Abstract
Parkinson’s disease has a large heritable component and genome-wide association studies have identified over 90 variants with disease-associated common variants, providing deeper insights into the disease biology. However, there have not been large-scale rare variant analyses for Parkinson’s disease.
To address this gap, we investigated the rare genetic component of Parkinson’s disease at minor allele frequencies <1%, using whole genome and whole exome sequencing data from 7184 Parkinson’s disease cases, 6701 proxy cases and 51 650 healthy controls from the Accelerating Medicines Partnership Parkinson's disease (AMP-PD) initiative, the National Institutes of Health, the UK Biobank and Genentech. We performed burden tests meta-analyses on small indels and single nucleotide protein-altering variants, prioritized based on their predicted functional impact.
Our work identified several genes reaching exome-wide significance. Two of these genes, GBA1 and LRRK2, have variants that have been previously implicated as risk factors for Parkinson’s disease, with some variants in LRRK2 resulting in monogenic forms of the disease. We identify potential novel risk associations for variants in B3GNT3, AUNIP, ADH5, TUBA1B, OR1G1, CAPN10 and TREML1 but were unable to replicate the observed associations across independent datasets. Of these, B3GNT3 and TREML1 could provide new evidence for the role of neuroinflammation in Parkinson’s disease. To date, this is the largest analysis of rare genetic variants in Parkinson’s disease.
Keywords: Parkinson’s disease, burden, GBA1, LRRK2, genetics, rare variant
Makarious et al. investigate the contribution of rare genetic variants, with minor allele frequencies <1%, to Parkinson’s disease risk. They confirm the known associations between rare variants in GBA1 and LRRK2 and Parkinson’s disease risk in individuals of European ancestry, and identify additional novel risk associations.
Introduction
Parkinson’s disease (PD) is a complex neurological disease likely caused by an interplay between ageing, environmental factors and genetics. While the role of common genetic variants in PD has been extensively studied using large genome-wide association studies (GWAS), rare variants can also contribute to familial and sporadic disease. To date, over 90 independent risk signals have been associated with PD, including common variants in close proximity to SNCA, TMEM175 and MAPT.1,2 Most of the risk alleles found by array-based GWAS have frequencies over 5% in the population of interest, often reside in non-coding regions of the genome, and typically have small effect sizes. In contrast, rare damaging and pathogenic variants implicated in PD, such as coding variants in SNCA3 and PRKN,4 have traditionally been identified using family-based approaches. One aspect of major interest in disease genetics is the large number of pleomorphic genes, where multiple variants of varying allele frequency present with a wide range of effect sizes.5 For example, in PD, GWAS identified common variants with moderate effects near GBA1, GCH1, LRRK2, SNCA and VPS13C,1 while familial studies identified rare variants in the same genes resulting in more damaging effects (e.g. LRRK2 p.G2019S and SNCA p.A53T).6–9
In contrast to common variants, there have been no large-scale efforts investigating the role of rare variants in PD on a genome-wide scale. Although rare variant associations for several PD genes (such as ARSA and ATP10B) have been reported in candidate gene studies,10,11 these genes remain controversial due to lack of replication in independent PD datasets.12–15 One of the main challenges that comes with analysing rare variants is that the quality and reliability of imputation procedures decreases with allele frequency. Since genome-wide genotyping methods are currently much cheaper than sequencing, most large datasets used for GWAS rely on imputed genotype data. A strength of the present study is that we focused on using whole genome (WGS) and whole exome sequencing (WES) to facilitate the analysis of rare variants. We performed the largest genome-wide analysis of rare variants in PD to date, investigating 7184 PD cases, 6701 proxy cases (defined as having a parent or sibling with PD) and 51 650 neurologically healthy controls of European ancestry from several large sequencing efforts. Using these data, we executed gene-level burden testing in order to understand how moderate- to large-effect rare variants contribute to the genetic aetiology of PD.
Materials and methods
Accelerating Medicines Partnership in Parkinson’s Disease and National Institutes of Health genome sequencing data
Whole genome sequencing data was obtained from multiple datasets including the Parkinson’s Progression Markers Initiative (PPMI), the Parkinson’s Disease Biomarkers Program (PDBP) and the Harvard Biomarker Study (HBS), BioFIND, SURE-PD3 and STEADY-PD3 as part of the Accelerating Medicines Partnership in Parkinson’s Disease (AMP-PD) initiative. Several other datasets were sequenced in parallel at the Laboratory of Neurogenetics (LNG) and the US Uniformed Services University (USHUS), including samples from the National Institutes of Health (NIH) PD clinic, the United Kingdom Brain Expression Consortium (UKBEC),16 the North American Brain Expression Consortium (NABEC)17 and Wellderly.18 All cohorts from AMP-PD (PPMI, PDBP, HBS, BioFIND, SURE-PD3 and STEADY-PD3) were processed using the Genome Analysis Toolkit (GATK) Best Practices guidelines set by the Broad Institute’s joint discovery pipeline and elaborated on elsewhere.19 All other cohorts were joint called separate from AMP-PD but in a similar manner, also from the processed WGS data following the GATK Best Practices using the Broad Institute’s workflow for joint discovery and Variant Quality Score Recalibration (VQSR).20 Data processing and quality control (QC) procedures have been described previously.19,21 Reported elsewhere, these sequencing metrics had a median/mean coverage between 33.3× and 35.0×.19 Additional quality control was performed to exclude closely related individuals (PI_HAT >0.125) by selecting one sample at random using PLINK (v1.922). All individuals were of European ancestry as confirmed by principal component analysis using HapMap3 European ancestry populations. Individuals recruited as part of a biased and/or genetic dataset, such as LRRK2 and GBA1 variant carriers within a specific effort of PPMI, were excluded from this analysis. Including all variants within the gene boundaries, a minimum allele count (MAC) threshold of 1 was applied. Exonic regions were subset from the whole genome sequencing data using the exome calling regions from gnomAD lifted over to hg38.23
UK Biobank
Exome sequencing data from a total of 200 643 individuals (OQFE dataset, field codes: 23151 and 23155) were downloaded from the UK Biobank in December of 2020.24 As described elsewhere, the UK Biobank Exome Sequencing Consortium sequenced these exomes with 95.8% of targeted bases covered at a depth of 20× or higher.25 Standard quality control was performed to exclude non-European outliers. Closely related individuals (PI_HAT >0.125) were excluded by selecting one sample at random using PLINK (v1.922). Standard exome sequencing data filtering was applied using suggested parameters as described in previous UK Biobank exome sequencing studies.25
UK Biobank phenotype data were obtained from ICD10 codes (field code: 41270), PD (field code: 131023), illnesses of father and mother (field codes: 20107 and 20110), parkinsonism (field code: 42031) or dementia (field code: 42018), genetic ethnic grouping (field code: 22006), year of birth (field code: 34) and age of recruitment (field code: 21022). Cases were defined as any individual identified as having PD using the above field code. Proxy cases were defined as having a parent or sibling with PD as previously reported.1 Controls were filtered to exclude any individuals with an age of recruitment <59 years, any reported nervous system disorders (Category 2406), a parent with PD or dementia (field codes: 20107 and 20110) and any reported neurological disorder (field codes: dementia/42018, vascular dementia/42022, frontotemporal dementia/42024, amyotrophic lateral sclerosis/42028, parkinsonism/42030, Parkinson’s disease/42032, progressive supranuclear palsy/42034, multiple system atrophy/42036).
Genentech
Whole genome sequencing data from Genentech included a total of 2710 PD cases and 8994 individuals used as controls. Cases of PD included 2318 individuals from 23andMe, a subset of those included in the analysis by Chang and colleagues26 who were contacted and provided consent for this analysis. An additional 392 PD cases were obtained from the Roche clinical trial TASMAR. Individuals included as controls were obtained from various Genentech clinical trials/studies and included cases for four diseases that do not share notable heritability with PD: age-related macular degeneration (n = 1735), asthma (n = 3398), idiopathic pulmonary fibrosis (n = 1532) and rheumatoid arthritis (n = 2329). Illumina HiSeq based 30× genome sequencing was performed on all samples using 150 bp paired-end reads. Genotypes with a genotype quality (GQ) < 20 were labelled as missing. The reads were then mapped to the GRCh38 reference genome with the Burrows–Wheeler Aligner (BWA),27 followed by application of GATK27,28 for base quality score recalibration, indel realignment and duplicate removal. This was followed by single nucleotide polymorphism (SNP) and insertion/deletion (indel) discovery and genotyping across all samples simultaneously using variant quality score recalibration according to GATK Best Practices recommendations.29–31 The 11 704 samples included in these analyses passed the following QC steps: genotype missing rate < 0.1, no sample pair had kinship coefficient (k0, i.e. probability of zero alleles shared identical-by-descent; or the value Z0 reported by PLINK’s–genome module) < 0.4; and no sample was an outlier in five iterations of outlier removal using principal component analysis (PCA).32
Variant annotation
Variants were annotated using the SnpEff and SnpSift annotation softwares (v4.3t33) as well as the Ensembl Variant Effect Predictor (VEP; v10434) package. Both the Combined Annotation Dependent Depletion (CADD; v1.435) and the Loss-of-Function (LoF) Transcript Effect Estimator (LOFTEE; v1.0223) VEP plugins were used. SnpEff is a toolbox based on 38 000 genomes that is designed to annotate genetic variants and predict their downstream functional consequences. SnpSift leverages multiple databases to filter SnpEff outputs and prioritize variants, and can predict amino acid changes as having ‘moderate’ or ‘high’ impact. The CADD plugin for VEP is a tool used to score the deleteriousness of single nucleotide variants, insertions and deletions. A CADD Phred score is a scaled measure of deleteriousness, with a score of 20 indicating that the variant is among the top 1% of deleterious variants in the genome.35 The LOFTEE plugin for VEP is uniquely designed to assess stop-gain, frameshift and splice-site disrupting variants and classify these as LoF with either low or high confidence. The following variant classes were used for gene burden analyses: (i) missense variants as defined by SnpEff; (ii) moderate or high impact variants as defined by SnpEff/SnpSift; (iii) high confidence LoF variants as defined by LOFTEE; and (iv) variants with either a CADD Phred score >20 or high confidence LoF variants as defined by LOFTEE.
