Abstract
Background
The root of Saposhnikovia divaricata (Turcz.) Schischk is a well-known traditional medicinal plant, containing various bioactive compounds with anti-inflammatory, antioxidant, and analgesic properties. However, no scientific studies have validated its clinical use as an anti-inflammatory agent against inflammatory bowel disease (IBD). This study aimed to investigate whether the root extract of S. divaricata ameliorates IBD and induces gut microbial alteration, using a RAW 264.7 cell line and a DSS-induced colitis mouse model.
Methods
To investigate the anti-inflammatory effects and alleviation of IBD, using a methanol extract of Saposhnikovia divaricata (Turcz.) Schischk. root (MESD), RAW 264.7, murine macrophages and a dextran sodium sulfate (DSS)-induced colitis mouse model were employed. 16S rRNA gene sequencing was conducted to determine the alterations in the gut microbiota of mice with DSS-induced colitis.
Results
MESD significantly decreased nitric oxide (NO) and inflammatory cytokine levels in lipopolysaccharide (LPS)-induced RAW 264.7 cells in vitro. Oral administration of MESD reduced the expression of inflammatory cytokines in the colons of mice with DSS-induced colitis. Additionally, MESD inhibited the abundance of Clostridium sensu stricto 1 and enhanced the predicted functional pathways, including l-glutamate degradation VIII (to propanoic acid). Seven compounds with anti-inflammatory properties were isolated from the MESD. Among them, 3′-O-acetylhamaudol and 3′-O-angeloylhamaudol exhibited strong anti-inflammatory effects in vitro.
Conclusion
Overall, MESD may be a potential natural product for the treatment of IBD by lowering inflammatory cytokine levels and altering gut microbiota composition.
Keywords: Saposhnikovia divaricata, DSS-induced colitis, Inflammatory bowel disease, Inflammatory response, Gut microbiota
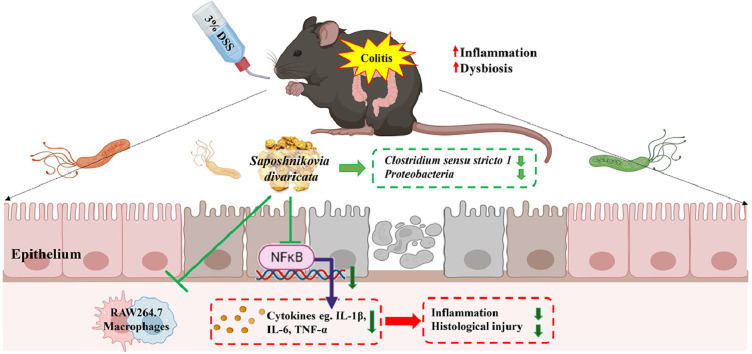
Graphical abstract
1. Introduction
Inflammatory bowel disease (IBD) includes two main types of inflammatory disorders, Crohn's disease (CD) and ulcerative colitis (UC), which are typically induced by multiple factors, including genetic, environmental, and immunological factors as well as abnormal immune responses related to the microbiota. However, the primary cause of IBD remains unknown.1 The most common symptoms of IBD include weight loss, fever, abdominal pain, diarrhea, and bloody stool.2 The incidence of IBD is increasing worldwide, along with the burden on patients’ healthcare systems.3 The estimated patient age-standardized prevalence rates of IBD increased from 79.5 per 100,000 in 1990 to 84.3 per 100,000 in 2017, with 6.8 million IBD cases reported globally.4
Currently, the most convenient therapies, including aminosalicylates (5-aminosalicylic acid), corticosteroids, immune modifiers (thiopurines), anti-tumor necrosis factor (anti-TNF) agents, and antibiotics (metronidazole), are administered as the primary treatment for patients with IBD.5,6 Despite advanced drug treatments, a significant percentage of patients do not benefit from these therapies owing to drug tolerance, lack of response, or loss of response to treatment. For instance, approximately 30 % of patients are primary non-responders, while up to 10 % lose their response during anti-TNF treatment (secondary non-response) every year.7 As an “alternative treatment strategy,” IBD patients have used herbal treatments owing to their perceived harmlessness and lack of side effects.8 Natural products, including animals, fungi, microorganisms, marine organisms, and plants, produce secondary metabolites with bioactivity and abundant structural diversity. As a result, natural products are a valuable source of drugs.1 Saposhnikovia divaricata (Turcz.) Schischk. (Umbelliferae) is distributed throughout