Abstract
We characterized the ability of a yeast to cleave the aromatic structure of the dioxin-like compound dibenzofuran. The yeast strain was isolated from a dioxin-contaminated soil sample and identified as Trichosporon mucoides. During incubation of glucose-pregrown cells with dibenzofuran, six major metabolites were detected by high-performance liquid chromatography. The formation of four different monohydroxylated dibenzofurans was proven by comparison of analytical data (gas chromatography-mass spectrometry) with that for authentic standards. Further oxidation produced 2,3-dihydroxydibenzofuran and its ring cleavage product 2-(1-carboxy methylidene)-2,3-dihydrobenzo[b]furanylidene glycolic acid, which were characterized by mass spectrometry and 1H nuclear magnetic resonance spectroscopy. These two metabolites are derived from 2-hydroxydibenzofuran and 3-hydroxydibenzofuran, as shown by incubation experiments using these monohydroxylated dibenzofurans as substrates.
Although many efforts have been made to reduce the emission of polychlorinated dibenzofurans and dioxins during combustion of dust or bleaching of pulp in paper mills, these compounds remain widespread environmental contaminants. This problem has given rise to an intensive search for microorganisms capable of metabolizing cyclic biaryl ether structures in order to evaluate their degradation potential in nature. Recently, some bacterial strains able to cometabolize (4, 13) or to grow with dibenzofuran (7, 16, 20) have been isolated and the mechanisms of oxygenation and ring cleavage have been described in detail (23). Degradation of halogenated dibenzofurans and dibenzo-p-dioxins by aerobic bacteria has so far been shown only for derivatives with up to two chlorine substituents (24). The mineralization potential for higher chlorinated compounds might be rather low. The biodegradation capacity of aerobic microorganisms could be important in the destruction of nonhalogenated or slightly halogenated intermediates arising from anaerobic dehalogenation of dibenzofurans, e.g., in sediments (1) or soils.
Although fungi constitute the majority of microbial biomass in soil, data on fungal catabolism of biaryl ether compounds are limited and restricted to hydroxylation processes (4, 11). Our objectives were (i) to identify fungal strains that could degrade dibenzofuran and (ii) to determine the pathway and the chemical intermediates through which the breakdown occurs.
MATERIALS AND METHODS
Isolation procedure.
We isolated yeast strains from a dioxin-contaminated soil sample from a copper works under selective conditions in a mineral salts medium (MM) (14) containing, per liter, 5 g of NH4H2PO4, 2.5 g of KH2PO4, 1 g of MgSO4 · 7H2O, 2 mg of FeCl3 · 6H2O, 20 mg of Ca(NO3)2 · 4H2O, and 10 ml of a trace element solution (containing, per liter, 50 mg of H3BO3, 40 mg of MgSO4 · 4H2O, 40 mg of ZnSO4 · 7H2O, 20 mg of Na2MoO4, 10 mg of CuSO4 · 5H2O, 10 mg of CoCl2, 10 mg of potassium iodide). Soil (2 g) was suspended in 100 ml of MM and supplemented with dibenzofuran (1 g/liter) as the sole source of carbon and chloramphenicol (30 μg/liter) to inhibit bacterial growth. After 7 days of shaking cultures at 30°C and 180 rpm, 10 ml was transferred to 90 ml of fresh medium and incubated under the same conditions. Identical transfers were performed 7 and 14 days later. Afterwards, microorganisms were obtained by plating 0.1 ml of the cultures on malt agar plates. Pure cultures were maintained on malt agar and stored at 4°C.
Yeast strains were characterized for their potential to degrade dibenzofuran by incubating glucose (1%)-grown cells in MM with dibenzofuran (500 μg/ml) for 7 days. Degradation product formation was determined by analyzing the culture supernatant by high-performance liquid chromatography (HPLC). All experiments described below were carried out with strain SBUG 801, which accumulated the largest quantity of metabolites in the culture medium.
Identification and characterization of strain SBUG 801.
