Abstract
Cryptosporidium species are a major cause of diarrhea and associated with growth failure. There is currently only limited knowledge of the parasite's genomic variability. We report a genomic analysis of Cryptosporidium parvum isolated from Bangladeshi infants and reanalysis of sequences from the United Kingdom. Human isolates from both locations shared 154 variants not present in the cattle-derived reference genome, suggesting host-specific adaptation of the parasite. Remarkably 34.6% of single-nucleotide polymorphisms unique to human isolates were nonsynonymous and 8.2% of these were in secreted proteins. Linkage disequilibrium decay indicated frequent recombination. The genetic diversity of C. parvum has potential implications for vaccine and therapeutic design.
Clinical Trials Registration. NCT02764918.
Keywords: Cryptosporidium, SNPs, genome sequences, parasite
A genomic analysis of C. parvum isolated from Bangladeshi infants and reanalysis of sequences from the United Kingdom were performed. Full genome sequences clustered by gp60 group and isolate source (human or animal) more than by geographic region.
Cryptosporidium parasites are a leading cause of death and disability due to diarrheal disease in infants in low- and middle-income countries [1–3]. The parasitic members of the eukaryotic single-celled Cryptosporidium genus can infect a broad range of hosts. While human cryptosporidiosis can be caused by at least 15 different species of Cryptosporidium, just 3 species cause the bulk of human disease: Cryptosporidium hominis, Cryptosporidium meleagridis, and Cryptosporidium parvum [4]. The aim of this work was to characterize the genetic diversity in the Cryptosporidium parasites infecting humans. No vaccine exists to prevent cryptosporidiosis and information on the population genetics of the parasite has important implications for the design and development of vaccines and therapeutics [5].
Fewer than 10 reference (formally assembled, annotated, and publicly available on the parasite database CryptoDB) genome sequences exist for these species [6]. Because only C. parvum can be cultured in vitro, C. parvum is the focus of many experimental models for vaccine and therapeutic development.
In previous work on cryptosporidiosis in Bangladesh and Africa, we and others discovered that extensive genomic diversity exists within C. hominis, including a high rate of sexual recombination and single nucleotide polymorphisms (SNPs) [7, 8]. Here, we extended this analysis by sequencing C. parvum isolates from children in Bangladesh and comparing these genome sequences to both the reference genome sequence, which was from cattle, and isolates collected from humans and cattle in the United Kingdom [9, 10]. We highlight the genomic variability among parasites collected in distinct geographic locations and between parasites isolated from humans versus cattle, as well as a high rate of recombination. These findings emphasize the need for further delineation of the variability in clinically relevant reference genome sequences to prioritize antigen selection and identify drug targets.
METHODS
Ethical Considerations
The study was approved by the Ethical and Research Review Committees of the International Centre for Diarrhoeal Disease Research, Bangladesh (icddr, b) and the Institutional Review Board of the University of Virginia. Informed written consent was obtained from the parents or guardians for the participation of their child in the study.
Infant Cohort
Starting in June 2014, 250 children born into an urban slum of Dhaka (Section 11 of Mirpur Thana) and 258 children from rural Mirzapur were enrolled in the first week after birth into a community-based prospective cohort study of enteric infections (“Cryptosporidiosis and Enteropathogens in Bangladesh”; ClinicalTrials.gov identifier NCT02764918). At the urban location, an additional 250 children from the same population were enrolled in a second companion community-based prospective cohort study focused on infant cryptosporidiosis (“Field Studies of Cryptosporidiosis and Enteropathogens in Bangladesh”). C. parvum was identified in 2% of all cryptosporidia infections (n = 3, 2 monoinfections) at the urban site and in 4% (total n = 4; 3 monoinfections) at the rural location [4]. Six of the oocyst isolates collected from both study sites had sufficient DNA for sequencing and were included in the analysis reported here, including 1 coinfected with C. hominis.
