeth attachment is attributed to the induction of innate immune responses rather than the presence of teeth on the mRNA strand. This finding is consistent with our previous result that mRNA preserves translational activity after hybridization with 17-nt complementary RNA (32).
The kinetics of antigen presentation and immunostimulation is critical in mRNA vaccines, as premature immune activation before antigen presentation may negatively impact the vaccination processes (39). In a kinetic analysis, we quantified the levels of immunostained OVA (ovalbumin) epitope (SIINFEKL)/MHC class I H-2Kb complexes on the surface of cultured dendritic cells (DC2.4 cells) to evaluate the antigen presentation. In parallel, the innate immune activation kinetics was evaluated by using reporter RAW-Lucia cell lines with and without RIG-I knockout. The cell lines express lLuc reporter under promoters responsive to interferon regulatory factors; thus, the kinetics of the lLuc level correlates with that of interferon responses. Notably, mRNA with a tooth provided similar kinetic profiles of epitope presentation on MHC class I and RIG-I-mediated immune responses, with both processes observed efficiently from 2 h to 6 h post-mRNA treatment (Fig. 6 J and K). These concurrent immune activation and antigen presentation processes may explain effective vaccination by comb-structured mRNA. Further, notably, mRNA with 1 tooth exhibited increased epitope presentation than mRNA without a tooth, although tooth attachment reduced protein expression efficiency in a reporter assay using gLuc mRNA (Fig. 6I). Immunostimulation by mRNA with 1 tooth might activate the epitope presentation process, overcoming the negative impact of immunostimulation on protein expression efficiency.
The feasibility of comb-structured mRNA was also studied for future clinical translation using human DCs. The introduction of 1, 3, and 5 teeth significantly improved the CD86 expression levels compared to mRNA without tooth and poly I:C (Fig. 6L). Furthermore, in a reporter assay using gLuc mRNA, comb-structured mRNA preserved approximately half of the translational activity of mRNA without tooth (Fig. 6M). These results indicate the availability of comb-structured mRNA in human antigen-presenting cells.
Comb-Structured mRNA Broadly Stimulates the Expression of Proinflammatory Molecules.
Further immunological characterization of comb-structured mRNA was performed using the multiplex immunoassay and enzyme-linked immunosorbent assay (ELISA) to measure protein expression levels of 25 types of cytokines, interferons, and chemokines. Twenty-four hours after mRNA introduction to mouse BMDCs, the concentration of these immune molecules in the culture medium was measured (Fig. 3 and SI Appendix, Fig. S7). The fold-change compared to untreated control is shown as a color gradient in Fig. 3 for 16 of the 25 immune molecules, while expression levels of the remaining nine molecules were below the detection limit.
Two proinflammatory cytokines [interleukin (IL)-1β, IL-6] and one chemokine [C-X-C motif chemokine ligand (CXCL)10] expressed more than twofold for mRNA without tooth compared to the untreated control and the control treated with Lipofectamine without mRNA (Fig. 3). This result revealed the immunostimulatory property of mRNA. Intriguingly, compared to mRNA without a tooth, comb-structured mRNA further increased the expression of IL-1β by approximately twofold, IL-6 by 17- to 33-fold, and CXCL10 by fivefold-to-sevenfold, in a manner dependent on the tooth number. Further notably, secretion of several proinflammatory molecules was enhanced only by comb-structured mRNA, not by mRNA without a tooth. Comb-structured mRNA enhanced the expression levels an approximately 100-fold for a type I interferon (IFN-β) and 2-10-fold for two proinflammatory cytokines [IL-1α, tumor necrosis factor (TNF)-α] and three chemokines [CXCL1, CC chemokine ligand (CCL)2, CCL5] compared to untreated control. In contrast, mRNA without teeth did not increase the expression of these immune molecules by more than twofold. These results show that the teeth tethering to the mRNA strand reinforces the immunostimulatory property of mRNA and broadens the types of secreted immune molecules. Such reinforcement was not observed for the mRNA tethering the teeth lacking a 5′ppp group, indicating the involvement of RIG-I in this immunostimulation process.
The separate introduction of mRNA and dsRNA also appears to induce relatively high levels of proinflammatory proteins in the heatmap. However, the levels were lower than observed after the introduction of comb-structured mRNA, even when the amount of dsRNA was equal between these two groups (SI Appendix, Fig. S7). This result is consistent with the quantification of IFN-β and IL-6 at the mRNA level (Fig. 2 E and F). Despite the properties of comb-structured mRNA to broadly stimulate the secretion of proinflammatory cytokines, i.e., type I interferon and chemokines, it provided a modest influence on the expression of anti-inflammatory cytokines (IL-10) and T helper 2 (Th2)-related cytokines (IL-4, IL-5, IL-9, IL-13) (40). The expression was undetected for IL-4, IL-5, and IL-10, low for IL-13, and unchanged for IL-9 after the comb-structure installation. These immunological profilings also indicate that comb-structured mRNA exerts much stronger immunostimulation than poly I:C. This result is consistent with its enhanced effect of increasing the expression of activation markers on the DC surface (Fig. 6).
