Abstract
Background
Remdesivir is approved for treatment of coronavirus disease 2019 (COVID-19) in nonhospitalized and hospitalized adult and pediatric patients. Here we present severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) resistance analyses from the phase 3 ACTT-1 randomized placebo-controlled trial conducted in adult participants hospitalized with COVID-19.
Methods
Swab samples were collected at baseline and longitudinally through day 29. SARS-CoV-2 genomes were sequenced using next-generation sequencing. Phenotypic analysis was conducted directly on participant virus isolates and/or using SARS-CoV-2 subgenomic replicons expressing mutations identified in the Nsp12 target gene.
Results
Among participants with both baseline and postbaseline sequencing data, emergent Nsp12 substitutions were observed in 12 of 31 (38.7%) and 12 of 30 (40.0%) participants in the remdesivir and placebo arms, respectively. No emergent Nsp12 substitutions in the remdesivir arm were observed in more than 1 participant. Phenotyping showed low to no change in susceptibility to remdesivir relative to wild-type Nsp12 reference for the substitutions tested: A16V (0.8-fold change in EC50), P323L + V792I (2.2-fold), C799F (2.5-fold), K59N (1.0-fold), and K59N + V792I (3.4-fold).
Conclusions
The similar rate of emerging Nsp12 substitutions in the remdesivir and placebo arms and the minimal change in remdesivir susceptibility among tested substitutions support a high barrier to remdesivir resistance development in COVID-19 patients.
Clinical Trials Registration. NCT04280705.
Keywords: COVID-19, RNA-dependent RNA polymerase, SARS-CoV-2, remdesivir, resistance
In participants with COVID-19 enrolled in a phase 3 clinical trial of remdesivir, similar rates of emergent amino acid substitutions were observed for remdesivir and placebo. Substitutions were associated with low to no change in viral susceptibility to remdesivir.
Remdesivir (GS-5734; Veklury) is a nucleotide analog prodrug that is metabolized into an active analog of adenosine triphosphate that inhibits the RNA-dependent RNA polymerase (RdRp) of viruses, including members of the Coronaviridae, Flaviviridae, Filoviridae, and Pneumoviridae families [1–3]. Remdesivir potently blocks severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) replication in vitro, improves pulmonary function, and prevents disease progression in ongoing SARS-CoV-2 infection in preclinical animal models [4–6]. Its clinical benefit in hospitalized adults with moderate-to-severe coronavirus disease 2019 (COVID-19) has been demonstrated in multiple phase 3 clinical trials, including the randomized, placebo-controlled Adaptive COVID-19 Treatment Trial-1 (ACTT-1; CO-US-540-5776) [7–9]. Recently, remdesivir treatment for 3 days in nonhospitalized participants showed a significant reduction in COVID-19–related hospitalization or all-cause death compared with placebo [10]. Remdesivir is approved for the treatment of COVID-19 in pediatric and adult hospitalized and nonhospitalized patients in the United States, the European Union, Japan, and elsewhere [11, 12].
RNA viruses have high mutation rates, allowing rapid adaptation to changing environments. Monitoring drug resistance development and impact on clinical outcomes is important to understand the benefits of direct-acting antiviral treatments. The target of remdesivir, the nonstructural protein 12 (Nsp12) RdRp, is highly conserved across coronaviruses, with near 100% identity in the enzyme active site [13, 14]. The low diversity and high genetic stability of the RNA replication complex suggests a minimal risk of preexisting SARS-CoV-2 resistance to remdesivir [15].
The most common Nsp12 substitution relative to ancestral SARS-CoV-2 observed in variants, such as Delta and Omicron, is P323L [16–18]. Phenotypic testing of a recombinant SARS-CoV-2 virus containing P323L showed no loss of remdesivir susceptibility; the 50% inhibition of virus replication (EC50) fold change was 0.95 versus wild type [18].
An in vitro resistance selection experiment with SARS-CoV-2 using the parent nucleoside analog of remdesivir, GS-441524, revealed emergent amino acid substitutions in Nsp12: V166A, N198S, S759A, V792I, C799F, and C799R [19]. Phenotypic testing of recombinant SARS-CoV-2 viruses containing the individual substitutions resulted in remdesivir EC50 fold changes from 1.7–3.3 versus wild-type SARS-CoV-2, indicating minimal to low-level reduced susceptibility [20]. In a second selection experiment using a SARS-CoV-2 isolate containing P323L, a single amino acid substitution (V166L) emerged. Phenotypic testing of recombinant SARS-CoV-2 viruses containing P323L alone or P323L + V166L resulted in remdesivir EC50 fold changes from 1.2–1.5 versus the reference strain, again indicating minimal to no loss of susceptibility [21]. Separately, another resistance selection study identified a single Nsp12 amino acid substitution, E802D, with low-level reduced susceptibility (2- to 6-fold change in remdesivir EC50) [22, 23].
ACTT-1, conducted early in the COVID-19 pandemic, was the pivotal randomized, placebo-controlled, phase 3 study demonstrating clinical benefit of remdesivir in hospitalized patients [7]. ACTT-1 demonstrated remdesivir's superiority to placebo in shortening time to recovery in adults hospitalized with COVID-19 (median, 10 vs 15 days). Here we conducted SARS-CoV-2 resistance analyses for ACTT-1 in the first description of such analyses conducted in a randomized placebo-controlled trial with remdesivir.
METHODS
Clinical Study
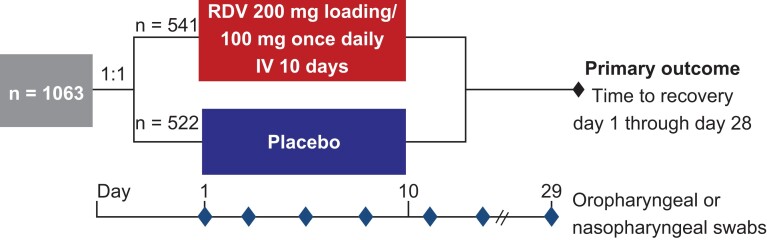
Details of ACTT-1 have been published previously [7]. Briefly, adults aged ≥18 years diagnosed with COVID-19 and hospitalized with evidence of lower respiratory tract involvement were randomized to receive either remdesivir (200 mg on day 1, followed by 100 mg/day for ≤9 days) or matching placebo for ≤10 days. The ACTT-1 protocol was approved by the institutional review board at each site (or by a centralized institutional review board as applicable) and was overseen by an independent data and safety monitoring board. Written informed consent was obtained from each patient or from the patient's legally authorized representative if the patient was unable to provide consent.
Oropharyngeal swabs were collected for virologic analyses from all participants at baseline (day 1) and longitudinally throughout the treatment and follow-up periods (days 3, 5, 8, 11, 15, and 29; Figure 1). Nasopharyngeal swabs were obtained where oropharyngeal swabs were not possible (eg, in intubated participants). Samples up to and including day 11 were collected while participants were hospitalized, and samples were collected on days 15 and 29 if participants could return to the clinic or remained hospitalized. Participants who received concomitant COVID-19 treatments during the study (including chloroquine, hydroxychloroquine, and lopinavir-ritonavir) were excluded.
