Abstract
Autosomal dominant Alzheimer’s disease (ADAD) offers a unique opportunity to study pathophysiological changes in a relatively young population with few comorbidities. A comprehensive investigation of proteome changes occurring in ADAD could provide valuable insights into AD-related biological mechanisms and uncover novel biomarkers and therapeutic targets. Furthermore, ADAD might serve as a model for sporadic AD, but in-depth proteome comparisons are lacking. We aimed to identify dysregulated CSF proteins in ADAD and determine the degree of overlap with sporadic AD. We measured 1472 proteins in CSF of PSEN1 or APP mutation carriers (n = 22) and age- and sex-matched controls (n = 20) from the Amsterdam Dementia Cohort using proximity extension-based immunoassays (PEA). We compared protein abundance between groups with two-sided t-tests and identified enriched biological pathways. Using the same protein panels in paired plasma samples, we investigated correlations between CSF proteins and their plasma counterparts. Finally, we compared our results with recently published PEA data from an international cohort of sporadic AD (n = 230) and non-AD dementias (n = 301). All statistical analyses were false discovery rate-corrected. We detected 66 differentially abundant CSF proteins (65 increased, 1 decreased) in ADAD compared to controls (q < 0.05). The most strongly upregulated proteins (fold change >1.8) were related to immunity (CHIT1, ITGB2, SMOC2), cytoskeletal structure (MAPT, NEFL) and tissue remodelling (TMSB10, MMP-10). Significant CSF-plasma correlations were found for the upregulated proteins SMOC2 and LILR1B. Of the 66 differentially expressed proteins, 36 had been measured previously in the sporadic dementias cohort, 34 of which (94%) were also significantly upregulated in sporadic AD, with a strong correlation between the fold changes of these proteins in both cohorts (rs = 0.730, P < 0.001). Twenty-nine of the 36 proteins (81%) were also upregulated among non-AD patients with suspected AD co-pathology. This CSF proteomics study demonstrates substantial biochemical similarities between ADAD and sporadic AD, suggesting involvement of the same biological processes. Besides known AD-related proteins, we identified several relatively novel proteins, such as TMSB10, MMP-10 and SMOC2, which have potential as novel biomarkers. With shared pathophysiological CSF changes, ADAD study findings might be translatable to sporadic AD, which could greatly expedite therapy development.
Keywords: dementia, familial, genetic, olink, proteome
Van der Ende et al. measured more than 1400 CSF proteins in familial Alzheimer's disease mutation carriers and found 66 significantly dysregulated proteins, including MAPT, NEFL, CHIT1 and MMP-10. Many of these proteins were similarly altered in sporadic Alzheimer's disease, suggesting involvement of the same biological processes.
Introduction
Autosomal dominant Alzheimer’s disease (ADAD) is a rare (<1%) form of Alzheimer’s disease (AD) caused by genetic mutations in amyloid precursor protein (APP), presenilin-1 (PSEN1) or presenilin-2 (PSEN2).1 ADAD has played a crucial role in identifying key pathological mechanisms underlying AD and the time course of AD-related brain changes.1,2 A thorough investigation of the biochemical changes occurring in ADAD could further improve our understanding of its complex pathophysiology, as well as help to identify novel biomarkers and therapeutic targets.
ADAD and sporadic AD are largely comparable in terms of phenotype, clinical course and neuropathology,1,3,4 and biochemical and structural measures have similar longitudinal profiles.3,5 Owing to the relatively young age at symptom onset in ADAD,6 age-related comorbidities which frequently confound sporadic AD studies are less common.7 Furthermore, the near complete penetrance of ADAD genetic mutations enables identification of individuals destined to develop AD on a predictable timescale, providing a valuable framework for prevention trials.1,8 Given their similarities, findings from ADAD studies might translate well to sporadic AD, but in-depth proteome comparisons are lacking.
In recent years, the field of proteomics has seen substantial technological improvements. Novel multiplexing immunoassays, such as proximity extension assays (PEA), enable highly sensitive measurements of thousands of proteins of interest simultaneously in minute sample volumes.9,10 These platforms have proven to be powerful tools to identify proteome changes and novel biomarkers in various diseases, including sporadic AD.11-15 In contrast, proteomics studies of ADAD are scarce and mostly small mass spectrometry-based studies focusing on presymptomatic mutation carriers,16-19 highlighting the need for an updated study of protein changes in ADAD.
While blood is gaining popularity in biomarker research for its convenience in clinical settings, CSF is probably a more suitable medium to investigate the biological mechanisms underlying AD.20 CSF is in direct contact with the brain, and proteins released from brain cells traffic freely to the CSF. Accordingly, 67% of CSF proteins have been detected in brain tissue.21 Furthermore, the CSF proteome is relatively unaffected by protein expression in other organ systems, facilitating a more brain-specific protein profile.20
In the present study, we investigated the abundance of over 1400 proteins in CSF of ADAD mutation carriers and controls using PEA. We aimed to delineate the biological processes involved in ADAD pathophysiology and uncover novel biomarkers and therapeutic targets. Using paired plasma samples, we determined which dysregulated CSF proteins correlate with their counterparts in blood and warrant further investigation as potential blood biomarkers. Finally, to determine the degree of similarity in ADAD and sporadic AD on the proteome level, we compared our results with recently published PEA data from a large cohort of sporadic AD and non-AD dementias.22
Materials and methods
Subjects
Autosomal dominant Alzheimer’s disease cohort
CSF samples were available for 22 carriers of a pathogenic mutation in APP or PSEN1, as well as 20 age- and sex-matched controls from the Amsterdam Dementia Cohort (ADC) biobank.23 A paired plasma sample, collected on the same day as CSF, was available for 21 mutation carriers. All participants underwent standardized neurological, cognitive and laboratory assessments, regardless of clinical diagnosis as previously described.23 The presence or absence of an ADAD-related pathogenic mutation was determined using PCR-based amplification of the appropriate exon followed by Sanger sequencing. Mutation carriers were considered presymptomatic or symptomatic [either mild cognitive impairment (MCI) or AD dementia] based on international consensus criteria.24,25 The control group consisted of individuals with subjective cognitive decline (SCD) in whom cognitive and laboratory investigations were normal. The Mini-Mental State Examination (MMSE) was used as a global measure of cognitive functioning.
Sporadic dementias cohort
The sporadic dementias cohort and corresponding PEA measurements, which were used for comparisons with our ADAD dataset, have been described in detail elsewhere.22 Here, we briefly highlight the most important methodological components relevant to the present study.
The sporadic dementias cohort included CSF samples from patients with sporadic AD (n = 230), non-AD dementia [either dementia with Lewy bodies (DLB, n = 110) or frontotemporal dementia (FTD, n = 191)] and controls (n = 195). Clinical diagnoses were assigned based on international consensus criteria.24-28 Controls had SCD with normal cognitive and laboratory investigations. Owing to the frequent occurrence of AD co-pathology among patients with non-AD dementias,29 this group was further separated into those with a CSF AD biomarker profile (see later), indicative of AD pathology, and those with normal CSF AD biomarkers. A subset of diagnoses was confirmed either by autopsy or, in the case of FTD, by the presence of an FTD-related genetic mutation.30 The majority of CSF samples were from the ADC; additional samples were collected through the Center for Neurodegenerative Disease Research at the University of Pennsylvania (‘UPenn’), Erasmus University Medical Center (‘Erasmus’) and the Goizueta Alzheimer’s disease Research Center at Emory University (‘Emory’).
Ethical approval
All studies described were approved by local medical ethical committees and all participants or their caregivers provided written informed consent according to the Declaration of Helsinki. Laboratory staff were blinded to the clinical and genetic status of samples.
Sample collection and protein profiling
CSF and plasma samples were collected by lumbar puncture and venipuncture, respectively, and were processed and stored in accordance with international consensus guidelines.31,32 For each sample, a fresh aliquot was used, i.e. without previous freeze-thaw cycles.
In the ADAD cohort, levels of CSF amyloid-β42 (Aβ42), total tau (t-tau) and phosphorylated tau-181 (p-tau) were analysed locally using commercially available kits [enzyme-linked immunosorbent assay (ELISA) (Innotest); Aβ42, hTau-Ag and phospho-tau(181P) (Fujirebio or Elecsys); Aβ42, t-tau and phospho-tau(181P) biomarker assays (Roche Diagnostics GmbH)]. Elecsys assay results were transformed to equivalent Innotest values using conversion formulas, which were previously developed in our centre using Passing-Bablok regression analyses.33 Innotest Aβ42 values were adjusted for drift over time.34 A positive AD biomarker profile was defined as a t-tau/Aβ42 ratio >0.46. In the sporadic dementias cohort, core AD biomarkers were measured locally using the aforementioned kits (for ADC and Erasmus samples) or using Luminex xMAP INNO-BIA AlzBio3 (Luminex Corp.) kits (for UPenn and Emory samples). A positive AD biomarker profile was defined as a t-tau/Aβ42 ratio >0.46 (ADC/Erasmus) or >0.30 (UPenn), or as an Aβ42/t-tau ratio <6 (Emory). The use of these ratios is consistent with the previous paper on PEA measurements in the sporadic dementias cohort,22 and their diagnostic accuracy to discriminate sporadic AD from other dementias and controls has been demonstrated.35
Proteomics analyses were performed using validated multiplex panels based on PEA technology (Olink Proteomics Inc.).9 Briefly, target proteins are bound by unique pairs of oligonucleotide-labelled antibodies. When in close proximity to one another, the oligonucleotides hybridize to form a PCR target sequence. The resulting DNA amplicon is quantified either on an Illumina NovaSeq platform using Next Generation Sequencing (Olink Explore) or on a Fluidigm BioMarkTM HD real-time PCR platform (Olink Target). Protein levels are reported on a log2 scale as normalized protein expression (NPX), which is derived from cycle threshold values and thus directly proportional to the concentration of target protein in the sample.