The yeast used was assigned to the genus Trichosporon by the urea hydrolysis test, sugar utilization (IC32; Biomerieux, Nürtingen, Germany), and morphology. Further identification to the species level using physiological characteristics, coenzyme Q type, and the D2 domain of 26S rRNA sequences was performed by the method of Gueho et al. (9, 10). Strain SBUG 801 has been deposited as strain DSM 12017 in the Deutsche Sammlung für Mikroorganismen, Braunschweig, Germany.
Growth and incubation conditions.
Cells were grown in 500-ml shake flasks with 100 ml of MM supplemented with 1 or 2% glucose and 1 ml of a vitamin solution (21). After incubation for 16 h at 30°C and 180 rpm on a rotary shaker, cells were harvested by centrifugation (3,100 × g, 5 min) and washed twice with sterile MM. The cell pellet was resuspended in MM to an optical density (600 nm) of 6. Dibenzofuran and 2- or 3-hydroxydibenzofuran were added in suitable concentrations of about 10 to 500 μg/ml (see Results) to the cell suspensions, and the cultures were incubated as described above. Cells in MM without substrate and dibenzofuran derivatives in MM without cells were used as controls.
Kinetics of dibenzofuran degradation and isolation of metabolites.
Cultures were incubated in 100-ml Erlenmeyer flasks containing 20 ml of the cell suspension in MM and 160 μg of dibenzofuran per ml at 30°C with agitation at 180 rpm. Flasks with autoclaved cells and dibenzofuran were used as additional controls. At each sampling period, cultures were removed and stored at −20°C. At the end of the experiment all samples and controls were thawed and extracted with 4 ml of trichloromethane for 5 min. The organic layer was separated from the aqueous phase by centrifugation in glass tubes (3,100 × g, 5 min). The contents of dibenzofuran and metabolites were estimated by gas-chromatographic analysis of 1 μl of the trichloromethane phase and HPLC analysis of 100 μl of the aqueous supernatant. The data are reported as means for two separate experiments with replicated batch cultures. Standard deviation was no more than 8%.
Additional cultures (500-ml flasks with 100 ml of the cell suspension and 50 to 250 μg of dibenzofuran or 2-hydroxydibenzofuran per ml) were incubated to enrich the yield of intermediates. Metabolites were extracted from the centrifuged culture supernatant twice with ethyl acetate at pH 7 and once again after acidification of the aqueous residue to pH 2. The organic phases were dried over anhydrous sodium sulfate, and the solvent was removed. The residues obtained were dissolved in methanol.
Chemical analysis and identification of metabolites.
Gas chromatography was carried out on a Shimadzu model GC-14A gas chromatograph with a flame ionization detector at 310°C equipped with a Permabond SE-54-DF-0.35 capillary column (50 m by 0.25 mm [inside diameter]; Macherey-Nagel, Düren, Germany). Helium was used as the carrier gas (180 kPa). The column temperature was programmed from 150 to 300°C at 10°C/min.
HPLC was performed on a Hewlett-Packard (Bad Homburg, Germany) HPLC apparatus 1050 M equipped with a quaternary pump system, a diode array detector 1040 M series I, and an HP Chemstation. The separation was achieved with a LiChroCart 125-4 RP-18 end-capped (5-μm) column (Merck, Darmstadt, Germany). The initial solvent composition was 30% methanol–70% phosphoric acid (0.1%), reaching 100% methanol after 14 min at a flow rate of 1 ml/min.
Metabolites were purified by semipreparative HPLC on a LiChrospher RP-18 end-capped column (16 by 250 mm, 10 μm; Knauer, Berlin, Germany) by using methanol–1% acetic acid (80:20 [vol/vol] for neutral extracts and 50:50 [vol/vol] for acidic extracts) as the mobile phase at a flow rate of 9 ml/min.
The UV-visible absorption spectra of degradation products were determined in a diode array detector.