Figure 1.
ACTT-1 study design. Abbreviations: ACTT-1, Adaptive COVID-19 Treatment Trial-1; IV, intravenous; RDV, remdesivir.
Virologic Resistance Analyses
Virologic resistance analyses aimed to determine whether amino acid substitutions in SARS-CoV-2 Nsp12 (RdRp) or in other parts of the replication-transcription complex (Nsp8, Nsp10, Nsp13, and Nsp14) emerged in participants, and whether identified substitutions altered susceptibility to remdesivir.
Sequencing analysis was conducted on samples from remdesivir-treated participants who were in the >80th percentile of cumulative viral shedding among study participants, as they might be at highest risk for resistance development. As a control, remdesivir-treated participants in who were in the <20th percentile of cumulative viral shedding and approximately half of the participants in the placebo arm were included. SARS-CoV-2 sequencing was limited to samples with >40 copies/polymerase chain reaction, which corresponds to the lower limit of detection for the sequencing assay (approximately 600 copies/mL). The whole genome of SARS-CoV-2 was amplified using the Swift Bioscience SARS-CoV-2 assay, and the nucleotide sequence was determined by Illumina NextSeq 500 or NextSeq 2000 (Illumina, Inc) at the University of Washington Virology Laboratory (Seattle, WA) [24, 25].
Sequencing data were received as FASTQ files (paired-end) that were split per sample and amplification pool. FASTQ files were aligned to hg38 reference using BWA version 0.7.15 [26] to exclude human RNA transcripts and to isolate viral reads for further processing. Viral reads were trimmed using Trimmomatic version 0.36 [27] for low quality (sliding window 4 bp, avg phred 15), and short reads (<50 base pairs) were filtered out. Reads were then aligned to the Wuhan-Hu-1 reference (NC_045512) using SMALT version 0.7.6 aligner (https://www.sanger.ac.uk/tool/smalt/). Base pairs from reads overlapping with primer regions were trimmed. Tabulated amino acid variants and insertion-deletion mutations (indels) were reported per genome position at frequency ≥15%, average phred score ≥20, and read depth ≥50. Consensus sequences were generated and included nucleotide mixtures when 1 base was present in ≥15% of the SARS-CoV-2 viral population and indels when present in ≥50% of the viral population. SARS-CoV-2 lineage was determined by Pangolin Software version 3.1.11 (pangoLEARN version 2021-08-09, https://github.com/cov-lineages/pangolin) using SARS-CoV-2 whole genome consensus sequences. A sequence coverage threshold of ≥70% of the SARS-CoV-2 coding region was used for lineage determination.
Amino acid substitutions in Nsp12 versus the reference sequence (Wuhan-Hu-1, NC_045512.2) that occurred in ≥3 participants at baseline were reported together with amino acid changes versus the baseline sequence. Postbaseline sequences were compared with participant-specific baseline sequences of the same sample type to determine if any amino acid substitutions emerged in Nsp12 during or after treatment. Substitutions detected as a mixture (15%–85%) at baseline that resolved to the consensus amino acid postbaseline were not considered to have emerged. Substitutions emerging in SARS-CoV-2 Nsp8, Nsp10, Nsp13, and Nsp14 were also reported as secondary analyses.
Phenotypic Analyses
Phenotypic analysis was attempted on postbaseline clinical isolates with identified treatment-emergent amino acid substitutions compared with their baseline sample in SARS-CoV-2 Nsp12.
Phenotyping was performed at the University of Washington Center for Innate Immunity and Immune Disease laboratories (Seattle, WA) and at Gilead Sciences (Foster City, CA). Details of the assays conducted are presented in the Supplementary Material. Briefly, SARS-CoV-2 from clinical isolates was propagated twice in Vero-TMPRSS2 cells. Remdesivir's antiviral activity against the clinical isolates was assessed at 44 hours postinfection and compared to its activity against the wild-type SARS-CoV-2 reference strain, WA1 (hCoV-19/USA-WA1/2020), using A549-hACE2-TMPRSS2 cells and antinucleoprotein enzyme-linked immunosorbent assay (ELISA). For each clinical isolate, experiments were performed twice with technical duplicates. Results of the phenotypic analysis were reported as fold change in the effective concentration of remdesivir to reach EC50 of the clinical isolates relative to the SARS-CoV-2 reference strain, WA1. SARS-CoV-2 whole-genome sequencing was performed using the same assay described previously, using the virus stocks to confirm homogeneity to the original virus.
Phenotyping of site-directed mutants (SDMs) of SARS-CoV-2 was conducted using an adapted replicon assay [28, 29]. Briefly, the 4 plasmids that encode SARS-CoV-2 genes for the nonstructural proteins and the nucleoprotein, with or without SDMs, were prepared. The 4 DNA fragments were isolated from SARS-CoV-2 SH01 (SARS-CoV-2/human/CHN/SH01/2020, GenBank MT121215), a plaque-purified Pango lineage B strain isolated from a patient in Shanghai. Huh7-1CN cells were mixed with RNA and immediately electroporated followed by luciferase assay at 48 hours postelectroporation. Relative luciferase signals were calculated by normalizing the luciferase signals of the compound-treated groups to that of the control (dimethylsulfoxide [DMSO]) groups (set as 100%). EC50 values were calculated using a nonlinear 4-parameter variable slope regression model as the concentration at which there was a 50% decrease in the luciferase reporter signal relative to DMSO vehicle alone (0% virus inhibition) and uninfected control culture (100% virus inhibition). Two experiments were performed with technical triplicates. Fold-change values were calculated by dividing the variant mean EC50 by the SH01 reference strain mean EC50.
Replication capacity was calculated by dividing the luciferase signal of the mutant replicon by the wild-type replicon at 48 hours posttransfection or by comparing the growth kinetics of the mutant in the recombinant virus to the wild type in A549-hACE2-TMPRSS2 cells at 48 hours postinfection using a plaque assay readout in TMPRSS2-expressing Vero cells. Student t test was performed to compare mutant and wild-type means.
RESULTS
Participants
Of 1048 participants enrolled, 94 in the remdesivir arm and 79 in the placebo arm met the resistance analysis criteria. In the remdesivir arm, this included 58 participants with >80th percentile cumulative viral shedding and 36 with <20th percentile viral shedding. In the placebo arm, this included 52 with >80th percentile viral shedding and 27 with <20th percentile viral shedding. For the postbaseline virologic analysis, 31 of 94 remdesivir-treated participants who qualified for sequencing (33.0%) had both baseline and postbaseline sequencing data available. In the placebo arm, 30 of 79 participants (38.0%) who qualified for sequencing had both baseline and postbaseline sequencing data available (Table 1). Consensus sequences were generated including substitutions observed at ≥15% of the SARS-CoV-2 viral population.
Table 1.