Comb-Structured mRNA Potentiates Lipoplex-Based mRNA Vaccines against Cancer.
To evaluate the potential of comb-structured mRNA in cancer vaccination, we utilized anionic lipoplex, comparable with that used in clinical trials (24). Its intravenous (i.v.) injection resulted in strong protein expression from mRNA in immune tissues such as the spleen, thereby providing efficient vaccination effects (23). Herein, anionic lipoplexes were prepared by mixing cationic liposome and mRNA possessing 0, 1, 3, and 5 teeth, with an excess mRNA in charge ratio to exert negatively charged properties. Regardless of the number of teeth, the lipoplexes showed an average size of around 200 nm with a polydispersity index below 0.12 in dynamic light scattering measurement (SI Appendix, Fig. S8A). ζ-potential of these lipoplexes was between −40 mV and −70 mV (SI Appendix, Fig. S8B).
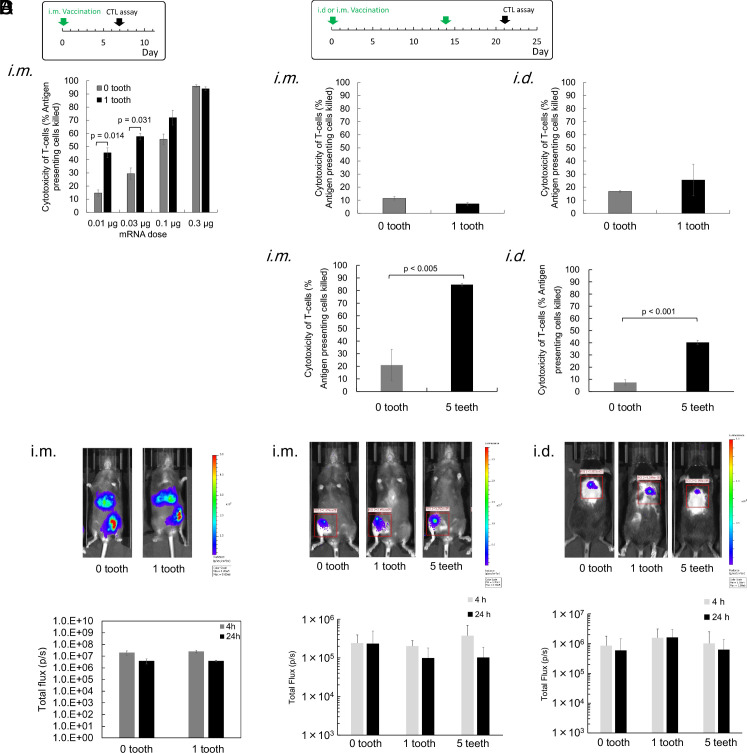
In this study, a lower mRNA dose (5 μg/mouse) was used compared to that used in a previous preclinical study of the comparable lipoplex (20 or 40 μg/mouse) (23), expecting that adjuvant functionalities of the comb-structured mRNA may reduce effective mRNA dose. In a reporter assay using comb-structured firefly luciferase (fLuc) mRNA, mRNA with 1 and 5 teeth exhibited fLuc expression efficiency in the spleen at a comparable level with mRNA without tooth at 24 h after injection (Fig. 4A). Flow cytometry analyses revealed that the expression level of CD86 in splenic CD11c-positive DCs tended to increase with an increase in tooth number, with statistical significance observed between mRNA without tooth and comb-structured mRNA with 5 teeth (Fig. 4B). This result demonstrates the appreciable potential of comb-structured mRNA for DC activation in vivo.
Vaccination effects were then assessed using mRNA expressing OVA as a model antigen by the in vivo CTL assay, which quantifies the killing of transplanted syngeneic splenic cells presenting the OVA antigen in vaccinated mice. Seven days after vaccination, comb-structured mRNA with 1, 3, and 5 teeth showed enhanced antigen-specific CTL response compared to mRNA without a tooth (Fig. 4C). Notably, OVA mRNA with 1 tooth, but not mRNA without a tooth, showed larger percentages of CD8- and CD4-positive splenocytes reactive with OVA peptide tetramers than an untreated control (SI Appendix, Fig. S9). The improved vaccination effect of comb-structured mRNA may be attributed to enhanced activation of DCs in immune tissues (Fig. 4B). Enhanced secretion of proinflammatory molecules (Fig. 3) may also contribute positively to the vaccination effect in comb-structured mRNA. In safety evaluation, we first measured serum levels of proinflammatory cytokines. Introducing one tooth to mRNA does not increase the serum level of IL-6 compared to mRNA without a tooth 6 h after vaccination, while mRNA with 5 teeth showed higher IL-6 levels than those in these two groups (Fig. 4D). Notably, the increase in IL-6 was modest in every group, and the levels returned to normal 24 h after vaccination. Serum levels of IFN-β 6 and 24 h after injection of all lipoplexes were below the detectable limit. In blood chemistry, all tested formulations did not significantly increase toxicity markers of the liver (AST, ALT), the kidney (BUN, Cre), and the whole body (LDH) compared to untreated control, except for a slight increase in ALT after injection of OVA mRNA with 5 teeth (SI Appendix, Fig. S10). Moreover, organ histology of the spleen, liver, lung, kidney, and heart was normal for all formulations (SI Appendix, Fig. S11). Based on these efficiency and safety analyses, we preferentially used mRNA with one tooth throughout the following experiments to minimize safety concerns. Meanwhile, when strong adjuvant effects were necessary, mRNA with 5 teeth was also used.
