Abstract
Within recent years, there has been a growing number of genes associated with amyotrophic lateral sclerosis (ALS), resulting in an increasing number of novel variants, particularly missense variants, many of which are of unknown clinical significance. Here, we leverage the sequencing efforts of the ALS Knowledge Portal (3864 individuals with ALS and 7839 controls) and Project MinE ALS Sequencing Consortium (4366 individuals with ALS and 1832 controls) to perform proteomic and transcriptomic characterization of missense variants in 24 ALS-associated genes.
The two sequencing datasets were interrogated for missense variants in the 24 genes, and variants were annotated with gnomAD minor allele frequencies, ClinVar pathogenicity classifications, protein sequence features including Uniprot functional site annotations, and PhosphoSitePlus post-translational modification site annotations, structural features from AlphaFold predicted monomeric 3D structures, and transcriptomic expression levels from Genotype-Tissue Expression. We then applied missense variant enrichment and gene-burden testing following binning of variation based on the selected proteomic and transcriptomic features to identify those most relevant to pathogenicity in ALS-associated genes.
Using predicted human protein structures from AlphaFold, we determined that missense variants carried by individuals with ALS were significantly enriched in β-sheets and α-helices, as well as in core, buried or moderately buried regions. At the same time, we identified that hydrophobic amino acid residues, compositionally biased protein regions and regions of interest are predominantly enriched in missense variants carried by individuals with ALS. Assessment of expression level based on transcriptomics also revealed enrichment of variants of high and medium expression across all tissues and within the brain. We further explored enriched features of interest using burden analyses and identified individual genes were indeed driving certain enrichment signals. A case study is presented for SOD1 to demonstrate proof-of-concept of how enriched features may aid in defining variant pathogenicity.
Our results present proteomic and transcriptomic features that are important indicators of missense variant pathogenicity in ALS and are distinct from features associated with neurodevelopmental disorders.
Keywords: amyotrophic lateral sclerosis, variants of uncertain significance, missense variants, protein features, transcriptomic features
Using large DNA sequencing datasets comprising more than 17 000 individuals, Dilliott et al. identify proteomic and transcriptomic features indicative of pathogenicity in missense variants associated with amyotrophic lateral sclerosis.
Introduction
Amyotrophic lateral sclerosis (ALS), also referred to as motor neuron disease, is a fatal neurodegenerative disorder characterized by adult-onset progressive degeneration of upper and lower motor neurons.1 The neuronal loss leads to progressive muscle weakness and eventual paralysis and death within 3–5 years, predominantly due to respiratory failure. Importantly, ALS displays high heritability, not only for individuals with familial forms of the disease, but also for seemingly sporadic or isolated cases with no apparent family history.2 The most common genetic causes of ALS, namely pathogenic variants in C9orf72, SOD1, TARDBP and FUS, have been reported in both patients with familial ALS (fALS) and sporadic ALS (sALS).3–5 As such, genetic testing has become an important tool in the clinical care of ALS patients, offering many advantages, including early, accurate diagnosis and access to emerging clinical trials targeted to specific genetic profiles.6–8 These benefits are now considered quite substantial and recent recommendations include the expansion of clinical genetic testing to a broader range of patients, not only to those with a known family history as has been the conventional approach.9
However, standardized clinical testing in ALS remains challenging. Over recent years, there has been a concerted effort to uncover novel genetic markers associated with ALS with a growing number of genes discovered,10 yet there are a large proportion of patients that remain genetically unexplained.5 As the compendium of ALS-associated genes has expanded, there has been a vast increase in the number of novel variants identified in those genes, and consequently, accurate pathogenicity classification remains challenging. Typically, variants are classified based on the American College of Medical Genetics and Genomics (ACMG) pathogenicity classification guidelines, which have become a standardized approach applied to all diseases and take into consideration a variant’s previous clinical reports, functional experimental evidence, segregation within a pedigree, minor allele frequency in healthy populations and relevant disease cohorts, and in silico predictions, among other evidence.11 But in most cases, the sum of the evidence is not sufficient to confidently classify a variant as pathogenic or benign, and the classification of variants of uncertain significance (VUS) in clinical testing is common. Previous genetic testing of known genes in ALS cohorts have identified VUS in 15–25% of patients,12–15 and in a recent study of >6000 sALS patients 47% of variants identified in ALS-associated genes were considered VUS.16 Additionally, data obtained from the variant database, ClinVar, demonstrate that even the most common, well studied ALS-associated genes—SOD1, TARDBP and FUS—have relatively high rates of VUS (Supplementary Fig. 1A). Not only is the identification of VUS from clinical testing frustrating for the patient and their families, but it can complicate genetic counselling and prevent patient enrolment in ongoing clinical trials.17,18
Missense variants are particularly common VUS,19 as the variant consequence can be difficult to predict without definitive experimental evidence to determine whether function of the encoded protein is gained, lost or altered. Importantly, missense variants are established drivers of neurodegenerative disease risk, as they can induce a gain of protein activity as observed in PSEN1 and PSEN2, which cause early onset Alzheimer’s disease and SOD1 in ALS.20–25 Although many in silico tools have been developed to aid in prediction of missense variant pathogenicity,26–29 the tools primarily rely on variant conservation and constraint, as well as broad biochemical properties. In silico tools are also not specific to any singular disease, such as ALS, and do not capture phenotype- and gene-specific nuances driving missense variant pathogenicity. Therefore, the tools and the ACMG guidelines may fail to capture evidence that could be derived from protein structure and function or from transcriptional expression.
Recently, enrichment analyses were applied to identify protein structural features characterizing likely pathogenic variation in neurodevelopmental disorders (NDD).30 Here, we aim to expand upon this approach to characterize the amino acid residue changes resulting from genetic missense variation in known ALS-associated genes, observed in large cohorts of ALS patients and controls, to identify signals of enrichment of specific protein structure, protein sequence, and transcriptomic features. We anticipate that our results will contribute to the improvement of interpretation of VUS pathogenicity in ALS clinical testing.
Materials and methods
ALS-associated gene selection
In total, 24 genes were selected that had previously been associated with ALS in the literature. Specifically, we only included genes with at least two publications describing rare coding single nucleotide variants in individuals with ALS. We also excluded any genes that display an unclear inheritance pattern in ALS (i.e. driven by risk alleles from genome-wide association studies) or had a limited or refuted gene-disease validity classification from the ALS ClinGen Gene Curation Expert Panel as of January 2022 (Clinical Genome ALS). A summary of the 24 ALS-associated genes, including the year of first association, inheritance pattern, method of first association, and protein function, is presented in Supplementary Table 1.
Study samples and sequencing
Sequencing data were obtained from the ALS Knowledge Portal and Project MinE ALS sequencing consortium.31,32 Briefly, the ALS Knowledge Portal contains summary data from whole exomes of 3864 individuals with ALS and 7839 controls. The dataset was subjected to rigorous and standard quality control, including variant and sample level assessment to account for depth, coverage, genetic ancestry matching using principal component analysis (PCA), and relatedness using identity by descent metrics. Carriers of the C9orf72 hexanucleotide expansion were excluded from the study. Further, ∼2000 of the case samples were screened for pathogenic variants in SOD1, FUS and TARDBP, and known variant carriers were excluded. Comprehensive details outlining sample acquisition, exome-sequencing and quality control were previously described.31 The Project MinE ALS sequencing consortium dataset contains whole-genome sequencing of 4366 individuals with ALS and 1832 controls. Again, details regarding sequencing methodology and quality control were previously described and largely compare to the steps undertaken by the ALS Knowledge Portal.32 Controls were matched to the individuals with ALS based on age, sex and geographical region. For the purposes of this study, Project MinE data were obtained through the Project MinE Data Browser (http://databrowser.projectmine.com/).
Variant genomic annotation and filtering
Variants from both datasets were annotated using Variant Effect Predictor (VEP; Ensemble v.105.0)33 with the following features: gene symbol; Human Genome Variation Society (HGVS) coding and protein sequence alteration; variant consequence; Genome Aggregation Database (gnomAD) v2.1.1 Non-Finnish European (NFE), non-neurological (n = 51 592) allele frequency34; and ClinVar pathogenicity classifications.35 Only variants within the 24 ALS-associated genes were retained for further analysis. Transcript identifiers used for each gene in the VEP annotation can be found in Supplementary Table 2. Variants were filtered to only include missense variants that were rare in the general population (allele frequency < 0.01 in the gnomAD v2.2.2 NFE non-neurological cohort). Variant reference and alternate amino acid biochemical properties were also annotated, including whether the amino acids were non-polar, polar, polar with a positive charge, or polar with a negative charge.
Variant protein feature annotation
Reference amino acid residues substituted in missense variants were annotated by two sets of protein sequence features and two sets of protein 3D structural features.
The protein sequence features included functional site annotations from the UniProt database36 and post-translational modification (PTM) site annotations from the PhosphoSitePlus database,37 referred to as functional and PTM features, respectively. Affected amino acid residues in the identified missense variants were annotated with 26 functional features including: active site, binding site, calcium binding site, coiled coil, compositional bias, cross link, disulphide bond, DNA binding site, domain, glycosylation, intramembrane, lipidation, metal binding site, modified residue, motif, nucleoprotein binding site, peptide, propeptide, region of interest, repeat, signal peptide, site, topological domain, transit peptide, transmembrane and zinc finger (Supplementary Table 3; https://www.uniprot.org/help/sequence_annotation). Similarly, affected amino acid residues in the missense variants were annotated with 10 PTM features including: acetylation, glycosylation (O-glycosylation), methylation, phosphorylation, sumoylation, ubiquitination, regulatory site, kinase-substrate, PTMvar (i.e. PTMs overlapping genetic variants) and disease-associated PTM (Supplementary Table 4).