Number of Participants with Sequencing Data in the As-Treated Population
| Mild/Moderate Disease | Severe Disease | Any Disease Severity | |||||
|---|---|---|---|---|---|---|---|
| RDV | Placebo | RDV | Placebo | RDV | Placebo | Total | |
| As-treated population | 55 | 49 | 477 | 467 | 532 | 516 | 1048 |
| Met resistance analysis criteria and sequencing attempteda | 9 | 5 | 85 | 74 | 94 | 79 | 173 |
| Sequencing data available | |||||||
| Baseline | 5 | 3 | 42 | 41 | 47 | 44 | 91 |
| Postbaseline | 5 | 2 | 31 | 32 | 36 | 34 | 70 |
| Baseline + postbaseline | 4 | 2 | 27 | 28 | 31 | 30 | 61 |
Abbreviations: COVID-19, coronavirus disease 2019; RDV, remdesivir.
aSome participants had no sequencing data available due to sample missing at baseline or postbaseline (n = 2), viral load below lower limit of detection of sequencing assay (n = 29), exclusion due to concomitant COVID-19 medication (chloroquine/hydroxychloroquine, n = 74), and assay failure (n = 7).
Baseline Sequence Characteristics
Among the 91 participants with available sequencing data, the most common SARS-CoV-2 lineages (≥3 participants) included B.1 (n = 33, 36.3%), B.1.333 (n = 9, 9.9%), and A.1 (n = 7, 7.7%; Table 2). Of the 91 participants with baseline sequencing data, P323L was the only amino acid substitution identified in ≥3 participants. It was observed in 70 participants (76.9%), including 35 of 47 participants (74.5%) treated with remdesivir and 35 of 44 participants (79.5%) treated with placebo. Phenotyping of Nsp12 P323L showed no loss of susceptibility to remdesivir [18].
Table 2.
Baseline Sequencing Data
| Participants, No. (% out of Participants With Baseline Sequencing Data Available)a | |||||||
|---|---|---|---|---|---|---|---|
| Mild/Moderate Disease | Severe Disease | Any Disease Severity | |||||
| RDV | Placebo | RDV | Placebo | RDV | Placebo | Total | |
| Participants with baseline sequencing datab | 5 | 3 | 42 | 41 | 47 | 44 | 91 |
| B.1 | 0 | 1 (33.3) | 13 (31.0) | 19 (46.3) | 13 (27.7) | 20 (45.5) | 33 (36.3) |
| B.1.333 | 0 | 0 | 4 (9.5) | 5 (12.2) | 4 (8.5) | 5 (11.4) | 9 (9.9) |
| A.1 | 1 (20.0) | 0 | 3 (7.1) | 3 (7.3) | 4 (8.5) | 3 (6.8) | 7 (7.7) |
| A | 1 (20.0) | 0 | 3 (7.1) | 2 (4.9) | 4 (8.5) | 2 (4.5) | 6 (6.6) |
| B.1.320 | 0 | 0 | 1 (2.4) | 2 (4.9) | 1 (2.1) | 2 (4.5) | 3 (3.3) |
| B.1.605 | 1 (20.0) | 0 | 1 (2.4) | 1 (2.4) | 2 (4.3) | 1 (2.3) | 3 (3.3) |
| Otherc | 2 (40.0) | 2 (66.7) | 17 (40.5) | 9 (22.0) | 19 (40.4) | 11 (25.0) | 30 (33.0) |
Abbreviations: RDV, remdesivir; SpO2, peripheral oxygen saturation.
aSevere disease was defined as requiring mechanical ventilation, requiring oxygen, SpO2 ≤94% on room air, or tachypnea (respiratory rate ≥24 breaths/minute). Mild/moderate disease was defined as having SpO2 >94% and respiratory rate <24 breaths/minute without supplemental oxygen.
bThe analysis included participants with viral shedding above the 80th and below the 20th percentiles.
cLineages observed in <3 participants included A.3, B.1.243, and B.1.540 observed in 2 participants each; and A.2.2, B, B.1.1, B.1.1.172, B.1.1.236, B.1.108, B.1.110, B.1.110.3, B.1.118, B.1.163, B.1.302, B.1.323, B.1.330, B.1.428, B.1.450, B.1.479, B.1.564, B.1.577, B.1.610, B.4.7, B.41, B.46, and B.57 observed in 1 participant each.
Emerging Substitutions in Nsp12
Emerging substitutions in Nsp12 were observed in 2.3% (24/1048) of total study participants in both the remdesivir (12/532) and placebo (12/516) arms. Among participants with baseline and postbaseline sequencing data, across any disease severity, emerging substitutions in Nsp12 were observed in 12 of 31 (38.7%) and 12 of 30 participants (40.0%) in the remdesivir and placebo arm, respectively (Table 3). This included emergent substitutions in 2 of 4 participants (50.0%) treated with remdesivir and 1 of 2 participants (50.0%) treated with placebo with mild or moderate disease and 10 of 27 participants (37.0%) treated with remdesivir and 11 of 28 participants (39.3%) treated with placebo with severe disease (Table 3). In the remdesivir arm, no emergent Nsp12 substitutions were observed in more than 1 participant and most were present as mixtures with wild-type SARS-CoV-2. Overall, across any disease severity, the emergent Nsp12 substitutions in the remdesivir arm were observed in 11 of 29 participants (37.9%) with >80th percentile of cumulative viral shedding and 1 of 2 participants (50.0%) with <20th percentile of cumulative viral shedding.
Table 3.