Then, the therapeutic potential of comb-structured mRNA was tested first using a mouse model bearing subcutaneous lymphoma expressing the OVA model antigen (E.G7-OVA). Mice were vaccinated before tumor inoculation in a prophylactic model and after the tumor grew to a specific size in a treatment model. In a prophylactic model, comb-structured mRNA and mRNA without a tooth suppressed tumor growth more efficiently than untreated control (Fig. 4E). Although comb-structured mRNA tends to be more effective than mRNA without a tooth, the difference between these groups is not statistically significant due to the effective prevention of tumor growth observed in mRNA without a tooth, which led us to use a more challenging treatment model. After three-time vaccination in this treatment model, comb-structured mRNA with a tooth significantly reduced tumor volume and prolonged survival compared to mRNA without a comb structure (Fig. 4F). To exclude the possible contribution of innate immune responses against comb-structured mRNA on the antitumor effect, a control experiment was performed using mRNA encoding luciferase, a nonantigen protein. As a result, luciferase mRNA with a tooth failed to reduce tumor volume (SI Appendix, Fig. S12), showing that antigen-specific immunity rather than innate immunity contributes to cancer treatment. From a safety viewpoint, 3-time sequential injection of anionic lipoplex loading mRNA with and without a tooth did not reduce mouse body weight compared with untreated control (SI Appendix, Fig. S13).
Furthermore, we evaluated the therapeutic potential of the comb-structured mRNA by targeting a tumor-associated antigen. In this clinically relevant model, mice bearing subcutaneous B16F0 melanoma received four-time injections with tyrosinase-related protein 2 (Trp2) mRNA using an anionic lipoplex after the tumor grew into a certain size. As a result, Trp2 mRNA with a tooth showed enhanced antitumor activity than Trp2 mRNA without a tooth (Fig. 4G). This result demonstrates the high therapeutic potential of the comb-structured mRNA vaccines, as targeting a tumor-associated antigen in this model requires overcoming immune tolerance.
Comb-Structured mRNA Is Versatile to Potentiate Other Two Types of mRNA Vaccines.
Recently, many studies employed iLNP via local injection for cancer vaccination (11, 13, 15), motivating us to evaluate the functionalities of comb-structured mRNA in this system. We encapsulated OVA mRNA with 0 and 1 tooth into iLNP used in an approved COVID-19 vaccine (BNT162b2), which efficiently induced cellular immunity in addition to humoral immunity (41). Vaccination was performed via an intramuscular (i.m.) route. Notably, the size was comparable between mRNA with 0 and 1 tooth (SI Appendix, Table S3). Then, anti-OVA cellular immunity was evaluated by the in vivo CTL assay one week after the vaccination. Among 4 tested doses of mRNA, mRNA with one tooth induced significantly enhanced antigen-specific CTL response compared to mRNA without a tooth, especially at low doses, showing substantial vaccination effects even at the mRNA dose of 0.01 μg (Fig. 7A).
Fig. 7.
Vaccines using iLNPs and PMs. (A and B) The in vivo CTL assay was performed at indicated schedules. iLNPs loading OVA mRNA with 0 or 1 tooth were injected into mice via i.m. route. n = 4. (B) PMs loading OVA mRNA with 0, 1, or 5 teeth were injected into mice via i.m. and i.d. routes. Tooth dose was 0.0047 μg for 0.01 μg mRNA, 0.0014 μg for 0.03 μg mRNA, 0.047 μg for 0.1 μg mRNA, and 0.014 μg for 0.3 μg mRNA, respectively. n = 4. (C and D) Luciferase expression was evaluated after i.m. injection of iLNP (C) and after i.m. and i.d. injection of PMs (D). Only expression at the muscle was quantified in (C). Representative images at 4 h are shown. n = 5 in (C) and n = 3 in (D). In i.m. injection, OVA mRNA dose for each injection was 20 μg, and tooth dose was 0.93 μg for 1 tooth and 4.7 μg for 5 teeth, respectively. In i.d. injection, OVA mRNA dose for each injection was 12 μg, and tooth dose was 0.56 μg for 1 tooth and 2.8 μg for 5 teeth, respectively.