In the ADAD cohort, 1472 CSF and plasma proteins were measured using four Olink Explore 384-plex panels (Cardiometabolic, Inflammation, Neurology and Oncology; Olink Proteomics). All samples were randomized on one plate. Samples were prepared according to the dilution blocks shown in Supplementary Table 1. Dilution factors were provided by the manufacturer based on validation experiments to estimate the linear range of quantification in different matrices. Quality control samples and negative controls were supplied by the manufacturer. Details on assay characteristics and quality control (QC) procedures are provided on the manufacturer’s website.36 For each protein, the lower limit of detection (LOD) was defined as the sample of blanks plus 3 standard deviations. Proteins were included in further analyses if they were above the LOD in >85% of samples and passed quality control. Extreme outliers, defined as NPX values above or below a z-score of 5, were capped at a z-score of ±5.
Of the 1472 measured proteins, 814 were above the LOD in >85% of CSF samples (Supplementary Table 2). Two proteins (IL6 and CXCL8) were measured in all four Olink panels; NPXs of these replicates were strongly correlated [rs (range): 0.77–0.98; Supplementary Fig. 1] and one replicate for each protein was selected at random for inclusion in subsequent analyses. Among the remaining 808 unique proteins measured in CSF samples of the 42 ADAD mutation carriers and controls, 21 extreme outlying NPX values (0.06%) were capped. Of the 808 proteins included in CSF analyses, 798 were detected in >85% of the 21 paired plasma samples and were included in correlative analyses between CSF and plasma.
In the sporadic dementias cohort, 979 CSF proteins were measured using the 11 Olink Target 96 panels that were available at the time (Cardiometabolic, Cardiovascular II and III, Cell regulation, Development, Immune response, Inflammation, Metabolism, Neurology, Oncology II and Organ damage; Olink Proteomics). CHI3L1 was excluded from our comparative analyses due to a presumed hook-effect in the version of the assay used in the sporadic dementias cohort.22
For validation purposes, we quantified the levels of NEFL, GFAP and THOP1 using ELISA techniques. NEFL and GFAP were measured with single molecule array technology (Simoa) using the N4PE kit [(502542; coefficient of variation of QCs: NEFL 5.3%, GFAP 4.7%). THOP1 was measured in triplicate using a homebrew assay developed on the EllaTM automated immunoassay system 48-Digoxigenin cartridges (lot 25519, coefficient of variation of QCs: 5.2%).37
Statistical analyses
All data processing and statistical analyses were performed using R version 4.0.3.
Demographic characteristics were compared between groups using Kruskal–Wallis tests for numerical data and chi-square tests for sex. Normality of NPX values was assessed using Shapiro–Wilk tests. As most (>80%) proteins were normally distributed, we used two-sided t-tests to compare CSF protein expression levels between ADAD mutation carriers and non-carriers. These comparisons were repeated after exclusion of presymptomatic mutation carriers. Fold changes were calculated based on the ratio of NPX values in mutation carriers to controls. Spearman’s rank correlation was used for correlative analyses with core AD biomarkers (CSF Aβ42, t-tau, p-tau), MMSE and proteins measured by ELISA for validation, as well as between CSF and plasma NPX values among mutation carriers. In the sporadic dementias cohort, cases and controls were not age- and sex-matched, and therefore, protein expression levels were analysed using nested linear models with age and sex as covariates.22 To eliminate the risk of misdiagnoses among non-AD dementias, we performed additional comparative analyses between ADAD and non-AD dementias, limiting non-AD dementias to subjects with pathological or genetic confirmation. All analyses were false discovery rate (FDR)-corrected using the Benjamini–Hochberg method. The statistical significance threshold was set at FDR-corrected q < 0.05.
Functional enrichment analysis was performed using Metascape,38 selecting Gene Ontology Biological Processes and Reactome as the ontology sources. All measured proteins included in the statistical analyses were used as the enrichment background. Default parameters were used for the analysis, in which terms with a P-value <0.01, a minimum count of three and an enrichment factor >1.5 (i.e. the ratio between observed counts and counts expected by chance) were collected and grouped into clusters based on their membership similarities. The most statistically significant term from each cluster was chosen to represent the cluster. In addition, we assessed the possible cellular origin of the differentially expressed CSF proteins by a cell type specificity assessment with the EWCE (expression weighted cell type enrichment) R package.39 Here, the input was the cell type expression of the DroNc-seq snRNAseq data set.40 Finally, we conducted pathway enrichment analyses with the KEGG database, inputting the differential expression and P-values of all measured proteins. We tailored the R package KEGGREST for the analyses, with the measured proteins as the background.41,42 A threshold was set at a minimum count of three proteins measured and P < 0.05. The enrichment is presented by the median beta estimate of up- or downregulation for the pathway.
Results
Subject characteristics are shown in Table 1. ADAD mutation carriers and controls did not differ significantly in terms of age or sex. Among the 22 mutation carriers, 18 were symptomatic and four were presymptomatic. All symptomatic mutation carriers as well as two presymptomatic carriers had a positive AD biomarker profile. Mutation types included various PSEN1 mutations in 16 mutation carriers from 15 pedigrees, and APP mutations in six mutation carriers from five pedigrees (Table 1). All symptomatic mutation carriers presented with an amnestic phenotype. A detailed overview of neuropsychological test results is provided in Supplementary Table 3.
Table 1.
Subject characteristics
| ADAD cohort | Sporadic dementias cohort | |||||||
|---|---|---|---|---|---|---|---|---|
| Mutation carriersa,b | Controls | P | Sporadic AD | Non-AD dementia | Controls | P | ||
| CSF+ | CSF− | |||||||
| n | 22 | 20 | – | 230 | 93 | 208 | 195 | – |
| Age, years | 48 (40–58) | 49 (45–52) | 0.668 | 66 (59–72) | 68 (63–74) | 66 (61–71) | 57 (52–62) | <0.001 |
| Disease duration if symptomatic, yearsc | 2 (1–3) | – | – | 3 (2–4) | 3 (1–4) | 3 (2–4) | – | 0.574 |
| Sex, female (%) | 11 (50%) | 8 (40%) | 0.734 | 95 (41%) | 39 (42%) | 71 (34%) | 79 (41%) | 0.377 |
| MMSEd | 23 (17–25) | 29 (28–29) | <0.001 | 21 (17–24) | 23 (19–27) | 25 (22–27) | 28 (27–29) | <0.001 |
| AD biomarker profile, n +/− (%)e | 20/2 (91%) | 0/20 (0%) | <0.001 | 226/3 (99%) | 93/0 (100%) | 0/208 (0%) | 1/193 (0.01%) | <0.001 |
| ApoE4, n +/− (%)f | 6/14 (27%) | 3/17 (15%) | 0.201 | 133/88 (60%) | 36/38 (49%) | 62/120 (34%) | 44/125 (26%) | <0.001 |
| Study site, n | ||||||||
| ADC | 22 | 20 | – | 210 | 51 | 151 | 194 | – |
| UPenn | 0 | 0 | – | 20 | 35 | 45 | 0 | – |
| Erasmus | 0 | 0 | – | 0 | 3 | 12 | 1 | – |
| Emory | 0 | 0 | – | 0 | 4 | 0 | 0 | – |
Numerical variables are shown as median (interquartile range). P-values were derived from chi-square tests and Mann–Whitney U or Kruskal–Wallis tests for categorical and numerical variables, respectively.
PSEN1 mutations: A79V (n = 3), A231V (n = 1), G384A (n = 1), L232F (n = 1), L262F (n = 5), Y115C (n = 4), G206D (n = 1); APP mutations: duplication (n = 4), H677R (n = 1), M722K (n = 1).
Clinical diagnoses: AD (n = 16), MCI (n = 2) or cognitively healthy (n = 4). All subjects with MCI and AD initially presented with memory complaints (i.e. amnestic phenotype).
Disease duration, defined as the difference in age at CSF collection and age at symptom onset as estimated by a caregiver, was missing in 119 subjects from the sporadic dementias cohort.
MMSE missing in two subjects from the ADAD cohort and in 39 subjects from the sporadic dementias cohort (six sporadic AD, 25 non-AD dementia, four controls).
A positive AD biomarker profile (CSF+) was defined based on site-specific cut-off values: t-tau/Aβ42 ratio >0.46 (ADC and Erasmus); t-tau/Aβ42 ratio >0.30 (UPenn); or Aβ42/t-tau ratio >6 (Emory).
At least one ApoE4 allele present.