Sampling and Specimen Testing
Diarrheal and monthly surveillance stools were evaluated for the presence of the major enteric parasitic pathogens, including Cryptosporidium spp. by use of a multiplexed quantitative polymerase chain reaction (qPCR) assay, which includes a broad-range assay that recognizes the common Cryptosporidium species that routinely infect humans (C. hominis, C. parvum, and C. meleagridis) as well as 2 other clinically relevant protozoan parasites (Giardia duodenalis and Entamoeba histolytica) [7, 11].
Genotyping Assay
The polymorphic region within the gp60 gene (cgd6_1080) was used to genotype Cryptosporidium-positive samples [12]. If the short-read genomic sequences were insufficient to type the region with confidence, a nested PCR reaction was performed as previously described and Sanger sequenced [7] (Supplementary Table 1). Mixed infections were identified by the presence of multiple gp60 genotypes.
Whole-Genome Resequencing and Analysis
The sequences of the Bangladesh isolates were obtained as previously described for C. hominis [7] and are deposited in the NCBI's Sequence Read Archive BioProject PRJEB14327 (SRA; Supplementary Table 1). Additional genome sequences were obtained from the SRA with the following selection criteria: availability of fastq files and sample collection methods presented in peer-reviewed publications. Furthermore, only samples that were collected from humans and purified to obtain oocysts were used, excluding mouse-derived parasites and metagenomic sequences. With these selection criteria, whole-genome nucleotide sequencing data from 12 isolates collected in the United Kingdom were acquired through their accession numbers (Supplementary Table 1) [9, 13]. Of note, 1 additional genome sequence (from a child in Uganda) passed these selection criteria; it was excluded because, with only 1 isolate, we could not compare multiple Ugandan isolates as we did with isolates from the United Kingdom and Bangladesh [7, 9].
For consistency in analysis, all genome sequences were analyzed in parallel and aligned to the Cryptosporidium parvum Iowa II reference genome sequence (CryptoDB version 53; [6]); the analytic code and additional detail are provided at https://github.com/maureencarey/cparvum_genomes_manuscript. First, sequences were downloaded from NCBI's SRA and trimmed to remove adaptors and restrict sequence length to 150 base pairs, and quality filtered with BBTools version 38.57 [14]. Read quality was evaluated with FastQC version 0.11.5 and MultiQC version 1.8 [15, 16]. Next, unmerged forward and reverse reads were aligned with BWA-mem version 0.7.17 [17] to the C. parvum Iowa II reference genome (accession number GCA_000165345.1), SAM files were converted and sorted to generate BAM files with SAMTools version 1.9 [18]. The mean depth of genome cover in each isolate was determined using (SAMTools; version 1.12) and multiallelic SNPs were identified using Freebayes (version 0.9.9) and tabulated using bcftools (version 1.9). Duplicate reads were marked with Picard version 2.20.6 (https://broadinstitute.github.io/picard). GATK's HaplotypeCaller (version 4.0.0.0) was then used to call SNPs [19]. For the combined population-based analyses the individual BAM files were then merged (Supplementary Table 1). For SNP analyses, oocyst genome sequences were treated as tetraploid; however, for linkage disequilibrium calculations, these genome sequences were treated as diploid for compatibility with available software [20]. Lastly, variants were filtered to remove low-quality SNPs using GATK's VariantFiltration (QUAL < 25.0, QD < 15.0, FS > 12.0, MQ < 58.0, MQRankSum < −3.0, ReadPosRankSum < −3.0 [19]); parameters were selected for consistency with our previous publication [7]. SnpEff version 4.3 was used to identify the variations that resulted in the substitution of a different amino acid in the encoded proteins [21].
Linkage disequilibrium and principal component analysis (PCA) were performed with PLINK version 1.90b6.16 [20, 22]. VCF files were analyzed and visualized in R 4.0.3 [23, 24]. The 32 Bangladesh C. hominis genome sequences [7] were rerun through an identical pipeline for comparison and randomly subset into groups of 16–18 to determine if the differences in the squared correlation coefficient between different alleles (r2) resulted from sample size bias [25].