Gas chromatography-mass spectrometry was carried out on a gas chromatograph GC 8000 linked to a mass selective detector MD 800 (Fisons Instruments, Mainz, Germany) operating at 70 eV, fitted with a 60-m DB5-ms column (0.25-mm by 0.33-μm film; J & W Scientific, Folsom, Calif.). Acid-extractable compounds were derivatized before analysis by methylation with diazomethane as described by De Boer and Backer (5) in a microapparatus (Aldrich-Chemie, Steinheim, Germany). High-resolution mass spectra were recorded on a Vacuum Generators (Manchester, United Kingdom) analytical instrument, model 70-250 SE.
The 1H and 13C nuclear magnetic resonance (NMR) spectra were recorded on a Bruker (Karlsruhe, Germany) WM 300 instrument at 300 MHz or on a DRX500 instrument at 500 MHz in CD3OD (99.99%).
Chemicals.
Dibenzofuran and 2-hydroxydibenzofuran were purchased from Aldrich-Chemie. 3-Hydroxydibenzofuran was synthesized from 3-aminodibenzofuran by the method of Wilkes (22). All chemicals and solvents were of the highest purity available.
RESULTS
From enrichment cultures with dibenzofuran various yeast strains were isolated, but none could grow with the biaryl compound as the sole source of carbon. Screening of the yeasts obtained for their potential to metabolize dibenzofuran following growth on glucose identified a strain that produced a broad pattern of degradation products when the culture supernatant was analyzed by HPLC.
Taxonomic characterization of the dibenzofuran-metabolizing yeast.
Microscopic observation showed formation of arthroconidia. This finding together with the results of the carbon utilization spectrum (IC32) and the hydrolysis of urea led us to classify the organism as a yeast belonging to the genus Trichosporon. This genus can be separated into two groups, one consisting of 15 species containing coenzyme Q-9 ubiquinone systems, and the other consisting of four species containing coenzyme Q-10 ubiquinone systems (10). Since this isolate had coenzyme Q-10, subsequent studies were confined to Trichosporon mucoides, T. moniliforme, T. cutaneum, and T. jirovecii. The physiological features of the isolate pointed to T. mucoides: assimilation of specific carbon sources (galactitol, glucono-δ-lactone, l-arabinitol, melibiose, and erythritol), no growth with nitrate or creatine, and growth at 37°C. However, all four species have very similar carbon and nitrogen compound assimilation patterns (2, 10). For final identification, we sequenced the taxonomically most informative 26S rRNA region from strain SBUG 801 and compared the data obtained with data for the type strains of the four species obtained by Gueho et al. (10). The sequence of strain SBUG 801 was the same as that of the CBS 7625 type culture of T. mucoides.
Metabolism of dibenzofuran.
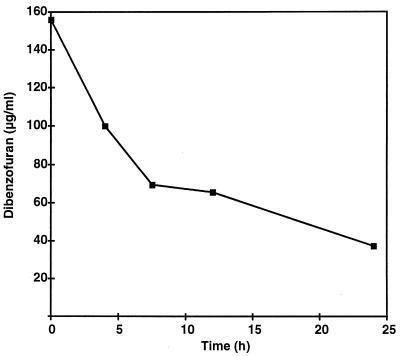
Glucose-grown cells of T. mucoides SBUG 801 metabolized dibenzofuran very rapidly. More than 50% of the substrate added was degraded within 8 h (Fig. 1) at a degradation rate of about 15 μg of dibenzofuran ml−1 h−1. After that time the degradation of the biaryl compound dropped drastically. The kinetics seem to be time dependent, because a similar decrease in the degradation rate occurred after 8 to 10 h when the initial dibenzofuran concentrations were 100 and 200 μg/ml. Nonsignificant change in the dibenzofuran concentration was observed in heat-killed cultures.
FIG. 1.
Degradation of dibenzofuran (160 μg/ml) by T. mucoides SBUG 801.
Detection and identification of intermediates.