Predicted monomeric 3D structures of the 24 ALS-associated proteins were collected from the AlphaFold database38,39 and were used to extract structural features of the reference amino acids. From the 3D structures, per-residue secondary structure type and residues’ solvent accessible surface area (ASA) were derived using the dictionary of protein secondary structure (DSSP) program.40 Each amino acid residue was annotated with one of nine different types of secondary structure. These nine different types of secondary structures were grouped into three broader categories: (i) β-strand/-sheet included β-strands and β-sheets; (ii) helices included 310-helices, α-helices, π-helices and polyproline II helices; and (iii) coils included loops, bends, and turns. For each residue, the ASA value (measured in units of square angstrom, Å2) was transformed to calculate the relative ASA (RSA) by normalizing the ASA of that residue by the surface area of the same type of residue in a reference state. We used the ASA normalizing values derived in Tien et al.41 using Gly-X-Gly tripeptide as the reference state for a given residue X. Based on the value of RSA, we labelled each amino acid with one of the following exposure levels: core, RSA < 5%; buried, 5% < RSA < 25%; medium-buried, 25% < RSA < 50%; medium-exposed, 50% < RSA < 75%; and exposed, RSA > 75%, as described by Iqbal et al.42 Note that for the structural feature analyses involving the annotations obtained from AlphaFold predicted structures, only variants at residues with a high quality [predicted local-distance difference test (pLDDT) > 70; per-residue estimates of reliability generated by the AlphaFold neural]38 were used.
Transcriptional variant expression annotation
Finally, the missense variants were annotated based on the proportion of expression across transcripts (pext) level, which were previously derived for all variant possibilities across all gene transcripts using data obtained from the Genotype-Tissue Expression (GTEx) portal.43,44 Briefly, isoform-level quantifications from 11 706 GTEx v7 tissue samples were used to estimate isoform expression levels if any given missense variant were present within a gene, producing a gene level expression-estimate annotation. Importantly, all values were normalized to the expected expression of the gene if it were to carry no missense variation. Average pext expression of each variant across all GTEx tissues were calculated and hereafter referred to as ‘all tissues’ expression levels, and average pext expression of each variant across all GTEx brain tissues were calculated and hereafter referred to as ‘brain’ expression levels. For consistency with the landmark manuscript, expression levels were binned based on the following: low, mean expression < 0.1; medium, mean expression ≥ 0.1 and ≤ 0.9; and high, mean expression > 0.9 as described by Cummings et al.44
Ethics approval
Only summary genetic data from the open-access ALS Knowledge Portal and the open-access Project MinE Data browser, were used for the analyses described herein. Appropriate and informed consent in accordance with each Research Ethics Board at each respective recruiting site was obtained as described in the primary publications.31,32
Statistical analyses
Missense variants were binned based on the above proteomic and transcriptomic features. To quantify the enrichment of missense variants with respect to these bins in the ALS Knowledge Portal and Project MinE ALS datasets, and identify features associated with ALS, we applied two-sided Fisher’s exact tests. Specifically, the number of individuals with ALS carrying each respective variant type was compared to the number of controls carrying the variant type. We also applied combined analyses between the two cohorts using the Cochran-Mantel-Haenszel (CMH) test. Significance was measured at an alpha-level of 0.05.
For proteomic and transcriptomic features demonstrating significant enrichment in individuals with ALS compared to the controls, variants were further binned by genes and the Fisher’s exact test was again applied, to determine if specific genes were driving the enrichment signals. Similarly, individual tests were applied to the ALS Knowledge Portal and Project MinE ALS datasets, followed by combined analyses between the two cohorts using the CMH test. Following Bonferroni multiple testing correction to account for the 24 genes, significance was measured at an alpha level of 2.08 × 10−3.
Statistical analyses were performed using the R statistical software v4.1.145 in R Studio v1.4.1717. Data visualization was performed using the ggplot2 R package (v3.3.6).46
SOD1 case study
The Genomics 2 Proteins (G2P) portal (https://g2p.broadinstitute.org/) was used to map SOD1 missense variants, collected from the ALS Knowledge Portal and Project MinE sequencing consortium and classified according to ClinVar pathogenicity ascertainment, onto the SOD1 AlphaFold predicted protein structure (AlphaFold ID: AF-P00441-F1).42 Similarly, the protein sequence and structural features, as well as transcriptomic features for each residue, were mapped onto the structure of SOD1 using the G2P portal. The structure annotated with variants and features was exported from the portal and visualized using the PyMOL software for downstream analyses (i.e. investigating the overlap between residues mutated in pathogenic and VUS in individuals with ALS and residues with features significantly associated with ALS variants in SOD1).
Results
Missense variants identified in ALS-associated genes
We first assessed the contribution of missense variation to the amount of uncertainty in pathogenicity assessment of ALS associated variation by interrogating the 24 selected ALS associated genes in the ClinVar database and identified all ALS associated variants within the genes (Supplementary Fig. 1A). DCTN1 had the greatest number of VUS reported in ClinVar (n = 364), although five other genes each had >100 VUS reported, including FUS, MATR3, SQSTM1, VAPB and VCP. In four out of these six genes with >100 VUS, the largest number of reported VUS in ClinVar were missense variants (Supplementary Fig. 1B).
Across 3864 individuals with ALS and 7839 controls from the ALS Knowledge Portal, 728 unique rare, missense variants were identified within 23 of the 24 ALS associated genes (Supplementary Table 5). No unique, rare variants were detected in CHCHD10. In 4366 individuals with ALS and 1832 controls from the Project MinE ALS sequencing consortium, 609 unique rare, missense variants were identified in 23 of the 24 ALS-associated genes (Supplementary Table 5). However, in this dataset no variants were reported in UBQLN2.
Again, the ClinVar database was interrogated, specifically for the rare variants identified in the ALS Knowledge Portal and Project MinE ALS sequencing consortium datasets (Supplementary Fig. 1C). Similarly, the largest number of VUS in both the individuals with ALS and controls within our dataset were observed in DCTN1.
Variant reference and alternate amino acids
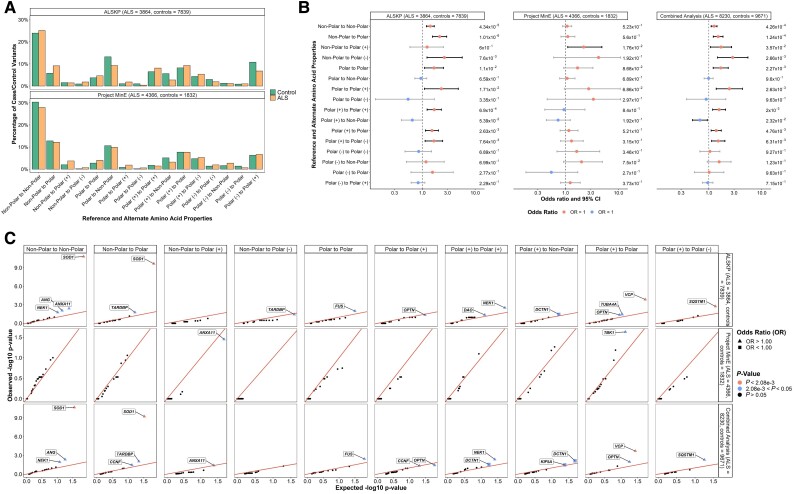
We assessed the distribution of the reference and alternate amino acid biochemical property changes involved in the rare missense variants identified in the individuals with ALS and controls of the ALS Knowledge Portal and Project MinE ALS sequencing consortium (Fig. 1A). To determine whether specific biochemical property changes were more frequent in missense variants in ALS, an enrichment analysis was performed on both datasets, followed by a combined analysis including variants observed in both datasets. From the combined analysis, we observed a significant enrichment of rare, missense variants resulting in a change from non-polar to non-polar amino acids, non-polar to polar amino acids, non-polar to polar positively charged amino acids, non-polar to polar negatively charged amino acids, polar to polar amino acids, polar to polar positively charged amino acids, polar positively charged to polar positively charged amino acids, polar positively charged to polar amino acids, and polar positively charged to polar negatively charged amino acids (P < 0.05; Fig. 1B). We also assessed whether specific genes were driving these enrichments of rare, missense variants affecting specific amino acid biochemical property changes and observed individuals with ALS to be significantly enriched for variants resulting in a change from a non-polar to another non-polar amino acid or a change from a non-polar to a polar amino acid in SOD1 [odds ratio, OR = 57.35 (7.36–447.07), P = 2.01 × 10−11 and OR = 31.49 (4.56–217.56), P = 6.04 × 10−10, respectively); and for variants resulting in a change from a polar positively charged to polar amino acid in VCP [OR = 13.02 (1.80–93.96), P = 2.04 × 10−4] across the combined datasets (Fig. 2C).
Figure 1.