Amino Acid Substitutions in Nsp12 Detected Postbaseline
| Participants, No. (% out of Participants With Baseline and Postbaseline Sequencing Data Available)a | |||||||
|---|---|---|---|---|---|---|---|
| Mild/Moderate Disease | Severe Disease | Any Disease Severity | Total | ||||
| RDV | Placebo | RDV | Placebo | RDV | Placebo | ||
| Participants with baseline and postbaseline sequencing datab | 4 | 2 | 27 | 28 | 31 | 30 | 61 |
| No substitution | 2 (50.0) | 1 (50.0) | 17 (63.0) | 18 (64.3) | 19 (61.3) | 18 (60.0) | 37 (60.7) |
| Any substitution | 2 (50.0) | 1 (50.0) | 10 (37.0) | 10 (35.7) | 12 (38.7) | 12 (40.0) | 24 (39.3) |
| A16V | 1 (25.0) | 0 | 0 | 0 | 1 (3.2) | 0 | 1 (1.6) |
| T20T/1 | 0 | 0 | 0 | 1 (3.6) | 0 | 1 (3.3) | 1 (1.6) |
| G44G/V | 0 | 0 | 1 (3.7) | 0 | 1 (3.2) | 0 | 1 (1.6) |
| F56F/S | 0 | 0 | 1 (3.7) | 0 | 1 (3.2) | 0 | 1 (1.6) |
| K59K/N | 0 | 0 | 1 (3.7) | 0 | 1 (3.2) | 0 | 1 (1.6) |
| D60D/H | 0 | 0 | 1 (3.7) | 0 | 1 (3.2) | 0 | 1 (1.6) |
| A97A/S | 0 | 1 (50.0) | 0 | 0 | 0 | 1 (3.3) | 1 (1.6) |
| A125A/T | 0 | 0 | 1 (3.7) | 0 | 1 (3.2) | 0 | 1 (1.6) |
| A176A/V | 0 | 0 | 1 (3.7) | 0 | 1 (3.2) | 0 | 1 (1.6) |
| P227P/Lc | 1 (25.0) | 0 | 0 | 1 (3.6) | 1 (3.2) | 1 (3.3) | 2 (3.3) |
| D291D/N | 0 | 0 | 0 | 1 (3.6) | 0 | 1 (3.3) | 1 (1.6) |
| A311A/E | 0 | 0 | 1 (3.7) | 0 | 1 (3.2) | 0 | 1 (1.6) |
| A311A/T | 0 | 0 | 0 | 1 (3.6) | 0 | 1 (3.3) | 1 (1.6) |
| V315V/F | 0 | 1 (50.0) | 0 | 0 | 0 | 1 (3.3) | 1 (1.6) |
| P323P/L | 0 | 0 | 1 (3.7) | 0 | 1 (3.2) | 0 | 1 (1.6) |
| T324T/I | 0 | 0 | 0 | 1 (3.6) | 0 | 1 (3.3) | 1 (1.6) |
| G345G/V | 0 | 1 (50.0) | 0 | 0 | 0 | 1 (3.3) | 1 (1.6) |
| V359V/I | 0 | 0 | 1 (3.7)d | 0 | 1 (3.2)d | 0 | 1 (1.6) |
| A382A/V | 0 | 0 | 0 | 1 (3.6) | 0 | 1 (3.3) | 1 (1.6) |
| N403N/I | 0 | 1 (50.0) | 0 | 0 | 0 | 1 (3.3) | 1 (1.6) |
| A423A/V | 0 | 0 | 0 | 1 (3.6) | 0 | 1 (3.3) | 1 (1.6) |
| S425S/P | 0 | 0 | 1 (3.7) | 0 | 1 (3.2) | 0 | 1 (1.6) |
| G427G/V | 0 | 0 | 1 (3.7) | 0 | 1 (3.2) | 0 | 1 (1.6) |
| Q444Q/H | 0 | 0 | 0 | 1 (3.6) | 0 | 1 (3.3) | 1 (1.6) |
| Y483Y/H | 0 | 0 | 0 | 1 (3.6) | 0 | 1 (3.3) | 1 (1.6) |
| S501L | 0 | 0 | 0 | 1 (3.6) | 0 | 1 (3.3) | 1 (1.6) |
| Q524Q/* | 1 (25.0) | 0 | 0 | 0 | 1 (3.2) | 0 | 1 (1.6) |
| T567T/I | 0 | 0 | 1 (3.7) | 0 | 1 (3.2) | 0 | 1 (1.6) |
| D684D/N | 1 (25.0) | 0 | 0 | 0 | 1 (3.2) | 0 | 1 (1.6) |
| T701T/M | 0 | 0 | 0 | 1 (3.6) | 0 | 1 (3.3) | 1 (1.6) |
| L707L/F | 0 | 0 | 1 (3.7) | 0 | 1 (3.2) | 0 | 1 (1.6) |
| Y719Y/C | 0 | 0 | 0 | 1 (3.6) | 0 | 1 (3.3) | 1 (1.6) |
| R733R/I | 0 | 0 | 1 (3.7) | 0 | 1 (3.2) | 0 | 1 (1.6) |
| V764V/L | 0 | 0 | 1 (3.7) | 0 | 1 (3.2) | 0 | 1 (1.6) |
| A771A/P | 0 | 0 | 1 (3.7) | 0 | 1 (3.2) | 0 | 1 (1.6) |
| V792I | 0 | 0 | 1 (3.7) | 0 | 1 (3.2) | 0 | 1 (1.6) |
| C799C/F | 0 | 0 | 1 (3.7) | 0 | 1 (3.2) | 0 | 1 (1.6) |
| G823G/C | 0 | 0 | 1 (3.7) | 0 | 1 (3.2) | 0 | 1 (1.6) |
| P830P/S | 0 | 0 | 0 | 1 (3.6) | 0 | 1 (3.3) | 1 (1.6) |
| P832P/S | 0 | 0 | 0 | 1 (3.6) | 0 | 1 (3.3) | 1 (1.6) |
| F843F/L | 0 | 0 | 1 (3.7) | 0 | 1 (3.2) | 0 | 1 (1.6) |
Abbreviations: RDV, remdesivir; SpO2, peripheral oxygen saturation.
aSevere disease was defined as requiring mechanical ventilation, requiring oxygen, SpO2 ≤94% on room air, or tachypnea (respiratory rate ≥24 breaths/minute). Mild/moderate disease was defined as having SpO2 >94% and respiratory rate <24 breaths/minute without supplemental oxygen.
bThis table lists Nsp12 substitutions emerged in the RDV and placebo arms with >80th percentile of cumulative viral shedding and <20th percentile of cumulative viral shedding.
cP227L was observed as a full mutant in 1 participant in the placebo arm and as a mixture with wild type in 1 participant in the RDV arm.
dIn the RDV arm, V359V/I and L707L/F were observed in 1 participant with <20th percentile of cumulative viral shedding. The other substitutions in the RDV arm were observed in participants with >80th percentile of cumulative viral shedding.
Clinical Outcomes
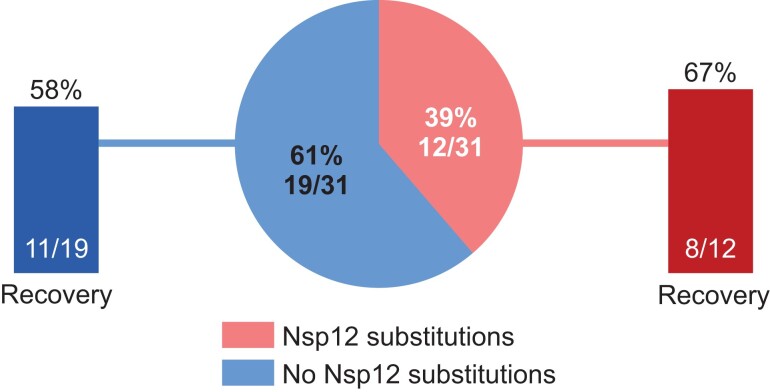
In ACTT-1, emergent Nsp12 substitutions in the remdesivir arm did not impact clinical recovery by day 28: 8 of 12 participants (66.7%) with emergent Nsp12 substitutions recovered and 11 of 19 participants (57.9%) without emergent Nsp12 substitutions recovered (Figure 2).
Figure 2.
Clinical recovery in participants treated with remdesivir with or without emergent Nsp12 amino acid substitutions. Recovery was defined as either discharged from the hospital or hospitalized but not requiring supplemental oxygen and no longer requiring ongoing medical care at day 28.