RESULTS
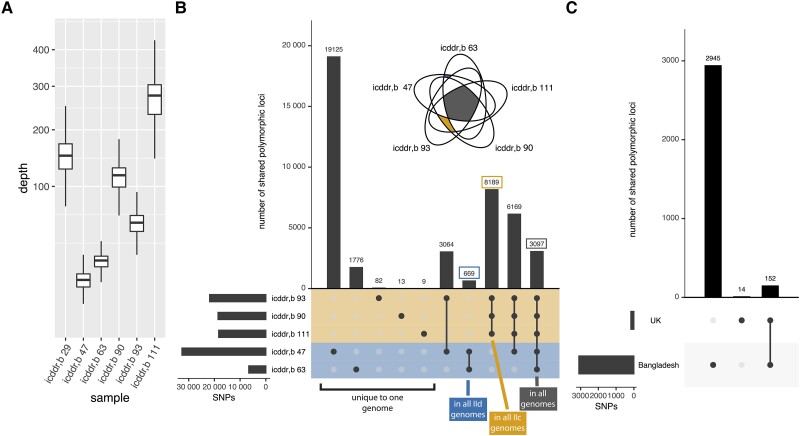
Oocysts purified by immunomagnetic separation from stool were sequenced to obtain 5 C. parvum genome sequence monoinfections (as determined by unique polymorphic gp60 region and only a small number of multiallelic SNPs [0.1% ± 0.08%]) and the mean depth of cover (DP) and the percentage of the reads that mapped to the C. parvum genome (mapped) for each isolate calculated (icddr, b 47, DP = 20.6, mapped 58.85%; icddr, b 63, DP = 32.25, mapped =97.97%; icddr, b 90, DP = 142.259, mapped = 88.74%, icddr, b 93, DP = 83.23, mapped = 88.32%; icddr, b 111, DP = 263.126, mapped = 99.40%). One additional isolate was also obtained from a child with a mixed C. parvum and C. hominis infection (icddr, b 29: DP = 194, mapped = 98.35%) (Figure 1A, Supplementalry Table 1). Parasite genome sequences contained between 6636 and 32 616 SNPs (total, 43 882) when compared to the reference genome, C. parvum Iowa II obtained from cattle (CryptoDB version 53) (Figure 1). No major structural variations (defined as insertions or deletions greater than 200 base pairs) were observed (see code, Supplementary Material). Only 3097 SNPs were common to all 5 monoinfection isolates (Figure 1B). For reference, 12 recently published genome sequences isolated in the United Kingdom sequenced using similar technology to a mean coverage of 101.5 reads per locus, were analyzed in parallel (Supplementary Figure 1A). In the UK isolates, 26 992 SNPs were detected, but only 166 of these SNPs were present in all of the UK isolates (Supplementary Figure 1B). There were 2945 SNPs found exclusively in the Bangladesh isolates (ie, in all Bangladesh isolates and in no UK isolates) whereas only 14 SNPs were found exclusively in the UK isolates (Figure 1C). There were 152 SNPs found to be shared with all of the human isolates, including 60 variants that resulted in a change in the amino acid sequences of the encoded proteins. Despite variability in the total number of SNPs and genes affected by SNPs across genome sequences (Supplementary Figure 2A and 2B), nonsynonymous variants were similarly represented in each genome (Supplementary Figure 2C); nonsynonymous SNPs yield functional differences in a protein by changing the amino acid sequence or adding or removing a stop codon. Such changes in the amino acid sequence in secreted proteins were of particular interest given the potential for interaction with the mammalian host and, therefore, these may be under selective pressure. Ninety-five genes in the C. parvum genome are predicted to encode secreted proteins (2.4% of all genes [6]). We found that the nonsynonymous SNPs occurred at a higher frequency in these genes (3% of the nonsynonymous SNPs were located in these genes) (Supplementary Figure 2C). Five genes encoding secreted proteins contained SNPs in all of these human isolates: cgd8_1740 (secreted GGC gene family protein), cgd3_10 (uncharacterized, SKSR gene family), cgd6_1180 (uncharacterized), cgd7_4340 (uncharacterized), cgd7_4500 (uncharacterized), and cgd8_3540 (uncharacterized, WYLE gene family). Three genes contained SNPs in only the Bangladesh isolates: cgd1_1680 (insulinase-like protease), cgd7_3390 (patatin-like phospholipase), and cgd8_3670 (uncharacterized); none of these proteins are known to trigger a host immune response.