Biochemical properties of reference and alternative amino acids involved in rare missense variants identified across 24 ALS-associated genes. Non-polar amino acids included alanine, glycine, isoleucine, leucine, methionine, phenylalanine, proline, tryptophan and valine; polar amino acids included asparagine, cysteine, glutamine, serine, threonine and tyrosine; polar positively charged amino acids [polar (+)] included arginine, histidine and lysine; polar negatively charged amino acids [polar(−)] included aspartic acid and glutamic acid. (A) Distribution of the biochemical property changes of the amino acids involved in missense variants of interest identified in the ALS Knowledge Portal (ALSKP) and Project MinE ALS sequencing consortium datasets. (B) Enrichment analyses were performed using Fisher’s exact testing to compare the number of variants carried by individuals with amyotrophic lateral sclerosis (ALS) and controls of the various potential biochemical property changes in the ALSKP and Project MinE datasets, followed by a Cochran–Mantel–Haenszel (CMH) test on the combined datasets. Significance was measured at an alpha-level of 0.05. (C) Quantile-quantile plots of rare variants of interest identified across 24 ALS-associated genes in ALS case-control sequencing datasets, defined by the biochemical property changes of the amino acids. The amino acid biochemical property changes significantly enriched for variants of interest in the individuals with ALS compared to the controls were further analysed to determine which genes were driving the enrichment of the features using Fisher’s exact testing. Enrichment of variants resulting in non-polar to non-polar amino acid changes and non-polar to polar amino acid changes were driven by SOD1 (ALSKP, P = 1.60 × 10−11; Project MinE, P = 1.13 × 10−1; combined analysis, P = 2.01 × 10−11 and ALSKP, P = 2.29 × 10−10; Project MinE, P = 1.15 × 10−1; combined analysis, P = 6.04 × 10−10, respectively). Enrichment of variants resulting in polar (+) to polar amino acid changes was driven by VCP (ALSKP, P = 1.41 × 10−4; Project MinE, P = 5.60 × 10−1; combined analysis, P = 2.04 × 10−4). An alpha-level of 2.08 × 10−3 was considered significant following Bonferroni correction accounting for the 24 genes analysed.
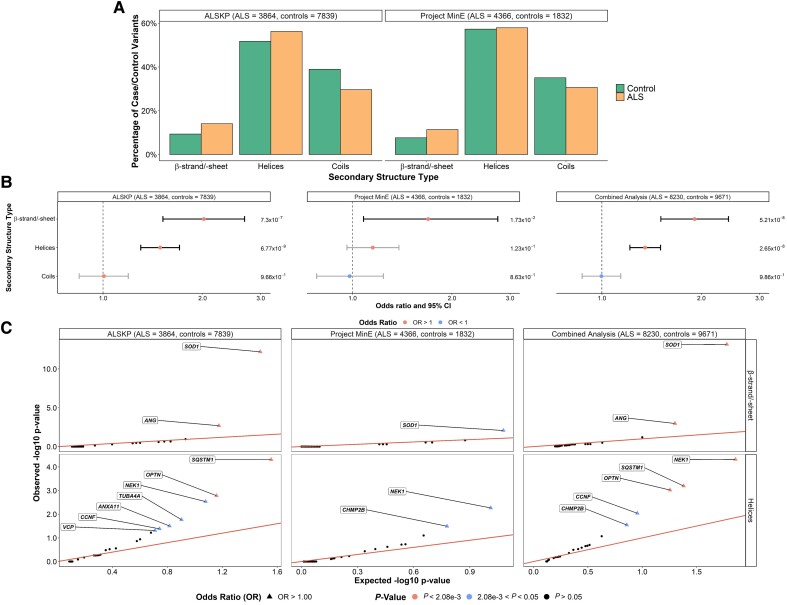
Figure 2.
Secondary structure types at which rare missense variants were identified across 24 ALS-associated genes. (A) Secondary structure types were obtained using the AlphaFold predicted structures and DSSP (dictionary of protein secondary structure) program for all residues at which variants of interest were observed in the ALS Knowledge Portal (ALSKP) and Project MinE ALS sequencing consortium. Secondary structure types were binned based on the following: β-strand/-sheet includes β-strands and β-sheets; helices includes 310-helices, α-helices, π-helices and polyproline II helices; and coils includes loops, bends and turns. (B) Enrichment analyses were performed using Fisher’s exact testing to compare the number of variants carried by individuals with amyotrophic lateral sclerosis (ALS) and controls at residues of each secondary structure type in the ALSKP and Project MinE datasets, followed by a Cochran–Mantel–Haenszel (CMH) test. Significance was measured at an alpha-level of 0.05. (C) Quantile-quantile plots of rare variants of interest identified across 24 ALS-associated genes in ALS case-control sequencing datasets, defined by their secondary structure type. The secondary structure types significantly enriched for variants of interest in the individuals with ALS compared to the controls were further analysed to determine which genes were driving the enrichment of the features using Fisher’s exact testing. Enrichment of variants in β-strands/-sheets was driven by SOD1 (ALSKP, P = 6.26 × 10−13; Project MinE, P = 8.38 × 10−3; combined analysis, P = 6.97 × 10−14) and ANG (ALSKP, P = 2.04 × 10−3; combined analysis, P = 1.05 × 10−3). Enrichment of variants in helices was driven by NEK1 (ALSKP, P = 2.92 × 10−3; Project MinE, P = 5.43 × 10−3; combined analysis, P = 4.81 × 10−5), SQSTM1 (ALSKP, P = 4.92 × 10−5; combined analysis, P = 6.41 × 10−4); and OPTN (ALSKP, P = 1.65 × 10−3; Project MinE, P = 5.43 × 10−3; combined analysis, P = 9.52 × 10−4). An alpha-level of 2.08 × 10−3 was considered significant following Bonferroni correction accounting for the 24 genes analysed.
We further examined the combinations of reference amino acids and alternate amino acids involved in the rare, missense variants in individuals with ALS versus controls. Across the combined datasets, the largest odds ratios were observed for missense variants involving glycine (Gly) to cystine (Cys), arginine (Arg) to leucine (Leu), or valine (Val) to Gly amino acid changes (OR = 14.11, 14.11 and 11.76, respectively; Supplementary Fig. 2).
Variant protein structural feature assessment
Quality of the predicted monomeric structures of the 24 ALS-associated proteins collected from the AlphaFold database are presented in Supplementary Table 6. For analyses involving the annotations obtained from AlphaFold predicted structures, only variants at residues with a high quality (pLDDT > 7038) were retained, including 74.2% of variants carried by individuals with ALS and 75.0% of variants carried by controls from the ALS Knowledge Portal; and 62.6% of variants carried by individuals with ALS and 64.5% of variants carried by controls from the Project MinE ALS sequencing consortium (Supplementary Fig. 3).
From the AlphaFold annotations, we assessed the secondary structure distribution of the reference amino acids involved in the rare, missense variants (Fig. 2A) to determine whether specific secondary structure types were more often affected in variants carried by individuals with ALS than controls (Fig. 2B). Following binning of the secondary structures into subtypes, the combined dataset analysis revealed that variants at residues involved in β-strands/sheets [OR = 1.91 (1.51–2.42), P = 5.21 × 10−8] and involved in helices [OR = 1.35 (1.21–1.50), P = 2.65 × 10−8] were significantly enriched in ALS. Notably, these subtype signals may have been driven by the significantly enriched secondary structure type signals identified for β-sheets [OR = 1.95 (1.53–2.47), P = 3.69 × 10−8] and α-helices [OR = 1.32 (1.18–1.48), P = 3.84 × 10−7; Supplementary Fig. 4). The β-strands/sheets signals were primarily driven by SOD1 [OR = 20.78 (7.14–60.47), P = 6.97 × 10−14] and ANG [OR = 3.57 (1.65–7.72), P = 1.05 × 10−3], whereas the signals in helices were driven by NEK1 [OR = 1.80 (1.36–2.38), P = 4.81 × 10−5], SQSTM1 [OR = 1.57 (1.21–2.05), P = 6.41 × 10−4], and OPTN [OR = 2.76 (1.49–5.09), P = 9.52 × 10−4] (Fig. 2C).
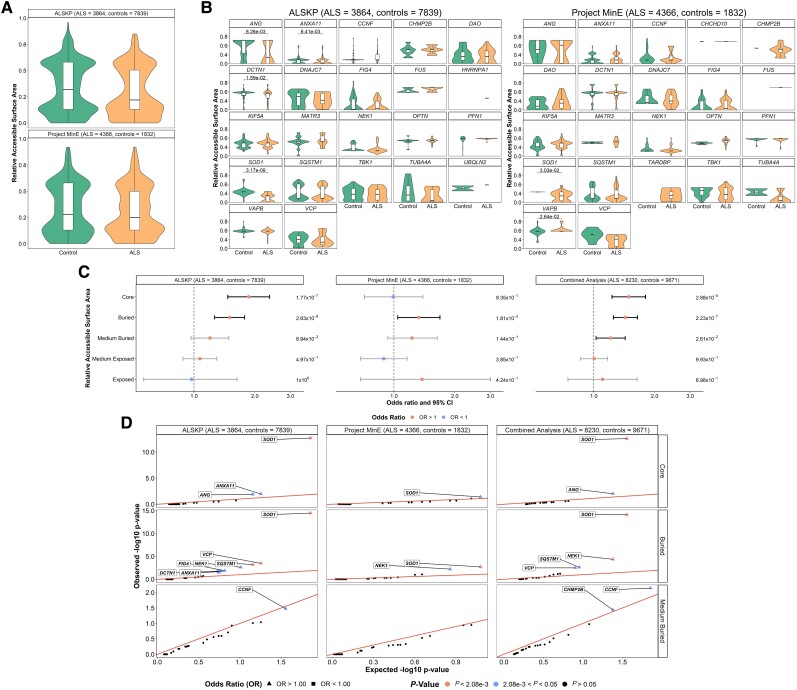
We next assessed the RSA of the reference amino acids involved in the rare, missense variants. Mean RSAs of the variants in any of the 24 genes carried by individuals with ALS and controls in the ALS Knowledge Portal and Project MinE ALS sequencing consortium were compared using Wilcoxon rank sum tests with continuity correction, but no significant differences were observed (ALS Knowledge Portal P-value = 0.142 and Project MinE ALS P-value = 0.1583; Fig. 3A). Gene specific comparisons of mean RSAs were also performed, and only SOD1 displayed a significant difference in both datasets with a lower mean RSAs in the variants carried by individuals with ALS compared to controls (ALS Knowledge Portal P = 3.17 × 10−9; Project MinE ALS P = 3.03 × 10−2; Fig. 3B).