Phenotypic Analyses
Phenotypic analysis was attempted on postbaseline clinical isolates from 11 of 12 participants in the remdesivir arm and 11 of 12 participants in the placebo arm (phenotypic analysis was not attempted on the clinical isolate with the P323P/L substitution because it does not confer reduced susceptibility to remdesivir in vitro [18]). Viable virus was cultured from 3 participants treated with remdesivir; no viable virus could be cultured from those receiving placebo. In the participant with the emergent Nsp12 A16V substitution at day 29, the EC50 fold change versus the wild-type SARS-CoV-2 strain was 0.75 (Table 4), suggesting no impact on remdesivir susceptibility. Presence of the emergent C799F substitution in 1 participant at day 11 (1 day after remdesivir treatment cessation) was associated with a 2.51-fold change in EC50 versus wild type. This participant had COVID-19 symptoms for 23 days prior to remdesivir treatment initiation and an ordinal scale clinical status of 5 at baseline (hospitalized, requiring supplemental oxygen), with no improvement in clinical status over time, and died at day 15. In another participant, emergent K59N and V792I substitutions in Nsp12 were observed at day 10 (last day of remdesivir treatment); at baseline, this participant had an ordinal scale clinical status of 5 (hospitalized, requiring supplemental oxygen) and recovered to clinical status of 3 (hospitalized, not requiring supplemental oxygen, no longer requires ongoing medical care) at day 22. This participant had COVID-19 symptoms for 5 days prior to remdesivir treatment initiation. Culturing of the virus from this participant was attempted twice and, in both cultures, K59N reverted to wild type. Phenotyping for remdesivir susceptibility was conducted on the cultured virus containing V792I, showing a 2.17-fold change in EC50. Further testing of K59N and V792I was conducted using replicons containing SDMs in Huh7-1CN cells. The K59N mutant was associated with a 0.98-fold change in EC50 versus wild type, suggesting no reduction in susceptibility to remdesivir. The K59N + V792I mutant was associated with a 3.41-fold change in EC50, suggesting low-level reduced susceptibility similar to the V792I mutant alone (3.15-fold change; Table 4). These results indicate low-level reduced susceptibility of C799F and V792I to remdesivir in vitro. The V792I and C799F substitutions are rare in the Global Initiative on Sharing All Influenza Data (GISAID) database (0.004% [549/15 000 071] and 0.001% [155/15 000 071] of SARS-CoV-2 sequences, respectively, as of 6 April 2023). The replicon containing the V792I substitution had a replication capacity of 9.1% compared with the wild-type replicon. The mutant recombinant virus with the C799F substitution grew to 0.5% of wild type at 48 hours postinfection (Supplementary Figure 1). These results indicate that these substitutions were associated with reduced fitness.
Table 4.
Remdesivir EC50 Phenotyping Against Clinical Isolates With Emergent Nsp12 Amino Acid Substitutions and SDMs
| Clinical Isolates | Remdesivir EC50 (μM) | EC50 Fold Change (SD) From Reference | ||
|---|---|---|---|---|
| Substitution | First Replicate | Second Replicate | Mean (SD) | |
| Clinical isolates (frequency of substitution, %) | ||||
| Wild-type SARS-CoV-2 (WA1)a | 0.26 | 0.21 | 0.24 (0.04) | 1.0 |
| A16V (88.6%) | 0.19 | 0.16 | 0.18 (0.02) | 0.8 (0.05) |
| C799F (22.4%) | 0.60 | 0.57 | 0.58 (0.03) | 2.5 (0.30) |
| V792I (99%)b | 0.50 | 0.50 | 0.50 (0.0) | 2.2 (0.36) |
| S759A + V792I positive controlc | 2.82 | 2.49 | 2.66 (0.23) | 11.4 (0.90) |
| SDMs | ||||
| Wild type, SH01d | 15.37 | 13.87 | 14.62 (1.06) | 1.0 |
| K59N | 14.7 | 14.04 | 14.37 (0.47) | 1.0 (0.04) |
| K59N + V792I | 49.82 | 49.61 | 49.72 (0.15) | 3.4 (0.24) |
| V792I | 45.9 | 46.01 | 45.96 (0.08) | 3.2 (0.23) |
| D684N | No replication | No replication | NA | NA |
| V764L | No replication | No replication | NA | NA |
Abbreviations: EC50, half maximal effective concentration; NA, not applicable; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; SD, standard deviation; SDM, site-directed mutant.
aWA1: clinical isolate with wild-type reference SARS-CoV-2 lineage A strain from January 2020 in Washington State.
bK59N and V792I substitutions in Nsp12 emerged at day 10 in participant COV.01517. P323L in Nsp12 was observed at baseline and at all time points sequenced including day 10. Culturing of the virus was attempted twice and, in both cultures, K59N reverted to wild type. Phenotyping was conducted on the cultured virus containing P323L and V792I.
cThe positive control used for these experiments contained the Nsp12 S759A and V792I substitutions and was generated using recombinant SARS-CoV-2 virus. These substitutions were previously observed in an in vitro resistance selection experiment using GS-441524 (the parent nucleoside of remdesivir [21]).
dSH01: wild-type reference SARS-CoV-2 replicon generated from clinical isolate from Shanghai (lineage B).
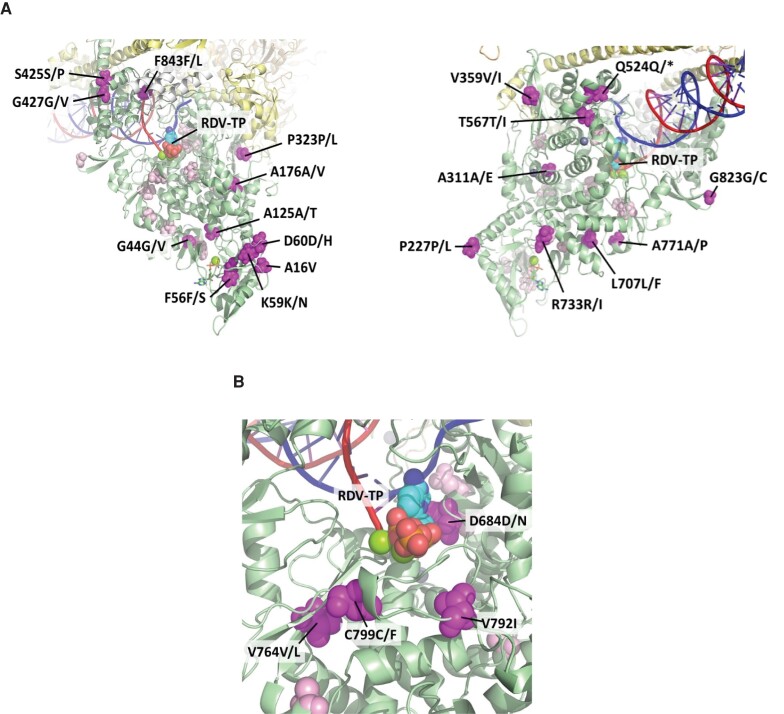
Based on analysis of prior cryoelectron microscopy (Cryo-EM) structures of the SARS-CoV-2 polymerase complex [30], most emerging Nsp12 substitutions in the remdesivir arm were located on the surface of the protein (Figure 3), distant from the polymerase active site and the viral RNA. Emerging substitutions D684D/N, V764V/L, V792I, and C799C/F are closer to the active site (within 15 Å); however, none have a direct interaction with the RNA or the incoming nucleotide.