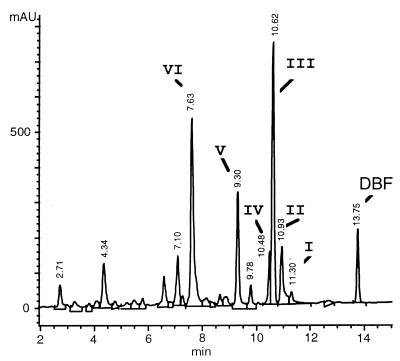
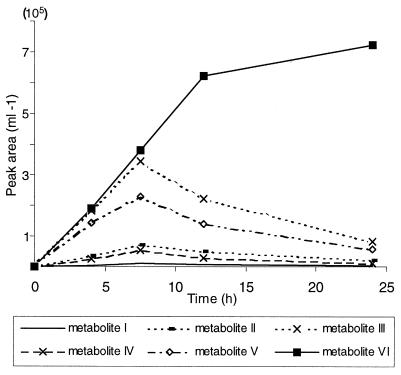
Dibenzofuran degradation was accompanied by the formation of at least six water-soluble intermediates. The major metabolites (designated I to VI) had retention times of 10 to 12 min (I to IV), 9.3 min (V), and 7.6 min (VI). The HPLC peak at 13.8 min was dibenzofuran (Fig. 2). Metabolites I to V accumulated over a period of about 10 h and seemed to be metabolized during subsequent incubation. The concentration of metabolite VI increased continuously (Fig. 3). Metabolites were characterized by their UV-visible spectra during detection with a diode array detector. None of these substances accumulated in the control flasks.
FIG. 2.
HPLC elution profile of an aqueous culture supernatant after incubation of T. mucoides SBUG 801 with dibenzofuran (500 μg/ml) for 12 h. At least six dibenzofuran (DBF) degradation products (I to VI) were detectable after separation on a reversed-phase column. AU, absorbance units.
FIG. 3.
Product accumulation and decomposition during incubation of glucose-grown cells of T. mucoides SBUG 801 with dibenzofuran (160 μg/ml). Amounts of intermediates were calculated from their absorbance at 220 nm.
Monohydroxylated compounds.
Compounds I to IV were identified as the four isomers of monohydroxylated dibenzofurans by comparing UV and mass spectra with those of available standards (2- and 3-hydroxydibenzofuran), with data from the literature (1-hydroxydibenzofuran [4]), or with our own data (4-hydroxydibenzofuran [11]). 2-Hydroxydibenzofuran was always the most common isomer (maximum concentration, approximately 20 μg/ml). In contrast, 3- and 4-hydroxydibenzofuran were detected at about 2 to 5 μg/ml, and 1-hydroxydibenzofuran was detected in much smaller quantities (no more than 0.5 μg/ml).
Metabolites V and VI.
Metabolites V and VI were rapidly released into the medium. Compound V reached its maximum concentration (6 μg/ml) at about 8 h, while compound VI accumulated throughout the entire incubation up to 35 μg/ml (Fig. 3). Incubation experiments with glucose-grown cells of T. mucoides SBUG 801 with 125 μg of 2-hydroxydibenzofuran or 3-hydroxydibenzofuran per ml as the substrate also resulted in the accumulation of metabolites V and VI in the culture fluid. Thus, 2- and 3-hydroxydibenzofuran were formed as intermediates during dibenzofuran degradation.
After incubation of the yeast strain with 2-hydroxydibenzofuran (125 μg/ml) for 30 h, about 90% of the substrate had been transformed to metabolite VI, while a high level (18 μg/ml) of metabolite V was achieved with higher amounts of 2-hydroxydibenzofuran (250 μg/ml and more). Therefore, all further incubations concerning enrichment and preparation of these compounds were carried out with 2-hydroxydibenzofuran as the substrate.
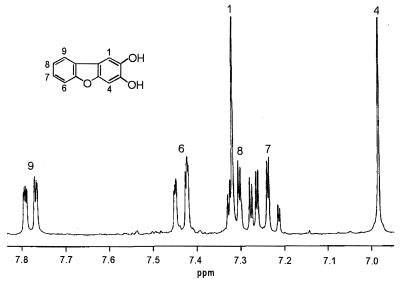
Metabolite V was extractable with ethyl acetate at a pH of 7 and yielded a light brown powder after purification by semipreparative HPLC. The UV absorption spectrum showed absorption maxima at 247, 298, and 313 nm. The mass spectrum revealed a molecular weight of m/z 200. The molecular ion of the methyl derivative was observed at m/z 228, suggesting the presence of two hydroxyl groups in the molecule. The molecular weight of the metabolite, the gas-chromatographic behavior of the methyl derivative, and the mass spectrum all were consistent with metabolite V being a dihydroxylated dibenzofuran. 1H NMR and 13C NMR showed the presence of six protons and 12 carbons in the aromatic area. Apart from the signals of a nonsubstituted ring (two doublets, two triplets), two singlets were observed by 1H NMR (Fig. 4). Because of the spin-spin coupling of isolated meta protons, the singlets in metabolite V must be in the para position, proving this intermediate to be 2,3-hydroxydibenzofuran.