In the sporadic dementias cohort, controls were significantly younger than subjects with sporadic AD and non-AD dementia, but no differences in sex were observed. Of 230 sporadic AD subjects, 226 had a positive AD biomarker profile, two had a negative biomarker profile and biomarker data were missing in two subjects. Six sporadic AD subjects were autopsy confirmed, including the two with a negative AD biomarker profile and one with missing biomarkers. The non-AD dementias group consisted of 93 subjects with a positive AD biomarker profile (53 FTD, 40 DLB), and 208 with a negative AD biomarker profile (138 FTD, 70 DLB). Thirty-one non-AD subjects (26 FTD, 5 DLB) in the biomarker-positive group (33%), as well as 64 (56 FTD, 8 DLB) in the biomarker-negative non-AD group (31%), were confirmed by autopsy or genetics. Twenty-two DLB diagnoses were supported by FPCIT single-photon emission computed tomography (DAT-SPECT).43
CSF proteomics analyses
Differential CSF protein abundance in ADAD
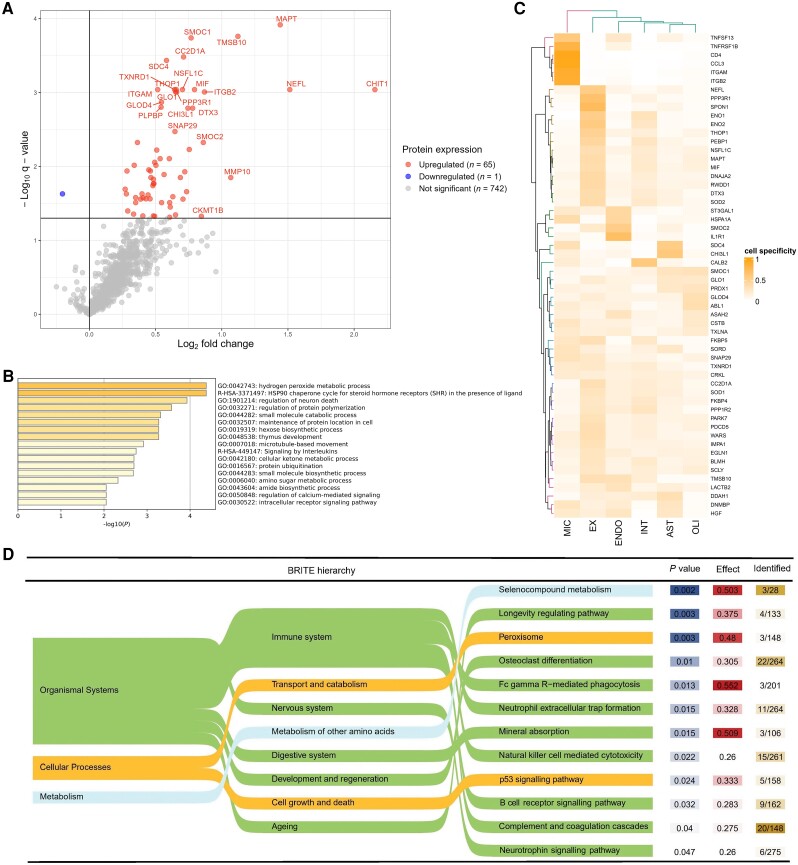
Results for all proteins included in the analyses of ADAD mutation carriers versus controls are shown in Supplementary Table 2. In total, 65 CSF proteins were significantly upregulated in ADAD mutation carriers compared to controls, and one protein, IL17A, was significantly downregulated (q < 0.05; Fig. 1A and Supplementary Table 4A). The largest fold change was seen for CHIT1 (fold change = 4.5), followed by NEFL, MAPT, TMSB10 and MMP-10 (fold change > 2). The most significantly dysregulated proteins (q < 0.001) included those involved in cytoskeletal structure (MAPT, NEFL), tissue remodelling (TMSB10), immunity (CHIT1, ITGAM, MIF, ITGB2), synapse function (CC2D1A, PPP3R1), vesicle transport (SDC4, NSFL1C) and energy metabolism (GLO1).
Figure 1.
Differential protein abundance and associated biological pathways in ADAD mutation carriers. (A) Differential protein abundance in ADAD mutation carriers versus controls. The log2 fold change, which is equivalent to the difference in log2-transformed normalized protein expression (NPX) levels, is plotted against the –log10-transformed q-value. The horizontal line indicates the statistical significance threshold, set at q < 0.05. Significantly dysregulated proteins are shown in red (upregulated) or blue (downregulated). The top 20 proteins with the smallest q-value and the top 10 proteins with the largest fold change are labelled. (B) Enriched biological pathways among ADAD mutation carriers. Functional enrichment was performed using Metascape,35 selecting Gene Ontology Biological Processes and Reactome as ontology sources. All analysed proteins were included as the enrichment background (n = 808). Terms with a P-value <0.01, a minimum count of three and an enrichment factor >1.5 (i.e. the ratio between observed counts and counts expected by chance) were collected and grouped into clusters based on their membership similarities. P-values were calculated based on the accumulative hypergeometric distribution. Kappa scores are used as the similarity metric when performing hierarchical clustering on the enriched terms, and subtrees with a similarity of >0.3 are considered a cluster. The most statistically significant term is chosen to represent the cluster. Stronger terms indicate more significant enrichment. (C) Cell-type specificity of dysregulation proteins. In this heat map, the dysregulated CSF proteins are clustered based on the proportion of cell-type expression. The columns list the cell types: microglia (MIC), excitatory neuron (EX), endothelial cells (ENDO), interneurons (INT), astrocytes (AST) or oligodendrocytes (OLI). (D) Enriched KEGG pathways according the BRITE hierarchy, with increasing granularity from left to right. For the analyses, the measured proteins from the background. A threshold was set at a minimum count of three proteins measured and P < 0.05. The enrichment is presented by the median beta estimate of up- or downregulation for the pathway, the P-value and the number of proteins detected with the current proteomics analyses relative to all proteins in the pathway.
After exclusion of presymptomatic mutation carriers (n = 4), we identified 89 significantly upregulated proteins (Supplementary Fig. 2 and Supplementary Table 4B), including 25 proteins which were significant in the original analyses. The top 20 most significant proteins remained unchanged except for one (LACT2B replacing DTX3).
In support of the validity and replicability of our findings, we found a strong correlation between CSF proteomics and ELISA techniques for measurement of NEFL (rs = 0.97, P < 0.001), GFAP (rs = 0.79, P < 0.001) and THOP1 (rs = 0.86, P < 0.001) (Supplementary Fig. 3).
Biological pathway analysis
In the functional enrichment analysis using Metascape, the most significantly enriched terms among ADAD mutation carriers included several terms related to cellular metabolism, such as small molecule catabolic processes, hydrogen peroxide metabolic process and carbohydrate metabolic processes (Fig. 1B). The terms associated with the largest number of proteins in our list included signalling by interleukins (13 proteins), regulation of neuron death (12 proteins) and small molecule catabolic process (nine proteins). The origin of the proteins based on cell type expression data was diverse, with clear clusters of predominant microglia (n = 6 proteins), excitatory neuron (n = 15 proteins), endothelial (n = 4 proteins) and astrocyte (n = 2 proteins) expressed proteins, as well as several protein clusters less clearly related to one cell type (Fig. 1C). In terms of KEGG pathway enrichment, significant enrichment of the systems neurology and immune response was found. On the deeper layer, the most significant enriched pathways were selenocompound metabolism, longevity regulation and peroxisome (Fig. 1D).
Correlation with core Alzheimer’s disease biomarkers and Mini-Mental State Examination
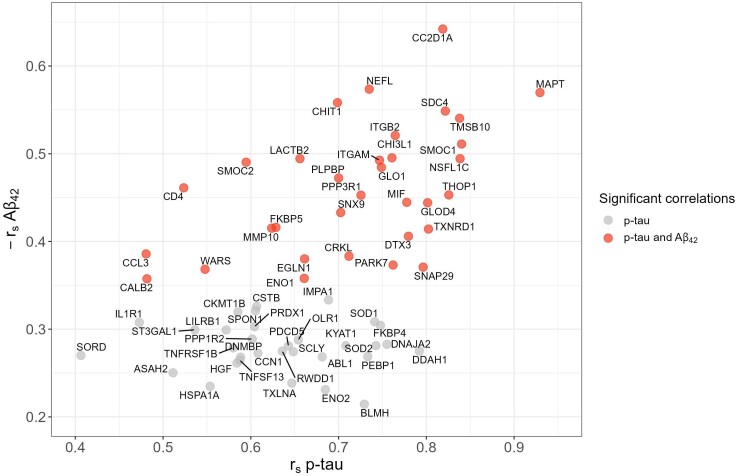
Moderate to strong positive correlations were found for all but one of the 66 differentially expressed proteins with CSF p-tau [median (range) rs = 0.68 (0.41–0.93)] and t-tau [rs = 0.72 (0.41–0.97)] (Fig. 2). In addition, 33 of these proteins correlated negatively with CSF Aβ42 [rs = −0.45 (−0.64; −0.35)]. One protein, IL17A, was negatively correlated with p-tau and t-tau and not significantly correlated with Aβ42.
Figure 2.
Correlations between differentially abundant CSF proteins and core CSF AD biomarkers. The correlation coefficient with Aβ42 and p-tau was determined for each protein using Spearman’s rho and considered significant at q < 0.05. All proteins shown (n = 65) correlated significantly with p-tau; those shown in red also correlated with Aβ42. For visualization purposes, correlation coefficients with Aβ42 are inverted (i.e. −rs Aβ42). One protein, IL-17A, was negatively correlated with p-tau (rs = −0.38; q = 0.015) and positively correlated with Aβ42 (rs = 0.11; q = 0.48) and is not shown here.
Significant negative correlations with MMSE in mutation carriers were found for CC2D1A (rs = −0.504, P = 0.020), PARK7 (rs = −0.446, P = 0.043) and MAPT (rs = −0.445, P = 0.043), but these were no longer significant after correction for multiple testing (Supplementary Table 5).