Polymer-based systems have been rarely reported for mRNA vaccines despite their vast potential in mRNA delivery, which may be partially attributed to the low immunostimulatory properties of polymers (42). Thus, increasing the mRNA adjuvant effect using comb-structured mRNA may open the opportunity to use polymer-based systems in vaccination. In this study, we used a polyplex micelle (PM), which provides efficient protein expression after in vivo local delivery with successful treatment outcomes in several disease models (26). Block copolymer composed of poly(ethylene glycol) (PEG) and polycation (poly(N-[N′-(2-aminoethyl)-2-aminoethyl]aspartamide): PAsp(DET)) segments was used to prepare PM by mixing mRNA and this block copolymer in an aqueous solution. Prepared PM possesses a core-shell structure with mRNA in the core surrounded by a PEG shell. In a vaccination experiment using OVA mRNA, PMs were injected twice at a 2-wk interval via i.m. and intradermal (i.d.) injection. Notably, the size was comparable between mRNA with 0 and 1 tooth (SI Appendix, Table S4). The in vivo CTL assay performed one week after the second injection showed that mRNA with and without a tooth induced only minimal vaccination effect. There was no significant difference in the effect between these two groups (Fig. 7B). Considering that this result is attributed to almost nonimmunogenic properties of PMs (26, 43), we increased the number of teeth from 1 to 5. The increase did not largely influence PM size (SI Appendix, Table S4). In both administration routes, mRNA with 5 teeth successfully enhanced the vaccination effect for inducing CTL response compared to mRNA without a tooth (Fig. 7B). Meanwhile, local administration of PM loading mRNA with 5 teeth did not significantly increase IL-6 and IFN-β (SI Appendix, Fig. S14). Worth to notice is that tethering teeth showed only modest effects on mRNA introduction efficiency in iLNPs (Fig. 7C) and PMs (Fig. 7D), which may contribute to enhanced vaccination effects after the teeth tethering in these systems (Fig. 7 A and B).
Discussion
Loading the antigen and adjuvant together into a single vaccine formulation ensures the codelivery of these two components into the same antigen-presenting cells, showing promise in vaccination (8–10). This strategy has demonstrated its feasibility in RIG-I adjuvants, i.e., dsRNA with 5′ triphosphate, wherein nanoparticles coloading RIG-I adjuvants and protein/peptide antigens provided efficient vaccination against cancer and influenza (20, 21, 44). In application of this strategy to mRNA vaccines, simple mixing of RIG-I adjuvants and antigen mRNA does not ensure coloading of these two components into delivery carriers at an intended ratio. Alternatively, a comb-structured mRNA attached with dsRNA adjuvants, developed in this study, allows coloading of mRNA and adjuvants into various delivery formulations with minimal influence on their physicochemical properties. In addition, attaching dsRNA into mRNA increased immunostimulating efficiency compared to delivering dsRNA tooth and mRNA without annealing with each other (Fig. 2 E and F and SI Appendix, Fig. S3), as discussed below. Notably, the comb-structured mRNA successfully potentiated three types of existing mRNA vaccine formulations possessing completely different immunogenic properties. iLNPs intrinsically have intense adjuvant functionalities (16–18), synergizing with comb-structured mRNA adjuvant to further improve vaccination effects (Fig. 7A). As a result, the effective dose of iLNP vaccines was able to be reduced. Meanwhile, comb-structured mRNA improved the vaccine effects of less immunostimulating anionic lipoplex (18) (Fig. 4) and even almost noninflammatory PM (26, 43) (Fig. 7B). Thus, our strategy of integrating adjuvancy into comb-structured mRNA provides more freedom in choosing formulation materials for mRNA vaccines focusing on essential properties other than immunostimulation, including the reduction of side reactions and the improvement in targetability of antigen-presenting cells. Among the three delivery systems showing the benefit of comb-structured mRNA in this study, an anionic lipoplex, a formulation proceeding into clinical trials, was selected in cancer vaccination studies. However, future studies are required to optimize delivery carriers with possible combinations with immune checkpoint inhibitors using several treatment schedules and doses with safety analyses for each formulation.
Interestingly, the optimal tooth number is dependent on the delivery formulations and routes. For example, a single tooth tethered to a small dose of mRNA (0.01 μg/mouse at minimum) was effective enough in iLNP formulation, presumably because of the intrinsic immunostimulatory properties of the iLNP. In contrast, 5 teeth-tethered mRNA at an increased dose (12, 20 μg/mouse) for the PM formulation was optimum for effective vaccination without a systemic increase in inflammatory molecules. Furthermore, one tooth at an mRNA dose of 5 μg/mouse was optimal for anionic lipoplex to exert an efficient antitumor effect without significantly increasing serum cytokine levels. These results highlight that the immunostimulation intensity of comb-structured mRNA systems is systematically adjusted to various mRNA vaccine formulations.
Comb-structured mRNA does not require additional exogenous materials for immunostimulation other than RNA composed of naturally existing nucleotides. This characteristic is a potential advantage of safety in clinical application. Remarkably, tethering of only one short dsRNA tooth (24 bp) to a long mRNA strand drastically induced the immunological response with a negligible increase in total RNA doses, as demonstrated in the immunostimulation effect of 783-nt gLuc mRNA (Figs. 2, 3, 5, and 6) and the vaccination effect of 1437-nt OVA mRNA (Figs. 4 and 7). Such short teeth introduction minimally affected physicochemical properties of mRNA formulations, as shown in SI Appendix, Fig. S8 for lipoplex, SI Appendix, Table S3 for iLNP, and SI Appendix, Table S4 for PM, respectively, indicating the broad applicability of this approach in various types of mRNA formulations.