FIG. 4.
1H NMR spectrum of metabolite V, 2,3-dihydroxydibenzofuran (300 Mhz; CD3OD). Trimethylsilane was the internal standard.
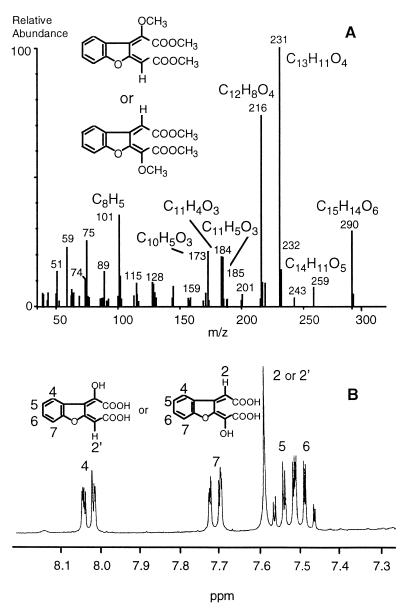
Extraction of culture supernatants at pH 2 and evaporation of the solvent resulted in residues highly enriched in metabolite VI. Because of the acid character of this intermediate, it was assumed to be a ring cleavage product. After purification by preparative HPLC, the compound was obtained as a yellow powder showing UV absorption maxima at 242 and 329 nm (Table 1). Gas chromatography-mass spectrometry was successful only after methylation. High-resolution mass spectrometry showed the molecular formula of the methylated compound to be C15H14O6. The base peak at m/z 231 was assigned to C13H11O4, resulting in the loss of COOCH3 from the molecular ion. Fragments at m/z 259 (C14H11O5) and m/z 216 (C12H8O4) were interpreted as loss of OCH3 and CH3-COOCH3 (Fig. 5A). 1H NMR and 13C NMR of the unmethylated substance showed five protons and 12 carbon atoms. Similar to the spectrum of metabolite V, the 1H NMR spectrum of metabolite VI showed the proton signals of an unsubstituted aromatic ring. In addition, only one singlet was obtained, representing one isolated proton (Fig. 5B). 13C-distortionless enhancement by polarization transfer (DEPT) 90 NMR data (five signals) indicated seven quaternary carbons and only one aliphatic methine proton. Because of the molecular weight of 290 and the presence of carboxyl groups, the compound was assumed to be a ring cleavage product showing a hydroxylated muconic acid as a partial structure. All NMR signals presented in Table 2 and additional NMR studies (500 MHz) involving long-range coupling experiments like heteronuclear multiple bond correlation and nuclear Overhauser enhancement spectroscopy were consistent with the structure of 2-(1-carboxy methylidene)-2,3-dihydrobenzo[b]furanylidene glycolic acid. Since coupling between the hydrogen of the hydroxyl group and hydrogen atoms on the aromatic-ring system could not be detected, it was impossible to unambiguously identify the location of the hydroxyl group to either of the side chains.
TABLE 1.