Figure 3.
Relative solvent accessible areas (RSAs) of rare missense variants identified across 24 ALS-associated genes. For all residues at which variants of interest were observed in the datasets, the AlphaFold predicted structures and DSSP (dictionary of protein secondary structure) program were used to derive RSAs. (A) A Wilcoxon rank sum test with continuity correction compared mean RSA of variants carried by the individuals with amyotrophic lateral sclerosis (ALS) and controls from the ALS Knowledge Portal (ALSKP) and Project MinE ALS sequencing consortium. Significant differences were not observed in either dataset (P-value = 0.142 and P-value = 0.1583, respectively). (B) Wilcoxon rank sum tests compared mean RSA of variants carried by the individuals with ALS and controls per gene. Significant P-values are displayed. Genes with <2 variant counts were excluded. (C) Enrichment analyses were performed using Fisher’s exact testing to compare the number of variants carried by individuals with ALS and controls of various RSA levels in the two datasets, followed by a Cochran–Mantel–Haenszel (CMH) test. Significance was measured at an alpha-level of 0.05. (D) Quantile-quantile plots of rare missense variants identified across 24 ALS-associated genes in the two datasets, defined by their RSA level. The RSA levels significantly enriched for missense variants in the individuals with ALS were further analysed to determine which genes were driving the enrichment of the features using Fisher’s exact testing. Enrichment of variants in the core was driven by SOD1 (ALSKP, P = 2.11 × 10−13; Project MinE, P = 3.96 × 10−2; combined analysis, P = 2.72 × 10−13) and ANG (ALSKP, P = 1.23 × 10−2; combined analysis, P = 9.73 × 10−3). Enrichment of variants in buried regions was driven by SOD1 (ALSKP, P = 3.38 × 10−15; Project MinE, P = 1.97 × 10−3; combined analysis, P = 7.71 × 10−15), NEK1 (ALSKP, P = 2.41 × 10−3; combined analysis, P = 4.39 × 10−5), SQSTM1 (ALSKP, P = 6.13 × 10−4; Project MinE, P = 1.97 × 10−3; combined analysis, P = 1.69 × 10−5) and VCP (ALSKP, P = 3.26 × 10−4; combined analysis, P = 3.12 × 10−3). An alpha-level of 2.08 × 10−3 was considered significant following Bonferroni correction accounting for the 24 genes analysed. Solvent area levels were defined as: core, RSA < 5%; buried, 5% < RSA < 25%; medium-buried, 25% < RSA < 50%; medium-exposed, 50% < RSA < 75%; and exposed, RSA > 75%.
RSAs were then binned into five exposure levels, including: core, RSA <5%; buried, 5% < RSA < 25%; medium-buried, 25% < RSA < 50%; medium-exposed, 50% < RSA < 75%; and exposed, RSA > 75%, to determine whether there was an enrichment of specific exposure levels. In the combined dataset analysis, variants across all 24 genes were enriched in individuals with ALS at core [OR = 1.49 (1.23–1.80), P = 2.88 × 10−5], buried state [OR = 1.43 (1.25–1.64), P = 2.23 × 10−7] and medium-buried state [OR = 1.21 (1.03–1.43), P = 2.61 × 10−2] (Fig. 3C).
Gene-based analysis revealed that ALS variant enrichment in residues at protein core was driven by SOD1 [OR = 68.29 (8.70–535.85), P = 2.72 × 10−13]; and that at buried level was also driven by SOD1 [OR = 119.41 (7.37–1935.76), P = 7.71 × 10−15] as well as NEK1 [OR = 1.93 (1.41–2.63), P = 4.39 × 10−5] (Fig. 3D). No individual genes were significantly enriched for variants of residues at medium-buried residues.
Variant protein sequence feature assessment
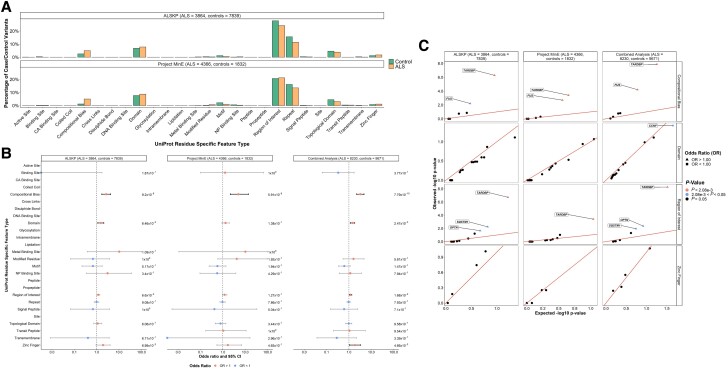
The variants were also annotated with UniProt-defined functional features of the reference amino acids (Fig. 4A). We observed a significant enrichment of variants in ALS in the combined dataset analysis at amino acids involved in compositional bias [OR = 3.22 (2.21–4.70), P = 7.79 × 10−10]; domains [OR = 1.44 (1.14–1.81), P = 2.47 × 10−3]; regions of interest [OR = 1.18 (1.03–1.35), P = 1.68 × 10−2]; and zinc fingers [OR = 1.84 (1.04–3.27), P = 4.66 × 10−2; Fig. 4B]. The specific genes driving the signal in compositionally biased regions were TARDBP [OR = 27.66 (5.77–132.52), P = 1.31 × 10−8] and FUS [OR = 2.93 (1.74–4.94), P = 6.28 × 10−5]; and in regions of interest, was also TARDBP [OR = 28.68 (6.02–136.63), P = 5.29 × 10−9] (Fig. 4C).
Figure 4.
UniProt protein sequence functional features of rare missense variants identified across 24 ALS-associated genes. (A) Distribution of UniProt defined protein sequence functional features of the residues at which variants of interest were identified carried by the individuals with amyotrophic lateral sclerosis (ALS) and controls from the ALS Knowledge Portal (ALSKP) and Project MinE ALS sequencing consortium. (B) Enrichment analyses were performed using Fisher’s exact testing to compare the number of variants carried by individuals with ALS and controls of various UniProt functional features in the ALSKP and Project MinE datasets, followed by a Cochran–Mantel–Haenszel (CMH) test. Significance was measured at an alpha-level of 0.05. (C) Quantile-quantile plots of rare variants of interest identified across 24 ALS-associated genes in the two datasets, defined by their UniProt functional features. The UniProt functional features significantly enriched for variants of interest in the individuals with ALS compared to the controls were further analysed to determine which genes were driving the enrichment of the features using Fisher’s exact testing. Enrichment of variants in compositionally biased regions was driven by TARDBP (ALSKP, P = 5.23 × 10−7; Project MinE, P = 3.94 × 10−4; combined analysis, P = 1.31 × 10−8) and FUS (ALSKP, P = 2.03 × 10−3; Project MinE, P = 6.19 × 10−3; combined analysis, P = 6.28 × 10−5). Enrichment of variants in regions of interest was driven by TARDBP (ALSKP, P = 1.91 × 10−7; Project MinE, P = 3.94 × 10−4; combined analysis, P = 5.29 × 10−9). An alpha-level of 2.08 × 10−3 was considered significant following Bonferroni correction accounting for the 24 genes analysed.
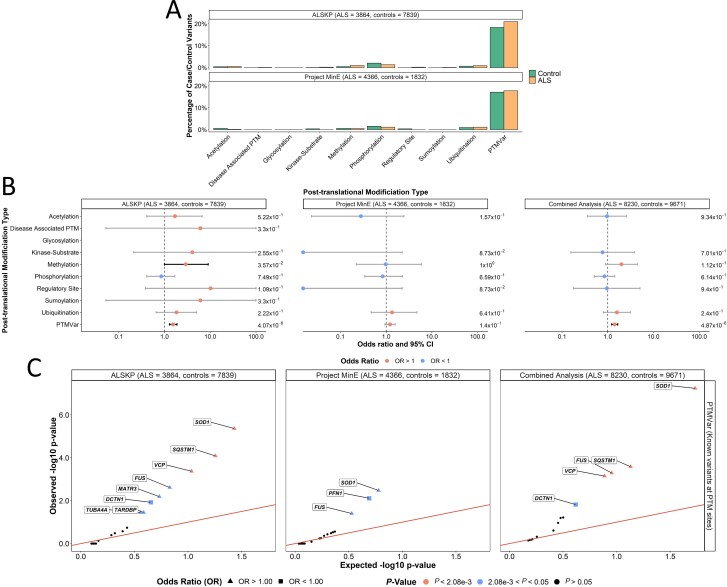
Similarly, we surveyed the PTM features of the reference amino acids (Fig. 5A). We observed a significant enrichment of variants in ALS in the combined dataset analysis at amino acids involved in any known PTM [OR = 1.42 (1.22–1.64), P = 4.87 × 10−6; Fig. 5B]. This was primarily driven by SOD1 [OR = 3.57 (2.21–5.76), P = 5.77 × 10−8]; SQSTM1 [OR = 1.65 (1.26–2.16), P = 2.61 × 10−4]; FUS [OR = 23.19 (2.49–216.23), P = 5.12 × 10−4]; and VCP [OR = 20.00 (1.15–346.50), P = 7.13 × 10−4] (Fig. 5C).