Figure 1.
Novel genome sequencing reveals conserved variants and variants associated with geographic location. A, Sequencing depth. Genome-wide sequencing depth for each Bangladesh isolate (including the mixed infection); analogous data for UK isolates is available in Supplementary Figure 1. Each box represents the median (inner line), 25th percentile, and 75th percentile. Upper whiskers extend from the top of the box to the largest value within 1.5 times the interquartile range, and the lower whisker extends to the smallest value within 1.5 times the interquartile range. B, Core set of conserved single-nucleotide polymorphisms (SNPs) and SNPs unique to each genome. SNPs in each monoinfection isolate from Bangladesh were compared using the UpSet visualization approach. Left barchart represents the total number of SNPs in each genome. Top bar chart represents the number of SNPs shared between the different genome sequences (indicated by the filled circles below). The mixed infection is not shown. Genome sequences in yellow are IIc (gp60 grouping), genome sequences in blue are IId. The inset Venn diagram is shown to highlight the SNPs shared between the genome sequences with the different gp60 genotype. C, SNPs found in all isolates. SNPs found in all Bangladesh isolates or all UK isolates were compared.
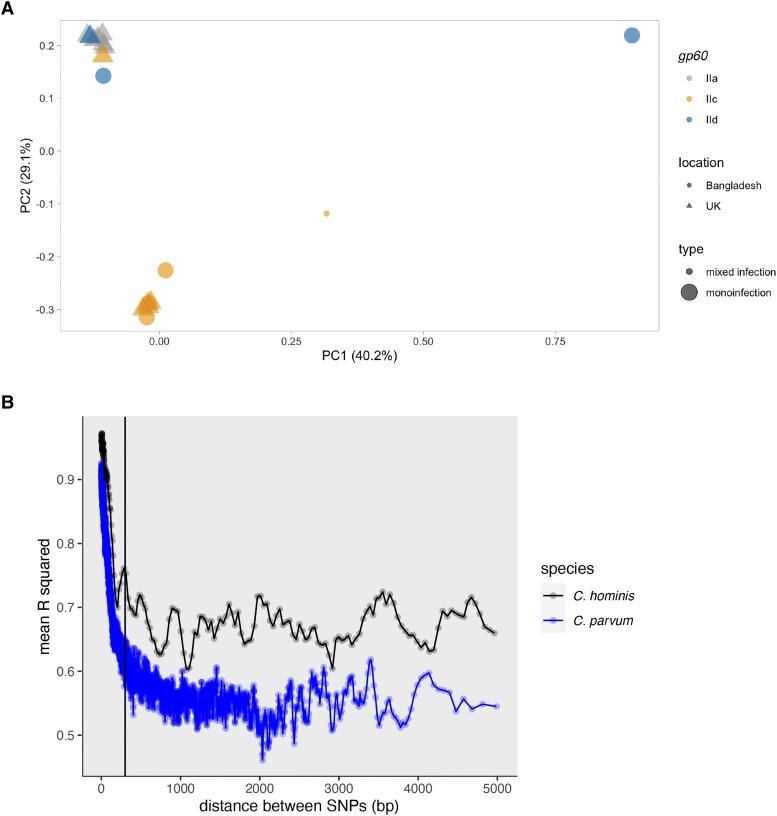
By comparing all SNP variations in each genome using a PCA, the full genome sequences clustered by gp60 group more so than geographic region (PERMANOVA, location P > .5; gp60 P < .04; Figure 2A). gp60 is the most frequently used Cryptosporidium genotyping system. The gp60 locus is evaluated by identifying individual SNPs to distinguish parasite types and the number of repetitions in a highly variable microsatellite region to identify subtypes [26,27]; the C. parvum gp60 group IIc is thought to have a strong preference for human hosts whereas C. parvum isolates with other gp60 genotypes are thought to more commonly infect other mammals [9]. The reference genome from C. parvum Iowa II, used in this study, belongs to the IIa group and therefore would be expected to have a broad range of mammalian hosts (GCA_015245375.1 VEupath Release 53) [28]. This trend in gp60 group clustering via PCA was conserved when focusing on only those nonsynonomous SNPs that resulted in changes in the amino acid sequences of the secreted proteins (Supplementary Figure 3A and 3B). Because PCA and associated statistical tests can be biased by outliers, we also confirmed that this trend was consistent when excluding 1 genome from Bangladesh that appeared to be an outlier (identifier icddr, b 47). Outlier status was based on SNP content after removing biases (base pairs were filtered for quality and normalized to read depth and root mean square mapping quality over all the reads at the site). The reanalysis via PCA of the data minus icddr, b 47 (Supplementary Figure 3C) did not alter our initial conclusion based on the number of shared SNPs (Figure 1B) or our conclusion after the performance of the first PCA that the genome sequences belonging to the gp60 group IIc in the United Kingdom and Bangladesh were more similar to each other than to the group IId genome sequences (Figure 2A).
Figure 2.