Characterization of substrates and intermediates of dibenzofuran degradation by T. mucoides SBUG 801
| Compound | Metabolite | UV absorption maxima (nm)a | Characteristic mass ions (m/z) |
|---|---|---|---|
| DBFb | 249, 279, 294(s) | 168 [M+], 139 [M]-CHO | |
| 1-Hydroxy-DBF | I | 224, 256, 268, 274, 298, 306 | 184 [M+]; 155 [M]-CHO, 128 [M]-COC2H4 |
| 2-Hydroxy-DBF | III | 251, 289, 317(s) | 184 [M+], 155 [M]-CHO, 128 [M]-COC2H4 |
| 3-Hydroxy-DBF | II | 213, 231, 253, 297 | 184 [M+], 155 [M]-CHO, 128 [M]-COC2H4 |
| 4-Hydroxy-DBF | IV | 224(s), 255, 279, 295/305(s) | 184 [M+], 155 [M]-CHO, 128 [M]-COC2H4 |
| 2,3-Dihydroxy-DBF | V | 216, 247, 298, 313 | 200 [M+], 171[M]-CHO, 126 [M]-CH2O-CH2O-CH2 |
| Ring cleavage product | VI | 242, 329 | 290 [M+], 259 [M]-OCH3, 231 [M]-COOCH3, 216 [M]-COOCH3-CH3c |
s, shoulder.
DBF, dibenzofuran.
Methyl derivative.
FIG. 5.
Mass spectrum of the methyl derivative (A) and 1H NMR spectrum of metabolite VI, 2-(1-carboxy methylidene)-2,3-dihydrobenzo[b]furanylidene glycolic acid, a ring cleavage product formed during incubation of T. mucoides SBUG 801 with dibenzofuran (B).
TABLE 2.
1H NMR data for 2-(1-carboxy methylidene)-2,3-dihydrobenzo[b]furanylidene glycolic acid (metabolite VI) recorded at 300 MHz
| Proton assignment | δ (ppm) | 3J (Hz) | 4J (Hz) |
|---|---|---|---|
| H4, dd | 8.03 | 7.3 (4, 5) | 1.7 (4, 6) |
| H5, m | 7.48 | 7.4 (5, 6) | 1.3 (5, 7) |
| H6, m | 7.53 | 7.8 (6, 7) | |
| H7, dd | 7.71 | ||
| H2′, s | 7.59 |
During incubation of glucose-grown cells of T. mucoides SBUG 801 with low amounts of 2,3-dihydroxydibenzofuran, metabolite VI was detected.
DISCUSSION
We investigated the ability of a yeast to biodegrade dibenzofuran, a model compound for intermediates arising from highly persistent environmental pollution with biaryl ether structures. These structures may be nonhalogenated and slightly halogenated compounds which result from anaerobic or chemical dehalogenation processes. Screening of soil samples for suitable organisms revealed a strain identified as T. mucoides which had been isolated only from human tissue previously. This strain was able to grow with aromatic compounds like phenol or benzoic acid and transformed dibenzofuran as well. The degradation of monocyclic aromatic compounds is common in yeasts (12, 15), and phenol degradation by T. cutaneum has been investigated in detail (8, 17). However, information on the ability of yeasts to cleave the aromatic ring in biaryl or polycyclic compounds is still lacking.
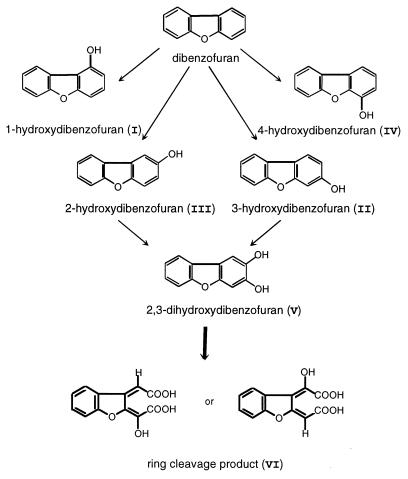
We suggest that T. mucoides SBUG 801 degrades dibenzofuran via 2,3-hydroxydibenzofuran (metabolite V) since ring cleavage of this intermediate leads to the formation of 2-(1-carboxy methylidene)-2,3-dihydrobenzo[b]-furanylidene glycolic acid (metabolite VI [Fig. 6]). The accumulation of high levels of monohydroxylated dibenzofurans and the formation of the dihydroxylated derivative from both 2-hydroxydibenzofuran and 3-hydroxydibenzofuran indicated the action of monooxygenases during the first degradation steps. Hydroxylation of biaryl compounds by monooxygenases in fungi has been frequently described (3, 4, 6, 11, 19); however oxidation of the resulting hydroxylated derivatives has not been well documented. The only previous report of this activity in yeasts showed that diphenyl ether could be transformed via hydroxylated derivatives to a lactone by Trichosporon sp. strain SBUG 752 (18).