eastern Siberia and northern Asia and is a well-known traditional medicine in China, Japan, and Korea. S. divaricata is used to treat rheumatism, generalized pain, headaches, fever, cold, and arthralgia.9 S. divaricata is also rich in bioactive constituents and contains various structural compounds, including polysaccharides, chromones, and coumarins.10 Previous studies have shown that the root of S. divaricata and its constituents can inhibit pro-inflammatory cytokines, inducible nitric oxide synthase (iNOS), and cyclooxygenase-2 (COX-2) in lipopolysaccharide (LPS)-induced murine macrophages RAW 264.7.11 In addition, recent studies have shown that the active compounds prim-O-glucosylcimifugin, 4′-O-β-D-glucosyl-5-O-methylvisamminol, sec‑O-glucosylhamaudol, and cimifugin have effectively inhibited stimulated pain in mice, while the antipyretic, analgesic, and anti-inflammatory properties inhibited the LPS induced interleukin-6 (IL-6) and NO production.12, 13, 14 According to Weidong Xu et al. ,15 a combination of four plants commonly used in Traditional Chinese Medicine, including S. divaricata, was shown to be effective in treating irritable bowel syndrome (IBS). These combinations reduced intestinal inflammation and reduced the expression of interleukin-1β (IL-1β), IL-6, IL-10, and tumor necrosis factor-alpha (TNF-α) in the colon.
Animal models of IBD have been widely used to evaluate the development of inflammation and the microbial composition. Over the past few decades, several murine colitis models have been developed to explore the mechanisms underlying human IBD. Several studies have used a dextran sodium sulfate (DSS)-induced colitis model to determine its similarity to human UC. Water-soluble (40–50 kDa) DSS induces intestinal inflammation while interrupting the configuration of the epithelial monolayer of the large intestine, overproducing pro-inflammatory cytokines, and leading to gut microbial dysbiosis.16
The gut microbiota has a symbiotic relationship with the host organism and plays critical roles in metabolism, energy balance, and the immune system. A healthy gut microbiota provides the first defense mechanism against microbial pathogens and contributes to the maturation of immune cells.17 In contrast, microbial dysbiosis mediates severe inflammation in the gut microbiota. Compared to healthy individuals, individuals with IBD exhibited decreased intestinal microbiota diversity, increased harmful metabolites, and enhanced content of pro-inflammatory factors.18 The microbiota of IBD patients is characterized by decreased diversity, reduced abundance of Firmicutes in CD patients or Bacteroidetes in UC patients, and increased abundance of Proteobacteria and Actinobacteria.19,20 However, whether the root of S. divaricata inhibits IBD symptoms in animal models or changes the gut microbiota remains unknown.
In this study, we investigated the ameliorating effects of S. divaricata root extract on gut microbiota composition in a DSS-induced colitis model. Furthermore, the anti-inflammatory activity of the methanolic extract of S. divaricata (Turcz.) Schischk. root (MESD), and compounds isolated from MESD were identified using the murine macrophage RAW 264.7 cell line.
2. Methods
2.1. Plant materials
The Saposhnikovia divaricata (Turcz.) Schischk (voucher specimen number – M23) was collected from Dornod, Mongolia (46°N, 56′29.3″; 116°E,12′08.4″) in September 2019, and was identified by the Institute of Chemistry and Chemical Technology. The roots of S. divaricata were dried and extracted with methanol (1:10, w/v) for 72 h at room temperature. The supernatant was collected after filtration, and the extraction procedure was repeated twice using root residues. The collected liquid was evaporated using Rotavapor R-100 (Büchi, Flawil, Switzerland).