Despite the rapid linkage disequilibrium decay observed in the Cryptosporidium parvum genome sequences the whole-genome SNP profile continued to be concordant with the gp60 genotype. A, gp60 summarizes genome-wide variation. Principal component (PC) analysis of genome-wide variation (as implemented in Plink 2.0) is shown. All quality-filtered single-nucleotide polymorphisms (SNPs) were used. Points represent genome sequences and are color coded by gp60 grouping. Shapes represent patient location. The small dot represents the mixed infection from Bangladesh. Significance as determined by a PERMANOVA: location, P > .5; gp60, P < .04; however, this statistical test is biased by outliers, and so please see Supplementary Figure 3. B, Similar rate of recombination in C. parvum when compared to Cryptosporidium hominis. Comparison between C. parvum (blue) and C. hominis (black) linkage disequilibrium decay, calculated with Plink 2.0. The region of linkage disequilibrium in C. hominis was previously calculated as being <300 and 300 bp is indicated by the vertical black line at 300 bp on the x-axis [7]. Although the threshold values were different in C. hominis and C. parvum the rate of linkage disequilibrium decay appeared similar. To confirm this observation the data were reanalyzed using comparable genome numbers (Supplementary Figure 4).
Previous work highlighted the unusually short regions of linkage disequilibrium in C. hominis (<300 bp) and, as a result, C. hominis genome sequences do not cluster by gp60 genotype [7]. In C. parvum the gp60 genotype is, however, routinely used to both track outbreaks and as a genetic marker to describe genomic lineages [9, 12]. Its use for this purpose has, however, been controversial [29–32]. Thus, we next asked if gp60 was more representative of whole-genome C. parvum diversity in Bangladesh than in C. hominis because C. parvum has less recombination. Our results, however, indicated that this is not the case. The C. parvum parasites also had a high rate of recombination (Figure 2B) similar to that occurring in the Bangladesh C. hominis genome sequences [25]. Any differences in observed linkage disequilibrium curves are explained by the reduced number of genome sequences included in the study when compared to the C. hominis study (Supplementary Figure 4) [25].
DISCUSSION
Here, we demonstrate that C. parvum isolates from human infections in geographically diverse locations have genetic differences from the reference genome line, isolated from cattle. We present high-quality sequence data from 5 C. parvum isolates collected from patients in Bangladesh (Figure 1A and 1B), compared them to both the animal-derived reference genome (C. parvum Iowa II) and 12 recently published human isolates from the United Kingdom, and identified a set of SNPs shared by all 17 human isolates (Figure 1C). Over 39% of these SNPs specific to Cryptosporidium genome sequences isolated from human hosts resulted in functional differences in the encoded proteins; these variants highlight key differences between human-derived isolates and the animal-derived parasite line that was used to generate a reference genome. Importantly, 3% of all SNPs in the Bangladesh and UK isolates were both nonsynonymous and located within the 2.4% of genes that encode for secreted proteins, which are potentially under selection pressure from the human host (Supplementary Figure 2).