Figure 5.
Post-translational modification (PTM) features rare missense variants identified across 24 ALS-associated genes. (A) Distribution of PTM features of the residues at which all missense variants were identified. (B) Enrichment analyses were performed using Fisher's exact testing to compare the number of variants carried by individuals with ALS and controls at which PTMs occur in the ALS Knowledge Portal (ALSKP) and Project MinE datasets, followed by a Cochran–Mantel–Haenszel (CMH) test. Significance was measured at an alpha-level of 0.05. (C) Quantile-quantile plots of rare missense variants identified across 24 ALS-associated genes in ALS case-control sequencing datasets, that are located at known post-translational modification sites (PTMVar). Known PTM sites, which were significantly enriched for variants of interest in the individuals with ALS compared to the controls from the two datasets, were further analysed to identify the genes driving the enrichment using Fisher’s exact testing. Enrichment of variants was driven by SOD1 (ALSKP, P = 4.32 × 10−6; Project MinE, P = 3.33 × 10−3; combined analysis, P = 5.77 × 10−8), SQSTM1 (ALSKP, P = 8.13 × 10−5; combined analysis, P = 2.61 × 10−4), FUS (ALSKP, P = 2.43 × 10−3; Project MinE, P = 3.96 × 10−2; combined analysis, P = 5.12 × 10−4) and VCP (ALSKP, P = 4.26 × 10−4; combined analysis, P = 7.13 × 10−4). An alpha-level of 2.08 × 10−3 was considered significant following Bonferroni correction accounting for the 24 genes analysed.
Variant transcriptional expression levels
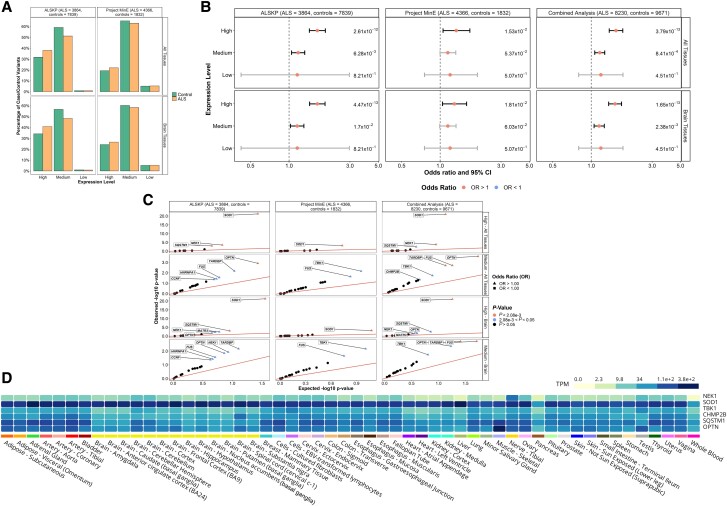
Finally, all variants were annotated with expression levels estimated using GTEx across all tissues and specifically within brain tissues (Fig. 6A). We compared the expression levels of the residues and observed a significant enrichment of variants in ALS at amino acids with high and medium expression levels in all tissues [OR = 1.56 (1.38–1.77), P = 3.79 × 10−13 and OR = 1.17 (1.07–1.28), P = 8.41 × 10−4, respectively]; and in brain specific tissues [OR = 1.54 (1.37–1.73), P = 1.65 × 10−13 and OR = 1.15 (1.05–1.27), P = 2.38 × 10−3, respectively; Fig. 6B]. For the high expression in both ‘all tissues’ and in ‘brain tissues’, SOD1 was the primary driver [OR = 10.49 (5.58–19.70), P = 9.70 × 10−22 and OR = 10.49 (5.58–19.70), P = 9.70 × 10−22, respectively; Fig. 6C]. Further, variants with medium expression in all tissues were significantly enriched in OPTN [OR = 2.52 (1.52–4.16), P = 2.60 × 10−4] and variants with medium expression both in all tissues and in brain tissues were significantly enriched in FUS [OR = 2.37 (1.39–4.03), P = 1.94 × 10−3 and OR = 2.37 (1.39–4.03), P = 1.94 × 10−3, respectively] as well as TARDBP [OR = 2.63 (1.42–4.87), P = 1.96 × 10−3 and OR = 2.63 (1.42–4.87), P = 1.96 × 10−3, respectively].
Figure 6.
Expression levels of rare missense variants identified across 24 ALS-associated genes. (A) Distribution of expression levels across all tissues and in brain tissues of all missense variants found in the ALS Knowledge Portal (ALSKP) and Project MinE sequencing consortium datasets were identified. (B) Enrichment analyses were performed using Fisher’s exact testing to compare expression levels across all tissues and in brain tissues of variants carried by individuals with ALS and controls in the two datasets, followed by a Cochran–Mantel–Haenszel (CMH) test. Expression levels were defined as: low, mean expression < 0.1; medium, mean expression ≥ 0.1 and ≤ 0.9; and high, mean expression > 0.9. Significance was measured at an alpha-level of 0.05. (C) Quantile-quantile plots of missense variants identified across the 24 ALS-associated genes, defined by their expression level in brain tissues and all tissues. The expression levels significantly enriched for variants of interest in the individuals with ALS compared to the controls from the ALSKP and Project MinE ALS datasets were further analysed to determine which genes were driving the enrichment of the features using Fisher’s exact testing. Enrichment of variants with high expression in all tissues was driven by SOD1 (ALSKP, P = 5.64 × 10−22; Project MinE, P = 6.60 × 10−4; combined analysis, P = 9.70 × 10−22), NEK1 (ALSKP, P = 3.22 × 10−4; combined analysis, P = 1.52 × 10−4) and SQSTM1 (ALSKP, P = 1.04 × 10−3; combined analysis, P = 3.30 × 10−3); and enrichment of variants with medium expression in all tissues was driven by OPTN (ALSKP, P = 1.22 × 10−3; combined analysis, P = 2.60 × 10−4), FUS (ALSKP, P = 2.79 × 10−2; Project MinE, P = 2.27 × 10−2; combined analysis, P = 1.94 × 10−3) and TARDBP (ALSKP, P = 6.80 × 10−3; combined analysis, P = 1.96 × 10−3). Similarly, enrichment of variants with high expression in brain tissues was driven by SOD1 (ALSKP, P = 5.64 × 10−22; Project MinE, P = 6.60 × 10−4; combined analysis, P = 9.70 × 10−22); and enrichment of variants with medium expression in brain tissues was driven by FUS (ALSKP, P = 2.79 × 10−2; Project MinE, P = 2.27 × 10−2; combined analysis, P = 1.94 × 10−3), TARDBP (ALSKP, P = 6.80 × 10−3; combined analysis, P = 1.96 × 10−3) and OPTN (ALSKP, P = 1.44 × 10−2; combined analysis, P = 4.36 × 10−3). An alpha-level of 2.08 × 10−3 was considered significant following Bonferroni correction accounting for the 24 genes analysed. (D) Average expression of the genes identified as driving enrichment of variants with medium and high expression in the brain and all tissues in individuals with ALS compared to controls based on data from the Genotype-Tissue Expression (GTEx) Portal.
Case study: SOD1
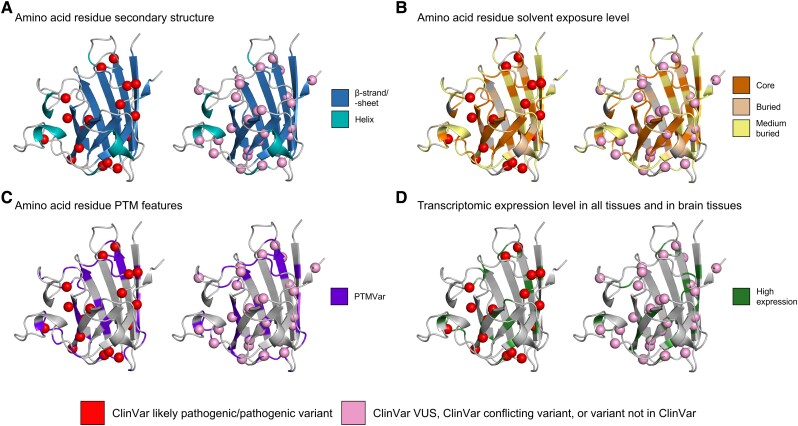
To highlight the strength of our approach, we present a case study visualizing our results regarding one of the most well established ALS-associated genes, SOD1 (Fig. 7). All missense variants identified within SOD1 in individuals with ALS were annotated with ClinVar pathogenicity classifications and the variant positions were mapped onto the AlphaFold predicted protein structure in the G2P Portal. Protein structure, protein sequence, and transcriptomic features were also mapped onto the SOD1 3D protein structure, including the significantly enriched features: β-sheets/-strands, core and buried regions, known sites of PTMs, and high expression in all tissues and in brain tissues.
Figure 7.