Figure 3.
Emergent Nsp12 amino acid substitutions in participants treated with remdesivir relatively (A) distant and (B) nearer to the RdRp active site or RNA. The structure is a model of RDV-TP incorporation in the SARS-CoV-2 polymerase active site based on the 6XEZ Cryo-EM structure [30]. A, Map of the observed postbaseline amino acid substitutions on Cryo-EM structure of SARS-CoV-2 polymerase complex in rotated view. Full Nsp12 protein (in green) with the locations of the observed postbaseline amino acid substitutions located in this view of the structure are shown in magenta. For context, all substitutions not highlighted in this particular view are shown in light pink. White is Nsp7, yellow is Nsp8 (2 subunits), and orange is Nsp13 (2 subunits). The template RNA strand is shown in blue and the nascent RNA strand is shown in red. B, Map of observed postbaseline amino acid substitutions closest to the active site of Nsp12 and on Cryo-EM structure of the SARS-CoV-2 polymerase complex. Abbreviations: Cryo-EM, cryoelectron microscopy; RdRp, RNA-dependent RNA polymerase; RDV-TP, triphosphate form of remdesivir; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Additional phenotyping of SDMs was performed using the replicon system in Huh7-1CN cells for Nsp12 substitutions observed in the remdesivir arm that were determined to be closest to the active site based on Cryo-EM modeling. This included D684N and V764L, which were observed in 1 participant each (Table 4). Transfection of the mutant replicons was attempted twice; however, phenotypic results were not generated due to lack of replication.
Emerging Substitutions in Nsp8, Nsp10, Nsp13, and Nsp14
Of the 31 participants in the remdesivir arm with both baseline and postbaseline sequencing data available, emergent substitutions in Nsp8, Nsp10, Nsp13, or Nsp14 were observed in 3 (9.7%), 2 (6.5%), 11 (35.5%), and 7 (22.6%) participants with any disease severity, respectively (Supplementary Table 1). Most substitutions occurred as a mixture with wild-type SARS-CoV-2, and none were observed in ≥1 participant. Of the 30 participants in the placebo arm with baseline and postbaseline sequencing data, substitutions in Nsp8, Nsp13, or Nsp14 were observed in 1 (3.3%), 10 (33.3%), and 6 (20.0%) participants, respectively. No substitutions were observed in Nsp10 in the placebo arm.
DISCUSSION
ACTT-1 was a pivotal, randomized, placebo-controlled, phase 3 study that demonstrated clinical benefit of remdesivir in hospitalized patients [7]. The resistance analyses reported here are the first published from a randomized placebo-controlled trial with remdesivir. From the sequencing analyses of oropharyngeal or nasopharyngeal swabs, a similar rate of emergent amino acid substitutions in Nsp12 and other proteins of the replication/transcription complex was observed in participants treated with remdesivir or placebo. This suggests that most substitutions were due to natural viral evolution and not related to remdesivir. These substitutions could be due to adaptation of the virus to evade the immune response [31–34].
In vitro resistance selection experiments with SARS-CoV-2 have demonstrated a high barrier to developing remdesivir resistance, with high passage numbers required to develop substitutions that have low impact on susceptibility [18, 19, 21, 22]. The viruses emerging from selection experiments displayed impaired replication, suggesting that development of resistance may result in a significant fitness cost [19]. Importantly, in this analysis, only 2 participants had the emergence of substitutions associated with low-level reduced remdesivir susceptibility (V792I and C799F; ≤3.4-fold change in EC50 compared with wild type and associated with reduced fitness). In these participants, the substitutions emerged either on the last day of treatment (day 10) or 1 day after treatment cessation (day 11). It is unclear whether these changes may meaningfully impact clinical outcomes given the low fold change values. Notably, clinical recovery was similar between participants with and without emergent Nsp12 substitutions in the remdesivir arm, suggesting minimal impact on clinical outcome.
The risk of drug resistant virus transmission is likely low because SARS-CoV-2 viral RNA typically declines to undetectable levels after 2–3 weeks [35, 36], preceded by an even more rapid reduction in infectious viral titer [37]. However, rare cases of prolonged shedding have been described in older or immunocompromised patients [38–40]. Fortunately, surveillance of Nsp12 substitutions continues to demonstrate high sequence conservation [14–16, 41], and the prevalence of substitutions associated with remdesivir resistance is exceedingly low [15, 20, 23, 42]. It will be important to continue to monitor substitutions as new variants emerge. Similarly, continued monitoring of resistance substitutions to antivirals targeting SARS-CoV-2 proteins other than Nsp12, which may have a lower barrier to resistance development compared with remdesivir, are also warranted. For example, in vitro resistance selection experiments using the oral protease inhibitor nirmatrelvir revealed that Nsp5 L50F, E166A, and L167F mutations conferred up to 80-fold resistance, although these mutations were exceedingly rare and were associated with a fitness cost [43]. Additional in vitro resistance selection studies found that E166V in Nsp5 conferred high-level resistance to nirmatrelvir with a substantial fitness cost; however, compensatory mutations arose, including T21I and L50F, that restored fitness while maintaining resistance [43, 44]. Several other hot spots for drug resistance mutations in Nsp5 have been identified from clinical isolates [45].
ACTT-1 enrolled participants early in the pandemic, between February 2020 and May 2020. At baseline, the only amino acid substitution in Nsp12 observed was P323L (76.9% of baseline sequences), consistent with the SARS-CoV-2 variants circulating at the time [16]. The most common lineage among the participants in our study was B.1; in the intervening period, SARS-CoV-2 has evolved greatly and multiple novel variants have emerged [46]. Currently, P323L, which does not confer reduced susceptibility to remdesivir, is present in most circulating variants and remains the most common defining substitution in Nsp12 [18, 21].
SARS-CoV-2 variants of concern primarily exhibit changes in the spike protein, which mediates virus entry and is the target of therapeutic monoclonal antibodies. Increasing evidence shows that therapeutic monoclonal antibodies lose efficacy against Omicron variants, such that there are no US Food and Drug Administration–authorized therapeutic monoclonal antibodies for treatment of COVID-19 at the time of writing [33, 47–49]. Remdesivir's inhibition of viral RNA synthesis has been demonstrated biochemically to occur via 2 mechanisms. Following incorporation of its triphosphate metabolite into nascent RNA, remdesivir's 1′-CN substitution clashes with the Nsp12 protein, causing either delayed chain termination [13] or template-dependent inhibition [50]. Thus, the 2 potential mechanisms for resistance to develop would be for the rate of incorporation of the inhibitor to be reduced or for the subsequent clashes to be alleviated. Remdesivir retains comparable potency against Delta and Omicron, as well as earlier variants, compared with the wild-type reference strain [18], which is anticipated to be maintained for future variants due to the low evolutionary rate of the RdRp.