Gene burden analysis and meta-analysis
The AMP-PD and NIH datasets were merged prior to gene burden analysis, with 3848 duplicates removed prior to analysis. Rare variant testing for this merged dataset, the UK Biobank case–control dataset, and the UK Biobank proxy control datasets were performed using the Sequence Kernel Association Test–Optimal (SKAT-O) and the Combined and Multivariate Collapsing (CMC) Wald algorithms.36,37 These algorithms were run using the RVtests package (v2.1.038). The CMC Wald test collapses and combines all rare variants and then performs a Wald test, where only an alternative model is fit and the effect size is estimated.39 SKAT-O is an optimized sequencing kernel association test designed to combat limitations introduced by the SKAT and burden tests. SKAT-O aggregates the associations between variants and the phenotype of interest while allowing for SNP–SNP interactions and has been proven to detect genes more reliably than a burden or SKAT test separately by adaptively selecting the best linear combination of both SKAT and burden tests to maximize test power.40 All analyses were stratified by the four variant classes described above and by maximum minor allele frequencies (MAF) levels of 1% and 0.1%. For Genentech data, SKAT-O and CMC-Wald tests were performed using the R package SKAT.41
The combined AMP-PD and NIH dataset was adjusted for sex, age and the first five principal components. The UK Biobank datasets were adjusted for sex, Townsend scores and the first five principal components. For the UK Biobank analyses, only neurologically healthy controls 60 years and older were included in analyses, and therefore age was not included as a covariate. Meta-analyses of the resulting summary statistics per gene were performed using custom Python (v3.7) scripts, which we have made available on our GitHub (https://github.com/neurogenetics/PD-BURDEN). In summary, the two meta-analysis approaches used in this study were: (i) a combined P-value approach using Fisher’s test; and (ii) a weighted Z-score approach. In previous studies, Fisher’s method was reported to detect >75% of causal effects (either deleterious or protective) that are in the same direction.42 Unless otherwise stated, all results reported in this manuscript correspond to the SKAT-O rare variant test, and all meta-analyses were performed using the combined P-values reported following Fisher’s test.
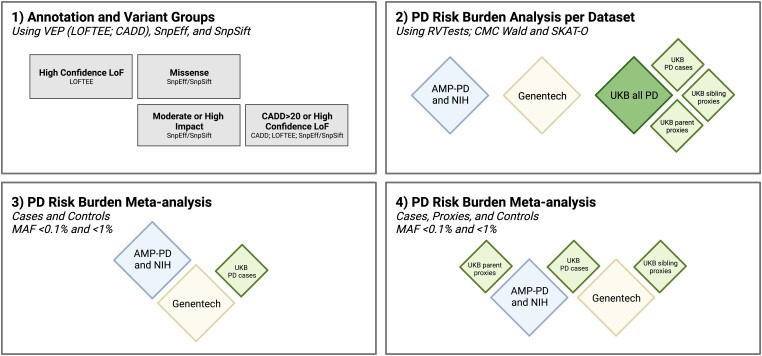
Rare variant analyses were performed on each dataset separately and all data is using genome build hg38. Two joint meta-analyses were performed as follows: (i) a case-control meta-analysis between the combined AMP-PD and NIH dataset, the Genentech dataset and the UK Biobank case-control dataset; and (ii) a meta-analysis of the case-control and proxy control results from the combined AMP-PD and NIH dataset, the Genentech dataset, the UK Biobank PD case-control dataset, the UK Biobank sibling proxy cases dataset and the UK Biobank parent proxy cases dataset. A summary of the analysis workflow is outlined in Fig. 1.
Figure 1.
Graphical representation of the analysis workflow. (1) Annotation was performed using Variant Effect Predictor (VEP) and four variant groups were selected: (i) missense variants as defined by SnpEff; (ii) moderate or high impact variants as defined by SnpEff/SnpSift; (iii) high confidence Loss-of-Function (LoF) variants as defined by LOFTEE; and (iv) variants with either a CADD Phred score >20 or high confidence LoF variants as defined by LOFTEE. (2) Burden analysis was performed on each dataset separately at rare (minor allele frequency, MAF < 1%) and ultra-rare (MAF < 0.1%) cut-offs. (3) Meta-analysis Strategy 1 using only Parkinson's disease (PD) cases and controls, otherwise referred to as the ‘case-control’ meta-analysis. (4) Meta-analysis Strategy 2 using PD cases, PD proxy cases (siblings and parent) and controls, otherwise referred to as the ‘case-control-proxies’ meta-analysis. UKB = UK Biobank.
Power calculations
One hundred gene simulations were run using the power calculation function with default European haplotypes made available in the SKAT R package (v2.0.140). The total sample size was estimated at 65 535, with 7184 PD cases, 6701 proxy cases down-weighted to one-fourth of a PD case (corresponding to 1675 cases), and 51 650 controls resulting in a case proportion of 13.5%. We estimated the disease prevalence of PD at 1% as previously estimated43 and used an exome-wide significance threshold calculated by assuming 20 000 protein-coding genes, resulting in a Bonferroni correction of 2.50 × 10−6. Since we used two different algorithms for burden testing, we set the final threshold of significance to 1 × 10−6. Power calculations based on varying percentages of causality (10%, 5%, 3%, 1% and 0.5%) and causal MAF (0.05%, 0.1%, 0.5%, 1%, 3% and 5%) are reported in Supplementary Table 5. Assuming at least 3% of the rare alleles tested are causal, this analysis has ≥80% power to detect associations at the tested MAF cut-offs (Supplementary Table 5).
Results
Study overview
A total of 7184 PD cases, 6701 sibling/parent proxy cases and 51 650 controls with whole genome (AMP-PD, NIH and Genentech) or exome (UK Biobank) sequencing were included in this analysis (Table 1). Rare variant gene-level burden tests were performed across all genes for four variant classes and two MAF cut-offs (Fig. 1). As expected, we observed that more deleterious variant classes resulted in fewer variants tested per gene. For a full overview of the frequency and number of variants within each gene in cases and controls, stratified by variant class and cohort (excluding Genentech), see Supplementary Tables 11–14.
Table 1.
Datasets overview after quality control
| Dataset | Sample size | Agea (Mean ± SD) | Sex (male; %) | |||
|---|---|---|---|---|---|---|
| Cases | Controls | Cases | Controls | Cases sex (male; %) | Controls sex (male; %) | |
| AMP-PD and NIH Genomes (includes: PPMI, PDBP, HBS, BioFIND, NIH PD clinic, UKBEC, NABEC) | 3369 | 4605 | 62.1 (11.8) | 71.9 (16.2) | 63.6 | 47.6 |
| UKB case-control (WES) | 1105 | 5643 | 62.9 (5.24) | 64.1 (2.84) | 62.4 | 47.6 |
| UKB sibling proxy-control (WES) | 668b | 3463 | 62.2 (5.59) | 64.1 (2.83) | 45.5 | 49.5 |
| UKB parent proxy-control (WES) | 6033b | 28 945 | 58.1 (7.23) | 64.1 (2.82) | 42.5 | 48.7 |
| Genentech case-control (WGS) | 2710 | 8994 | 64.7 (10.4) | 59.2 (15.6) | 59.2 | 40.7 |
| Total | 7184 cases; 6701 proxies | 51 650 controls | – | – | – | – |
AMP-PD = Accelerating Medicines Partnership Parkinson’s disease; HBS = Harvard Biomarker Study; NABEC = North American Brain Expression Consortium; NIH = National Institutes of Health; PDBP = Parkinson’s disease Biomarkers Project; PPMI = Parkinson's Progression Markers Initiative; UKB = UK Biobank; UKBEC = UK Brain Expression Consortium; WES = whole-exome sequences; WGS = whole-genome sequences.
