Abstract
This scientific commentary refers to ‘Genetic topography and cortical cell loss in Huntington’s disease link development and neurodegeneration’ by Estevez-Fraga et al. (https://doi.org/10.1093/brain/awad275).
This scientific commentary refers to ‘Genetic topography and cortical cell loss in Huntington’s disease link development and neurodegeneration’ by Estevez-Fraga et al. (https://doi.org/10.1093/brain/awad275).
Evidence continues to accumulate demonstrating that the huntingtin protein (HTT) plays a critical role in neurodevelopment.1 Huntington’s disease is a neurodegenerative disorder caused by an expanded CAG repeat in the HTT gene leading to a mutated huntingtin protein (mHTT). This mutation results in significant neuronal loss, especially in the striatum and cortex, culminating in a gamut of motor, cognitive and psychiatric symptoms. Although several hypotheses exist, the precise mechanism through which mHTT instigates neurodegeneration remains elusive. Given the relationship between HTT and neurodevelopment, it is reasonable to hypothesize that the neurodegeneration witnessed in Huntington’s disease might be a delayed fallout of early neurodevelopmental changes triggered by the presence of mHTT. In this issue of Brain, Estevez-Fraga and colleagues2 venture into this intricate terrain, probing the ties between genetic topography, cortical cell loss and the overarching trajectory of disease progression. The team leveraged volumetric and diffusion MRI methodologies to craft Huntington’s disease-specific brain maps, juxtaposing them against control data to discern cortical regions of significant cell loss. The subsequent pairing of these imaging insights with gene expression data from the Allen Human Brain Atlas painted a detailed panorama of the Huntington’s disease genetic landscape and its potential influence on both neurodevelopment and neurodegeneration.
A standout revelation from this work was the robust positive correlation between cortical cell loss and the expression of developmental genes. Specifically, areas of the cortex that demonstrated significant atrophy were also areas with the greatest expression of genes implicated in development. In stark contrast, synaptic and metabolic genes, previously implicated in neurodegeneration, showcased a negative correlation, such that greater expression of these genes was associated with less cortical cell loss.2 This dichotomy hints at a sophisticated balance between the genetic foundations of Huntington’s disease and the observed brain morphological alterations.
Based on these observations, the authors are left with the difficult task of trying to interpret the complex interaction between HTT and neurodevelopment in the context of a growing body of literature on this topic. They ascribe to the notion that because patients with Huntington’s disease have one mutant allele and one wild-type allele, they will therefore have reduced levels of wild-type HTT (wtHTT), which may be insufficient to support normal development. Support for this loss-of-function theory comes from preclinical animal models showing that a reduction in wtHTT during normal development leads to cortical and striatal degeneration.3 However, this theory seems to contrast with another hypothesis for the role of HTT in neurodevelopment and Huntington’s disease pathology that is well-supported by the literature. According to this hypothesis, there are three aspects of HTT function that are important to consider when evaluating the role of HTT in neurodevelopment: (i) HTT seems to act by an exclusively dominant function of its longest allele; (ii) the effects of HTT are CAG-dependent throughout the entire range of repeats from below to above disease threshold, across an allelic continuum; and (iii) HTT’s role in brain development is likely to create a functional advantage early in life and may have been positively selected for in human brain evolution. Importantly, it is the creation of this early functional advantage that may lead to vulnerability to degeneration later in life.
In regard to the first point, preclinical studies have shown that HTT affects energy metabolism and gene expression in a fully dominant fashion.4 Moreover, the well-known phenomenon whereby the length of the CAG repeat is highly correlated with the age of disease onset also occurs in a fully dominant manner.5 In studies of human brain development, HTT has been shown to affect brain structure and cognitive function—including when the number of CAG repeats is below disease threshold—with the effects driven only by the longest allele.6 Finally, in individuals with Huntington’s disease, the phenotype of those who are homozygous for mHTT appears no different from that of individuals who are heterozygous for mHTT.7 This comparison supports the theory that mHTT acts in a dominant fashion since the additional mHTT allele seems to have no clinical effect. In addition, it refutes the notion that the absence of wtHTT negatively affects neurodevelopment and, thereby, neurodegeneration, as homozygous carriers would be expected to have significant neurodevelopmental changes that would predispose them to a more severe disease course.