With only 2 publicly available C. parvum reference genome sequences [6] and fewer than 10 published whole-genome sequencing studies, this study adds considerably to the field of parasite genomics by increasing the number and diversity of available genome sequences, specifically those derived from human infections. As such, 4 of these novel genome sequences obtained from monoinfections were gp60 group IIc, the genotype group described as the human C. parvum (C. parvum anthroponosum; Figure 1B). Importantly, the Bangladesh isolates had more total SNPs and more shared SNPs than the UK isolates (Figure 1B and Supplementary Figures 1B and 2A), emphasizing the diversity of parasites in this region [7].
Furthermore, we show that gp60 is somewhat representative of full genomic diversity in C. parvum in this set of genome sequences. Genome sequences with the same gp60 genotype are more similar than genome sequences from a single geographic location (Figure 1B and Figure 2A). The (partial) utility of gp60 genotyping in this species is contrary to what was observed in C. hominis [7]. Lastly, we show that this difference between species is not due to a significantly decreased rate of recombination in C. parvum (Figure 2B). The species difference may be the result of differential selective pressure on the gp60 protein in each species. As the function of gp60 is uncharacterized and the host range varies between the 2 parasite species, functional differences across species in the protein or in the host's response to the protein are plausible.
This study has several limitations, most notably the small number of C. parvum genome sequences obtained from human and cattle samples and the limited geographic range of infection locations sampled. However, the study also has notable strengths including the acquisition of the C. parvum genome sequences isolated from a Bangladesh population where this parasite is endemic and the identification of a core set of SNPs apparently unique to human isolates of C. parvum.
CONCLUSIONS
Clinically derived isolates of C. parvum collected in Bangladesh were highly divergent from one another with over 30% of SNPs in these isolates resulting in changes in protein open reading frames. Genetic variability within this population is unexpected given the low prevalence of this Cryptosporidium species and the homogenous host population [4]. Furthermore, frequent recombination also occurred in C. parvum, consistent with a previous study on C. hominis. Unlike C. hominis, however, the gp60 genotyping system remained representative of genomic variation in C. parvum. Bangladeshi human-derived isolates share many SNPs with clinically derived isolates from the United Kingdom, indicating divergence from the animal-derived reference genome and emphasizing the need for high-quality reference genome sequences from both human and animal infections. The functional significance of these genomic changes in the parasite remains to be discovered, but highlights that such genetic diversity may need to be accounted for in identifying targetable antigens for vaccine development.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Contributor Information
Maureen Carey, Departments of Medicine, University of Virginia, Charlottesville, Virginia, USA.
Tuhinur Arju, International Centre for Diarrhoeal Diseases Research, Dhaka, Bangladesh.
James A Cotton, Wellcome Sanger Institute, Cambridge, United Kingdom.
Masud Alam, International Centre for Diarrhoeal Diseases Research, Dhaka, Bangladesh.
Mamun Kabir, International Centre for Diarrhoeal Diseases Research, Dhaka, Bangladesh.
Abu S G Faruque, International Centre for Diarrhoeal Diseases Research, Dhaka, Bangladesh.
Rashidul Haque, International Centre for Diarrhoeal Diseases Research, Dhaka, Bangladesh.
William A Petri, Jr, Departments of Medicine, University of Virginia, Charlottesville, Virginia, USA.
Carol A Gilchrist, Departments of Medicine, University of Virginia, Charlottesville, Virginia, USA.
Notes
Author contributions. T. A. processed samples and J. A. C. provided sequencing data. M. A. C., C. A. G., and W. A. P. conceived the analysis plan and M. A. C. performed bioinformatic analyses with J. A. C. providing consultation. W. A. P., R. H., and A. S. G. F. founded the birth cohort and directed the study. Field work and data collection at the International Centre for Diarrhoeal Disease Research, Bangladesh (icddr, b) were performed by M. A. and M. K., with supervision from A. S. G. F. and R. H. Drafting of the manuscript was performed by M. A. C. and C. A. G. All authors edited and approved the final manuscript.