CSF-plasma correlations
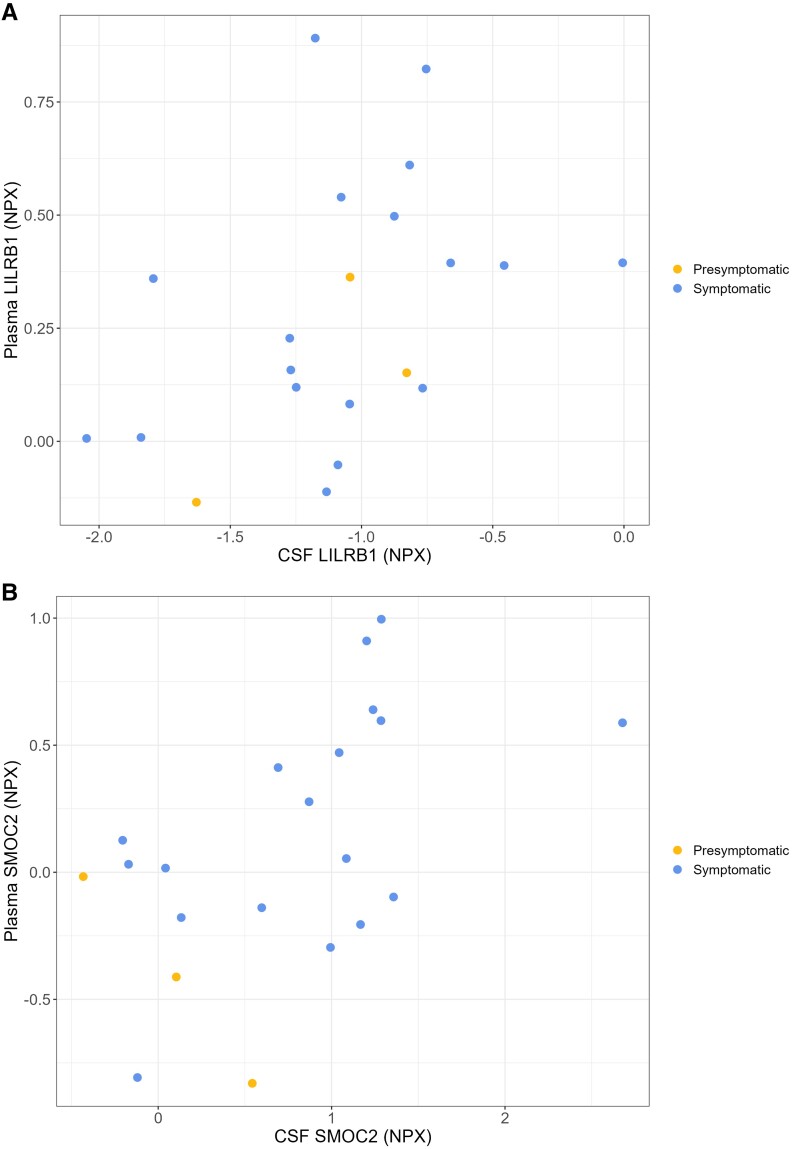
Across all 798 proteins analysed in both CSF and plasma samples of 21 mutation carriers, a significant positive correlation was found for 11 proteins (Supplementary Table 6A). Of the 66 significantly dysregulated CSF proteins, 63 were also measured in plasma; among these, SMOC2 (rs = 0.522, P = 0.016) and LILRB1 (rs = 0.514, P = 0.018) correlated with their plasma counterparts, although these correlations were no longer significant after FDR-correction (Fig. 3 and Supplementary Table 6B).
Figure 3.
Correlations between CSF and plasma (A) LILRB1 and (B) SMOC2 among ADAD mutation carriers. CSF and plasma protein abundance are expressed on a log2 scale as normalized protein expression (NPX). Correlation coefficients and P-values are derived from Spearman’s rho. Blue and yellow symbols represent symptomatic (n = 18) and presymptomatic mutation carriers (n = 4), respectively.
Comparison with sporadic dementias
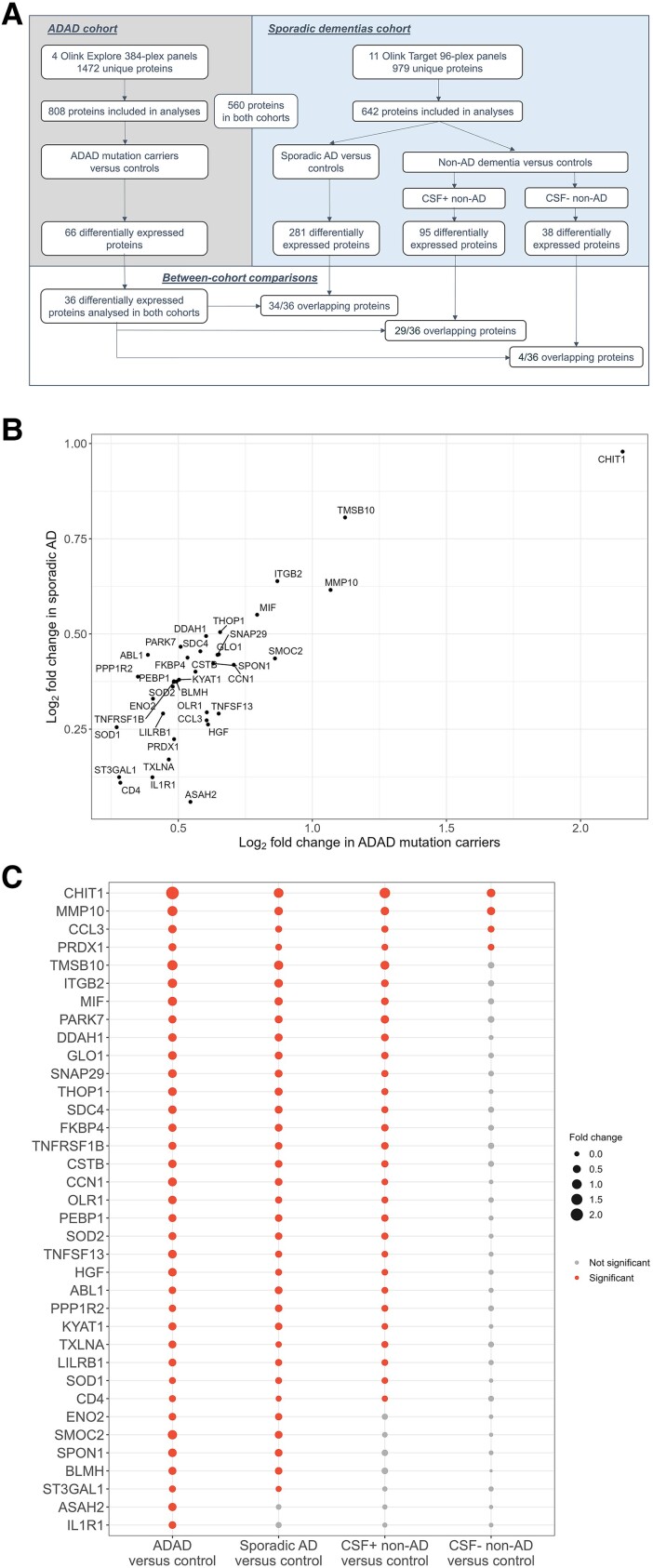
Overall, 560 proteins measured in the ADAD cohort had previously been measured in the sporadic dementias cohort, including 36 of the 66 significantly dysregulated proteins in ADAD (Supplementary Table 4A).
Among these 36 proteins, 34 (94%) were also significantly upregulated in sporadic AD (Fig. 4A). A strong correlation was found between the fold changes of these proteins in both cohorts (rs = 0.730, P < 0.001) (Fig. 4B). Of these 36 proteins, 29 (81%) were also upregulated in non-AD dementias with suspected AD co-pathology. In contrast, in the comparison with non-AD dementias with a negative AD biomarker profile (i.e. without suspected AD co-pathology), just 4 of the 36 proteins (MMP-10, CHIT1, CCL3, PRDX1; 11%) were dysregulated (Fig. 4C). Finally, ASAH2 and IL1R1 were upregulated in ADAD but not in any of the sporadic dementia groups.
Figure 4.
Comparisons of differentially abundant CSF proteins in ADAD with sporadic dementias. (A) Flow chart showing design of between-cohort comparisons. In total, 560 proteins were measured in both cohorts and included in the comparisons, including 36 of the 66 differentially regulated proteins among ADAD mutation carriers. CSF+ non-AD and CSF− non-AD indicate non-AD dementia patients with a positive or negative AD biomarker profile, respectively. (B) Scatter plot showing log2-fold changes in ADAD mutation carriers and sporadic AD patients versus controls for the proteins (n = 34) which were significantly upregulated in both cohorts. (C) Bubble chart displaying overlap of differentially abundant proteins in the various comparisons. Red circles indicate significantly dysregulated proteins; the circle size is proportional to the fold change. Of the 36 proteins upregulated in ADAD and included in the comparison study, 34 were also upregulated in sporadic AD compared to controls; 29 proteins were upregulated in both sporadic AD and CSF+ non-AD dementia, whereas just four were also upregulated among CSF− non-AD dementia. Two proteins were uniquely dysregulated in ADAD. ADAD = autosomal dominant Alzheimer’s disease.
Among pathologically or genetically confirmed non-AD dementias with a positive AD biomarker profile, 13 of the 36 proteins were found to overlap with ADAD (Supplementary Fig. 4).
Discussion
We performed a systematic and detailed investigation of CSF proteome changes in ADAD mutation carriers using state-of-the-art PEA technology. Our results demonstrate large and significant changes in the CSF proteome in ADAD, with involvement of inflammatory, cytoskeletal and metabolic proteins, as well as a very large overlap in proteome changes with sporadic AD.