2.2. Isolation and identification of the major compounds of the Saposhnikovia divaricata (Turcz.) Schischk. root extract
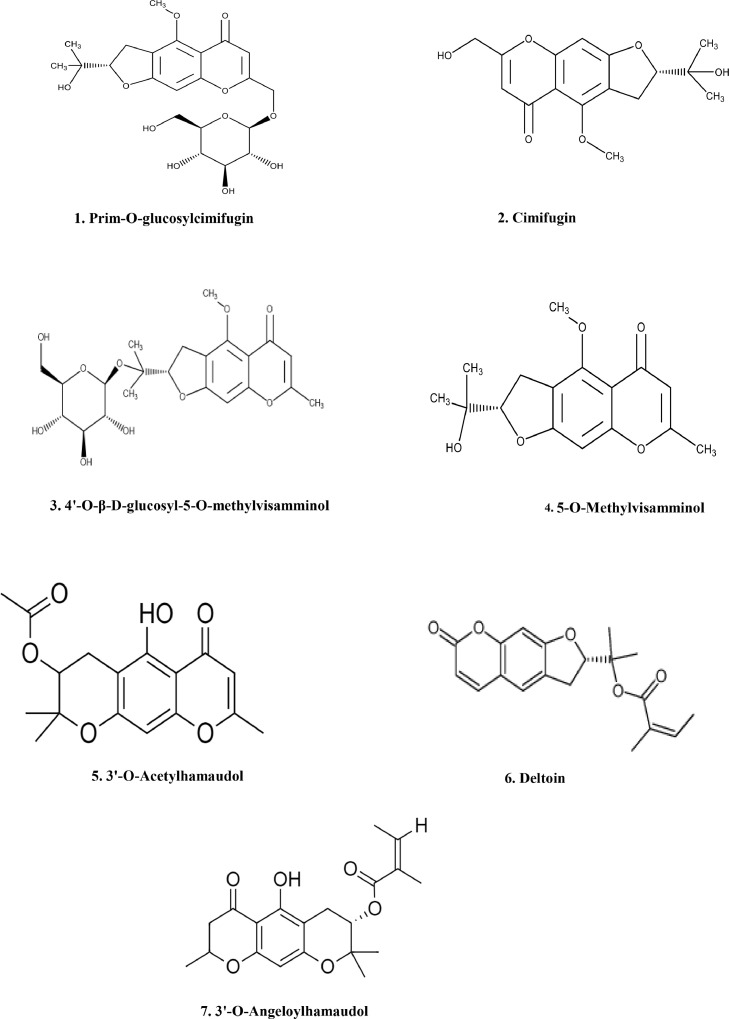
MESD was separated using reversed-phase preparative LC (Gilson, 321 Pump, Middleton, Wisconsin, USA) and a semi-preparative Luna C18 column (00G-4252-N0, 250 × 10 mm, 5 μm; Phenomenex, Torrance, CA, USA) at a flow rate of 4 mL/min. Seven compounds were isolated via preparative LC, and the purity and molecular mass of the separated compounds were first checked using Agilent LC/MS (Agilent Technologies 1260 Infinity Quaternary LC and Agilent Technologies 1200 Series, Santa Clara, California, USA). Acetonitrile (solvent A) and water (solvent B) were used for reverse-phase HPLC analysis under the following gradient conditions:0–10 min, solvent A, 20–60 %; 10–25 min, solvent A, 60–70 %; 25–40 min, solvent A, 70–80 %; 40–50 min, solvent A; 80–90 %; 50–60 min, solvent A, 90–20 %. The flow rate was 1 mL/min and signals were obtained at 280 nm. The mass spectra of each compound were analyzed in the mass scan mode (100–2000 m/z) under positive and negative conditions. The retention times and mass signals of the chemical compounds were compared with those reported previously,21, 22, 23 and a total of seven chemical compounds were identified (prim-O-glucosylcimifugin, cimifugin, 4′-O-β-d-glucosyl-5-O-methylvisamminol, and 5-O-methylvisamminol, 3′-O-acetylhamaudol, deltoin, and 3′-O-angeloylhamaudol).
2.3. Cell cultures
The murine macrophage cell line RAW 264.7, was purchased from American Type Culture Collection (Manassas, VA, USA). RAW 264.7 cells were grown in minimum essential medium (MEM) supplemented with 10 % fetal bovine serum (Sigma-Aldrich, St. Louis, MO, USA) and 1 % penicillin-streptomycin. The cells were incubated in a 5 % CO2 incubator at 37 °C.