Acknowledgement. We are grateful to Dr Matthew Berriman for advice and assistance in the sequencing and for the willing participation of the parents and children at the icddr, b study sites. We also thank the field workers, nurses, laboratory staff of the Parasitology Laboratory of icddr, b who worked for this project, and Gladys Andino, at the University of Virginia Research Computing Core without whom we could not have completed this research. We also acknowledge the helpful feedback from members of the Petri laboratory, University of Virginia Trans-University Microbiome Initiative's Data Science team, and Pankaj Kumar, as well as Uma Nayak for her curation of the clinical database.
Disclaimer. The funders had no role in study design, data collection and analysis, or decision to submit for publication.
Financial support. This work was supported by the National Institute of Allergy and Infectious Diseases (grant numbers R21 AI-109118 to C. A. G., and R01 AI-043596 to C. A. G. and W. A. P.); the Bill and Melinda Gates Foundation (grant number OPP1100514 to A. S. G. F.); University of Virginia Engineering-in-Medicine seed grant to M. A. C. and W. A. P.; the Henske Family Foundation; and the Wellcome Trust (grant number 206194). M. A. C. was supported by the PhRMA Foundation Postdoctoral Fellowship in Translational Medicine and Therapeutics and the University of Virginia Trans-University Microbiome Initiative. TechLab, Inc donated the Giardia/Cryptosporidium QUIK CHEKS used in the parent study. Work at icddr, b was supported by the core donors (Government of the People’s Republic of Bangladesh, Global Affairs Canada, the Swedish International Development Cooperation Agency, and UKAid).
Potential conflict of interests. W. A. P. is a consultant for TechLab, a company that makes diagnostic tests for cryptosporidiosis. All other authors report no potential conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Haque R, Mondal D, Karim A, et al. . Prospective case-control study of the association between common enteric protozoal parasites and diarrhea in Bangladesh. Clin Infect Dis 2009; 48:1191–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Liu J, Platts-Mills JA, Juma J, et al. . Use of quantitative molecular diagnostic methods to identify causes of diarrhoea in children: a reanalysis of the GEMS case-control study. Lancet 2016; 388:1291–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Platts-Mills JA, Babji S, Bodhidatta L, et al. . Pathogen-specific burdens of community diarrhoea in developing countries: a multisite birth cohort study (MAL-ED). Lancet Glob Health 2015; 3:e564–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Steiner KL, Ahmed S, Gilchrist CA, et al. . Species of cryptosporidia causing subclinical infection associated with growth faltering in rural and urban Bangladesh: a birth cohort study. Clin Infect Dis 2018; 67:1347–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sparks H, Nair G, Castellanos-Gonzalez A, White AC. Treatment of Cryptosporidium: what we know, gaps, and the way forward. Curr Trop Med Rep 2015; 2:181–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Heiges M. CryptoDB: a Cryptosporidium bioinformatics resource update. Nucleic Acids Res 2006; 34:D419–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gilchrist CA, Cotton JA, Burkey C, et al. . Genetic diversity of Cryptosporidium hominis in a Bangladeshi community as revealed by whole-genome sequencing. J Infect Dis 2018; 218:259–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tichkule S, Jex AR, van Oosterhout C, et al. . Comparative genomics revealed adaptive admixture in Cryptosporidium hominis in Africa. Microb Genom 2021; 7:mgen000493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nader JL, Mathers TC, Ward BJ, et al. . Evolutionary genomics of anthroponosis in Cryptosporidium. Nat Microbiol 2019; 4:826–36. [DOI] [PubMed] [Google Scholar]
- 10. Abrahamsen MS, Templeton TJ, Enomoto S, et al. . Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science 2004; 304:441–5. [DOI] [PubMed] [Google Scholar]
- 11. Liu J, Kabir F, Manneh J, et al. . Development and assessment of molecular diagnostic tests for 15 enteropathogens causing childhood diarrhoea: a multicentre study. Lancet Infect Dis 2014; 14:716–24. [DOI] [PubMed] [Google Scholar]
- 12. Chalmers RM, Robinson G, Elwin K, Elson R. Analysis of the Cryptosporidium spp. and gp60 subtypes linked to human outbreaks of cryptosporidiosis in England and Wales, 2009 to 2017. Parasit Vectors 2019; 12:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hadfield SJ, Pachebat JA, Swain MT, et al. . Generation of whole genome sequences of new Cryptosporidium hominis and Cryptosporidium parvum isolates directly from stool samples. BMC Genomics 2015; 16:650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bushnell B.BBMap: BBTools: a suite of fast, multithreaded bioinformatics tools designed for analysis of DNA and RNA sequence data. https://sourceforge.net/projects/bbmap/. Accessed 8 August 2022.