The remarkable similarities in CSF protein dysregulation between ADAD and sporadic AD provide convincing evidence of shared pathways in both diseases and confirm that mechanistic insights gained from ADAD CSF studies may apply to sporadic AD as well. These findings warrant replication in future proteomics studies that directly compare ADAD and sporadic AD protein profiles (i.e. generated in the same study), ideally also in paired brain tissue and blood samples.19 The ability to identify preclinical ADAD mutation carriers, who are certain to develop the disease, uniquely offers the opportunity to study very early pathological brain changes and test drugs aimed at secondary prevention.1,5 Our results support a framework in which early-stage biomarkers and potential therapies are identified in ADAD and replicated in sporadic AD, which could greatly expedite the journey towards effective treatment. The power of such an approach is demonstrated by the oft-cited example of statins, which were initially trialled in familial hypercholesterolaemia and are now widely used to treat the much more common sporadic form.44
In order to extrapolate ADAD findings to sporadic AD, some differences between both diseases should be considered. First, a fundamental difference exists in the aetiology of amyloid pathology. While mutations in APP, PSEN1 and PSEN2 directly alter APP metabolism to produce more aggregation-prone Aβ species or more total Aβ, Aβ aggregation in sporadic AD probably results from a chronic imbalance in Aβ production and clearance following a complex interaction of (epi)genetic and environmental factors.45,46 Nonetheless, our results suggest common disease mechanisms downstream of Aβ pathology, and potential efficacy of drugs targeting these processes may be independent of the initial underlying aetiology. Second, symptom onset in ADAD is on average 30 years earlier than in sporadic AD,6,47 and the effects of ageing-related comorbidities as well as the more frequent occurrence of multiple neuropathologies in sporadic AD7,48,49 must be taken into account. Third, given the more rapid rate of decline in ADAD reported in some studies,50 treatment effects in sporadic AD might be more subtle than expected based on ADAD studies. Fourth, mutation-specific differences in molecular biomarkers,51,52 pathology53 and clinical features54,55 suggest divergent neurotoxic effects depending on the nature and position of the genetic mutation.56,57 As such, certain ADAD variants might align less well with sporadic AD. The mutations included in the present study were diverse, precluding identification of mutation-specific proteome changes. A better understanding of the molecular consequences of each mutation might enable biochemically-driven mutation groupings and reveal which genetic subtypes match best with sporadic AD.
Many of the dysregulated proteins identified here have previously been linked to AD, providing strong support for our approach and findings. These include extensively studied proteins, such as NEFL, CHIT-1, MAPT and CHI3L1,58-60 as well as proteins which have emerged more recently in the dementia field, such as SMOC2, MIF and MMP-10.13,22,61-66 CSF proteins not previously linked to AD, such as GLOD4 and DTX3, require replication in independent CSF data, as well as brain tissue studies, to further examine their relevance. Here, we discuss a selection of proteins based on their relevance to and novelty in AD research.
SMOC1 and SMOC2 were among the most strongly upregulated proteins in ADAD mutation carriers (respective fold changes = 1.7 and 1.8). Although their function in the brain is unclear, the closely related SPARC protein is highly expressed in glia and implicated in microglial response to damaging stimuli.67-69 Robust elevations in CSF SMOC115,16,19,21,70-72 and SMOC222,63 have been reported in AD across various proteomics platforms, and SMOC1 accurately discriminates AD from controls (area under the receiver operating characteristic curve = 0.84).70 We and others found strong correlations with CSF Aβ42,15 and post-mortem studies show co-localization of SMOC1 and/or SMOC2 with Aβ plaques,73,74 suggesting that these proteins may be directly related to amyloid pathology. Accordingly, CSF SMOC1 was not elevated in non-AD dementias in our study nor in other cohorts of patients with non-AD neurodegenerative diseases.15 Further studies are required to determine the differential diagnostic potential of SMOC1 and SMOC2. The correlations between CSF and plasma SMOC2, as well as elevated levels in AD plasma reported elsewhere15 are promising, and replication is warranted to explore its potential as a peripheral biomarker.
CSF MMP-10 was strongly upregulated in our ADAD dataset (fold change = 2.1), replicating previous findings in MCI and sporadic AD.13,63-66 MMP-10 belongs to a family of matrix metalloproteinases (MMPs) which degrade extracellular matrix proteins. In the brain, MMPs are overexpressed by astrocytes and microglia in response to Aβ accumulation and are capable of degrading Aβ.75,76 In line with the MMP-10 increases in our non-AD dementias groups, elevated levels of MMPs have been reported in other neurodegenerative diseases13,22,77,78 and therefore might reflect a non-specific inflammatory response in an effort to clear toxic protein aggregation. While its non-specific nature precludes its use as a differential diagnostic biomarker, MMP-10 was recently shown to have additional prognostic value besides core AD biomarkers to predict conversion from MCI to AD79; this prognostic potential is supported by the strong correlations with p-tau in our cohort (rs = 0.84, P < 0.001). The lack of correlation with MMSE in our study, which was similarly absent for the other differentially expressed proteins, might be due to limited statistical power in the relatively small sample of 18 symptomatic mutation carriers and requires replication in larger cohorts. Taken together, our results strongly support further investigation into MMP-10 as an inflammatory biomarker in neurodegenerative diseases.
We also observed a large fold change for TMBS10 (fold change = 2.2) a member of the thymosin beta family, which is implicated in cell proliferation, angiogenesis and tissue repair.80 CSF TMSB10 has only recently come to light in relation to dementia, with strong increases reported in sporadic AD.22 Its more extensively studied paralog, TMSB4, is elevated in CSF in AD and Creutzfeldt-Jakob disease81 and thought to be neuroprotective in various neurological diseases.80 Interestingly, TMSB4 overexpression in mouse models with APP/PSEN1 mutations reduced Aβ accumulation and attenuated AD phenotypes.82 The elevated TMSB10 levels found in our study might reflect an attempt to mitigate CNS damage secondary to AD. If confirmed, thymosin upregulation might be considered as a potential therapeutic strategy for dementia, as has been suggested for stroke and multiple sclerosis.80
Of note, four (MMP-10, CCL3, PRDX1, CHIT1) of the ADAD-associated proteins were upregulated in non-AD dementias in the absence of AD co-pathology. These probably reflect non-specific inflammation-related changes, corroborating elevated levels of these proteins found in other neurodegenerative diseases.13,22,60,77,78,83-85 However, we caution against over-interpreting lack of statistically significant results in the remaining ADAD-associated proteins in these subjects, which might be due to a lack of statistical power in a heterogeneous population of FTD and DLB patients, and emphasize that we cannot draw conclusions regarding the AD-specificity of proteins. In the same vein, the proteins which were upregulated in ADAD but not in sporadic AD and non-AD dementias need not necessarily be ADAD-specific: ASAH2 is implicated in lipid metabolism by its cleavage of ceramides, the metabolism of which is disrupted in (sporadic) AD.86,87 IL1R1, a member of the interleukin-1 family, plays a central role in the pro-inflammatory response seen in sporadic AD and various other diseases.88,89
We identified a larger number of differentially expressed proteins after exclusion of presymptomatic mutation carriers, suggesting that certain proteins change around after disease onset or fluctuate depending on the disease stage.17 A better understanding of the biochemical changes occurring in presymptomatic stages will be essential for identification of early biomarkers and therapeutic targets. Future studies that include enough presymptomatic carriers to analyse protein dynamics over the course of ADAD, for example by relating protein changes with estimated years to onset, will be highly insightful.
The protein dysregulation profiles identified here are broadly in line with recent proteomics studies of sporadic AD, both those using Olink technology11,63 and other strategies such as (targeted) mass spectrometry.90-92 We examined the biological significance of our findings using two pathway enrichment analyses, which revealed functional enrichment of terms related to immunity, energy metabolism, ageing and cell death. Our cell type expression analyses confirm a prominent role for microglia in AD protein dysregulation. The clear clusters of proteins expressed by endothelial cells is in line with previously reported dysregulation of proteins implicated in angiogenesis and blood–brain barrier function.63,93 Together, these findings provide further evidence of the similarities between ADAD and sporadic AD on the protein level.
Strengths of this study include the well-characterized cohort of ADAD mutation carriers and age-matched controls, as well as the very extensive protein panels which cover a wide range of biological processes. The large number of dysregulated proteins identified in this study, which remained significant after multiple testing correction, demonstrates the large-scale CSF proteome disruptions occurring in ADAD, and highlights the potential of CSF proteomics to study disease pathophysiology in vivo, even in rare diseases with modest sample sizes. The comparison of ADAD and sporadic AD on the CSF proteome level is novel and represents an important step in furthering our understanding of the similarities and differences between the two. The PEA platform employed here guarantees excellent sensitivity by using PCR-based protein quantification; at the same time, cross-reactive binding is minimal due to its reliance on dual-recognition immunoassays, which require binding of two antibodies and hybridization of oligonucleotides to produce a signal.9,15,94 The fact that we were able to replicate numerous previous findings underlines its reproducibility, which remains a major issue in the field of proteomics.95,96 Finally, the vast majority of samples from both cohorts was collected in the ADC as part of the same biobanking study, and the other centres adhered to consensus protocols for CSF biobanking, facilitating homogeneity in pre-analytical protocols.
This study must be viewed in light of some limitations. First, certain proteins of interest, such as neurogranin and SNAP-25, were not included in this targeted proteomics approach. However, given the very large number of proteins included, there is still ample room to identify novel protein dysregulations. Second, the large number of statistical tests inherent to proteomics studies necessitates multiple testing correction. While we believe the FDR correction applied in this discovery study, with a 5% threshold of false-positive results, offers an appropriate balance between the risks of false-negative versus false-positive results, the need for independent validation studies is evident. Third, our comparisons between ADAD and sporadic dementias might have been affected by the inevitable age differences between cohorts. Furthermore, the protein panels and assays used in both cohorts were not identical, and minor assay changes might have led to improved detectability of certain proteins in the ADAD cohort, which used more recently developed panels. A potential risk of overfitting exists given the use of the 560 proteins measured in both cohorts, which we mitigated by expressing the overlap in protein expression between both cohorts as a fraction of the 36 dysregulated proteins in ADAD. Future replication studies would ideally measure the same protein panels in both cohorts. Finally, we must acknowledge the risk of misdiagnoses among FTD and DLB cases, especially in those with an AD biomarker profile. However, diagnoses were made in specialized memory clinics, and almost one-third of FTD and DLB cases was either pathologically or genetically confirmed. Moreover, the analysis including only these confirmed cases revealed similar protein dysregulations, albeit with a smaller number of significant proteins (possibly due to a lack of statistical power).