Age for AMP-PD and NIH datasets reported at recruitment or baseline, ages reported for UK Biobank datasets at recruitment, ages reported for Genentech at recruitment.
Indicates proxy cases.
Genetic burden testing in large Parkinson's disease case–control datasets
Initial gene burden analyses per dataset (AMP-PD and NIH Genomes, Genentech, UK Biobank cases, UK Biobank sibling proxies and UK Biobank parent proxies) resulted in several known PD genes (e.g. GBA1 and LRRK2) reaching significance exome-wide (P < 1 × 10−6; Tables 2 and 3 and Supplementary Tables 15 and 16), confirming the validity of our approach. Lambda values per dataset showed minimal genomic inflation when adjusted for the number of cases, proxy cases and controls (λ1000; Supplementary Table 3). As expected, datasets with smaller sample sizes, such as the UK Biobank sibling proxy control dataset, resulted in increased genomic deflation when analysed separately (λ1000 < 0.9).
Table 2.
Genes reaching exome-wide significance (P < 1 × 10−6) in MAF <1% in meta-analyses and individual datasets following SKAT-O
| Variant class (MAF <1%) | Gene | Case only meta P-value | Case proxies meta P-value | AMP-PD and NIH P-value | GNE P-value | UKB case P-value | UKB sibling P-value | UKB parent P-value |
|---|---|---|---|---|---|---|---|---|
| Missense | GBA1 ** | 3.27 × 10−14 | 1.46 × 10−21 | 1.05 × 10−5 | 1.32 × 10−8 | 3.14 × 10−4 | 0.247 | 2.15 × 10−10 |
| LRRK2 * | 7.15 × 10−7 | 9.46 × 10−6 | 1.96 × 10−7 | 0.047 | 0.372 | 0.615 | 0.482 | |
| Moderate or high impact | GBA1 ** | 9.10 × 10−15 | 1.32 × 10−22 | 1.05 × 10−5 | 5.70 × 10−8 | 1.89 × 10−5 | 0.073 | 2.15 × 10−10 |
| LRRK2 * | 7.23 × 10−7 | 9.85 × 10−6 | 2.09 × 10−7 | 0.040 | 0.413 | 0.584 | 0.527 | |
| TUBA1B | 0.69 | 9.02 × 10−5 | NA | 0.647 | 0.501 | 0.352 | 9.48 × 10−7 | |
| LoF | B3GNT3 ** | 4.40 × 10−9 | 3.36 × 10−9 | NA | 4.40 × 10−9 | NA | NA | 0.032 |
| CAPN10 ** | 3.60 × 10−7 | 7.84 × 10−7 | NA | 0.005 | 3.75 × 10−6 | 0.053 | 0.394 | |
| CADD > 20 or LoF | GBA1 ** | 3.72 × 10−14 | 9.12 × 10−22 | 1.24 × 10−5 | 6.99 × 10−8 | 5.77 × 10−5 | 0.130 | 2.15 × 10−10 |
| LRRK2 * | 2.49 × 10−7 | 4.22 × 10−6 | 2.23 × 10−7 | 0.012 | 0.409 | 0.735 | 0.485 | |
| ADH5 | 4.62 × 10−6 | 6.15 × 10−5 | 0.512 | 0.170 | 3.13 × 10−7 | 0.491 | 0.768 | |
| OR1G1 | 0.215 | 6.56 × 10−6 | 0.848 | 0.029 | 0.620 | 6.58 × 10−7 | 0.063 |
AMP-PD = Accelerating Medicines Partnership in Parkinson’s Disease; GNE = Genentech; LoF = loss-of-function; MAF = minor allele frequency; NIH = National Institutes of Health; UKB = UK Biobank.
Denotes genes that pass exome-wide significance (P < 1 × 10−6) in one meta-analysis.
Denotes genes that pass exome-wide significance (P < 1 × 10−6) in both meta-analyses.
Table 3.
Genes reaching exome-wide significance (P < 1 × 10−6) in MAF < 0.1% in meta-analyses and individual datasets following SKAT-O
| Variant class (MAF <1%) | Gene | Case only meta P-value | Case proxies meta P-value | AMP-PD and NIH P-value | GNE P-value | UKB case P-value | UKB sibling P-value | UKB parent P-value |
|---|---|---|---|---|---|---|---|---|
| Missense | GBA1 * | 1.86 × 10−5 | 4.48 × 10−12 | 0.022 | 2.30 × 10−2 | 2.55 × 10−4 | 5.41 × 10−3 | 6.86 × 10−8 |
| Moderate or high impact | GBA1 * | 1.71 × 10−6 | 4.87 × 10−16 | NA | 0.088 | 1.13 × 10−6 | 0.001 | 5.13 × 10−10 |
| AUNIP ** | 1.54 × 10−8 | 2.70 × 10−7 | NA | 0.023 | 3.04 × 10−8 | 0.170 | 1 | |
| TUBA1B | 0.690 | 9.02 × 10−5 | NA | 0.647 | 0.501 | 0.352 | 9.48 × 10−7 | |
| TREML1 * | 0.048 | 3.58 × 10−7 | NA | 0.010 | 0.858 | 0.001 | 1.41 × 10−5 | |
| LoF | B3GNT3** | 4.40 × 10−9 | 3.36 × 10−9 | NA | 4.40 × 10−9 | NA | NA | 0.032 |
| AUNIP ** | 1.64 × 10−8 | 2.04 × 10−7 | NA | 0.024 | 3.13 × 10−8 | 0.116 | 1 | |
| CAPN10 ** | 3.60 × 10−7 | 7.84 × 10−7 | NA | 0.005 | 3.75 × 10−6 | 0.053 | 0.394 | |
| CADD > 20 or LoF | GBA1 ** | 2.33 × 10−7 | 1.20 × 10−14 | 0.017 | 0.127 | 4.56 × 10−7 | 8.93 × 10−4 | 7.89 × 10−8 |
| LRRK2 | 3.46 × 10−6 | 2.65 × 10−6 | 0.727 | 6.15 × 10−7 | 0.044 | 0.771 | 0.014 | |
| AUNIP * | 2.12 × 10−7 | 1.53 × 10−6 | 0.886 | 0.032 | 3.15 × 10−8 | 0.125 | 1 | |
| OR1G1 | 0.215 | 6.56 × 10−6 | 0.848 | 0.029 | 0.620 | 6.58 × 10−7 | 0.063 |
AMP-PD = Accelerating Medicines Partnership in Parkinson’s Disease; GNE = Genentech; LoF = loss-of-function; MAF = minor allele frequency; NA = not applicable; NIH = National Institutes of Health; UKB = UK Biobank.
Denotes genes that pass exome-wide significance (P < 1 × 10−6) in one meta-analysis.
Denotes genes that pass exome-wide significance (P < 1 × 10−6) in both meta-analyses.
Rare variant burden analysis of both GBA1 and LRRK2 reached significance exome-wide in the initial analysis of missense, moderate/high impact, and LoF or highly deleterious (CADD Phred > 20) variants. In our analyses, we focused on LoF variants to limit the scope of burden testing to rare variants that are the most likely to be highly deleterious. GBA1 was significant for these variant categories in both the Genentech (P = 1.32 × 10−8, P = 5.70 × 10−8 and P = 6.99 × 10−8, respectively) and UK Biobank parent proxies (P = 2.15 × 10−10, P = 2.15 × 10−10 and 2.15 × 10−10, respectively) datasets. LRRK2 was significant for these categories in the combined AMP-PD and NIH dataset (P = 1.96 × 10−7, P = 2.09 × 10−7 and P = 2.23 × 10−7, respectively); note that the ‘LRRK2 LoF variants only’ variant class was not significant, which is in line with previously reported data.44 LoF variants in B3GNT3 were significant exome-wide in the Genentech dataset (P = 4.40 × 10−9) and replicated at nominal significance in the UK Biobank parent proxies dataset [P = 0.032; Supplementary Figs 3–9 for Genentech (hg38: chr19:17807816:T:G; chr19:17807816:T:G; chr19:17807816:T:G) and UK Biobank (hg38: chr19:17807982:GC:G; chr19:17808033:C:T; chr19:17812105:C:CA)]. Moderate and high impact variants in TUBA1B were significant in the UK Biobank parent proxies dataset (P = 9.48 × 10−7). LoF or highly deleterious variants in ADH5 were significant in the UK Biobank cases-control dataset (P = 3.13 × 10−7), and LoF or highly deleterious variants in OR1G1 were significant in the UK Biobank sibling proxies dataset (P = 6.58 × 10−7; Table 2 and Supplementary Table 16).