- 15. Ewels P, Magnusson M, Lundin S, Käller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016; 32:3047–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. 08/2022Andrews S. FastQC: a quality control tool for high throughput sequence data. Babraham Bioinformatics, Babraham Institute, 2010. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/. Accessed 8 August 2022. [Google Scholar]
- 17. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009; 25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li H, Handsaker B, Wysoker A, et al. . The sequence alignment/map format and SAMtools. Bioinformatics 2009; 25:2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Poplin R, Ruano-Rubio V, DePristo MA, et al. . Scaling accurate genetic variant discovery to tens of thousands of samples. bioRxiv, doi: 10.1101/201178, 24July2018, preprint: not peer reviewed. [DOI] [Google Scholar]
- 20. Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 2015; 4:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cingolani P, Platts A, Wang LL, et al. . A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff. Fly (Austin) 2012; 6:80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Purcell S, Neale B, Todd-Brown K, et al. . PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81:559–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. RStudio Team . RStudio: integrated development for R. https://posit.co/download/rstudio-desktop/. Accessed 7 August 2022.
- 24. Wickham H, Chang W, Henry L, et al. ggplot2: Create Elegant Data Visualisations Using the Grammar of Graphics. CRAN R-Project.http://CRAN.R-Project.Org/package=ggplot2.
- 25. Tenesa A, Navarro P, Hayes BJ, et al. . Recent human effective population size estimated from linkage disequilibrium. Genome Res 2007; 17:520–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cama VA, Bern C, Roberts J, et al. . Cryptosporidium species and subtypes and clinical manifestations in children, Peru. Emerg Infect Dis 2008; 14:1567–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Baptista RP, Li Y, Sateriale A, et al. . Long-read assembly and comparative evidence-based reanalysis of Cryptosporidium genome sequences reveal expanded transporter repertoire and duplication of entire chromosome ends including subtelomeric regions. Genome Res 2022; 32:203–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tanrıverdi S, Grinberg A, Chalmers RM, et al. . Inferences about the global population structures of Cryptosporidium parvum and Cryptosporidium hominis. Appl Environ Microbiol 2008; 74:7227–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Widmer G, Lee Y. Comparison of single- and multilocus genetic diversity in the protozoan parasites Cryptosporidium parvum and C. hominis. Appl Environ Microbiol 2010; 76:6639–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Robinson G, Chalmers RM. Assessment of polymorphic genetic markers for multi-locus typing of Cryptosporidium parvum and Cryptosporidium hominis. Exp Parasitol 2012; 132:200–15. [DOI] [PubMed] [Google Scholar]
- 31. Abal-Fabeiro JL, Maside X, Bello X, Llovo J, Bartolomé C. Multilocus patterns of genetic variation across Cryptosporidium species suggest balancing selection at the gp60 locus. Mol Ecol 2013; 22:4723–32. [DOI] [PubMed] [Google Scholar]
- 32. Afgan E, Nekrutenko A, Grüning BA, et al. . The galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2022 update. Nucleic Acids Res 2022; 50:W345–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.