2.4. Determination of nitric oxide level
RAW 264.7 cells were seeded on 96-well plates (2.5 × 104 cells/well) and incubated for 24 h in a 5 % CO2 incubator at 37 °C. Fresh media with three different concentrations (2.5, 5, and 10 μg/mL) of MESD extract and compounds (40 μM and 80 μM) were added to each well following removal of the culture medium and rinsing with Dulbecco's phosphate-buffered saline (DPBS). After 1 h, 50 µL of medium with or without LPS (final concentration of 1 μg/mL) was added and incubated for 24 h. The next day, the culture supernatant was transferred to a new plate, and the nitric oxide (NO) concentration was detected using Griess reagent (1:1 ratio) (Sigma-Aldrich, USA). The absorbance was measured at 540 nm using a Synergy HTX multimode microplate reader (BioTek, Winooski, VT, USA).
2.5. Animal study
Seven-week-old C57BL/6 J male mice (n = 48) were purchased from the Central Lab. Animal Inc. (Seoul, Korea) and acclimated for five days before treatment. All mice were housed in a specific pathogen-free room with filter-top cages under a 12 h light and 12 h dark cycle and provided free access to an AIN-76A (Envigo, Indianapolis, USA) diet and water. Based on the body weight, the mice were arbitrarily divided into six groups (N = 8 per group). To induce acute colitis in mice, 3 % DSS was provided in drinking water (autoclaved) for nine days, except for mice in the control group. The body weight and food intake were measured daily. Carboxymethyl cellulose (CMC) solution (0.5 %) was used as the vehicle solution and was administered to the control (CON) and 3 % DSS groups. The positive control group was administered 100 mg/kg 5-aminosalicylic acid (ASA), and the MESD group was administered three different concentrations of the MESD extract (50, 100, and 200 mg/kg) in vehicle solution. All treatment solutions were administered via oral gavage once daily during the colitis-induction period. After completion of the experiment, the mice were euthanized via an intraperitoneal injection of a ketamine and xylazine mixture (10 mL/kg). For further investigation, blood, colon, and cecum samples were collected and stored at −80 C.
2.6. Enzyme-Linked immunosorbent assay (ELISA) and colorimetric assay
Commercial colorimetric and ELISA kits were used according to the manufacturers’ instructions. Interleukin-1β (IL-1β), interleukin 6 (IL-6), and tumor necrosis factor-alpha (TNF-α) ELISA kits were purchased from RayBiotech (Peachtree Corners, GA, USA), and prostaglandin E2 (PGE2) ELISA kits were obtained from R&D Systems (Minneapolis, Minnesota, US). Aspartate aminotransferase (AST) and alanine aminotransferase (ALT) colorimetric kits were purchased from ElabScience (Houston, TX).
2.7. Histological analysis
To assess the severity of colitis inflammation, mouse colons were collected and cut into segments. Colon tissue was fixed with 10 % formaldehyde, embedded in paraffin blocks, sectioned, and stained with hematoxylin and eosin (H&E) for histological evaluation. Images were acquired using an AXIO Zoom.V16 fluorescence microscope (ZEISS, Jena, Germany).
2.8. Quantitative real-time polymerase chain reaction (qRT-PCR) analysis
Total mRNA was extracted from cells and colon tissues using a commercial kit (GeneAll Biotechnology, South Korea). Total isolated mRNA was reverse-transcribed into complementary DNA (cDNA) using a TaKaRa cDNA Synthesis kit (Kusatsu, Shiga, Japan) according to the manufacturer's protocol. The mRNA expression levels of the samples were analyzed using qRT-PCR (Light Cycler 480 Real-Time PCR System; Roche, Basel, Switzerland), with mouse β-actin as the endogenous control. The relative gene expression was calculated using the comparative Ct value (2−ΔΔCt). The primer sequences used in this study are listed in Table 1.
Table 1.