Mapping of missense variants carried by individuals with ALS onto the AlphaFold-predicted structure of SOD1. The Genomics2Protein portal was used to map missense variants carried by the individuals with amyotrophic lateral sclerosis (ALS) from the ALS Knowledge Portal and Project MinE ALS sequencing consortium onto the predicted monomeric 3D structures of SOD1 collected from the AlphaFold database. Missense variants were annotated using the ClinVar database to determine whether they had been previously classified based on their pathogenicity. For the purposes of visualization, all variants classified as a variant of uncertain significance (VUS) or conflicting interpretation by ClinVar or not classified by ClinVar were binned. Following the application of a burden analysis approach to determine whether individual genes were driving protein structure, protein sequence, or transcriptomic feature enrichment, we identified the following features were significantly enriched in SOD1 missense variation carried by individuals with ALS compared to controls: (A) β-strand/-sheets secondary structures; (B) core and buried solvent exposure levels; (C) known post-translational modification sites (PTM); and (D) high transcriptomic expression levels in all tissues and in brain tissues. PTMVar = any known post-translational modification sites.
Of the 42 unique missense variants identified in SOD1 in individuals with ALS, 16 (38.1%) were classified as pathogenic or likely pathogenic in ClinVar, all of which were located within at least one of the protein structure, protein sequence, or transcriptomic features found to be enriched in SOD1 missense variants in the individuals with ALS. Ten (23.8%) of these variants were within a β-strand/-sheet (Fig. 7A), 13 (31.0%) were within a core or buried region (Fig. 7B), 11 (26.2%) were located at known PTM sites (Fig. 7C), all 16 exhibited high expression levels (Fig. 7D), and six (14.3%) were located at a position annotated with all four enriched features.
Twenty-six (61.9%) of the unique missense variants in SOD1 carried by individuals with ALS were classified as either a VUS in ClinVar, as having conflicting interpretations in ClinVar, or were not reported in ClinVar. Of these variants with unknown pathogenicity, 23 (54.8%) were located within at least one of the enriched features, and seven (16.7%) were located at a position annotated as all four enriched features.
Further, of the 42 missense variants identified within SOD1 in individuals with ALS, 11 had experimental structures available in the Protein Data Bank (PDB) database. We collected 31 PDB entries corresponding to the missense variants (Supplementary Table 7) and analysed the changes in the protein’s 3D structure. In Supplementary Fig. 5A, we compared the RSA of the mutated residue and the reference residue in the wild-type protein. Consistent with our findings (Fig. 3), reference residues affected by ALS variants in the wild-type protein were primarily in core, buried and medium-buried states; however, most variants (n = 9; 81.8%) resulted in an increase in the RSA value of the residue. This suggests that pathogenic SOD1 variants tend to cause the mutated residues to become more solvent-exposed. We also assessed the secondary structure of affected residues in SOD1 wild-type and mutant structures. Supplementary Fig. 5B–D displays the content of α-helix, β-strand and coil within the protein structure, defined as the fraction of residues with helix, strand or coil conformations out of total residues. We observed that SOD1 mutant structures tend to have lower β-strand content and higher coil content compared to the wild-type SOD1 structures, which is in line with our results showing that β-strand is a significant ALS variant associated structural feature (Fig. 2).
Interestingly, two missense variants were identified in SOD1 exclusively in control samples, namely p.Thr136Ile and p.Asp110Tyr. Although both variants were considered to have high expression, neither variant were located within an enriched secondary structure type, an enriched solvent exposure level, nor a known PTM site.
Discussion
Despite the rapid advancement in gene discovery in ALS, our understanding of the pathogenicity of the variants identified within those genes remains challenging, particularly when faced with missense variation. Here, we have used large-scale sequencing and a case-control enrichment analysis approach to identify patterns of protein structure, protein sequence, and transcriptomic features of missense variants in 24 known ALS-associated genes.
While it could be suggested that the features we found enriched in individuals with ALS would be relevant to pathogenic missense variation across a range of diseases, we hypothesize that our results are specific to ALS. We observed a strong, significant signal of enrichment for missense variants in compositionally biases protein regions (Fig. 4). In contrast, a recent assessment of UniProt functional features enriched in 189 NDD-associated genes across >360 000 samples found a depletion of NDD variants in compositionally biased protein regions compared to controls.30 Similarly, missense variants in ‘regions of interest’ that mediate protein-protein interactions or another biological processes were significantly enriched individuals with ALS herein, but not in individuals with NDD. One contributing factor to these differences may be that NDD pathogenic variants often display loss-of-function properties,47–49 while ALS variants often display gain-of-function properties.22,50–52 But additional investigations will be required to determine whether the pathological mechanisms translate to the protein features involved in disease-associated missense variation. Additionally, it has been previously shown that functional sites, such as active sites, metal, nucleotide-phosphate, and co-factor binding sites, are enriched for missense variations across 1330 Mendelian disease-associated genes,42 yet we observed no significant association between these functional features and ALS missense variants (Fig. 4). These differences suggest that our results are not generalizable to other Mendelian phenotypes.
Upon identification of features enriched for ALS case variants in all 24 genes, we applied a burden analysis approach to determine whether individual genes were driving feature enrichment. The burden signals observed in SOD1 were of particular interest, as further demonstrated by the presented case study. Notably, SOD1 missense variants in individuals with ALS were significantly found to affect non-polar residues, β-sheets/-strands, protein core or buried regions, and known sites of PTMs, as well as have high expression relative to wild-type SOD1 in all tissues and brain specific tissues. Upon further investigation of the missense variants in SOD1 by visualization of the protein structure, we observed that of the total 42 unique missense variants in individuals with ALS, 38.1% were classified as pathogenic or likely pathogenic in the ClinVar database and had at least one protein feature found to be enriched in SOD1 missense variants in ALS (Fig. 7). Further, assessment of experimentally solved structures of 11 out of the 42 SOD1 missense variants showed that the variants largely resulted in greater solvent exposure of residues and loss of β-sheet secondary structures, demonstrating complementarity with the statistical signals of enrichment we observed regarding potentially pathogenic protein structural features. Similarly, previous studies have found mutations within core β-sheets are more likely to alter the packing of the protein resulting in out-of-register β-sheet oligomers, thought to be important to the cytotoxicity of SOD1,53,54 and loss of stability and increased susceptibility to aggregation.55 Further evidence suggests variants affecting core and buried residues of SOD1 result in destabilization of interactions imperative for proper folding and in exposure of hydrophobic residues, which again induces aggregation.56–59 Finally, aggregation of SOD1 is prompted by loss of and alteration in PTMs, such as copper and zinc binding, disulphide formation, phosphorylation, and ubiquitination, among others.60–62 Collectively, the consistency between our findings, ClinVar, structural and functional evidence for SOD1 present a proof-of-concept that the identified proteomic and transcriptomic features enriched in ALS may improve the definition of pathogenic variants across all 24 ALS-associated genes.
In addition to SOD1, seven genes displayed significant enrichment of at least one proteomic or transcriptomic feature that could be incorporated into a gene- and disease-specific variant pathogenicity classification structure (Table 1). This may be especially beneficial for the pathogenicity classification of variants in genes with exceptionally high frequencies of missense VUS, such as SQSTM1 (Supplementary Fig. 1A and B).63 Based on our findings, extra consideration should be taken for SQSTM1 missense variants located within helices, specifically α-helices, or at known PTM sites. Interestingly, α-helices within the SQSTM1 encoded protein, p62, are main structural components of two of its primary functional domains, namely the PB1 domain and the UBA ubiquitin-binding domain.64,65 Further, many post-translational modifications have been observed for p62, including acetylation, disulphide bridging, phosphorylation and ubiquitination, which are imperative for the protein’s role in autophagy.66–68 Current ACMG pathogenicity classification guidelines do not account for these gene-specific features, unless the regions are also predetermined mutational hot spots.11 Although we are not recommending that variant pathogenicity classification could be based entirely on any one of the enriched features, we do suspect that incorporating the evidence into classification frameworks will improve accuracy of missense variant pathogenicity classification particularly within genes with large rates of VUS, in a similar manner to the incorporation of in silico prediction tool-based evidence.11
Table 1.
Proteomic and transcriptomic features significantly enriched in ALS-associated genes, which may be useful in gene-specific variant pathogenicity interpretation
| Gene | Significantly enriched feature | Odds ratio | P-value |
|---|---|---|---|
| ANG | β-strand/sheet | 3.57 | 1.05 × 10−3 |
| FUS | Compositional bias | 2.93 | 6.28 × 10−5 |
| PTM site at known variant | 23.19 | 5.12 × 10−4 | |
| Medium expression, all tissues | 2.37 | 1.94 × 10−3 | |
| Medium expression, brain tissues | 2.37 | 1.94 × 10−3 | |
| NEK1 | Helices | 1.80 | 4.81 × 10−5 |
| Buried RSA | 1.93 | 4.39 × 10−5 | |
| High expression, all tissues | 1.98 | 1.52 × 10−4 | |
| OPTN | Helices | 2.76 | 9.52 × 10−4 |
| Medium expression, all tissues | 2.52 | 2.60 × 10−4 | |
| SOD1 | Non-polar to non-polar amino acid change | 57.35 | 2.01 × 10−11 |
| Non-polar to polar amino acid change | 31.49 | 6.04 × 10−10 | |
| β-strand/sheet | 20.78 | 6.97 × 10−14 | |
| Core RSA | 59.34 | 2.11 × 10−13 | |
| Buried RSA | 44.26 | 3.38 × 10−15 | |
| PTM site at known variant | 3.57 | 5.77 × 10−8 | |
| High expression, all tissues | 10.49 | 9.70 × 10−22 | |
| High expression, brain tissues | 10.49 | 9.70 × 10−22 | |
| SQSTM1 | Helices | 1.57 | 6.41 × 10−4 |
| PTM site at known variant | 1.65 | 2.61 × 10−4 | |
| TARDBP | Compositional bias | 27.66 | 1.31 × 10−8 |
| Region of interest | 28.68 | 5.29 × 10−9 | |
| Medium expression, all tissues | 2.63 | 1.96 × 10−3 | |
| Medium expression, brain tissues | 2.63 | 1.96 × 10−3 | |
| VCP | Polar positively charged to polar amino acid change | 13.02 | 2.04 × 10−4 |
| PTM site at known variant | 10.58 | 4.26 × 10−4 |
Results of the combined Cochran–Mantel–Haenszel (CMH) test of the ALS Knowledge Portal and Project MinE ALS sequencing consortium were used to determine significant proteomic and transcriptomic features. An alpha-level of 2.08 × 10−3 was considered significant following Bonferroni correction accounting for the 24 genes analysed. Gene-specific significantly enriched proteomic or transcriptomic features were not observed for the genes: ANXA11, CCNF, CHCHD10, CHMP2B, DAO, DCTN1, DNAJC7, FIG4, HNRNPA1, KIF5A, MATR3, PFN1, TBK1, TUBA4A, UBQLN2 and VAPB. For all gene-based enrichment results see Supplementary Table 8. PTM = post-translational modification; RSA = relative solvent accessible area.