In conclusion, the marked similarities in CSF protein profiles between ADAD and sporadic AD provide convincing evidence of dysregulation of the same biological processes. Although the two cannot be directly equated, these exciting results indicate that findings from ADAD studies might be extrapolated to sporadic AD, which could greatly benefit therapeutics development. In this light, further exploration of plasma protein dysregulation among ADAD mutation carriers may help identify valuable peripheral biomarkers of AD.
Supplementary Material
Contributor Information
Emma L van der Ende, Neurochemistry Laboratory, Department of Clinical Chemistry, Amsterdam Neuroscience, Amsterdam UMC, Vrije Universiteit Amsterdam, 1081 HV Amsterdam, The Netherlands.
Sjors G J G In ‘t Veld, Neurochemistry Laboratory, Department of Clinical Chemistry, Amsterdam Neuroscience, Amsterdam UMC, Vrije Universiteit Amsterdam, 1081 HV Amsterdam, The Netherlands.
Iris Hanskamp, Neurochemistry Laboratory, Department of Clinical Chemistry, Amsterdam Neuroscience, Amsterdam UMC, Vrije Universiteit Amsterdam, 1081 HV Amsterdam, The Netherlands.
Sven van der Lee, Alzheimer Center Amsterdam, Department of Neurology, Amsterdam UMC, Vrije Universiteit Amsterdam, 1081 HV Amsterdam, The Netherlands; Genomics of Neurodegenerative Diseases and Aging, Human Genetics, Amsterdam UMC, Vrije Universiteit Amsterdam, 1081 HV Amsterdam, The Netherlands.
Janna I R Dijkstra, Alzheimer Center Amsterdam, Department of Neurology, Amsterdam UMC, Vrije Universiteit Amsterdam, 1081 HV Amsterdam, The Netherlands; Genomics of Neurodegenerative Diseases and Aging, Human Genetics, Amsterdam UMC, Vrije Universiteit Amsterdam, 1081 HV Amsterdam, The Netherlands.
Yanaika S Hok-A-Hin, Neurochemistry Laboratory, Department of Clinical Chemistry, Amsterdam Neuroscience, Amsterdam UMC, Vrije Universiteit Amsterdam, 1081 HV Amsterdam, The Netherlands.
Elena R Blujdea, Neurochemistry Laboratory, Department of Clinical Chemistry, Amsterdam Neuroscience, Amsterdam UMC, Vrije Universiteit Amsterdam, 1081 HV Amsterdam, The Netherlands.
John C van Swieten, Alzheimer Center and Department of Neurology, Erasmus University Medical Center, 3015 GD Rotterdam, The Netherlands.
David J Irwin, Department of Neurology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA.
Alice Chen-Plotkin, Department of Neurology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA.
William T Hu, Department of Neurology, Emory University School of Medicine, Atlanta, GA 30307, USA.
Afina W Lemstra, Alzheimer Center Amsterdam, Department of Neurology, Amsterdam UMC, Vrije Universiteit Amsterdam, 1081 HV Amsterdam, The Netherlands.
Yolande A L Pijnenburg, Alzheimer Center Amsterdam, Department of Neurology, Amsterdam UMC, Vrije Universiteit Amsterdam, 1081 HV Amsterdam, The Netherlands.
Wiesje M van der Flier, Alzheimer Center Amsterdam, Department of Neurology, Amsterdam UMC, Vrije Universiteit Amsterdam, 1081 HV Amsterdam, The Netherlands; Department of Epidemiology and Data Science, Amsterdam Neuroscience, Amsterdam UMC, Vrije Universiteit Amsterdam, 1081 HV Amsterdam, The Netherlands.
Marta del Campo, Neurochemistry Laboratory, Department of Clinical Chemistry, Amsterdam Neuroscience, Amsterdam UMC, Vrije Universiteit Amsterdam, 1081 HV Amsterdam, The Netherlands; Departamento de Ciencias Farmacéuticas y de la Salud, Facultad de Farmacia, Universidad San Pablo-CEU, CEU Universities, 28003 Madrid, Spain; Barcelonabeta Brain Research Center (BBRC), Pasqual Maragall Foundation, 08005 Barcelona, Spain.
Charlotte E Teunissen, Neurochemistry Laboratory, Department of Clinical Chemistry, Amsterdam Neuroscience, Amsterdam UMC, Vrije Universiteit Amsterdam, 1081 HV Amsterdam, The Netherlands.
Lisa Vermunt, Neurochemistry Laboratory, Department of Clinical Chemistry, Amsterdam Neuroscience, Amsterdam UMC, Vrije Universiteit Amsterdam, 1081 HV Amsterdam, The Netherlands; Alzheimer Center Amsterdam, Department of Neurology, Amsterdam UMC, Vrije Universiteit Amsterdam, 1081 HV Amsterdam, The Netherlands.
Data availability
The data from the sporadic dementias cohort are available at https://www.synapse.org/PRIDE_AD. At the time of writing, submission of the data from the ADAD cohort to https://www.synapse.org has been initiated and is awaiting approval.
Funding
Research of the Alzheimer Center Amsterdam is part of the neurodegeneration research program of Amsterdam Neuroscience. Alzheimer Center Amsterdam is supported by Stichting Alzheimer Nederland and Stichting Steun Alzheimercentrum Amsterdam. The clinical database structure was developed with funding from Stichting Dioraphte. The chair (W.M.F.) is supported by the Pasman stichting. A.W.L. has received funding from Stichting Dioraphte, Alzheimer Nederland and ZonMW (project no. 733050509). M.C. is supported by the attraction talent fellowship of Comunidad de Madrid (2018-T2/BMD-11885) and ‘PROYECTOS I+D+I—2020’- Retos de investigación from the Ministerio Español de Ciencia e innovación (PID2020-115613RA-I00). Research programs of W.M.F. have been funded by ZonMW, Nederlandse Organisatie voor Wetenschappelijk Onderzoek, Seventh Framework Programme, EU Joint Programme – Neurodegenerative Disease Research, Alzheimer Nederland, Hersenstichting, Health∼Holland, stichting Dioraphte, Gieskes-Strijbis Fonds, stichting Equilibrio, Stichting Edwin Bouw fonds, Pasman stichting, stichting Alzheimer & Neuropsychiatrie Foundation, Philips, Biogen, Novartis, Roche, and Fujifilm. She is recipient of ABOARD, which is a public-private partnership receiving funding from ZonMW (#73305095007) and Health∼Holland, (PPP-allowance; #LSHM20106). C.E.T. is supported by the European Commission [Marie Curie International Training Network, grant agreement No. 860197 (MIRIADE)], Innovative Medicines Initiative (Horizon 2020, grant no 831434), European Platform for Neurodegenerative Research Consortium [IMI 2 Joint Undertaking (JU), grant No. 101034344] and JPND (bPRIDE), National MS Society (Progressive MS alliance), Alzheimer Association, Health~Holland, Nederlandse Organisatie voor Wetenschappelijk Onderzoek (ZonMW), Alzheimer Drug Discovery Foundation, Selfridges Group Foundation and Alzheimer Nederland. C.E.T. is the recipient of ABOARD, which is a public-private partnership receiving funding from ZonMW (#73305095007) and Health∼Holland (PPP-allowance; #LSHM20106). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Competing interests
A.W.L. has performed contract research with Axovant, EIP Pharma and Combinostics. All funding is paid to her institution. W.M.F. has performed contract research for Biogen MA Inc. and Boehringer Ingelheim, and has been an invited speaker at Boehringer Ingelheim, Biogen MA Inc, Danone, Eisai, WebMD Neurology (Medscape), NovoNordisk, Springer Healthcare and the European Brain Council. W.M.F. is consultant to Oxford Health Policy Forum CIC, Roche and Biogen MA Inc. She participated in the advisory boards of Biogen MA Inc., Roche and Eli Lilly. All funding is paid to her institution. W.M.F. is a member of the steering committee of PAVE and Think Brain Health. W.M.F. was associate editor of Alzheimer, Research & Therapy in 2020/2021 and is an associate editor at Brain. C.E.T. has a collaboration contract with ADx Neurosciences, Quanterix and Eli Lilly and performed contract research or received grants from AC-Immune, Axon Neurosciences, Bioconnect, Bioorchestra, Brainstorm Therapeutics, Celgene, EIP Pharma, Eisai, Fujirebio, Grifols, Instant Nano Biosensors, Merck, Novo Nordisk, PeopleBio, Roche, Siemens, Toyama and Vivoryon. She serves on the editorial boards of Medidact Neurologi, Springer, Alzheimer’s Research & Therapy and Neurology: Neuroimmunology & Neuroinflammation. She had speaker contracts for Roche, Grifols and Novo Nordisk. L.V. received a grant for the CORAL consortium by Olink. The remaining authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Bateman RJ, Aisen PS, De Strooper B, et al. . Autosomal-dominant Alzheimer’s disease: a review and proposal for the prevention of Alzheimer’s disease. Alzheimers Res Ther. 2011;3:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bateman RJ, Xiong C, Benzinger TL, et al. . Clinical and biomarker changes in dominantly inherited Alzheimer’s disease. N Engl J Med. 2012;367:795–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schindler SE, Fagan AM. Autosomal dominant Alzheimer disease: a unique resource to study CSF biomarker changes in preclinical AD. Front Neurol. 2015;6:142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lippa CF, Saunders AM, Smith TW, et al. . Familial and sporadic Alzheimer’s disease: neuropathology cannot exclude a final common pathway. Neurology. 1996;46:406–412. [DOI] [PubMed] [Google Scholar]