Simple rationality in adjuvant design is also merit for this dsRNA tooth-based approach. Fine-tuning the structure of just a short dsRNA tooth (24 bp) allows sufficient adjuvant functionalities. Interestingly, subtle changes in the dsRNA structure largely influence the immunostimulation intensity (Fig. 2). Although detailed insights into the mechanisms underlying this phenomenon are yet to be clarified, it would be worth discussing the possible scheme of RIG-I recognition. According to previous reports, the 5′ triphosphate structure at the blunt end is a binding motif of RIG-I recognition, and a slight disturbance of the blunt structure, i.e., the presence of 5′ or 3′ overhang with a few base-pairs, drastically decreases immune activation (22, 27). In addition, RIG-I requires at least 20-nt dsRNA structure for its activation. Thus, unintended hybridization products, including loop structure or dsRNA with 5′ or 3′ overhang, might decrease the RIG-I recognition, depending on the tooth dsRNA sequence and length. Another possible determinant in RIG-I recognition is the dsRNA microenvironment. Indeed, the gap sequence length between dsRNA tooth and mRNA and the binding of a tooth to mRNA largely influenced immunostimulation with a tooth with a 2-nt gap sequence inducing the expression of inflammatory molecules more efficiently than that with 10-nt gap sequence and without attaching to mRNA (Figs. 2 and 3 and SI Appendix, Fig. S7). It would be meaningful to propose some potential mechanisms explaining the influence of the local environment surrounding dsRNA. Compared to a tooth with a 10-nt gap and a nonannealed tooth, a tooth with a 2-nt gap sequence located closer to mRNA induced more robust innate immune responses. Therefore, the proximity between dsRNA tooth and mRNA might play a critical role in immunostimulation. In one hypothesis, the 17-nt cRNA/mRNA dsRNA along mRNA strands might facilitate the binding of the nearby dsRNA region from 5′ppp-RNA and cRNA in a tooth to RIG-I or be recognized simultaneously with the tooth by RIG-I. In another hypothesis, dsRNA might become more resistant to nuclease attack when proximal to mRNA. Indeed, RNA steric structure largely influences the susceptibility to enzymatic degradation (45, 46). Practically, even if dsRNA teeth were unexpectedly detached from mRNA after administration into the body, the detached teeth would induce minimal off-target inflammation in cells and tissues that do not show antigen expression from mRNA.
Previously, we reported the preparation of immunostimulatory mRNA by hybridizing poly U sequence in its polyA tail region, which was the first attempt to integrate adjuvancy directly into antigen-encoding mRNA for vaccine formulation (47). The approach in the present study has several advantages over this previous approach. First, only the present comb-structured mRNA can control the immunostimulation intensity, providing advantages in safety and versatility as aforementioned. Second, the current comb-structured approach uses chemically synthesized cRNA for precise preparation of the blunt-ended dsRNA with 5′ triphosphate, a binding motif for RIG-I activation. In contrast, preparation of such a structure was unachievable in the previous approach, which entirely relies on IVT for poly U preparation. More critically, IVT preparation of poly U in the previous approach may produce contaminant RNA strands complementary to poly U via an uncontrolled process of RNA-templated RNA transcription during IVT (28–30), and dsRNA from poly U and the contaminant RNA may influence the immunostimulating activity of mRNA hybridized with poly U. The current approach circumvents the formation of such by-products by justifying the sequences of 5′ppp-RNA in combination with the proper chemical synthesis of cRNA.
The present system drastically increased the secretion of IFN-β in vitro, although the influence of type I interferon on mRNA vaccines is still controversial. Previous studies showed either positive or negative effects of type I interferon on cellular immunity induction by mRNA vaccines (23, 39, 48–50). Although the precise mechanisms underlying positive outcomes of comb-structured mRNA in the present study need further investigation, comb-structured mRNA induced the secretion of various proinflammatory molecules (Fig. 3). Accordingly, some of the molecules might influence positively to override the negative effect of type I interferon. Notably, comb-structured mRNA controls the intensity of type I interferon induction, potentially helping avoid its negative influences. In addition, concurrent antigen presentation and interferon responses, shown in Fig. 6 J and K, might explain the positive influence of interferon on vaccinations, as suggested by a previous study (39).
In conclusion, our system is a simple and robust platform to supply a desired intensity of immunostimulation to various mRNA cancer vaccine systems and improve their efficiency only by adding a small amount of RNA. However, vaccination efficiency and antitumor activity need improvement for future clinical translation. The current study focuses on system development and fundamental analyses using existing delivery systems, i.e., anionic lipoplex, iLNP, and PMs. In a future study, we will co-optimize the delivery systems along with tuning the immunostimulatory intensity of mRNA to improve vaccination efficiency. Furthermore, antitumor activity will be strengthened by combination with immune checkpoint inhibitors, which provide a synergistic effect with mRNA cancer vaccines (13, 51).