There were also significant signals of enrichment that were not driven by specific ALS-associated genes. While the lack of significant results in these burden analyses may have resulted from lack of statistical power due to sample size, the ALS Knowledge Portal and Project MinE ALS sequencing consortium are considered two of the largest ALS sequencing datasets currently available. Moreover, the number of missense variants that were annotated with these specific features were not exceptionally small; for example, across the two datasets there were a greater number of medium buried missense variants (n = 644) identified than core missense variants (n = 482) for which specific gene burdens were identified. Therefore, it is not unreasonable to propose that features that are not driven by single genes may indeed be relevant across all, or many, ALS-associated genes and warrant further exploration.
There are important caveats to our presented analyses that we acknowledge within our study. The individuals included within the ALS Knowledge Portal and Project MinE ALS sequencing consortium cohorts were largely of European ancestry,31,32 but it is well established that rates of VUS are much higher in under-represented populations.69,70 As larger, more diverse cohorts of ALS patients become available, it will be important to replicate our analyses using those datasets to ensure that our results remain applicable across all populations. The results presented also have yet to be functionally validated using in vitro or in vivo methodologies; however, as the success of case-control analyses have demonstrated in the identification of novel disease-associated genes, the application of large-scale sequencing data is highly opportunistic in the identification of genetic signatures, and our study demonstrates how integrating other ‘-omics’ data types may expand our understanding of known disease-associated genes. As previously stated, we do not believe that the features we identified can be used as sole evidence for pathogenicity, rather our results suggest that the incorporation of proteomic and transcriptomic evidence may offer additional insights for missense variation.
In conclusion, our results demonstrate that rare, missense variants identified in two of the largest ALS case-control cohorts to date exhibit enrichment of protein structure, protein sequence and transcriptomic features that may be useful in defining variant pathogenicity. Incorporating gene-specific features into variant classification may offer the ability to better identify true pathogenic missense variants in ALS clinical cohorts, thereby improving diagnoses and making clinical trials more accessible to a larger proportion of patients.
Supplementary Material
Acknowledgements
A.A.D. is supported by the Canadian Institute of Health Research Banting Postdoctoral Fellowship program. S.M.K.F. is supported by grants from Fondation Brain Canada, ALS Society of Canada, and the Tanenbaum Open Science Institute (TOSI) at The Montreal Neurological Institute and Hospital, McGill University. S.K. and S.I. are supported by the Merkin Institute of Transformative Technologies in Healthcare, the Broad Institute of MIT and Harvard.
Contributor Information
Allison A Dilliott, Department of Neurology and Neurosurgery, McGill University, Montreal, Quebec H3A 0G4, Canada; Montreal Neurological Institute-Hospital, McGill University, Montreal, Quebec H3A 2B4, Canada.
Seulki Kwon, The Center for the Development of Therapeutics, Broad Institute of MIT and Harvard, Cambridge, MA 02142, USA.
Guy A Rouleau, Department of Neurology and Neurosurgery, McGill University, Montreal, Quebec H3A 0G4, Canada; Montreal Neurological Institute-Hospital, McGill University, Montreal, Quebec H3A 2B4, Canada; Department of Human Genetics, McGill University, Montreal, Quebec, Canada.
Sumaiya Iqbal, The Center for the Development of Therapeutics, Broad Institute of MIT and Harvard, Cambridge, MA 02142, USA; Analytic and Translational Genetics Unit, Massachusetts General Hospital, Boston, MA 02114, USA.
Sali M K Farhan, Department of Neurology and Neurosurgery, McGill University, Montreal, Quebec H3A 0G4, Canada; Montreal Neurological Institute-Hospital, McGill University, Montreal, Quebec H3A 2B4, Canada; Department of Human Genetics, McGill University, Montreal, Quebec, Canada.
Data availability
ALS Knowledge Portal data are available through the Neurodegenerative Disease Knowledge Portal (https://ndkp.hugeamp.org/). Project MinE sequencing consortium data are available through the Project MinE Databrowser (http://databrowser.projectmine.com/). The GTEx-derived pext expression data (https://gnomad.broadinstitute.org/downloads/), AlphaFold Protein Structure Database (https://alphafold.ebi.ac.uk/), and Genomics 2 Protein Portal (https://g2p.broadinstitute.org/) are open access and were used for these analyses.
Competing interests
The authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Brown RH, Al-Chalabi A. Amyotrophic lateral sclerosis. N Engl J Med. 2017;377:162–172. [DOI] [PubMed] [Google Scholar]
- 2. Al-Chalabi A, Fang F, Hanby MF, et al. . An estimate of amyotrophic lateral sclerosis heritability using twin data. J Neurol Neurosurg Psychiatry. 2010;81:1324–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Majounie E, Renton AE, Mok K, et al. . Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: A cross-sectional study. Lancet Neurol. 2012;11:323–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Byrne S, Elamin M, Bede P, et al. . Cognitive and clinical characteristics of patients with amyotrophic lateral sclerosis carrying a C9orf72 repeat expansion: A population-based cohort study. Lancet Neurol. 2012;11:232–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mejzini R, Flynn LL, Pitout IL, Fletcher S, Wilton SD, Akkari PA. ALS Genetics, mechanisms, and therapeutics: Where are we now? Front Neurosci. 2019;13:1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dharmadasa T, Scaber J, Edmond E, et al. . Genetic testing in motor neurone disease. Pract Neurol. 2022;22:107–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Richards D, Morren JA, Pioro EP. Time to diagnosis and factors affecting diagnostic delay in amyotrophic lateral sclerosis. J Neuro Sci. 2020;417:117054. [DOI] [PubMed] [Google Scholar]
- 8. Singh N, Ray S, Srivastava A. Clinical mimickers of amyotrophic lateral sclerosis-conditions we cannot afford to miss. Ann Indian Acad Neurol. 2018;21:173–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Salmon K, Kiernan MC, Kim SH, et al. . The importance of offering early genetic testing in everyone with amyotrophic lateral sclerosis. Brain. 2022;145:1207–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dilliott AA, Al Nasser A, Elnageeb M, et al. . Clinical testing panels for ALS: Global distribution, consistency, and challenges. Amyotroph Lateral Scler Frontotemporal Degener. 2023;24:420–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Richards S, Aziz N, Bale S, et al. . Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American college of medical genetics and genomics and the association for molecular pathology. Genet Med. 2015;17:405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shepheard SR, Parker MD, Cooper-Knock J, et al. . Value of systematic genetic screening of patients with amyotrophic lateral sclerosis. J Neurol Neurosurg Psychiatry. 2021;92:510–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Roggenbuck J, Palettas M, Vicini L, Patel R, Quick A, Kolb SJ. Incidence of pathogenic, likely pathogenic, and uncertain ALS variants in a clinic cohort. Neurol Genet. 2020;6:e390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pensato V, Magri S, Bella ED, et al. . Sorting rare ALS genetic variants by targeted Re-sequencing panel in Italian patients: OPTN, VCP, and SQSTM1 variants account for 3% of rare genetic forms. J Clin Med. 2020;9:412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lattante S, Marangi G, Doronzio PN, et al. . High-Throughput genetic testing in ALS: The challenging path of variant classification considering the ACMG guidelines. Genes (Basel). 2020;11:1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mehta PR, Iacoangeli A, Opie-Martin S, et al. . The impact of age on genetic testing decisions in amyotrophic lateral sclerosis. Brain. 2022;145:4440–4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chio A, Battistini S, Calvo A, et al. . Genetic counselling in ALS: Facts, uncertainties and clinical suggestions. J Neurol Neurosurg Psychiatry. 2014;85:478–485. [DOI] [PubMed] [Google Scholar]
- 18. Klepek H, Goutman SA, Quick A, Kolb SJ, Roggenbuck J. Variable reporting of C9orf72 and a high rate of uncertain results in ALS genetic testing. Neurol Genet. 2019;5:e301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Federici G, Soddu S. Variants of uncertain significance in the era of high-throughput genome sequencing: A lesson from breast and ovary cancers. J Exp Clin Cancer Res. 2020;39:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Winklhofer KF, Tatzelt J, Haass C. The two faces of protein misfolding: Gain- and loss-of-function in neurodegenerative diseases. EMBO J. 2008;27:336–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ruffo P, Perrone B, Conforti FL. SOD-1 Variants in amyotrophic lateral sclerosis: Systematic Re-evaluation according to ACMG-AMP guidelines. Genes (Basel). 2022;13:537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sau D, De Biasi S, Vitellaro-Zuccarello L, et al. . Mutation of SOD1 in ALS: A gain of a loss of function. Hum Mol Genet. 2007;16:1604–1618. [DOI] [PubMed] [Google Scholar]