Ultra-rare variant (MAF < 0.1%) burden analysis of missense, moderate/high impact, and LoF or highly deleterious variants in GBA1 were significant exome-wide in the UK Biobank parent proxies dataset (P = 6.88 × 10−8, P = 5.13 × 10−10 and P = 7.89 × 10−8, respectively). LoF or highly deleterious variants in GBA1 were also significant in the UK Biobank case-control dataset (P = 4.56 × 10−7). LoF or highly deleterious variants in LRRK2 were significant in the Genentech dataset (P = 6.15 × 10−7). Moderate/high impact variants in AUNIP were significant in the UK Biobank case-control dataset (P = 3.04 × 10−8) and TUBA1B in the UK Biobank parent proxies dataset (P = 9.48 × 10−7). LoF variants in B3GNT3 were significant in the Genentech dataset (P = 4.40 × 10−9) and AUNIP in the UK Biobank case-control dataset (P = 3.13 × 10−8). LoF or highly deleterious variants in AUNIP were significant in the UK Biobank case-control dataset (P = 3.15 × 10−8), and LoF or highly deleterious variants in OR1G1 were significant in the UK Biobank sibling proxies dataset (P = 6.58 × 10−7). Ultra-rare variant burden analysis identified no significant genes exome-wide in any of the four variant classes within the AMP-PD and NIH genomes (P < 1 × 10−6; Table 3 and Supplementary Table 15).
Meta-analyses of large Parkinson's disease datasets
The first meta-analysis (herein called the case-control meta-analysis) excluded any UK Biobank proxy cases. The second meta-analysis (herein called the case-control-proxies meta-analysis) included UK Biobank proxy cases in addition to cases and controls. No significant divergence from expected lambda values (range: 0.97–1.00) were detected in any of the meta-analyses performed (Supplementary Table 4). Rare variant burden analysis of missense, moderate/high impact, and LoF or highly deleterious variants in GBA1 were significant exome-wide across both meta-analyses (case-control P = 3.27 × 10−14, P = 9.10 × 10−15 and P = 3.722 × 10−14, respectively; and case-control-proxies P = 1.46 × 10−21, P = 1.32 × 10−22 and P = 9.12 × 10−22, respectively). High confidence LoF variants in CAPN10 (case-control P = 3.60 × 10−7, case-control-proxies P = 7.84 × 10−7) and B3GNT3 (case-control P = 4.40 × 10−9, case-control-proxies P = 3.36 × 10−9) were also significant exome-wide (Table 2).
Ultra-rare variant burden analysis of moderate/high impact variants and high confidence LoF variants in AUNIP were significant exome-wide across both meta-analyses (case-control P = 1.54 × 10−8 and P = 1.64 × 10−8, respectively; and case-control-proxies P = 2.70 × 10−7 and P = 2.04 × 10−7, respectively). Moderate/high impact variants in TREML1 were significant with the inclusion of proxy cases. As in the rare variant burden analysis, ultra-rare LoF variants in CAPN10 (case-control P = 3.60 × 10−7, case-control-proxies P = 7.84 × 10−7) and B3GNT3 (case-control P = 4.40 × 10−9, case-control-proxies P = 3.36 × 10−9) were also significant. Notably, both rare (MAF < 1%) and ultra-rare (MAF < 0.1%) GBA1 variants showed significant associations with PD risk (Tables 2 and 3).
B3GNT3 was identified in the high confidence LoF variant class group, with P-values of 4.40 × 10−9 in the Genentech dataset and P = 0.032 in the UK Biobank parent proxies. However, no variants meeting this criteria were present in the AMP-PD and NIH genomes, so the association of rare LoF variants in B3GNT3 could not be confirmed. The majority of novel candidate genes identified in this study (B3GNT3, AUNIP, ADH5, TUBA1B, OR1G1, CAPN10 and TREML1) only reached significance exome-wide using the SKAT-O test (Supplementary Table 7). Full results from the SKAT-O and CMC Wald burden tests performed for each variant class, MAF cut-off, and meta-analysis group can be found on our GitHub repository (https://github.com/neurogenetics/PD-BURDEN).
Conditional LRRK2 analysis
Since LRRK2 p.G2019S is a relatively common risk factor for PD, we explored whether the rare variant association at LRRK2 is driven primarily by this variant. For these analyses, LRRK2 p.G2019S status per individual was coded as 0, 1 or 2 depending on the allelic dosage, allowing us to condition on LRRK2 p.G2019S status without removing carriers. Allelic status was then included as a covariate for the burden analyses. The observed association at LRRK2 was lost (P > 0.05) after conditioning on the allelic status of LRRK2 p.G2019S for all of the tested variant categories and MAF thresholds in the discovery datasets (excluding Genentech; Supplementary Table 8). Besides LRRK2 p.G2019S, no other substantial coding risk in LRRK2 was detected. However, it is important to note that other previously identified rare coding variants that have been shown to increase risk to PD were not detected in this study, including LRRK2 p.R1441H.45
Assessment of previously reported Parkinson's disease causal or high risk genes and GWAS regions
We next assessed a large number of genes that showed rare variant associations with PD in previous studies (for a full list, see Supplementary Table 9; for frequencies and number of variants for each gene, variant class and dataset, see Supplementary Tables 11–14). Besides the previously discussed GBA1 and LRRK2, none of these genes met exome-wide significance (P > 1 × 10−6) in our analysis. However, we did observe sub-significant association signals for LoF or highly deleterious variants in ARSA (P = 8.73 × 10−5) and DNAJC6 (P = 8.08 × 10−4; Supplementary Tables 9, 11 and 12). Since we did not detect a P-value of interest in PRKN (P = 0.30), which has been robustly associated with predominantly early onset PD in previous studies, we investigated the enrichment of possible homozygous and potentially compound heterozygous PRKN mutations in PD. In the most stringent variant class (LoF or highly deleterious variants), we found a frequency of 0.41% in cases and 0.07% in controls in the combined AMP-PD and NIH dataset (Supplementary Table 6). We also did not detect P-values of interest in well-established autosomal dominant genes including: VPS35 (present only in Genentech dataset, nvariants = 4; lowest meta P-valuecase-control meta-analysis = 0.235; MAF = 0.001; high confidence LoF variants; Supplementary Tables 11 and 13), or SNCA (lowest meta P-valuecase-proxy-control meta-analysis = 0.274; MAF = 0.001; CADD > 20 or LoF variants; Supplementary Tables 11 and 13), with mutations in the gene previously associated with an earlier onset (<50 years) and more severe form of PD. This is likely due to the very low frequency of pathogenic mutations in these genes and it is therefore difficult to detect signficant burden test signals.
We also attempted to determine whether known PD loci identified by GWAS present rare variant associations, as has been shown previously near SNCA, GBA1, GCH1, VPS13C and LRRK2.6–9 We assessed a total of 82 PD GWAS regions, 78 of which were identified in the largest GWAS of Europeans,1 two of which were identified in the largest PD GWAS of East Asians2 and two of which were identified in the largest PD GWAS investigating progression46 (Supplementary Table 10). Looking broadly at each meta-analysis group, only two genes, GBA1 and LRRK2, were significant after Bonferroni correction for 2361 unique genes within 1 megabase of known PD loci, suggesting that rare coding variants do not play a large role in these GWAS regions but rather that signals are driven by non-coding variants in these regions.
Discussion
We report the results of rare variant gene burden tests of PD using the largest sample size to date, including 7184 PD cases, 6701 proxy cases and 51 650 healthy controls. A meta-analysis of gene burden results reaffirms that rare variants in GBA1 and LRRK2 are associated with PD risk in individuals with European ancestry. However, we also observed several novel PD-associated genes (B3GNT3, AUNIP, ADH5, TUBA1B, OR1G1, CAPN10 and TREML1) that met exome-wide significance (P < 1 × 10−6) in our analysis. Although these genes were not significant across all of the datasets tested (Supplementary Table 7) and we were unable to replicate the associations at exome-wide significance in independent datasets, this may be due to varied power in the different datasets due to sample size and/or geographical population differences between the datasets that influence the presence or absence of rare variants of interest. We observed the strongest evidence of a novel rare variant association at B3GNT3, where LoF variants showed a significant meta-analysis P-value (P = 4.40 × 10−9) primarily driven by the Genentech (P = 4.40 × 10−9) and UK Biobank (parent proxies P = 0.032) datasets. Variants meeting this criteria were not present in the combined AMP-PD and NIH genomes, requiring additional data to confirm association with PD risk. Upon investigation, we found that three LoF variants in B3GNT3 are associated with increased risk of PD. Of the four individuals carrying these B3GNT3 variants in the Genentech dataset, three were PD cases and one was a control. The three PD cases reported a family history of PD, which is not uncommon in this cohort and not necessarily indicative of familial PD (up to 30% self-report a family history of PD). While these three PD cases reported earlier age at onset than typical PD (manifestation in their thirties and below), no enrichment for tremors, gait disturbances, REM sleep disturbances or anosmia were reported. Additionally, no evidence of excess identity-by-descent (IBD) between these three PD cases were found (average k0 = 0.91). These variants in B3GNT3 are rare, with three variants driving the association in both the Genentech and UK Biobank parent proxies datasets and are therefore likely to be absent in the remaining datasets analysed.