Primer sequences used for qRT-PCR.
| Gene | Forward primer | Reverse primer |
|---|---|---|
| Il-1β | GGTACATCAGCACCTCACAA | TTAGAAACAGTCCAGCCCATAC |
| Il-6 | CCCAACAGACCTGTCTATACC | CAGCTTATCTGTTAGGAGAGC |
| Tnf-α | TCCCCAAAGGGATGAGAAGTTC | GGGAGTAGACAAGGTACAAC |
| Nos2 | CCA AGCCCTCACCTACTTCC | CTCTGAGGGCTGACACAAGG |
| Cox-2 | GAAGTCTTTGGTCTGGTGCCTG | GTCTGCTGGTTTGGAATAGTTGC |
| Adgre1 | CTTTGGCTATGGGCTTCCAGT | GCAAGGACAGAGTTTATCGTG |
| β-actin | CATTGCTGACAGGATGCAGAAGG | TGCTGGAAGGTGGACAGTGAGG |
2.9. Western blotting
Protein expression in cell lysates and colon tissues was determined by western blotting. Mouse colon tissue was homogenized in RIPA lysis buffer containing 1 % protease inhibitor cocktail (Thermo Fisher Scientific, USA) and 1 % phenylmethylsulfonyl fluoride (PMSF). Total protein concentration was determined using the Bradford protein assay. All primary and anti-rabbit secondary antibodies were obtained from Cell Signaling Technology (Danvers, Massachusetts, USA), and anti-mouse secondary antibodies were purchased from Santa Cruz Biotechnology (Dallas, Texas, USA). The concentration of the primary antibodies used was 1:500 and that of the secondary antibodies was 1:2000.
2.10. Cecum DNA extraction and 16S rRNA gene sequencing
After the necropsy, cecum samples were collected and stored at −80 C until analysis. The cecum samples were homogenized and DNA was isolated using a PowerFecal Pro-DNA kit (Qiagen, Hilden, Germany) with bead-beating. DNA samples were collected and eluted in nuclease-free water. The DNA concentration and quality were verified using a NanoDrop spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). To amplify the V3-V4 region of the 16S rRNA gene, PCR was performed using the universal primer set 341F and barcoded 806R. PCR products were confirmed by denaturing gradient gel electrophoresis and gel-purified using AMPure XT beads (Beckman Coulter Genomics, Danvers, MA, USA) and Qubit dsDNA high-sensitivity reagent (Invitrogen, Carlsbad, CA, USA). The samples were sequenced on a MiSeq platform using a paired-end 2 × 300-bp reagent kit (Illumina, San Diego, CA, USA).
2.11. 16S analysis
Raw reads were processed, filtered, and analyzed using the QIIME2-DADA2 pipeline24,25 to determine amplicon sequence variants (ASV). ASVs were aligned using the Mafft aligner26 and q2-alignment plug-in. The q2-phylogeny plugin was used to reconstruct the phylogeny with FastTree.27 Taxonomic classification was assigned using classify-sklearn28 against SILVA database release 138.29 Alpha diversity (Shannon diversity index, Faith's phylogenetic diversity, observed ASVs, and Plelou's evenness) analyses were performed at a rarefaction depth of 9000 sequences per sample. Subsequent processing and analyses were performed using PhyloSeq to import the data generated from QIIME2 into R (v.4.1.2).30
For functional inference of the ASVs identified in mice, PICRUSt2 (v2.2.0-b)31 was used. Phylogenetic placement in PICRUSt2 was based on the results of three analysis tools:1) HMMER (http://www.hmmer.org) for ASV placement; 2) EPA-ng32 to determine the optimal position of the placed ASVs in a reference phylogeny; and 3) GAPPA33 to create a new tree incorporating the ASV placements. Finally, a phylogenetic tree that included both the reference genome and the environmentally sampled organisms was generated and used to predict the copy numbers of individual gene families for each ASV. The metabolic pathway database (MetaCyc) pathway abundance, which is the main high-level prediction output, was calculated in PICRUSt2 by structured mapping of the gene families for enzyme commission (EC) gene families to pathways.34 ANCOM was used to identify the differentially abundant MetaCyc pathways.
2.12. Statistical analysis
All data are expressed as mean ± standard error of the mean (SEM). A one-way analysis of variance (ANOVA), followed by Duncan's multiple range test, was performed using IBM SPSS Statistics for Windows, version 25 (IMB Corp. Armonk, NY, USA). Differences were considered statistically significant at p < 0.05. The PERMDISP2 function of the Vegan R Package35 was used to determine the homogeneity difference in the dispersion of microbial compositions between the groups with 999 permutations. A nonparametric PERMANOVA statistical test36 was used to compare the microbiota composition using the Vegan R package.35 Finally, a differential abundance analysis of genera was performed using ANCOM 2.137 in R (v.4.1.2) (R Core Team). Significance was defined as an ANCOM W>0.7.