- 5. Morris JC, Weiner M, Xiong C, et al. . Autosomal dominant and sporadic late onset Alzheimer’s disease share a common in vivo pathophysiology. Brain. 2022;145:3594–3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ryman DC, Acosta-Baena N, Aisen PS, et al. . Symptom onset in autosomal dominant Alzheimer disease: a systematic review and meta-analysis. Neurology. 2014;83:253–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cairns NJ, Perrin RJ, Franklin EE, et al. . Neuropathologic assessment of participants in two multi-center longitudinal observational studies: the Alzheimer Disease Neuroimaging Initiative (ADNI) and the Dominantly Inherited Alzheimer Network (DIAN). Neuropathology. 2015;35:390–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mills SM, Mallmann J, Santacruz AM, et al. . Preclinical trials in autosomal dominant AD: implementation of the DIAN-TU trial. Rev Neurol (Paris). 2013;169:737–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Assarsson E, Lundberg M, Holmquist G, et al. . Homogenous 96-plex PEA immunoassay exhibiting high sensitivity, specificity, and excellent scalability. PLoS One. 2014;9:e95192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cui M, Cheng C, Zhang L. High-throughput proteomics: a methodological mini-review. Lab Invest. 2022;102:1170–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vromen EM, Del Campo Milán M, Scheltens P, Teunissen CE, Visser PJ, Tijms BM. CSF Proteomic signature predicts progression to Alzheimer’s disease dementia. Alzheimers Dement (NY). 2022;8:e12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gaetani L, Bellomo G, Parnetti L, Blennow K, Zetterberg H, Di Filippo M. Neuroinflammation and Alzheimer’s disease: a machine learning approach to CSF proteomics. Cells. 2021;10:1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Boström G, Freyhult E, Virhammar J, et al. . Different inflammatory signatures in Alzheimer’s disease and frontotemporal dementia cerebrospinal fluid. J Alzheimers Dis. 2021;81:629–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jiang Y, Zhou X, Ip FC, et al. . Large-scale plasma proteomic profiling identifies a high-performance biomarker panel for Alzheimer’s disease screening and staging. Alzheimers Dement. 2022;18:88–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dammer EB, Ping L, Duong DM, et al. . Multi-platform proteomic analysis of Alzheimer’s disease cerebrospinal fluid and plasma reveals network biomarkers associated with proteostasis and the matrisome. Alzheimers Res Ther. 2022;14:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ringman JM, Schulman H, Becker C, et al. . Proteomic changes in cerebrospinal fluid of presymptomatic and affected persons carrying familial Alzheimer disease mutations. Arch Neurol. 2012;69:96–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Muenchhoff J, Poljak A, Thalamuthu A, et al. . Changes in the plasma proteome at asymptomatic and symptomatic stages of autosomal dominant Alzheimer’s disease. Sci Rep. 2016;6:29078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Qin W, Li F, Jia L, et al. . Phosphorylated tau 181 serum levels predict Alzheimer’s disease in the preclinical stage. Front Aging Neurosci. 2022;14:900773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cruchaga C, Sung Y, Yang C, et al. . Multi-tissue proteomics identifies molecular signatures for sporadic and genetically defined Alzheimer disease cases. Preprint (Version 1). Res Sq. doi:1021203/rs3rs-923492/v1
- 20. Hansson O. Biomarkers for neurodegenerative diseases. Nat Med. 2021;27:954–963. [DOI] [PubMed] [Google Scholar]
- 21. Higginbotham L, Ping L, Dammer EB, et al. . Integrated proteomics reveals brain-based cerebrospinal fluid biomarkers in asymptomatic and symptomatic Alzheimer’s disease. Sci Adv. 2020;6:eaaz9360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. del Campo M, Peeters CFW, Johnson ECB, et al. . CSF Proteome profiling across the Alzheimer’s disease spectrum reflects the multifactorial nature of the disease and identifies specific biomarker panels. Nature Aging. 2022;2:1040–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. van der Flier WM, Scheltens P. Amsterdam Dementia cohort: performing research to optimize care. J Alzheimers Dis. 2018;62:1091–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Albert MS, DeKosky ST, Dickson D, et al. . The diagnosis of mild cognitive impairment due to Alzheimer’s disease: recommendations from the national institute on aging-Alzheimer’s association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7:270–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McKhann GM, Knopman DS, Chertkow H, et al. . The diagnosis of dementia due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7:263–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gorno-Tempini ML, Hillis AE, Weintraub S, et al. . Classification of primary progressive aphasia and its variants. Neurology. 2011;76:1006–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rascovsky K, Hodges JR, Knopman D, et al. . Sensitivity of revised diagnostic criteria for the behavioural variant of frontotemporal dementia. Brain. 2011;134(9):2456–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McKeith IG, Boeve BF, Dickson DW, et al. . Diagnosis and management of dementia with Lewy bodies: fourth consensus report of the DLB Consortium. Neurology. 2017;89:88–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Elahi FM, Miller BL. A clinicopathological approach to the diagnosis of dementia. Nat Rev Neurol. 2017;13:457–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lashley T, Rohrer JD, Mead S, Revesz T. Review: an update on clinical, genetic and pathological aspects of frontotemporal lobar degenerations. Neuropathol Appl Neurobiol. 2015;41:858–881. [DOI] [PubMed] [Google Scholar]
- 31. Teunissen CE, Petzold A, Bennett JL, et al. . A consensus protocol for the standardization of cerebrospinal fluid collection and biobanking. Neurology. 2009;73:1914–1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hok AHYS, Willemse EAJ, Teunissen CE, Del Campo M. Guidelines for CSF processing and biobanking: Impact on the identification and development of optimal CSF protein biomarkers. Methods Mol Biol. 2019;2044:27–50. [DOI] [PubMed] [Google Scholar]
- 33. Willemse EAJ, van Maurik IS, Tijms BM, et al. . Diagnostic performance of Elecsys immunoassays for cerebrospinal fluid Alzheimer’s disease biomarkers in a nonacademic, multicenter memory clinic cohort: the ABIDE project. Alzheimers Dement (Amst). 2018;10:563–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tijms BM, Willemse EAJ, Zwan MD, et al. . Unbiased approach to counteract upward drift in cerebrospinal fluid amyloid-β 1-42 analysis results. Clin Chem. 2018;64:576–585. [DOI] [PubMed] [Google Scholar]
- 35. Duits FH, Teunissen CE, Bouwman FH, et al. . The cerebrospinal fluid “Alzheimer profile”: easily said, but what does it mean? Alzheimers Dement. 2014;10:713–23.e2. [DOI] [PubMed] [Google Scholar]
- 36. Olink. https://olink.com/
- 37. Hok-A-Hin YS, Bolsewig K, Ruiters DN, et al. . Thimet oligopeptidase as a potential CSF biomarker for Alzheimer’s disease: a cross-platform validation study. Alzheimers Dement (Amst).2023;15:e12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhou Y, Zhou B, Pache L, et al. . Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10:1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Skene NG, Grant SG. Identification of vulnerable cell types in major brain disorders using single cell transcriptomes and expression weighted cell type enrichment. Front Neurosci. 2016;10:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Habib N, Avraham-Davidi I, Basu A, et al. . Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat Methods. 2017;14:955–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tenenbaum D, Maintainer B. KEGGREST: Client-side REST access to the Kyoto Encyclopedia of Genes and Genomes (KEGG). R package version 1.38.0. 2022.
- 42. Carlson M. org.Hs.eg.db: Genome wide annotation for Human. R package version 3.15.0. 2019.