Previously suggested PD GWAS loci also harbour rare variants of interest, such as SYT11, FGF20 and GCH1.47 We identified no significant P-values in these genes, consistent with a similar, albeit smaller, analysis performed in the East Asian population.47 Therefore, it is tempting to speculate what the exact mechanism is that underlies these PD GWAS loci. While likely that some risk variants will affect gene expression differences, it is, however, unclear if all risk variants contribute to risk via this mechanism.
The vast majority of previously PD-associated genes were not nominated by our analysis, including PINK1 and PRKN (PARK2), which are the most common genetic cause of early onset PD.48 This is somewhat expected since burden testing algorithms are most well-powered to detect dominant and high-risk variants such as those in GBA1 and LRRK2 and are less sensitive to recessive and ultra-rare mutations. It is also important to note that PD patients who carry PRKN, PINK1 and SNCA mutations often have a slightly different PD phenotype (e.g. earlier onset, varying progression rates, rapid dementia onset) compared to the general PD population.49 Since most PD cases included in this analysis showed onset of symptoms in their sixties, it is less likely that they will harbour pathogenic PRKN mutations than those with early onset PD (Table 1). Additionally, it is also worth noting that certain known disease causing variants are extremely rare, for example SNCA pathogenic missense variants have so far been identified in ∼25 reports and therefore are likely too rare to be identified in the current dataset. It is therefore likely that such mutation carriers are underrepresented in the datasets included in this study.
Immune involvement including adaptive T-lymphocyte response in PD is well described and reviewed elsewhere.50B3GNT3 encodes an enzyme involved in the synthesis of L-selectin required for lymphocyte homing, particularly for rolling of leucocytes on endothelial cells, facilitating their migration into inflammatory sites. TUBA1B encodes the 1B chain of alpha-tubulin, the main constituent of cytoskeleton. Growing evidence suggests the role of microtubule defects in progressive neuronal loss in PD.51,52 Alpha-tubulin has previously been shown to aggregate as a result of mutations in genes encoding proteins well known to be implicated in PD, including parkin53 and alpha-synuclein.54TREML1 is one of the TREM receptors that are increasingly being implicated in neurodegenerative disorders like Alzheimer’s disease, PD and multiple sclerosis.55–57ADH5 encodes for one of the alcohol dehydrogenases, which have been studied in the past for association with PD risk with conflicting results.58–60 There is no clear, discernible connection between known PD biology and the function of the remaining three genes: AUNIP, OR1G1 and CAPN10. Further studies providing genetic support and functional data for these and related genes will be necessary to uncover their potential role in PD.
There are several limitations of this study. First, our analysis was restricted to individuals of European ancestry. It is important to expand rare variant analyses of PD to non-European populations, as well as varying age-at-onset ranges, as more whole genome and whole exome sequencing data becomes available. While our analysis was constrained to assessing four variant classes, we acknowledge that by creating these variant classes we are, in turn, testing specific types of mechanisms. For example, in the case of LoF variants, we are assessing mutations that impair protein function and its impact on disease risk, which is a limitation if the disease mechanism is gain-of-function. Although the sample size is large compared to previous rare variant analyses of PD, we lack power to detect associations in genes where ≤3% of the variants tested are putatively functional or causal, as some rare variant tests weigh rarer variants with increased penetrance and effect size differently or not at all (Supplementary Table 5). Since our literature search for previously reported rare variant associations was comprehensive and not limited to late-onset PD, it is possible that failure to replicate these associations is due to our analysis focusing on associations in late-onset PD compared to controls. Another limitation is, since not all the datasets included in the meta-analysis were not jointly called from an alignment of raw reads, it is possible that batch effects in sites, sequencing and data processing may bias the results. The meta-analysis model of these analyses to leverage the power of the datasets without combining them should however limit these biases. Further follow-up of candidate genes via segregation in multiplex families or resequencing in large case-control datasets, particularly those enriched for early onset and familial cases, is warranted. Additionally, our analysis included parent and sibling proxy cases from the UK Biobank to increase statistical power. Although PD proxy cases have shown to be valuable in large-scale studies investigating common variants1 and we have demonstrated their utility at detecting rare variant associations in known PD genes such as GBA1 (Supplementary Table 7), we acknowledge that caution should be used when searching for recessive forms of disease. Finally, the vast majority of PD patients included in this study are from the ‘general’ PD population, of which typically less than ∼10% have a positive family history. Future rare variant studies will benefit from recruitment efforts that prioritize PD patients who are highly suspected to have a monogenic form of disease since these individuals are more likely to harbour highly pathogenic or causal mutations that have not previously been associated with PD. This strategy is being actively used for recruitment of PD patients by the Global Parkinson's Genetics Program (GP2).61
Clinical heterogeneity within PD cases has been well documented, and further validation is needed to confirm the pathogenicity of rare or ultra-rare variants and their impact on disease.62–64 Analysis of rare variants restricted to subtypes of PD may identify genes important in PD subtypes but not PD as a whole. Our analysis was also restricted to SNVs and small indels, as we did not look at copy number variants generated by short- or long-read sequencing since we did not have access to all raw data to perform such analyses. Future analyses will benefit from including copy number variants which have been shown to be important and causal for PD4,65,66 and especially using of long-read sequencing, as long-read sequencing is able to identify more and more robustly copy number variants in comparison to short-read sequencing.67
Overall, we performed the largest PD genetic burden test to date. We identified GBA1 and LRRK2 as two genes harbouring rare variants associated with PD and nominated several other previously unidentified genes. While we have identified mutations in B3GNT3 and TREML1 potentially associated with increased risk of PD to be previously linked with neuroinflammation, further research into the biological mechanisms are critical to confirm the role of these genes in PD. Further replication in larger datasets that prioritize familial PD cases and individuals of non-European ancestry will provide greater insight into the nominated genes.
Supplementary Material
Acknowledgements
We would like to thank all of the subjects who donated their time and biological samples to be part of this study. This study used the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health (http://hpc.nih.gov). Figure 1 was designed at Biorender.com. Data used in the preparation of this article were obtained from the AMP-PD Knowledge Platform. For up-to-date information on the study, visit https://www.amp-pd.org. AMP-PD—a public-private partnership—is managed by the FNIH and funded by Celgene, GSK, the Michael J. Fox Foundation for Parkinson’s Research, the National Institute of Neurological Disorders and Stroke, Pfizer and Verily. We would like to thank AMP-PD for the publicly available whole-genome sequencing data, including cohorts from the Fox Investigation for New Discovery of Biomarkers (BioFIND), the Parkinson’s Progression Markers Initiative (PPMI) and the Parkinson’s Disease Biomarkers Program (PDBP). The Parkinson's Disease Biomarker Program (PDBP) consortium is supported by the National Institute of Neurological Disorders and Stroke (NINDS) at the National Institutes of Health. A full list of PDBP investigators can be found at https://pdbp.ninds.nih.gov/policy. Harvard Biomarker Study (HBS) is a collaboration of HBS investigators (full list of HBS investigators found at https://www.bwhparkinsoncenter.org/biobank) and funded through philanthropy and NIH and Non-NIH funding sources. The HBS Investigators have not participated in reviewing the data analysis or content of the manuscript. We also thank all of our Genentech colleagues involved in the Human Genetics Initiative involved in generating the sequence data including Natalie Bowers, Julie Hunkapiller, Jens Reeder and Suresh Selvaraj. We are grateful to the Banner Sun Health Research Institute Brain and Body Donation Program of Sun City, Arizona, for the provision of human brain tissue and data. UKBEC: Consortium members include; Juan A. Botía, University of Murcia & UCL Great Ormond Street Institute of Child Health; Karishma D’Sa, Crick Institute; Paola Forabosco, Istituto di Ricerca Genetica e Biomedica, Italy; Sebastian Guelfi, Verge Genomics & UCL Great Ormond Street Institute of Child Health; Adaikalavan Ramasamy, Singapore Institute for Clinical Sciences; Regina H. Reynolds, UCL Great Ormond Street Institute of Child Health; Colin Smith, The University of Edinburgh; Daniah Trabzuni, UCL Queen Square Institute of Neurology; Robert Walker, The University of Edinburgh; Michael E. Weale, Genomics Plc, Oxford UK. This work was supported by the UK Dementia Research Institute which receives its funding from DRI Ltd, funded by the UK Medical Research Council, Alzheimer’s Society and Alzheimer’s Research UK.Medical Research Council (award number MR/N026004/1) and Medical Research Council (award number MR/N026004/1). The Brain and Body Donation Program is supported by the National Institute of Neurological Disorders and Stroke (U24 NS072026 National Brain and Tissue Resource for Parkinson’s Disease and Related Disorders), the National Institute on Aging (P30 AG19610 Arizona Alzheimer’s Disease Core Center), the Arizona Department of Health Services (contract 211002, Arizona Alzheimer’s Research Center), the Arizona Biomedical Research Commission (contracts 4001, 0011, 05-901 and 1001 to the Arizona Parkinson’s Disease Consortium) and the Michael J. Fox Foundation for Parkinson’s Research. We thank the NIH NeuroBioBank (https://neurobiobank.nih.gov) for providing human brain tissue samples and data. Wellderly: This work is supported by Scripps Research Translational Institute, an NIH-NCATS Clinical and Translational Science Award (CTSA; 5 UL1TR002550). LNG Path confirmed: We are grateful to the Banner Sun Health Research Institute Brain and Body Donation Program of Sun City, Arizona for the provision of human biological materials (or specific description, e.g. brain tissue, cerebrospinal fluid). The Brain and Body Donation Program has been supported by the National Institute of Neurological Disorders and Stroke (U24 NS072026 National Brain and Tissue Resource for Parkinson’s Disease and Related Disorders), the National Institute on Aging (P30 AG19610 Arizona Alzheimer’s Disease Core Center), the Arizona Department of Health Services (contract 211002, Arizona Alzheimer’s Research Center), the Arizona Biomedical Research Commission (contracts 4001, 0011, 05-901 and 1001 to the Arizona Parkinson’s Disease Consortium) and the Michael J. Fox Foundation for Parkinson’s Research. We thank the NIH NeuroBioBank for the provision of tissue samples. NABEC: We thank members of the North American Brain Expression Consortium (NABEC) for providing samples derived from brain tissue. Brain tissue for the NABEC cohort were obtained from the Baltimore Longitudinal Study on Aging at the Johns Hopkins School of Medicine, the NICHD Brain and Tissue Bank for Developmental Disorders at the University of Maryland, the Banner Sun Health Research Institute Brain and Body Donation Program and the University of Kentucky Alzheimer’s Disease Center Brain Bank.