Materials and Methods
Preparation of Comb-Structured mRNA and mRNA Nanoparticles.
mRNA and 5′ppp-RNA were prepared by IVT, while cRNA was chemically synthesized. For hybridization, the mixture of mRNA, 5′ppp-RNA, and cRNA at an equimolar ratio was heated at 65 °C for 5 min and gradually cooled down to 30 °C in 10 min before settling down to 4 °C. Anionically charged mRNA lipoplex was prepared according to the previous report (23). Briefly, a cationic liposome prepared from DOTMA and DOPE (1:1 molar ratio) using a thin-film method was mixed with mRNA in NaCl solution at a nitrogen/phosphate (N/P) ratio of 0.5. For iLNP preparation, ethanol solution containing ionizable lipid, ALC-0315, phospholipid, 1,2-distearoyl-sn-glycero-3-phosphocholine, cholesterol, and PEG-lipid, ALC-0159 at the molar ratio of 46.3:9.4:42.7:1.6 was mixed with mRNA in citrate buffer (pH3) using microfluidics followed by buffer replacement with PBS. The PM was prepared at an N/P ratio of 5, as previously described (43), using a block copolymer of 44-kDa PEG and PAsp(DET) with a polymerization degree 59. For detail, see SI Appendix.
Cell Culture Experiments.
mRNA was introduced into DC2.4 cells in Fig. 2, reporter cell lines expressing Lucia luciferase or SEAP responding to proinflammatory stimuli in Fig. 5, mouse BMDCs in Figs. 3 and 6 A–K, and human DCs in Fig. 6 L and M. Lipofectamine LTX was used for mRNA introduction unless otherwise described. The concentration of mRNA and poly IC in the cultured medium was adjusted to 1,000 ng/mL in Figs. 2, 3, and 6 A–K, and 500 ng/mL in Figs. 5 and 6 L and M. In Fig. 2, proinflammatory transcript levels were measured using quantitative PCR. In Fig. 5, proinflammatory responses were quantified based on Lucia luciferase or SEAP levels. In Fig. 6, cell surface expression of DC activation marker proteins was measured by flow cytometry after immunocytochemical staining. In Fig. 3, Luminex® Multiplex Assays (ThermoFisher Scientific) were performed to quantify expression levels of 24 molecules, including G-CSF, IFN-γ, IL-1α, 1β, 2, 4, 5, 6, 7, 9, 10, 12p40, 12p70, 13, 15, 17, TNF-α, CXCL1, 2, 10, and CCL2, 3, 4, 5. Expression of IFN-β was quantified using a Mouse IFN-β Duoset ELISA kit. Out of 25 inflammatory molecules, 16 were above detection limits, and the levels of these 16 molecules are shown in a heatmap. For detail, see SI Appendix.
Vaccination Experiments.
C57BL/6J mice (7 wk, female) were used. For the in vivo CTL assay, CFSE-labeled splenocytes added with a SIINFEKL epitope were intravenously injected into vaccinated mice. One day later, the viability of the injected splenocytes in the spleen was evaluated by flow cytometry. For tumor challenge, E.G7-OVA cells or B16F0 cells were inoculated subcutaneously. The mRNA doses for each vaccination are 5 μg for anionic lipoplex loading OVA mRNA, 10 μg for anionic lipoplex loading Trp2 mRNA, 0.01, 0.03, 0.1, and 0.3 μg for i.m. injection of iLNP loading OVA mRNA, 20 μg for i.m. injection of PM loading OVA mRNA, and 12 μg for i.d. injection of PM loading OVA mRNA. All animal experiments were conducted under the approval of the animal care and use committees in the Innovation Center of NanoMedicine, Kawasaki Institute of Industrial Promotion (Kanagawa, Japan), and Kyoto Prefectural University of Medicine (Kyoto, Japan). For detail, see SI Appendix.
Data Presentation.
Data are presented as mean ± SEM. Statistical analyses were performed by unpaired two-tailed Student's t test for comparing two groups and ANOVA followed by Tukey’s post hoc test for comparing three or more groups. For detail, see SI Appendix.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
This work was supported by the Center of Innovation Program from Japan Science and Technology Agency [JPMJCE1305 to K.K.], Grants-in-Aid for Challenging Research (Pioneering) [18H05378 to K.K.], for Scientific Research (A) [21H04962 to S.U., 21H04967 to K.K.] and (B) [18K03529 to S.U.] from the Ministry of Education, Culture, Sports, Science and Technology, Japan, Leading Advanced Projects for Medical Innovation [21gm0010008s0101 to S.U.], and Research Program on Emerging and Re-emerging Infectious Diseases [21fk0108620h0001 to S.U.] from Japan Agency for Medical Research and Development. We would like to thank Hiroaki Kinoh, Xueying Liu, Joachim Van Guyse, Keisuke Nagao, Hikaru Saitoh, Yuki Sato, Yuki Tada (iCONM), Satomi Nakagahara, and Yumi Fujii (NanoCarrier Ltd.) for their technical assistance.
Author contributions
S.U. designed research; T.A.T., S.A., M.M.-M., A.H., N.Y., E.B., Z.W., S.F., and S.U. performed research; T.A.T., S.A., N.Y., and S.U. analyzed data; and T.A.T., S.A., K.K., and S.U. wrote the paper.