FIG. 6.
Proposed pathway for the first steps of dibenzofuran degradation by T. mucoides SBUG 801.
Monohydroxylated and dihydroxylated dibenzofuran derivatives were metabolized very rapidly by cells of T. mucoides SBUG 801, leading to the formation of a derivative of hydroxymuconic acid with a benzofuran substructure. Cleavage of the dihydroxylated ring presumably utilizes a mechanism similar to that described for the degradation of diphenyl ether by Trichosporon sp. strain SBUG 752 (18). In both species a third hydroxyl group must be introduced into dihydroxylated intermediates before ortho cleavage of the aromatic structure can take place. In contrast to T. mucoides (coenzyme Q-10), strain SBUG 752 belongs to the group of coenzyme Q-9-containing Trichosporon species.
This is the first report of a fungus capable of cleaving the aromatic structure of dibenzofuran. The results together with those for strain SBUG 752 show that Trichosporon strains not only degrade monoaromatic compounds but also can degrade biaryl ether substrates. Further studies, particularly with chlorinated dibenzofurans, will be required to determine if these yeasts can effectively degrade halogenated environmental pollutants.
ACKNOWLEDGMENTS
This study was supported by grants 0319519 A0 and 1460638 R9 from the Bundesministerium für Bildung und Forschung.
We thank R. Kunze for help with obtaining and interpreting the rDNA/rRNA sequence data and M. Kindermann and S. Siegert for recording NMR data.
REFERENCES
- 1.Ballerstedt H, Kraus A, Lechner U. Reductive dechlorination of 1,2,3,4-tetrachlorodibenzo-p-dioxin and its products by anaerobic mixed cultures from Saale river sediment. Environ Sci Technol. 1997;31:1749–1753. [Google Scholar]
- 2.Barnett J A, Payne R W, Yarrow D. Yeasts: characteristics and identification. Cambridge, United Kingdom: Cambridge University Press; 1983. [Google Scholar]
- 3.Cerniglia C E, Hebert R L, Szaniszlo P J, Gibson D T. Fungal transformation of naphthalene. Arch Microbiol. 1978;117:135–143. doi: 10.1007/BF00402301. [DOI] [PubMed] [Google Scholar]
- 4.Cerniglia C E, Morgan J C, Gibson D T. Bacterial and fungal oxidation of dibenzofuran. Biochem J. 1979;180:175–185. doi: 10.1042/bj1800175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Boer T D, Backer H J. Diazomethane. In: Leonard N J, editor. Organic synthesis. Vol. 36. New York, N.Y: John Wiley & Sons; 1956. pp. 14–16. [Google Scholar]
- 6.Dodge R H, Cerniglia C E, Gibson D T. Fungal metabolism of biphenyl. Biochem J. 1979;178:223–230. doi: 10.1042/bj1780223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fortnagel P, Harms H, Wittich R-M, Francke W, Krohn S, Meyer H. Cleavage of dibenzofuran and dibenzo-p-dioxin ring systems by a Pseudomonas bacterium. Naturwissenschaften. 1989;76:222–223. doi: 10.1007/BF00627694. [DOI] [PubMed] [Google Scholar]
- 8.Gaal A, Neujahr H Y. Metabolism of phenol and resorcinol in Trichosporon cutaneum. J Bacteriol. 1979;137:13–21. doi: 10.1128/jb.137.1.13-21.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gueho E, Kurtzman C P, Peterson S W. Evolutionary affinities of heterobasidiomycetous yeasts estimated from 18S and 25S ribosomal RNA sequence divergence. Syst Appl Microbiol. 1989;12:230–236. [Google Scholar]