- 23. Van Giau V, Pyun JM, Suh J, Bagyinszky E, An SSA, Kim SY. A pathogenic PSEN1 Trp165Cys mutation associated with early-onset Alzheimer's disease. BMC Neurol. 2019;19:188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fraser PE, Yang DS, Yu G, et al. . Presenilin structure, function and role in Alzheimer disease. Biochim Biophys Acta. 2000;1502:1–15. [DOI] [PubMed] [Google Scholar]
- 25. Weggen S, Beher D. Molecular consequences of amyloid precursor protein and presenilin mutations causing autosomal-dominant Alzheimer's disease. Alzheimers Res Ther. 2012;4:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Samocha KE, Kosmicki JA, Karczewski KJ, et al. . Regional missense constraint improves variant deleteriousness prediction. bioRxiv. [Preprint]. 10.1101/148353 [DOI]
- 27. Sim NL, Kumar P, Hu J, Henikoff S, Schneider G, Ng PC. SIFT Web server: Predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012;40(Web Server issue):W452–W457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet. 2013;Chapter 7:Unit7 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kircher M, Witten DM, Jain P, O'Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Iqbal S, Brunger T, Perez-Palma E, et al. . Delineation of functionally essential protein regions for 242 neurodevelopmental disorders. Brain. 2022;146:519–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Farhan SMK, Howrigan DP, Abbott LE, et al. . Exome sequencing in amyotrophic lateral sclerosis implicates a novel gene, DNAJC7, encoding a heat-shock protein. Nat Neurosci. 2019;22:1966–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Project MinE ALS Sequencing Consortium . Project MinE: Study design and pilot analyses of a large-scale whole-genome sequencing study in amyotrophic lateral sclerosis. Eur J Hum Genet. 2018;26:1537–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. McLaren W, Gil L, Hunt SE, et al. . The ensembl variant effect predictor. Genome Biol. 2016;17:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Karczewski KJ, Francioli LC, Tiao G, et al. . The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Landrum MJ, Lee JM, Riley GR, et al. . Clinvar: Public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42(Database issue):D980–D985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. UniProt Consortium . Uniprot: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021;49(D1):D480–D489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hornbeck PV, Zhang B, Murray B, Kornhauser JM, Latham V, Skrzypek E. Phosphositeplus, 2014: Mutations, PTMs and recalibrations. Nucleic Acids Res. 2015;43(Database issue):D512–D520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jumper J, Evans R, Pritzel A, et al. . Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tunyasuvunakool K, Adler J, Wu Z, et al. . Highly accurate protein structure prediction for the human proteome. Nature. 2021;596:590–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kabsch W, Sander C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. [DOI] [PubMed] [Google Scholar]
- 41. Tien MZ, Meyer AG, Sydykova DK, Spielman SJ, Wilke CO. Maximum allowed solvent accessibilites of residues in proteins. PLoS One. 2013;8:e80635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Iqbal S, Perez-Palma E, Jespersen JB, et al. . Comprehensive characterization of amino acid positions in protein structures reveals molecular effect of missense variants. Proc Natl Acad Sci U S A. 10 2020;117:28201–28211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. GTEx Consortium . The genotype-tissue expression (GTEx) project. Nat Genet. 2013;45:580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cummings BB, Karczewski KJ, Kosmicki JA, et al. . Transcript expression-aware annotation improves rare variant interpretation. Nature. 2020;581:452–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. 2021. https://www.R-project.org.
- 46. Wickham H. Ggplot2: Elegant graphics for data analysis. 2nd edn. Springer; 2009. [Google Scholar]
- 47. Kosmicki JA, Samocha KE, Howrigan DP, et al. . Refining the role of de novo protein-truncating variants in neurodevelopmental disorders by using population reference samples. Nat Genet. 2017;49:504–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kour S, Rajan DS, Fortuna TR, et al. . Loss of function mutations in GEMIN5 cause a neurodevelopmental disorder. Nat Commun. 2021;12:2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chen G, Han L, Tan S, et al. . Loss-of-function of KMT5B leads to neurodevelopmental disorder and impairs neuronal development and neurogenesis. J Genet Genomics. 2022;49:881–890. [DOI] [PubMed] [Google Scholar]
- 50. Baron DM, Fenton AR, Saez-Atienzar S, et al. . ALS-associated KIF5A mutations abolish autoinhibition resulting in a toxic gain of function. Cell Rep. 2022;39:110598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. White MA, Kim E, Duffy A, et al. . TDP-43 gains function due to perturbed autoregulation in a tardbp knock-in mouse model of ALS-FTD. Nat Neurosci. 2018;21:552–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sama RR, Fallini C, Gatto R, et al. . ALS-linked FUS exerts a gain of toxic function involving aberrant p38 MAPK activation. Sci Rep. 2017;7:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chan PK, Chattopadhyay M, Sharma S, et al. . Structural similarity of wild-type and ALS-mutant superoxide dismutase-1 fibrils using limited proteolysis and atomic force microscopy. Proc Natl Acad Sci U S A. 2013;110:10934–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sangwan S, Zhao A, Adams KL, et al. . Atomic structure of a toxic, oligomeric segment of SOD1 linked to amyotrophic lateral sclerosis (ALS). Proc Natl Acad Sci U S A. 2017;114:8770–8775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Monhasery N, Moll J, Cusman C, et al. . Transcytosis of IL-11 and apical redirection of gp130 is mediated by IL-11alpha receptor. Cell Rep. 2016;16:1067–1081. [DOI] [PubMed] [Google Scholar]
- 56. Munch C, Bertolotti A. Exposure of hydrophobic surfaces initiates aggregation of diverse ALS-causing superoxide dismutase-1 mutants. J Mol Biol. 2010;399:512–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tompa DR, Kadhirvel S. Changes in hydrophobicity mainly promotes the aggregation tendency of ALS associated SOD1 mutants. Int J Biol Macromol. 2020;145:904–913. [DOI] [PubMed] [Google Scholar]
- 58. Ghosh DK, Kumar A, Ranjan A. T54r mutation destabilizes the dimer of superoxide dismutase 1(T54R) by inducing steric clashes at the dimer interface. RSC Adv. 2020;10:10776–10788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Teilum K, Smith MH, Schulz E, et al. . Transient structural distortion of metal-free cu/zn superoxide dismutase triggers aberrant oligomerization. Proc Natl Acad Sci U S A. 2009;106:18273–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Furukawa Y, Kaneko K, Yamanaka K, O'Halloran TV, Nukina N. Complete loss of post-translational modifications triggers fibrillar aggregation of SOD1 in the familial form of amyotrophic lateral sclerosis. J Biol Chem. 2008;283:24167–24176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tsang CK, Liu Y, Thomas J, Zhang Y, Zheng XF. Superoxide dismutase 1 acts as a nuclear transcription factor to regulate oxidative stress resistance. Nat Commun. 2014;5:3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Trist BG, Genoud S, Roudeau S, et al. . Altered SOD1 maturation and post-translational modification in amyotrophic lateral sclerosis spinal cord. Brain. 2022;145:3108–3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Fecto F, Yan J, Vemula SP, et al. . SQSTM1 Mutations in familial and sporadic amyotrophic lateral sclerosis. Arch Neurol. 2011;68:1440–1446. [DOI] [PubMed] [Google Scholar]
- 64. Jakobi AJ, Huber ST, Mortensen SA, et al. . Structural basis of p62/SQSTM1 helical filaments and their role in cellular cargo uptake. Nat Commun. 2020;11:440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ciani B, Layfield R, Cavey JR, Sheppard PW, Searle MS. Structure of the ubiquitin-associated domain of p62 (SQSTM1) and implications for mutations that cause paget's disease of bone. J Biol Chem. 2003;278:37409–37412. [DOI] [PubMed] [Google Scholar]
- 66. Berkamp S, Mostafavi S, Sachse C. Structure and function of p62/SQSTM1 in the emerging framework of phase separation. FEBS J. 2021;288:6927–6941. [DOI] [PubMed] [Google Scholar]
- 67. Lee Y, Chou TF, Pittman SK, Keith AL, Razani B, Weihl CC. Keap1/cullin3 modulates p62/SQSTM1 activity via UBA domain ubiquitination. Cell Rep. 2017;19:188–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Matsumoto G, Wada K, Okuno M, Kurosawa M, Nukina N. Serine 403 phosphorylation of p62/SQSTM1 regulates selective autophagic clearance of ubiquitinated proteins. Mol Cell. 2011;44:279–289. [DOI] [PubMed] [Google Scholar]
- 69. Hoffman-Andrews L. The known unknown: The challenges of genetic variants of uncertain significance in clinical practice. J Law Biosci. 2017;4:648–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Appelbaum PS, Burke W, Parens E, et al. . Is there a way to reduce the inequity in variant interpretation on the hbasis of ancestry? Am J Hum Genet. 2022;109:981–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ALS Knowledge Portal data are available through the Neurodegenerative Disease Knowledge Portal (https://ndkp.hugeamp.org/). Project MinE sequencing consortium data are available through the Project MinE Databrowser (http://databrowser.projectmine.com/). The GTEx-derived pext expression data (https://gnomad.broadinstitute.org/downloads/), AlphaFold Protein Structure Database (https://alphafold.ebi.ac.uk/), and Genomics 2 Protein Portal (https://g2p.broadinstitute.org/) are open access and were used for these analyses.