- 43. Thomas AJ, Attems J, Colloby SJ, et al. . Autopsy validation of 123I-FP-CIT dopaminergic neuroimaging for the diagnosis of DLB. Neurology. 2017;88:276–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mabuchi H, Haba T, Tatami R, et al. . Effect of an inhibitor of 3-hydroxy-3-methyglutaryl coenzyme A reductase on serum lipoproteins and ubiquinone-10-levels in patients with familial hypercholesterolemia. N Engl J Med. 1981;305:478–482. [DOI] [PubMed] [Google Scholar]
- 45. Selkoe DJ, Hardy J. The amyloid hypothesis of Alzheimer’s disease at 25 years. EMBO Mol Med. 2016;8:595–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Paterson RW, Gabelle A, Lucey BP, et al. . SILK studies—capturing the turnover of proteins linked to neurodegenerative diseases. Nat Rev Neurol. 2019;15:419–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Global, regional, and national burden of neurological disorders, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Neurol. 2019;18:459–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Boyle PA, Yu L, Wilson RS, Leurgans SE, Schneider JA, Bennett DA. Person-specific contribution of neuropathologies to cognitive loss in old age. Ann Neurol. 2018;83:74–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schneider JA, Arvanitakis Z, Bang W, Bennett DA. Mixed brain pathologies account for most dementia cases in community-dwelling older persons. Neurology. 2007;69:2197–2204. [DOI] [PubMed] [Google Scholar]
- 50. Buckles VD, Xiong C, Bateman RJ, et al. . Different rates of cognitive decline in autosomal dominant and late-onset Alzheimer disease. Alzheimers Dement. 2022;18:1754–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chhatwal JP, Schultz SA, McDade E, et al. . Variant-dependent heterogeneity in amyloid β burden in autosomal dominant Alzheimer’s disease: cross-sectional and longitudinal analyses of an observational study. Lancet Neurol. 2022;21:140–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sun L, Zhou R, Yang G, Shi Y. Analysis of 138 pathogenic mutations in presenilin-1 on the in vitro production of Aβ42 and Aβ40 peptides by γ-secretase. Proc Natl Acad Sci U S A. 2017;114:E476–EE85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mann DM, Pickering-Brown SM, Takeuchi A, Iwatsubo T. Amyloid angiopathy and variability in amyloid beta deposition is determined by mutation position in presenilin-1-linked Alzheimer’s disease. Am J Pathol. 2001;158:2165–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ryan NS, Nicholas JM, Weston PSJ, et al. . Clinical phenotype and genetic associations in autosomal dominant familial Alzheimer’s disease: a case series. Lancet Neurol. 2016;15:1326–1335. [DOI] [PubMed] [Google Scholar]
- 55. Ryan NS, Rossor MN. Correlating familial Alzheimer’s disease gene mutations with clinical phenotype. Biomark Med. 2010;4:99–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Haapasalo A, Kovacs DM. The many substrates of presenilin/γ-secretase. J Alzheimers Dis. 2011;25:3–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Roher AE, Maarouf CL, Kokjohn TA. Familial presenilin mutations and sporadic Alzheimer’s disease pathology: is the assumption of biochemical equivalence justified? J Alzheimers Dis. 2016;50:645–658. [DOI] [PubMed] [Google Scholar]
- 58. Molinuevo JL, Ayton S, Batrla R, et al. . Current state of Alzheimer’s fluid biomarkers. Acta Neuropathol. 2018;136:821–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Rosén C, Andersson CH, Andreasson U, et al. . Increased levels of chitotriosidase and YKL-40 in cerebrospinal fluid from patients with Alzheimer’s disease. Dement Geriatr Cogn Dis Extra. 2014;4:297–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Abu-Rumeileh S, Steinacker P, Polischi B, et al. . CSF Biomarkers of neuroinflammation in distinct forms and subtypes of neurodegenerative dementia. Alzheimers Res Ther. 2019;12:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bacher M, Deuster O, Aljabari B, et al. . The role of macrophage migration inhibitory factor in Alzheimer’s disease. Mol Med. 2010;16(3–4):116–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Oikonomidi A, Tautvydaitė D, Gholamrezaee MM, Henry H, Bacher M, Popp J. Macrophage migration inhibitory factor is associated with biomarkers of Alzheimer’s disease pathology and predicts cognitive decline in mild cognitive impairment and mild dementia. J Alzheimers Dis. 2017;60:273–281. [DOI] [PubMed] [Google Scholar]
- 63. Whelan CD, Mattsson N, Nagle MW, et al. . Multiplex proteomics identifies novel CSF and plasma biomarkers of early Alzheimer’s disease. Acta Neuropathol Commun. 2019;7:169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bjerke M, Zetterberg H, Edman Å, Blennow K, Wallin A, Andreasson U. Cerebrospinal fluid matrix metalloproteinases and tissue inhibitor of metalloproteinases in combination with subcortical and cortical biomarkers in vascular dementia and Alzheimer’s disease. J Alzheimers Dis. 2011;27:665–676. [DOI] [PubMed] [Google Scholar]
- 65. Craig-Schapiro R, Kuhn M, Xiong C, et al. . Multiplexed immunoassay panel identifies novel CSF biomarkers for Alzheimer’s disease diagnosis and prognosis. PLoS One. 2011;6:e18850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Duits FH, Hernandez-Guillamon M, Montaner J, et al. . Matrix metalloproteinases in Alzheimer’s disease and concurrent cerebral microbleeds. J Alzheimers Dis. 2015;48:711–720. [DOI] [PubMed] [Google Scholar]
- 67. Lloyd-Burton SM, York EM, Anwar MA, Vincent AJ, Roskams AJ. SPARC regulates microgliosis and functional recovery following cortical ischemia. J Neurosci. 2013;33:4468–4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Liu X, Ying G, Wang W, et al. . Entorhinal deafferentation induces upregulation of SPARC in the mouse hippocampus. Brain Res Mol Brain Res. 2005;141:58–65. [DOI] [PubMed] [Google Scholar]
- 69. Vincent AJ, Lau PW, Roskams AJ. SPARC is expressed by macroglia and microglia in the developing and mature nervous system. Dev Dyn. 2008;237:1449–1462. [DOI] [PubMed] [Google Scholar]
- 70. Zhou M, Haque RU, Dammer EB, et al. . Targeted mass spectrometry to quantify brain-derived cerebrospinal fluid biomarkers in Alzheimer’s disease. Clin Proteomics. 2020;17:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Dayon L, Núñez Galindo A, Wojcik J, et al. . Alzheimer disease pathology and the cerebrospinal fluid proteome. Alzheimers Res Ther. 2018;10:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Li Y, Chen Z, Wang Q, et al. . Identification of hub proteins in cerebrospinal fluid as potential biomarkers of Alzheimer’s disease by integrated bioinformatics. J Neurol. 2022;270:1487–1500. [DOI] [PubMed] [Google Scholar]
- 73. Xiong F, Ge W, Ma C. Quantitative proteomics reveals distinct composition of amyloid plaques in Alzheimer’s disease. Alzheimers Dement. 2019;15:429–440. [DOI] [PubMed] [Google Scholar]
- 74. Drummond E, Kavanagh T, Pires G, et al. . The amyloid plaque proteome in early onset Alzheimer’s disease and Down syndrome. Acta Neuropathol Commun. 2022;10:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Rosenberg GA. Matrix metalloproteinases and their multiple roles in neurodegenerative diseases. Lancet Neurol. 2009;8:205–216. [DOI] [PubMed] [Google Scholar]
- 76. Yin KJ, Cirrito JR, Yan P, et al. . Matrix metalloproteinases expressed by astrocytes mediate extracellular amyloid-beta peptide catabolism. J Neurosci. 2006;26:10939–10948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Jeppsson A, Bjerke M, Hellström P, et al. . Shared CSF biomarker profile in idiopathic normal pressure hydrocephalus and subcortical small vessel disease. Front Neurol. 2022;13:839307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Minta K, Brinkmalm G, Al Nimer F, et al. . Dynamics of cerebrospinal fluid levels of matrix metalloproteinases in human traumatic brain injury. Sci Rep. 2020;10:18075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Adami PV M, Orellana A, García P, et al. . Matrix metalloproteinase 10 is linked to the risk of progression to dementia of the Alzheimer’s type. Brain. 2022;145:2507–2517. [DOI] [PubMed] [Google Scholar]
- 80. Zhang G, Murthy KD, Binti Pare R, Qian Y. Protective effect of Tβ4 on central nervous system tissues and its developmental prospects. Eur J Inflamm. 2020;18:2058739220934559. [Google Scholar]
- 81. Le Pera M, Urso E, Sprovieri T, et al. . Contribution of cerebrospinal fluid thymosin β4 levels to the clinical differentiation of Creutzfeldt-Jakob disease. Arch Neurol. 2012;69:868–872. [DOI] [PubMed] [Google Scholar]
- 82. Wang M, Feng LR, Li ZL, et al. . Thymosin β4 reverses phenotypic polarization of glial cells and cognitive impairment via negative regulation of NF-κB signaling axis in APP/PS1 mice. J Neuroinflammation. 2021;18:146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Sogorb-Esteve A, Swift IJ, Woollacott IOC, Warren JD, Zetterberg H, Rohrer JD. Differential chemokine alteration in the variants of primary progressive aphasia—A role for neuroinflammation. J Neuroinflammation. 2021;18:224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Miedema SSM, Mol MO, Koopmans FTW, et al. . Distinct cell type-specific protein signatures in GRN and MAPT genetic subtypes of frontotemporal dementia. Acta Neuropathol Commun. 2022;10:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Szeliga M. Peroxiredoxins in neurodegenerative diseases. Antioxidants (Basel). 2020;9:1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Filippov V, Song MA, Zhang K, et al. . Increased ceramide in brains with Alzheimer’s and other neurodegenerative diseases. J Alzheimers Dis. 2012;29:537–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Parveen F, Bender D, Law SH, Mishra VK, Chen CC, Ke LY. Role of ceramidases in sphingolipid metabolism and human diseases. Cells. 2019;8:1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Luís JP, Simões CJV, Brito RMM. The therapeutic prospects of targeting IL-1R1 for the modulation of neuroinflammation in central nervous system disorders. Int J Mol Sci. 2022;23:1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Italiani P, Puxeddu I, Napoletano S, et al. . Circulating levels of IL-1 family cytokines and receptors in Alzheimer’s disease: new markers of disease progression? J Neuroinflammation. 2018;15:342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Bader JM, Geyer PE, Müller JB, et al. . Proteome profiling in cerebrospinal fluid reveals novel biomarkers of Alzheimer’s disease. Mol Syst Biol. 2020;16:e9356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Johnson ECB, Carter EK, Dammer EB, et al. . Large-scale deep multi-layer analysis of Alzheimer’s disease brain reveals strong proteomic disease-related changes not observed at the RNA level. Nat Neurosci. 2022;25:213–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Wang H, Dey KK, Chen PC, et al. . Integrated analysis of ultra-deep proteomes in cortex, cerebrospinal fluid and serum reveals a mitochondrial signature in Alzheimer’s disease. Mol Neurodegener. 2020;15:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Visser PJ, Reus LM, Gobom J, et al. . Cerebrospinal fluid tau levels are associated with abnormal neuronal plasticity markers in Alzheimer’s disease. Mol Neurodegener. 2022;17:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Carlyle BC, Kitchen RR, Mattingly Z, et al. . Technical performance evaluation of olink proximity extension assay for blood-based biomarker discovery in longitudinal studies of Alzheimer’s disease. Front Neurol. 2022;13:889647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Pedrero-Prieto CM, García-Carpintero S, Frontiñán-Rubio J, et al. . A comprehensive systematic review of CSF proteins and peptides that define Alzheimer’s disease. Clin Proteomics. 2020;17:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Wesenhagen KEJ, Teunissen CE, Visser PJ, Tijms BM. Cerebrospinal fluid proteomics and biological heterogeneity in Alzheimer’s disease: a literature review. Crit Rev Clin Lab Sci. 2020;57:86–98. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data from the sporadic dementias cohort are available at https://www.synapse.org/PRIDE_AD. At the time of writing, submission of the data from the ADAD cohort to https://www.synapse.org has been initiated and is awaiting approval.