3. Result
3.1. MESD inhibits inflammatory response in LPS-induced RAW 264.7 cells
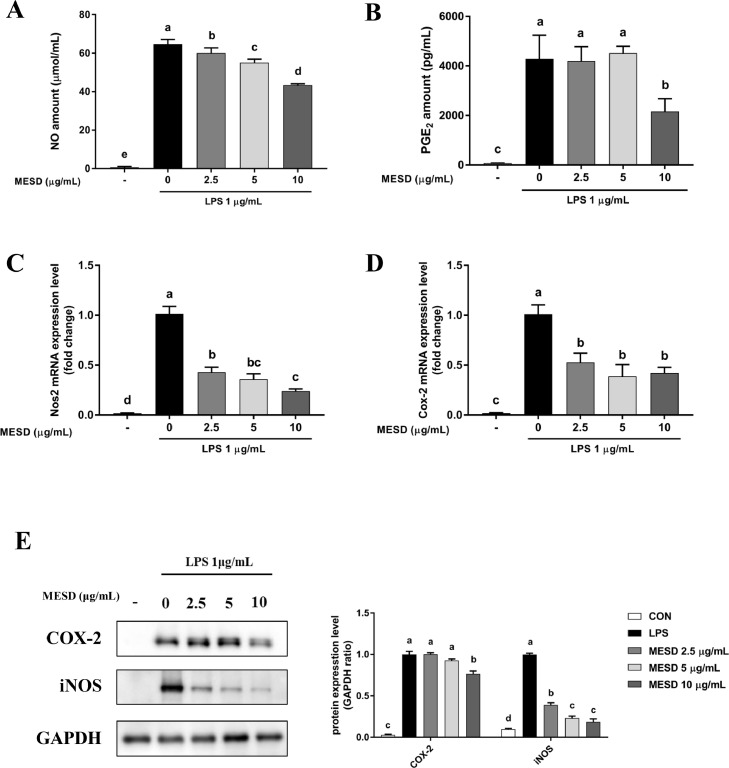
To determine the anti-inflammatory effect of MESD in vitro, the mRNA expression of inflammatory markers, such as nitric oxide synthase 2 (Nos2) and Cox-2, were measured in LPS-induced RAW 264.7 cells following PGE2 and NO production. MESD significantly decreased the NO and PGE2 levels in LPS-stimulated cells (Fig. 1A and 1B). The LPS-induced mRNA expression levels of Nos2 and Cox-2 were markedly reduced by MESD (Fig. 1C and 1D). MESD decreased COX-2 and iNOS protein expression in a dose-dependent manner (Fig. 1E). Overall, MESD exhibited anti-inflammatory effects by inhibiting the expression of inflammatory factors in macrophages.
Fig 1.
Anti-inflammatory effects of the methanol extract of Saposhnikovia divaricata root (MESD) in LPS-induced RAW 264.7 cells.
(A, B) Nitric oxide (NO) and Prostaglandin E2 (PGE2) content in cell culture media. (C, D) mRNA levels of nitric oxide synthase 2 (Nos2) and cyclooxygenase-2 (Cox-2). (E) the protein levels of COX-2 and iNOS. All data are presented as mean ± SEM. Different superscripts indicate significant differences at p <0.05.