Competing interests
K.K. is a Founder and a Member of the Board of NanoCarrier Ltd. M.M.-M. is an employee of NanoCarrier Ltd. N.Y., K.K., and S.U. have filed a patent application (Publication No. WO/2018/124181), and NanoCarrier Ltd. (M.M.-M.) holds a right to the patent.
Footnotes
This article is a PNAS Direct Submission. D.J.I. is a guest editor invited by the Editorial Board.
Contributor Information
Kazunori Kataoka, Email: kkataoka@g.ecc.u-tokyo.ac.jp.
Satoshi Uchida, Email: uchida.anme@tmd.ac.jp.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Pardi N., Hogan M. J., Porter F. W., Weissman D., mRNA vaccines–A new era in vaccinology. Nat. Rev. Drug Discov. 17, 261–279 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miao L., Zhang Y., Huang L., mRNA vaccine for cancer immunotherapy. Mol. Cancer 20, 41 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbier A. J., Jiang A. Y., Zhang P., Wooster R., Anderson D. G., The clinical progress of mRNA vaccines and immunotherapies. Nat. Biotechnol. 40, 840–854 (2022). [DOI] [PubMed] [Google Scholar]
- 4.Sahin U., et al. , Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature 547, 222–226 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Cafri G., et al. , mRNA vaccine–induced neoantigen-specific T cell immunity in patients with gastrointestinal cancer. J. Clin. Invest. 130, 5976–5988 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weide B., et al. , Direct injection of protamine-protected mRNA: Results of a phase 1/2 vaccination trial in metastatic melanoma patients. J. Immunother. 32, 498–507 (2009). [DOI] [PubMed] [Google Scholar]
- 7.Van Lint S., et al. , Preclinical evaluation of TriMix and antigen mRNA-based antitumor therapy. Cancer Res. 72, 1661–1671 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Schlosser E., et al. , TLR ligands and antigen need to be coencapsulated into the same biodegradable microsphere for the generation of potent cytotoxic T lymphocyte responses. Vaccine 26, 1626–1637 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Fischer N. O., et al. , Colocalized delivery of adjuvant and antigen using nanolipoprotein particles enhances the immune response to recombinant antigens. J. Am. Chem. Soc. 135, 2044–2047 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Kuai R., Ochyl L. J., Bahjat K. S., Schwendeman A., Moon J. J., Designer vaccine nanodiscs for personalized cancer immunotherapy. Nat. Mater. 16, 489–496 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oberli M. A., et al. , Lipid nanoparticle assisted mRNA delivery for potent cancer immunotherapy. Nano Lett. 17, 1326–1335 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haabeth O. A. W., et al. , mRNA vaccination with charge-altering releasable transporters elicits human T cell responses and cures established tumors in mice. Proc. Natl. Acad. Sci. U.S.A. 115, E9153–E9161 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miao L., et al. , Delivery of mRNA vaccines with heterocyclic lipids increases anti-tumor efficacy by STING-mediated immune cell activation. Nat. Biotechnol. 37, 1174–1185 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Zhang H., et al. , Delivery of mRNA vaccine with a lipid-like material potentiates antitumor efficacy through Toll-like receptor 4 signaling. Proc. Natl. Acad. Sci. U.S.A. 118, e2005191118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Islam M. A., et al. , Adjuvant-pulsed mRNA vaccine nanoparticle for immunoprophylactic and therapeutic tumor suppression in mice. Biomaterials 266, 120431 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alameh M. G., et al. , Lipid nanoparticles enhance the efficacy of mRNA and protein subunit vaccines by inducing robust T follicular helper cell and humoral responses. Immunity 54, 2877–2892.e7 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ndeupen S., et al. , The mRNA-LNP platform’s lipid nanoparticle component used in preclinical vaccine studies is highly inflammatory. iScience 24, 103479 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tahtinen S., et al. , IL-1 and IL-1ra are key regulators of the inflammatory response to RNA vaccines. Nat. Immunol. 23, 532–542 (2022). [DOI] [PubMed] [Google Scholar]
- 19.Abbasi S., Uchida S., Multifunctional immunoadjuvants for use in minimalist nucleic acid vaccines. Pharmaceutics 13, 644 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heidegger S., et al. , RIG-I activating immunostimulatory RNA boosts the efficacy of anticancer vaccines and synergizes with immune checkpoint blockade. EBioMedicine 41, 146–155 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koerner J., et al. , PLGA-particle vaccine carrying TLR3/RIG-I ligand Riboxxim synergizes with immune checkpoint blockade for effective anti-cancer immunotherapy. Nat. Commun. 12, 2935 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schlee M., et al. , Recognition of 5’ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity 31, 25–34 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kranz L. M., et al. , Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature 534, 396–401 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Sahin U., et al. , An RNA vaccine drives immunity in checkpoint-inhibitor-treated melanoma. Nature 585, 107–112 (2020). [DOI] [PubMed] [Google Scholar]