There is abundant evidence to support the idea that HTT acts in a CAG-dependent fashion across an allelic continuum. In a study of children with non-pathologic ranges of CAG repeats (15–34), a greater number of CAG repeats was associated with advantageous changes in brain structure and with better cognitive function.6 In children who carry mHTT, this pattern continues, with each increase in the length of the CAG repeat leading to greater cognitive skills and higher brain volumes.8,9 Taken together, this supports the theory that the function of HTT creates phenotypic variation in a linear fashion across the allelic continuum.
Evolutionary biologists have long postulated that genes with triplet repeats can generate a range of phenotypes, creating the variation needed for adaptive evolution. Recent work showed that the polyglutamine tract encoded by CAGs in HTT is under strong selective evolutionary pressure. Moreover, increases in CAG repeat lengths significantly correlated with changes in gene transcriptional networks governing neuronal function, corroborating the hypothesis that longer CAG repeats could generate more mature, more connected and more functional neurons.10 This is born out in the clinical studies that show increasing CAG repeats lead to larger brain volumes and superior cognitive skills. Importantly, the studies of children with CAG repeats in the pathologic range found their cognitive skills to be significantly better than those of children with repeats in the non-pathologic range, supporting the idea that not only does mHTT drive brain function, it creates a functional advantage early in life.8
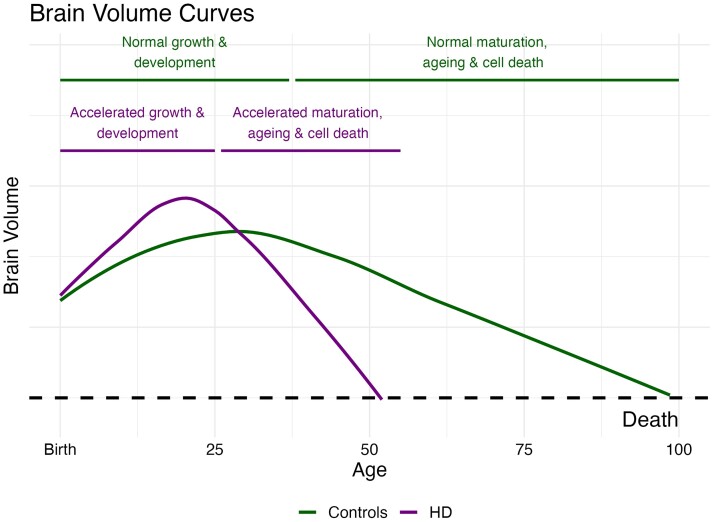
The findings from Estevez-Fraga and colleagues2 provide further evidence that HTT-induced neurodevelopmental changes likely contribute significantly to future neurodegeneration in Huntington’s disease. But how might an early functional advantage set the stage for later degeneration? In children with mHTT, the trajectory of striatal volume from early in life to age 18 was driven by mHTT and modified by the length of the CAG repeat.9 Initial striatal hypertrophy was followed by a linear decline in volume, in sharp contrast to the volume trajectory of children without the mutant gene. Moreover, the greater the number of repeats, the steeper the decline in striatal volume. This highlights the connection between early advantage and later degeneration and supports the theory that neural circuits that developed to be functionally superior may also have developed with structural liabilities that make them especially vulnerable to degeneration. One possibility is that the course of development and degeneration in Huntington’s disease is simply an accelerated trajectory of normal processes. Specifically, the growth and development of a superior brain early in life may lead to an accelerated process of ageing (Fig. 1).
Figure 1.
Neurodevelopment and neurodegeneration in Huntington’s disease. Control subjects (green line) show normal brain development that peaks around the age of 30 with slow volumetric decline associated with normal ageing. Subjects with Huntington’s disease (purple line) experience brain hypertrophy earlier in life that may be associated with cognitive advantages. However, this advantageous neurodevelopment produces vulnerabilities leading to accelerated ageing processes and neurodegeneration.