Contributor Information
Mary B Makarious, Laboratory of Neurogenetics, National Institute on Aging, National Institutes of Health, Bethesda, MD 20814, USA; Department of Clinical and Movement Neurosciences, UCL Queen Square Institute of Neurology, London WC1N 3BG, UK; UCL Movement Disorders Centre, University College London, London WC1N 3BG, UK.
Julie Lake, Laboratory of Neurogenetics, National Institute on Aging, National Institutes of Health, Bethesda, MD 20814, USA.
Vanessa Pitz, Integrative Neurogenomics Unit, Laboratory of Neurogenetics, National Institute on Aging, National Institutes of Health, Bethesda, MD 20814, USA.
Allen Ye Fu, Laboratory of Neurogenetics, National Institute on Aging, National Institutes of Health, Bethesda, MD 20814, USA; Department of Cell Biology and Neuroscience, Rutgers University, Piscataway, NJ 08854, USA.
Joseph L Guidubaldi, Integrative Neurogenomics Unit, Laboratory of Neurogenetics, National Institute on Aging, National Institutes of Health, Bethesda, MD 20814, USA; Center for Alzheimer's and Related Dementias (CARD), National Institute on Aging and National Institute of Neurological Disorders and Stroke, National Institutes of Health, Bethesda, MD 20814, USA.
Caroline Warly Solsberg, Memory and Aging Center, Department of Neurology, University of California San Francisco, San Francisco, CA 94158, USA; Pharmaceutical Sciences and Pharmacogenomics, University of California San Francisco, San Francisco, CA 94143, USA.
Sara Bandres-Ciga, Center for Alzheimer's and Related Dementias (CARD), National Institute on Aging and National Institute of Neurological Disorders and Stroke, National Institutes of Health, Bethesda, MD 20814, USA.
Hampton L Leonard, Laboratory of Neurogenetics, National Institute on Aging, National Institutes of Health, Bethesda, MD 20814, USA; Center for Alzheimer's and Related Dementias (CARD), National Institute on Aging and National Institute of Neurological Disorders and Stroke, National Institutes of Health, Bethesda, MD 20814, USA; Data Tecnica International, Washington, DC 20812, USA.
Jonggeol Jeffrey Kim, Integrative Neurogenomics Unit, Laboratory of Neurogenetics, National Institute on Aging, National Institutes of Health, Bethesda, MD 20814, USA; Preventive Neurology Unit, Wolfson Institute of Preventive Medicine, Queen Mary University of London, London EC1M 6BQ, UK.
Kimberley J Billingsley, Laboratory of Neurogenetics, National Institute on Aging, National Institutes of Health, Bethesda, MD 20814, USA; Center for Alzheimer's and Related Dementias (CARD), National Institute on Aging and National Institute of Neurological Disorders and Stroke, National Institutes of Health, Bethesda, MD 20814, USA.
Francis P Grenn, Laboratory of Neurogenetics, National Institute on Aging, National Institutes of Health, Bethesda, MD 20814, USA.
Pilar Alvarez Jerez, Laboratory of Neurogenetics, National Institute on Aging, National Institutes of Health, Bethesda, MD 20814, USA; Center for Alzheimer's and Related Dementias (CARD), National Institute on Aging and National Institute of Neurological Disorders and Stroke, National Institutes of Health, Bethesda, MD 20814, USA.
Chelsea X Alvarado, Center for Alzheimer's and Related Dementias (CARD), National Institute on Aging and National Institute of Neurological Disorders and Stroke, National Institutes of Health, Bethesda, MD 20814, USA; Data Tecnica International, Washington, DC 20812, USA.
Hirotaka Iwaki, Laboratory of Neurogenetics, National Institute on Aging, National Institutes of Health, Bethesda, MD 20814, USA; Center for Alzheimer's and Related Dementias (CARD), National Institute on Aging and National Institute of Neurological Disorders and Stroke, National Institutes of Health, Bethesda, MD 20814, USA; Data Tecnica International, Washington, DC 20812, USA.
Michael Ta, Center for Alzheimer's and Related Dementias (CARD), National Institute on Aging and National Institute of Neurological Disorders and Stroke, National Institutes of Health, Bethesda, MD 20814, USA; Data Tecnica International, Washington, DC 20812, USA.
Dan Vitale, Center for Alzheimer's and Related Dementias (CARD), National Institute on Aging and National Institute of Neurological Disorders and Stroke, National Institutes of Health, Bethesda, MD 20814, USA; Data Tecnica International, Washington, DC 20812, USA.
Dena Hernandez, Laboratory of Neurogenetics, National Institute on Aging, National Institutes of Health, Bethesda, MD 20814, USA.
Ali Torkamani, Department of Integrative Structural and Computational Biology, Scripps Research Institute, La Jolla, CA 92037, USA.
Mina Ryten, NIHR Great Ormond Street Hospital Biomedical Research Centre, University College London, London WC1N 1EH, UK; Department of Genetics and Genomic Medicine, Great Ormond Street Institute of Child Health, University College London, London WC1N 1EH, UK.
John Hardy, UK Dementia Research Institute and Department of Neurodegenerative Disease and Reta Lila Weston Institute, UCL Queen Square Institute of Neurology and UCL Movement Disorders Centre, University College London, London WC1N 3BG, UK; Institute for Advanced Study, The Hong Kong University of Science and Technology, Hong Kong SAR, China.
Sonja W Scholz, Neurodegenerative Diseases Research Unit, National Institute of Neurological Disorders and Stroke, Bethesda, MD 20814, USA; Department of Neurology, Johns Hopkins University Medical Center, Baltimore, MD 21287, USA.
Bryan J Traynor, Laboratory of Neurogenetics, National Institute on Aging, National Institutes of Health, Bethesda, MD 20814, USA; Department of Neurology, Johns Hopkins University Medical Center, Baltimore, MD 21287, USA.
Clifton L Dalgard, The American Genome Center, Uniformed Services University of the Health Sciences, Bethesda, MD 20814, USA.
Debra J Ehrlich, Parkinson’s Disease Clinic, Office of the Clinical Director, National Institute of Neurological Disorders and Stroke, Bethesda, MD 20814, USA.
Toshiko Tanaka, Translational Gerontology Branch, National Institute on Aging, NIH, Baltimore, MD 21224, USA.
Luigi Ferrucci, Translational Gerontology Branch, National Institute on Aging, NIH, Baltimore, MD 21224, USA.
Thomas G Beach, Civin Laboratory for Neuropathology, Banner Sun Health Research Institute, Sun City, AZ 85351, USA.
Geidy E Serrano, Civin Laboratory for Neuropathology, Banner Sun Health Research Institute, Sun City, AZ 85351, USA.
Raquel Real, Department of Clinical and Movement Neurosciences, UCL Queen Square Institute of Neurology, London WC1N 3BG, UK; UCL Movement Disorders Centre, University College London, London WC1N 3BG, UK.
Huw R Morris, Department of Clinical and Movement Neurosciences, UCL Queen Square Institute of Neurology, London WC1N 3BG, UK; UCL Movement Disorders Centre, University College London, London WC1N 3BG, UK.