Limitations to this study include that ACTT-1 was implemented 3 weeks before the World Health Organization declared COVID-19 a global pandemic and completed enrollment in 60 days. Logistical issues hindered consistent sample collection due to supply shortages (including personal protective equipment, swabs, and viral transport media) and sample shipping restrictions. These factors led to more missing samples than might be expected under normal circumstances, and few samples were obtained after participants were discharged from the hospital. Additionally, only 30% of participants who met the viral load criteria were successfully sequenced, and fewer had culturable virus. This was likely because ACTT-1 enrolled hospitalized patients early in the pandemic; therefore, patients were enrolled at a later stage in the disease and had relatively low viral loads. The population size of the virus when exposed to remdesivir was less than may be seen in an outpatient setting.
In conclusion, virologic analyses showed a similar rate of emergent Nsp12 substitutions in participants treated with remdesivir compared with placebo. These were associated with low to no change in remdesivir susceptibility among the treatment-emergent Nsp12 substitutions, supporting a high barrier to remdesivir resistance development in COVID-19 patients.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Contributor Information
Charlotte Hedskog, Gilead Sciences, Inc, Foster City, California, USA.
Lauren Rodriguez, Gilead Sciences, Inc, Foster City, California, USA.
Pavitra Roychoudhury, Center for Innate Immunity and Immune Disease, Department of Immunology, University of Washington, Seattle, Washington, USA; Fred Hutchinson Cancer Center, Seattle, Washington, USA.
Meei-Li Huang, Center for Innate Immunity and Immune Disease, Department of Immunology, University of Washington, Seattle, Washington, USA.
Keith R Jerome, Center for Innate Immunity and Immune Disease, Department of Immunology, University of Washington, Seattle, Washington, USA; Fred Hutchinson Cancer Center, Seattle, Washington, USA.
Linhui Hao, Center for Innate Immunity and Immune Disease, Department of Immunology, University of Washington, Seattle, Washington, USA.
Renee C Ireton, Center for Innate Immunity and Immune Disease, Department of Immunology, University of Washington, Seattle, Washington, USA.
Jiani Li, Gilead Sciences, Inc, Foster City, California, USA.
Jason K Perry, Gilead Sciences, Inc, Foster City, California, USA.
Dong Han, Gilead Sciences, Inc, Foster City, California, USA.
Gregory Camus, Gilead Sciences, Inc, Foster City, California, USA.
Alexander L Greninger, Center for Innate Immunity and Immune Disease, Department of Immunology, University of Washington, Seattle, Washington, USA; Fred Hutchinson Cancer Center, Seattle, Washington, USA.
Michael Gale, Jr, Center for Innate Immunity and Immune Disease, Department of Immunology, University of Washington, Seattle, Washington, USA.
Danielle P Porter, Gilead Sciences, Inc, Foster City, California, USA.
Notes
Acknowledgments. Medical writing and editorial support were provided by Michelle Preston, MSc, of Lumanity Communications Inc., and were funded by Gilead Sciences, Inc. We thank Tomas Cihlar and Romas Geleziunas for critical review of the manuscript.
Disclaimer . The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Financial support. This work was funded by Gilead Sciences, Inc and supported by the Infectious Diseases Clinical Research Consortium through the National Institute for Allergy and Infectious Diseases of the National Institutes of Health (grant number UM1AI148684). Funding to pay the Open Access publication charges for this article was provided by Gilead Sciences, Inc.
Potential conflicts of interest. C. H., L. R., J. L., J. K. P., D. H., and D. P. P. are employees and shareholders of Gilead Sciences, Inc. A. L. G. reports contract testing from Abbott, Cepheid, Novavax, Pfizer, Janssen, and Hologic and research support from Gilead outside of the described work. M. G. is a consultant for Merck, Biogen, and HDT Bio. G. C. is a shareholder and former employee of Gilead Sciences, Inc and a current employee of Vir Biotechnology. All other authors report no potential conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Lo MK, Jordan R, Arvey A, et al. . GS-5734 and its parent nucleoside analog inhibit filo-, pneumo-, and paramyxoviruses. Sci Rep 2017; 7:43395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sheahan TP, Sims AC, Graham RL, et al. . Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci Transl Med 2017; 9:eaal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brown AJ, Won JJ, Graham RL, et al. . Broad spectrum antiviral remdesivir inhibits human endemic and zoonotic deltacoronaviruses with a highly divergent RNA dependent RNA polymerase. Antiviral Res 2019; 169:104541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang M, Cao R, Zhang L, et al. . Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res 2020; 30:269–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pruijssers AJ, George AS, Schafer A, et al. . Remdesivir inhibits SARS-CoV-2 in human lung cells and chimeric SARS-CoV expressing the SARS-CoV-2 RNA polymerase in mice. Cell Rep 2020; 32:107940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Williamson BN, Feldmann F, Schwarz B, et al. . Clinical benefit of remdesivir in rhesus macaques infected with SARS-CoV-2. Nature 2020; 585:273–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Beigel JH, Tomashek KM, Dodd LE, et al. . Remdesivir for the treatment of Covid-19—final report. N Engl J Med 2020; 383:1813–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Spinner CD, Gottlieb RL, Criner GJ, et al. . Effect of remdesivir vs standard care on clinical status at 11 days in patients with moderate COVID-19: a randomized clinical trial. JAMA 2020; 324:1048–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Goldman JD, Lye DCB, Hui DS, et al. . Remdesivir for 5 or 10 days in patients with severe Covid-19. N Engl J Med 2020; 383:1827–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gottlieb RL, Vaca CE, Paredes R, et al. . Early remdesivir to prevent progression to severe Covid-19 in outpatients. N Engl J Med 2022; 386:305–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. VEKLURY (remdesivir) [prescribing information]. Forest City, CA: Gilead Sciences, Inc, 2022. [Google Scholar]
- 12. Gilead Sciences, Ltd. Veklury 100 mg powder for concentrate for solution for infusion [summary of product characteristics]. London, UK: Gilead Sciences, Ltd, 2022. [Google Scholar]