The role of mHTT in brain development has significant implications for current drug development strategies targeting mHTT reduction. The ultimate goal is to knock down the disease-causing gene early enough to prevent the disease from occurring altogether. However, the human brain takes nearly 30 years to fully mature, and an abundance of caution is required before attempting to lower the concentration of a protein known to be advantageous for early brain function. Unfortunately, therapies aimed at lowering mHTT levels later in life may be ineffective as they fail to address the vulnerabilities that were created by mHTT during neurodevelopment. In contrast, therapeutics designed to protect neurons that are vulnerable as a result of the observed neurodevelopmental changes may prove to be more effective. For example, mitochondrial dysfunction is known to occur in patients with Huntington’s disease. If the presence of mHTT leads to the development of neuronal circuits that are functionally superior but demand more energy, these will be especially susceptible to bioenergetic deficits and may therefore degenerate under the pressures of even normal processes such as ageing. Therapies aimed at overcoming this bioenergetic deficit may slow neurodegeneration by protecting vulnerable neurons.
The study by Estevez-Fraga and colleagues2 bridges many gaps in understanding the dynamic between genetic topography and cortical cell loss in Huntington’s disease. While it elucidates the potential developmental underpinnings of Huntington’s disease-associated neurodegeneration, the study also raises a myriad of new questions. The onus now lies on future research to integrate these findings with other studies, crafting a comprehensive narrative of the role of HTT in neurodevelopment.
Contributor Information
Jordan L Schultz, Department of Psychiatry, Carver College of Medicine at the University of Iowa, Iowa City, IA, USA; Department of Neurology, Carver College of Medicine at the University of Iowa, Iowa City, IA, USA; University of Iowa College of Pharmacy, Iowa City, IA, USA.
Mohit Neema, Department of Psychiatry, Carver College of Medicine at the University of Iowa, Iowa City, IA, USA.
Peg C Nopoulos, Department of Psychiatry, Carver College of Medicine at the University of Iowa, Iowa City, IA, USA; Department of Neurology, Carver College of Medicine at the University of Iowa, Iowa City, IA, USA; Stead Family Children’s Hospital at the University of Iowa, Iowa City, IA, USA.
Funding
This work is supported by the National Institute of Neurological Disorders and Stroke (K23-NS117736 to J.L.S. and U01-NS055903 to P.C.N.).
Competing interests
The authors report no competing interests.
References
- 1. Barnat M, Capizzi M, Aparicio E, et al. . Huntington’s disease alters human neurodevelopment. Science. 2020;369:787–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Estevez-Fraga C, Altmann A, Parker CS, et al. . Genetic topography and cortical cell loss in Huntington’s disease link development and neurodegeneration. Brain. 2023;146:4532-4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Arteaga-Bracho EE, Gulinello M, Winchester ML, et al. . Postnatal and adult consequences of loss of huntingtin during development: Implications for Huntington’s disease. Neurobiol Dis. 2016;96:144–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jacobsen JC, Gregory GC, Woda JM, et al. . HD CAG-correlated gene expression changes support a simple dominant gain of function. Hum Mol Genet. 2011;20:2846–2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lee J-M, Ramos EM, Lee J-H, et al. . CAG repeat expansion in Huntington disease determines age at onset in a fully dominant fashion. Neurology. 2012;78:690–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lee JK, Ding Y, Conrad AL, et al. . Sex-specific effects of the Huntington gene on normal neurodevelopment. J Neurosci Res. 2017;95:398–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cubo E, Martinez-Horta S-I, Santalo FS, et al. . Clinical manifestations of homozygote allele carriers in Huntington disease. Neurology. 2019;92:e2101–e2108. [DOI] [PubMed] [Google Scholar]
- 8. Schultz JL, van der Plas E, Langbehn DR, Conrad AL, Nopoulos PC. Age-related cognitive changes as a function of CAG repeat in child and adolescent carriers of mutant huntingtin. Ann Neurol. 2021;89:1036–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. van der Plas E, Langbehn DR, Conrad AL, et al. . Abnormal brain development in child and adolescent carriers of mutant huntingtin. Neurology. 2019;93:e1021–e1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Iennaco R, Formenti G, Trovesi C, et al. . The evolutionary history of the polyQ tract in huntingtin sheds light on its functional pro-neural activities. Cell Death Differ. 2022;29:293–305. [DOI] [PMC free article] [PubMed] [Google Scholar]