Jinhui Ding, Laboratory of Neurogenetics, National Institute on Aging, National Institutes of Health, Bethesda, MD 20814, USA.
J Raphael Gibbs, Laboratory of Neurogenetics, National Institute on Aging, National Institutes of Health, Bethesda, MD 20814, USA.
Andrew B Singleton, Laboratory of Neurogenetics, National Institute on Aging, National Institutes of Health, Bethesda, MD 20814, USA; Center for Alzheimer's and Related Dementias (CARD), National Institute on Aging and National Institute of Neurological Disorders and Stroke, National Institutes of Health, Bethesda, MD 20814, USA.
Mike A Nalls, Laboratory of Neurogenetics, National Institute on Aging, National Institutes of Health, Bethesda, MD 20814, USA; Center for Alzheimer's and Related Dementias (CARD), National Institute on Aging and National Institute of Neurological Disorders and Stroke, National Institutes of Health, Bethesda, MD 20814, USA; Data Tecnica International, Washington, DC 20812, USA.
Tushar Bhangale, Department of Human Genetics, Genentech, Inc., South San Francisco, CA 94080, USA.
Cornelis Blauwendraat, Laboratory of Neurogenetics, National Institute on Aging, National Institutes of Health, Bethesda, MD 20814, USA; Integrative Neurogenomics Unit, Laboratory of Neurogenetics, National Institute on Aging, National Institutes of Health, Bethesda, MD 20814, USA; Center for Alzheimer's and Related Dementias (CARD), National Institute on Aging and National Institute of Neurological Disorders and Stroke, National Institutes of Health, Bethesda, MD 20814, USA.
Data availability
Accelerating Medicines Partnership in Parkinson’s Disease (AMP PD data) and quality control notebooks are access-controlled (https://amp-pd.org/) and require individual sign-up to access the data. United Kingdom Biobank (UK Biobank) data are access-controlled and require an application for access (https://www.ukbiobank.ac.uk/). The remaining cohorts were obtained through collaborations with the National Institutes of Health (NIH) and Genentech. Each contributing study abided by the ethics guidelines set out by their institutional review boards, and all participants gave informed consent for inclusion in both their initial cohorts and subsequent studies. Each contributing study abided by the ethics guidelines set out by their institutional review boards, and all participants gave informed consent for inclusion in both their initial cohorts and subsequent studies. The research using data from the NIH Parkinson’s Disease clinic cohort was approved by the NIH Intramural institutional review board (IRB) under protocol number 01-N-0206. The research with the remaining cohorts was deemed ‘not human subjects research’ by the NIH Office of IRB Operations and stated that no IRB approval is required. The NIH Intramural IRB has waived ethical approval for the overall study (IRB #001161). All data produced in the present work are contained in the manuscript. All authors and the public can access the statistical programming code used in this project for the analyses and results generation on GitHub at https://github.com/neurogenetics/PD-BURDEN, as well as Supplementary tables and full results. M.B.M. and C.B. take final responsibility for the decision to submit the paper for publication. NABEC is available from NCBI dbGaP, study accession phs001300.v2.p1.
Funding
This research was supported in part by the Intramural Research Program of the National Institutes of Health (National Institute on Aging and National Institute of Neurological Disorders and Stroke; project numbers: 1ZIA-NS003154, Z01-AG000949-02, Z01-ES101986 and UK ADC NIA P30 AG072946). This research has been conducted using the UK Biobank Resource under Application Number 33601.
Competing interests
H.L.L., H.I., M.T., D.V. and M.A.N. declare that they are consultants employed by Data Tecnica International, whose participation in this is part of a consulting agreement between the US National Institutes of Health and said company. M.A.N. also currently serves on the scientific advisory board for Clover Therapeutics and is an advisor to Neuron23 Inc. H.R.M. is employed by UCL and in the last 24 months he reports paid consultancy from Biogen, Biohaven, Lundbeck; lecture fees/honoraria from Wellcome Trust, Movement Disorders Society. Research Grants from Parkinson’s UK, Cure Parkinson’s Trust, PSP Association, CBD Solutions, Drake Foundation, Medical Research Council and Michael J. Fox Foundation. H.R.M. is also a co-applicant on a patent application related to C9ORF72—Method for diagnosing a neurodegenerative disease (PCT/GB2012/052140). T.B. is employed by Genentech, Inc., a member of the Roche group. C.B. takes final responsibility for the decision to submit the paper for publication.
Supplementary material
Supplementary material is available at Brain online.
Appendix 1
UK Brain Expression Consortium members
John Hardy, Mike Weale, Daniah Trabzuni, Sebastian Guelfi, Juan Botia, Karishma D'Sa, Paola Forabosco, Colin Smith, Adaikalavan Ramasamy, Mina Ryten, Regina H. Reynolds, and Robert Walker. For a full list of UK Brain Expression Consortium (UKBEC) authors, see https://ukbec.wordpress.com/.
References
- 1. Nalls MA, Blauwendraat C, Vallerga CL, et al. Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet Neurol. 2019;18:1091–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Foo JN, Chew EGY, Chung SJ, et al. Identification of risk loci for Parkinson disease in Asians and comparison of risk between Asians and Europeans: a genome-wide association study. JAMA Neurol. 2020;77:746–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Polymeropoulos MH, Lavedan C, Leroy E, et al. Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science. 1997;276:2045–2047. [DOI] [PubMed] [Google Scholar]
- 4. Kitada T, Asakawa S, Hattori N, et al. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. [DOI] [PubMed] [Google Scholar]
- 5. Singleton A, Hardy J. A generalizable hypothesis for the genetic architecture of disease: pleomorphic risk loci. Hum Mol Genet. 2011;20:R158–R162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jansen IE, Gibbs JR, Nalls MA, et al. Establishing the role of rare coding variants in known Parkinson’s disease risk loci. Neurobiol Aging. 2017;59:220.e11–e220.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gaare JJ, Nido G, Dölle C, et al. Meta-analysis of whole-exome sequencing data from two independent cohorts finds no evidence for rare variant enrichment in Parkinson disease associated loci. PLoS One. 2020;15:e0239824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rudakou U, Yu E, Krohn L, et al. Targeted sequencing of Parkinson’s disease loci genes highlights SYT11, FGF20 and other associations. Brain. 2021;144:462–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mencacci NE, Isaias IU, Reich MM, et al. Parkinson’s disease in GTP cyclohydrolase 1 mutation carriers. Brain. 2014;137:2480–2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lee JS, Kanai K, Suzuki M, et al. Arylsulfatase A, a genetic modifier of Parkinson’s disease, is an α-synuclein chaperone. Brain. 2019;142:2845–2859. [DOI] [PubMed] [Google Scholar]
- 11. Martin S, Smolders S, Van den Haute C, et al. Mutated ATP10B increases Parkinson’s disease risk by compromising lysosomal glucosylceramide export. Acta Neuropathol. 2020;139:1001–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Makarious MB, Diez-Fairen M, Krohn L, et al. ARSA variants in α-synucleinopathies. Brain. 2019;142:e70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fan Y, Mao CY, Dong YL, et al. ARSA gene variants and Parkinson’s disease. Brain. 2020;143:e47. [DOI] [PubMed] [Google Scholar]
- 14. Tesson C, Lohmann E, Devos D, Bertrand H, Lesage S, Brice A. Segregation of ATP10B variants in families with autosomal recessive parkinsonism. Acta Neuropathol. 2020;140:783–785. [DOI] [PubMed] [Google Scholar]
- 15. Real R, Moore A, Blauwendraat C, Morris HR, Bandres-Ciga S; International Parkinson’s Disease Genomics Consortium (IPDGC) . ATP10B And the risk for Parkinson’s disease. Acta Neuropathol. 2020;140:401–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Trabzuni D, Thomson PC; United Kingdom Brain Expression Consortium (UKBEC) . Analysis of gene expression data using a linear mixed model/finite mixture model approach: Application to regional differences in the human brain. Bioinformatics. 2014;30:1555–1561. [DOI] [PubMed] [Google Scholar]
- 17. Gibbs JR, van der Brug MP, Hernandez DG, et al. Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLoS Genet. 2010;6:e1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Erikson GA, Bodian DL, Rueda M, et al. Whole-Genome sequencing of a healthy aging cohort. Cell. 2016;165:1002–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Iwaki H, Leonard HL, Makarious MB, et al. Accelerating medicines partnership: Parkinson’s disease. Genetic resource. Mov Disord. 2021;36:1795–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Poplin R, Ruano-Rubio V, DePristo MA, et al. Scaling accurate genetic variant discovery to tens of thousands of samples. bioRxiv. [Preprint] 10.1101/201178 [DOI] [Google Scholar]