- 10.Gueho E, Smith M T, de Hoog G S, Billon-Grand G, Christen R, Batenburg-van der Vegte W H. Contributions to a revision of the genus Trichosporon. Antonie Leeuwenhoek. 1992;61:289–316. doi: 10.1007/BF00713938. [DOI] [PubMed] [Google Scholar]
- 11.Hammer E, Schauer F. Fungal hydroxylation of dibenzofuran. Mycol Res. 1997;101:433–436. [Google Scholar]
- 12.Hofmann K H, Schauer F. Utilization of phenol by hydrocarbon assimilating yeasts. Antonie Leeuwenhoek. 1988;54:179–188. doi: 10.1007/BF00419204. [DOI] [PubMed] [Google Scholar]
- 13.Klecka G M, Gibson D T. Metabolism of dibenzo-p-dioxin and chlorinated dibenzo-p-dioxins by Beijerinckia species. Appl Environ Microbiol. 1980;39:228–236. doi: 10.1128/aem.39.2.288-296.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kreisel H, Schauer F. Methoden des mykologischen Laboratoriums. Jena, Germany: Gustav Fischer Verlag; 1987. p. 38. [Google Scholar]
- 15.Middelhoven W J. Catabolism of benzene compounds by ascomycetous and basidiomycetous yeasts and yeastlike fungi. Antonie Leeuwenhoek. 1993;63:125–144. doi: 10.1007/BF00872388. [DOI] [PubMed] [Google Scholar]
- 16.Monna L, Omori T, Kodama T. Microbial degradation of dibenzofuran, fluorene, and dibenzo-p-dioxin by Staphylococcus auriculans DBF63. Appl Environ Microbiol. 1993;59:285–289. doi: 10.1128/aem.59.1.285-289.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neujahr H Y, Gaal A. Phenolhydroxylase from yeast: purification and properties of the enzyme from Trichosporon cutaneum. Eur J Biochem. 1973;35:386–400. doi: 10.1111/j.1432-1033.1973.tb02851.x. [DOI] [PubMed] [Google Scholar]
- 18.Schauer F, Henning K, Pscheidl H, Wittich R M, Fortnagel P, Wilkes H, Sinnwell V, Francke W. Biotransformation of diphenyl ether by the yeast Trichosporon beigelii SBUG 752. Biodegradation. 1995;6:173–180. doi: 10.1007/BF00695348. [DOI] [PubMed] [Google Scholar]
- 19.Smith R V, Davis P J, Clark A M, Glover-Milton S. Hydroxylations of biphenyl by fungi. J Appl Bacteriol. 1980;49:65–73. doi: 10.1111/j.1365-2672.1980.tb01044.x. [DOI] [PubMed] [Google Scholar]
- 20.Strubel V, Rast H G, Fietz W, Knackmuss H-J, Engesser K-H. Enrichment of dibenzofuran utilizing bacteria with high co-metabolic potential toward dibenzodioxin and other anellated aromatics. FEMS Microbiol Lett. 1989;58:233–238. doi: 10.1016/0378-1097(89)90044-x. [DOI] [PubMed] [Google Scholar]
- 21.van der Walt J P. Criteria and methods used in classification. In: Lodder J, editor. The yeasts, a taxonomic study. Amsterdam, The Netherlands: North-Holland Publishing Co.; 1970. pp. 34–113. [Google Scholar]
- 22.Wilkes H. Untersuchungen zur Aufklärung von Stoffwechselwegen beim mikrobiellen Abbau von Diarylethern. Ph.D. thesis. Germany: University of Hamburg; 1993. [Google Scholar]
- 23.Wilkes H, Francke W, Harms H, Schmidt S, Fortnagel P. Mechanistic investigations on microbial degradation of diaryl ethers. Naturwissenschaften. 1992;79:269–271. [Google Scholar]
- 24.Wilkes H, Wittich R M, Timmis K N, Fortnagel P, Francke W. Degradation of chlorinated dibenzofurans and dibenzo-p-dioxins by Sphingomonas sp. strain RW1. Appl Environ Microbiol. 1996;62:367–371. doi: 10.1128/aem.62.2.367-371.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]