- 13. Gordon CJ, Tchesnokov EP, Woolner E, et al. . Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J Biol Chem 2020; 295:6785–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shannon A, Le NT, Selisko B, et al. . Remdesivir and SARS-CoV-2: structural requirements at both nsp12 RdRp and nsp14 exonuclease active-sites. Antiviral Res 2020; 178:104793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Martin R, Li J, Parvangada A, et al. . Genetic conservation of SARS-CoV-2 RNA replication complex in globally circulating isolates and recently emerged variants from humans and minks suggests minimal pre-existing resistance to remdesivir. Antiviral Res 2021; 188:105033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Showers WM, Leach SM, Kechris K, Strong M. Longitudinal analysis of SARS-CoV-2 spike and RNA-dependent RNA polymerase protein sequences reveals the emergence and geographic distribution of diverse mutations. Infect Genet Evol 2022; 97:105153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tsuchiya K, Yamamoto N, Hosaka Y, et al. . Molecular characterization of SARS-CoV-2 detected in Tokyo, Japan during five waves: identification of the amino acid substitutions associated with transmissibility and severity. Front Microbiol 2022; 13:912061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pitts J, Li J, Perry JK, et al. . Remdesivir and GS-441524 retain antiviral activity against Delta, Omicron, and other emergent SARS-CoV-2 variants. Antimicrob Agents Chemother 2022; 66:e00222-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stevens LJ, Pruijssers AJ, Lee HW, et al. . Mutations in the SARS-CoV-2 RNA-dependent RNA polymerase confer resistance to remdesivir by distinct mechanisms. Sci Transl Med 2022; 14:eabo0718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Du Pont V, Stevens L, Pruijssers A, et al. . Characterization of the susceptibility of specific SARS-CoV-2 nsp12 substitutions to remdesivir using a coronavirus reverse genetic system. Poster 160 V. 35th International Conference on Antiviral Research; 21–25 March 2022; Seattle, WA. [Google Scholar]
- 21. Checkmahomed L, Carbonneau J, Pont VD, et al. . In vitro selection of remdesivir-resistant SARS-CoV-2 demonstrates high barrier to resistance. Antimicrob Agents Chemother 2022; 66:e00198-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Szemiel AM, Merits A, Orton RJ, et al. . In vitro selection of remdesivir resistance suggests evolutionary predictability of SARS-CoV-2. PLoS Pathog 2021; 17:e1009929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gandhi S, Klein J, Robertson AJ, et al. . De novo emergence of a remdesivir resistance mutation during treatment of persistent SARS-CoV-2 infection in an immunocompromised patient: a case report. Nat Commun 2022; 13:1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shrestha L, Lin MJ, Xie H, et al. . Clinical performance characteristics of the Swift Normalase Amplicon Panel for sensitive recovery of severe acute respiratory syndrome coronavirus 2 genomes. J Mol Diagn 2022; 24:963–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Addetia A, Lin MJ, Peddu V, Roychoudhury P, Jerome KR, Greninger AL. Sensitive recovery of complete SARS-CoV-2 genomes from clinical samples by use of Swift Biosciences’ SARS-CoV-2 multiplex amplicon sequencing panel. J Clin Microbiol 2020; 59:e02226-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010; 26:589–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014; 30:2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang Y, Song W, Chen S, Yuan Z, Yi Z. A bacterial artificial chromosome (BAC)-vectored noninfectious replicon of SARS-CoV-2. Antiviral Res 2021; 185:104974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xie X, Muruato AE, Zhang X, et al. . A nanoluciferase SARS-CoV-2 for rapid neutralization testing and screening of anti-infective drugs for COVID-19. Nat Commun 2020; 11:5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen J, Malone B, Llewellyn E, et al. . Structural basis for helicase-polymerase coupling in the SARS-CoV-2 replication-transcription complex. Cell 2020; 182:1560–73.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gupta A M, Mandal S, Mandal S, Chakrabarti J. Immune escape facilitation by mutations of epitope residues in RdRp of SARS-CoV-2. J Biomol Struct Dyn 2022; 41:3542–52. [DOI] [PubMed] [Google Scholar]
- 32. Kumar S, Kumari K, Azad GK. Emerging genetic diversity of SARS-CoV-2 RNA dependent RNA polymerase (RdRp) alters its B-cell epitopes. Biologicals 2022; 75:29–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Harvey WT, Carabelli AM, Jackson B, et al. . SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol 2021; 19:409–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Garcia-Beltran WF, Lam EC, St Denis K, et al. . Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell 2021; 184:2372–83.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cevik M, Tate M, Lloyd O, Maraolo AE, Schafers J, Ho A. SARS-CoV-2, SARS-CoV, and MERS-CoV viral load dynamics, duration of viral shedding, and infectiousness: a systematic review and meta-analysis. Lancet Microbe 2021; 2:e13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kumar N, AbdulRahman A, AlAli S, Otoom S, Atkin SL, AlQahtani M. Time till viral clearance of severe acute respiratory syndrome coronavirus 2 is similar for asymptomatic and non-critically symptomatic individuals. Front Med (Lausanne) 2021; 8:616927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Killingley B, Mann AJ, Kalinova M, et al. . Safety, tolerability and viral kinetics during SARS-CoV-2 human challenge in young adults. Nat Med 2022; 28:1031–41. [DOI] [PubMed] [Google Scholar]
- 38. Owusu D, Pomeroy MA, Lewis NM, et al. . Persistent SARS-CoV-2 RNA shedding without evidence of infectiousness: a cohort study of individuals with COVID-19. J Infect Dis 2021; 224:1362–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhou C, Zhang T, Ren H, et al. . Impact of age on duration of viral RNA shedding in patients with COVID-19. Aging 2020; 12:22399–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xu K, Chen Y, Yuan J, et al. . Factors associated with prolonged viral RNA shedding in patients with coronavirus disease 2019 (COVID-19). Clin Infect Dis 2020; 71:799–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nesterenko PA, McLaughlin J, Tsai BL, et al. . HLA-A(*)02:01 restricted T cell receptors against the highly conserved SARS-CoV-2 polymerase cross-react with human coronaviruses. Cell Rep 2021; 37:110167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Focosi D, Maggi F, McConnell S, Casadevall A. Very low levels of remdesivir resistance in SARS-COV-2 genomes after 18 months of massive usage during the COVID19 pandemic: a GISAID exploratory analysis. Antiviral Res 2022; 198:105247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jochmans D, Liu C, Donckers K, et al. . The substitutions L50F, E166A and L167F in SARS-CoV-2 3CLpro are selected by a protease inhibitor in vitro and confer resistance to nirmatrelvir . mBio 2023; 14:e0281522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhou Y, Gammeltoft KA, Ryberg LA, et al. . Nirmatrelvir resistant SARS-CoV-2 variants with high fitness in an infectious cell culture system . Sci Adv 2022; 8:eadd7197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hu Y, Lewandowski EM, Tan H, et al. . Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir . bioRxiv, doi: 10.1101/2022.06.28.497978, 6September2022, preprint: not peer reviewed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chen Z, Azman AS, Chen X, et al. . Global landscape of SARS-CoV-2 genomic surveillance and data sharing. Nat Genet 2022; 54:499–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Takashita E, Kinoshita N, Yamayoshi S, et al. . Efficacy of antibodies and antiviral drugs against Covid-19 Omicron variant. N Engl J Med 2022; 386:995–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Planas D, Saunders N, Maes P, et al. . Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature 2022; 602:671–5. [DOI] [PubMed] [Google Scholar]
- 49. Cao Y, Yisimayi A, Jian F, et al. . BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection. Nature 2022; 608:593–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tchesnokov EP, Gordon CJ, Woolner E, et al. . Template-dependent inhibition of coronavirus RNA-dependent RNA polymerase by remdesivir reveals a second mechanism of action. J Biol Chem 2020; 295:16156–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.