- 21. Bandres-Ciga S, Saez-Atienzar S, Kim JJ, et al. Large-scale pathway specific polygenic risk and transcriptomic community network analysis identifies novel functional pathways in Parkinson disease. Acta Neuropathol. 2020;140:341–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Karczewski KJ, Francioli LC, Tiao G, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bycroft C, Freeman C, Petkova D, et al. The UK biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Backman JD, Li AH, Marcketta A, et al. Exome sequencing and analysis of 454,787 UK biobank participants. Nature. 2021;599:628–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chang D, Nalls MA, Hallgrímsdóttir IB, et al. A meta-analysis of genome-wide association studies identifies 17 new Parkinson’s disease risk loci. Nat Genet. 2017;49:1511–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McKenna A, Hanna M, Banks E, et al. The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Van der Auwera GA, Carneiro MO, Hartl C, et al. From FastQ data to high confidence variant calls: the genome analysis toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43:11.10.1–11.10.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Van der Auwera GA, O’Connor BD. Genomics in the cloud: Using docker, GATK, and WDL in terra. O’Reilly Media; 2020. [Google Scholar]
- 32. Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. [DOI] [PubMed] [Google Scholar]
- 33. Cingolani P, Platts A, Wang LL, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin). 2012;6:80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. McLaren W, Gil L, Hunt SE, et al. The ensembl variant effect predictor. Genome Biol. 2016;17:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rentzsch P, Witten D, Cooper GM, Shendure J, Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019;47:D886–D894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lee S, Wu MC, Lin X. Optimal tests for rare variant effects in sequencing association studies. Biostatistics. 2012;13:762–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lee S, Fuchsberger C, Kim S, Scott L. An efficient resampling method for calibrating single and gene-based rare variant association analysis in case-control studies. Biostatistics. 2016;17:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zeggini E, Morris A. Assessing rare variation in Complex traits: Design and analysis of genetic studies. Springer; 2015. [Google Scholar]
- 39. Lee S, Abecasis GR, Boehnke M, Lin X. Rare-variant association analysis: study designs and statistical tests. Am J Hum Genet. 2014;95:5–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lee S, Emond MJ, Bamshad MJ, et al. Optimal unified approach for rare-variant association testing with application to small-sample case-control whole-exome sequencing studies. Am J Hum Genet. 2012;91:224–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wu MC, Lee S, Cai T, Li Y, Boehnke M, Lin X. Rare-variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet. 2011;89:82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Derkach A, Lawless JF, Sun L. Robust and powerful tests for rare variants using Fisher’s method to combine evidence of association from two or more complementary tests. Genet Epidemiol. 2013;37:110–121. [DOI] [PubMed] [Google Scholar]
- 43. Tysnes OB, Storstein A. Epidemiology of Parkinson’s disease. J Neural Transm. 2017;124:901–905. [DOI] [PubMed] [Google Scholar]
- 44. Blauwendraat C, Reed X, Kia DAet al. Frequency of loss of function variants in LRRK2 in Parkinson disease. JAMA Neurol. 2018;75:1416–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liao J, Wu CX, Burlak C, et al. Parkinson disease-associated mutation R1441H in LRRK2 prolongs the “active state” of its GTPase domain. Proc Natl Acad Sci U S A. 2014;111:4055–4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tan MMX, Lawton MA, Jabbari E, et al. Genome-Wide association studies of cognitive and motor progression in Parkinson’s disease. Mov Disord. 2021;36:424–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pu JL, Lin ZH, Zheng R, et al. Association analysis of SYT11, FGF20, GCH1 rare variants in Parkinson’s disease. CNS Neurosci Ther. 2022;28:175–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kasten M, Hartmann C, Hampf J, et al. Genotype-phenotype relations for the Parkinson’s disease genes parkin, PINK1, DJ1: MDSGene systematic review. Mov Disord. 2018;33:730–741. [DOI] [PubMed] [Google Scholar]
- 49. Klein C, Westenberger A. Genetics of Parkinson’s disease. Cold Spring Harb Perspect Med. 2012;2:a008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mosley RL, Hutter-Saunders JA, Stone DK, Gendelman HE. Inflammation and adaptive immunity in Parkinson’s disease. Cold Spring Harb Perspect Med. 2012;2:a009381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Calogero AM, Mazzetti S, Pezzoli G, Cappelletti G. Neuronal microtubules and proteins linked to Parkinson’s disease: a relevant interaction? Biol Chem. 2019;400:1099–1112. [DOI] [PubMed] [Google Scholar]
- 52. Pellegrini L, Wetzel A, Grannó S, Heaton G, Harvey K. Back to the tubule: Microtubule dynamics in Parkinson’s disease. Cell Mol Life Sci. 2017;74:409–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ren Y, Zhao J, Feng J. Parkin binds to alpha/beta tubulin and increases their ubiquitination and degradation. J Neurosci. 2003;23:3316–3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cartelli D, Aliverti A, Barbiroli A, et al. α-Synuclein is a novel microtubule dynamase. Sci Rep. 2016;6:33289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Dardiotis E, Siokas V, Pantazi E, et al. A novel mutation in TREM2 gene causing nasu-hakola disease and review of the literature. Neurobiol Aging. 2017;53:194.e13–e194.e22. [DOI] [PubMed] [Google Scholar]
- 56. Feng CW, Chen NF, Sung CS, et al. Therapeutic effect of modulating TREM-1 via anti-inflammation and autophagy in Parkinson’s disease. Front Neurosci. 2019;13:769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Piccio L, Buonsanti C, Cella M, et al. Identification of soluble TREM-2 in the cerebrospinal fluid and its association with multiple sclerosis and CNS inflammation. Brain. 2008;131:3081–3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kim JJ, Bandres-Ciga S, Blauwendraat C, Gan-Or Z; International Parkinson’s Disease Genomics Consortium . No genetic evidence for involvement of alcohol dehydrogenase genes in risk for Parkinson’s disease. Neurobiol Aging. 2020;87:140.e19–e140.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Buervenich S, Carmine A, Galter D, et al. A rare truncating mutation in ADH1C (G78Stop) shows significant association with Parkinson disease in a large international sample. Arch Neurol. 2005;62:74–78. [DOI] [PubMed] [Google Scholar]
- 60. García-Martín E, Diez-Fairen M, Pastor P, et al. Association between the missense alcohol dehydrogenase rs1229984T variant with the risk for Parkinson’s disease in women. J Neurol. 2019;266:346–352. [DOI] [PubMed] [Google Scholar]
- 61. Global Parkinson’s Genetics Program . GP2: the global Parkinson’s genetics program. Mov Disord. 2021;36:842–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Campbell MC, Myers PS, Weigand AJ, et al. Parkinson disease clinical subtypes: key features & clinical milestones. Ann Clin Transl Neurol. 2020;7:1272–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Mu J, Chaudhuri KR, Bielza C, de Pedro-Cuesta J, Larrañaga P, Martinez-Martin P. Parkinson’s disease subtypes identified from cluster analysis of motor and non-motor symptoms. Front Aging Neurosci. 2017;9:301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sauerbier A, Jenner P, Todorova A, Chaudhuri KR. Non motor subtypes and Parkinson’s disease. Parkinsonism Relat Disord. 2016;22:S41–S46. [DOI] [PubMed] [Google Scholar]
- 65. Singleton AB, Farrer M, Johnson J, et al. Alpha-synuclein locus triplication causes Parkinson’s disease. Science. 2003;302:841. [DOI] [PubMed] [Google Scholar]
- 66. Scott AJ, Chiang C, Hall IM. Structural variants are a major source of gene expression differences in humans and often affect multiple nearby genes. Genome Res. 2021;31:2249–2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Billingsley KJ, Ding J, Jerez PA, et al. Genome-wide analysis of structural variants in Parkinson disease. Ann Neurol. 2023;93:1012–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Accelerating Medicines Partnership in Parkinson’s Disease (AMP PD data) and quality control notebooks are access-controlled (https://amp-pd.org/) and require individual sign-up to access the data. United Kingdom Biobank (UK Biobank) data are access-controlled and require an application for access (https://www.ukbiobank.ac.uk/). The remaining cohorts were obtained through collaborations with the National Institutes of Health (NIH) and Genentech. Each contributing study abided by the ethics guidelines set out by their institutional review boards, and all participants gave informed consent for inclusion in both their initial cohorts and subsequent studies. Each contributing study abided by the ethics guidelines set out by their institutional review boards, and all participants gave informed consent for inclusion in both their initial cohorts and subsequent studies. The research using data from the NIH Parkinson’s Disease clinic cohort was approved by the NIH Intramural institutional review board (IRB) under protocol number 01-N-0206. The research with the remaining cohorts was deemed ‘not human subjects research’ by the NIH Office of IRB Operations and stated that no IRB approval is required. The NIH Intramural IRB has waived ethical approval for the overall study (IRB #001161). All data produced in the present work are contained in the manuscript. All authors and the public can access the statistical programming code used in this project for the analyses and results generation on GitHub at https://github.com/neurogenetics/PD-BURDEN, as well as Supplementary tables and full results. M.B.M. and C.B. take final responsibility for the decision to submit the paper for publication. NABEC is available from NCBI dbGaP, study accession phs001300.v2.p1.