- 25.Hou X., Zaks T., Langer R., Dong Y., Lipid nanoparticles for mRNA delivery. Nat. Rev. Mater. 6, 1078–1094 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uchida S., Kataoka K., Design concepts of polyplex micelles for in vivo therapeutic delivery of plasmid DNA and messenger RNA. J. Biomed. Mater. Res. A 107, 978–990 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Schmidt A., et al. , 5’-triphosphate RNA requires base-paired structures to activate antiviral signaling via RIG-I. Proc. Natl. Acad. Sci. U.S.A. 106, 12067–12072 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cazenave C., Uhlenbeck O. C., RNA template-directed RNA synthesis by T7 RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 91, 6972–6976 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Triana-Alonso F. J., Dabrowski M., Wadzack J., Nierhaus K. H., Self-coded 3’-extension of run-off transcripts produces aberrant products during in vitro transcription with T7 RNA polymerase. J. Biol. Chem. 270, 6298–6307 (1995). [DOI] [PubMed] [Google Scholar]
- 30.Kariko K., Muramatsu H., Ludwig J., Weissman D., Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA. Nucleic Acids Res. 39, e142 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gholamalipour Y., Karunanayake Mudiyanselage A., Martin C. T., 3’ end additions by T7 RNA polymerase are RNA self-templated, distributive and diverse in character-RNA-seq analyses. Nucleic Acids Res. 46, 9253–9263 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoshinaga N., et al. , Induced packaging of mRNA into polyplex micelles by regulated hybridization with a small number of cholesteryl RNA oligonucleotides directed enhanced in vivo transfection. Biomaterials 197, 255–267 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Kariko K., Buckstein M., Ni H., Weissman D., Suppression of RNA recognition by Toll-like receptors: The impact of nucleoside modification and the evolutionary origin of RNA. Immunity 23, 165–175 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Kato H., et al. , Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J. Exp. Med. 205, 1601–1610 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Botos I., Liu L., Wang Y., Segal D. M., Davies D. R., The Toll-like receptor 3:dsRNA signaling complex. Biochim. Biophys. Acta 1789, 667–674 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heil F., et al. , Species-specific recognition of single-stranded RNA via Toll-like receptor 7 and 8. Science 303, 1526–1529 (2004). [DOI] [PubMed] [Google Scholar]
- 37.Pollard C., et al. , Type I IFN counteracts the induction of antigen-specific immune responses by lipid-based delivery of mRNA vaccines. Mol. Ther. 21, 251–259 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uchida S., Kataoka K., Itaka K., Screening of mRNA chemical modification to maximize protein expression with reduced immunogenicity. Pharmaceutics 7, 137–151 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Hoecke L., et al. , The opposing effect of type I IFN on the T cell response by non-modified mRNA-lipoplex vaccines is determined by the route of administration. Mol. Ther. Nucleic Acids 22, 373–381 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kostinov M. P., Akhmatova N. K., Khromova E. A., Kostinova A. M., Cytokine profile in human peripheral blood mononuclear leukocytes exposed to immunoadjuvant and adjuvant-free vaccines against influenza. Front. Immunol. 11, 1351 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vogel A. B., et al. , BNT162b vaccines protect rhesus macaques from SARS-CoV-2. Nature 592, 283–289 (2021). [DOI] [PubMed] [Google Scholar]
- 42.Blakney A. K., et al. , Polymeric and lipid nanoparticles for delivery of self-amplifying RNA vaccines. J. Control. Release 338, 201–210 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Uchida S., et al. , In vivo messenger RNA introduction into the central nervous system using polyplex nanomicelle. PLoS One 8, e56220 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Toy R., et al. , TLR7 and RIG-I dual-adjuvant loaded nanoparticles drive broadened and synergistic responses in dendritic cells in vitro and generate unique cellular immune responses in influenza vaccination. J. Control. Release 330, 866–877 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Han D., Park Y., Kim H., Lee J. B., Self-assembly of free-standing RNA membranes. Nat. Commun. 5, 4367 (2014). [DOI] [PubMed] [Google Scholar]
- 46.Yoshinaga N., et al. , Bundling mRNA strands to prepare nano-assemblies with enhanced stability towards RNase for in vivo delivery. Angew. Chem. Int. Ed. Engl. 58, 11360–11363 (2019). [DOI] [PubMed] [Google Scholar]
- 47.Uchida S., et al. , Designing immunostimulatory double stranded messenger RNA with maintained translational activity through hybridization with poly A sequences for effective vaccination. Biomaterials 150, 162–170 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Broos K., et al. , Particle-mediated intravenous delivery of antigen mRNA results in strong antigen-specific T-cell responses despite the induction of type I interferon. Mol. Ther. Nucleic Acids 5, e326 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Udhayakumar V. K., et al. , Arginine-rich peptide-based mRNA nanocomplexes efficiently instigate cytotoxic T cell immunity dependent on the amphipathic organization of the peptide. Adv. Healthc. Mater. 6, 1601412 (2017). [DOI] [PubMed] [Google Scholar]
- 50.Li C., et al. , Mechanisms of innate and adaptive immunity to the Pfizer-BioNTech BNT162b2 vaccine. Nat. Immunol. 23, 543–555 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen J., et al. , Lipid nanoparticle-mediated lymph node-targeting delivery of mRNA cancer vaccine elicits robust CD8(+) T cell response. Proc. Natl. Acad. Sci. U.S.A. 119, e2207841119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All study data are included in the article and/or SI Appendix.