3.2. Oral administration of MESD alleviates colitis symptoms and pro-inflammatory cytokine levels in the plasma of mice with DSS-induced colitis
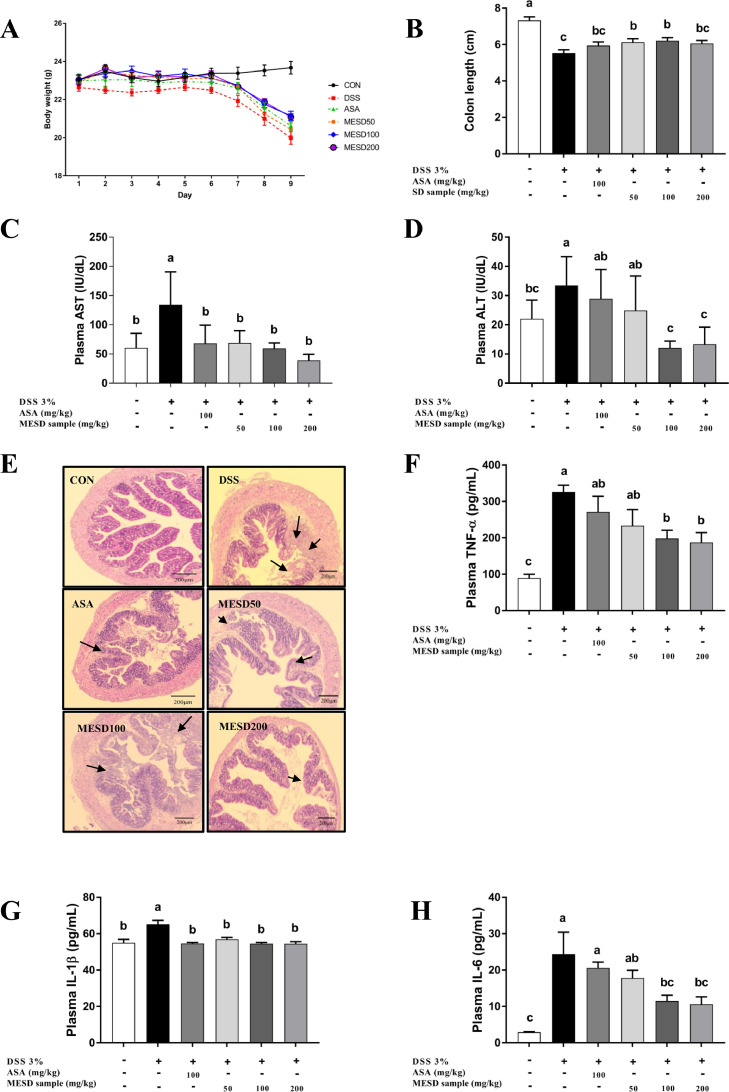
To confirm that the in vitro anti-inflammatory effects were validated in vivo, we investigated whether MESD could effectively inhibit colitis in mice with inflammatory diseases. The anti-colitis effects of MESD were verified by the induction of chronic inflammation in a mouse model of DSS-induced colitis. As shown in Fig. 2A and 2B, DSS decreased body weight and colon length. However, MESD alleviated body weight loss and colon shortening. Compared with the control (CON) group, the DSS group showed ulceration of the epithelial layer of the bowel wall, and the histological indications were improved in MESD-treated mice (Fig. 2E). The CON group had clear crypts and mucosal layers, whereas the DSS group had severe damage. However, the epithelial layer was restored in the MESD treatment group and crypt formation was observed. MESD significantly reduced the levels of plasma aspartate aminotransferase (AST) and alanine aminotransferase (ALT) (p < 0.05; Fig. 2C and 2D) and liver injury markers known to be elevated in colitis.38 To evaluate the anti-inflammatory effects of MESD in the DSS-induced colitis model, the production of pro-inflammatory cytokines, including TNF-α, IL-1β, and IL-6, were measured using plasma. Compared with the DSS group, the MESD group displayed reduced production of pro-inflammatory cytokines in the plasma (Fig. 2F-H).
Fig 2.
MESD alleviated colitis-related phenotypes in the DSS-induced colitis mouse model. (A, B) Changes in body weight and colon length. 5-aminosalicylic acid (ASA) was used as a positive control. (C, D) Aspartate aminotransferase (AST) and Alanine aminotransferase (ALT) levels in plasma in DSS-induced mice. (E) Hematoxylin and Eosin (H&E) staining of the cross-section of the colon tissue, magnification (x200 μm). Arrows indicate damage to colonic tissues, including inflammatory cell infiltrates and epithelial erosions. (F) The expression of inflammatory cytokines, tumor necrosis factor-α (TNF-α), (G) interleukin-1β (IL-1β), and (H) interleukin-6 (IL-6), in plasma based on ELISA assay. All data are presented as mean ± SEM, with significant differences at p <0.05.
3.3. MESD suppresses the mRNA expression of inflammatory markers in DSS-induced colitis mice
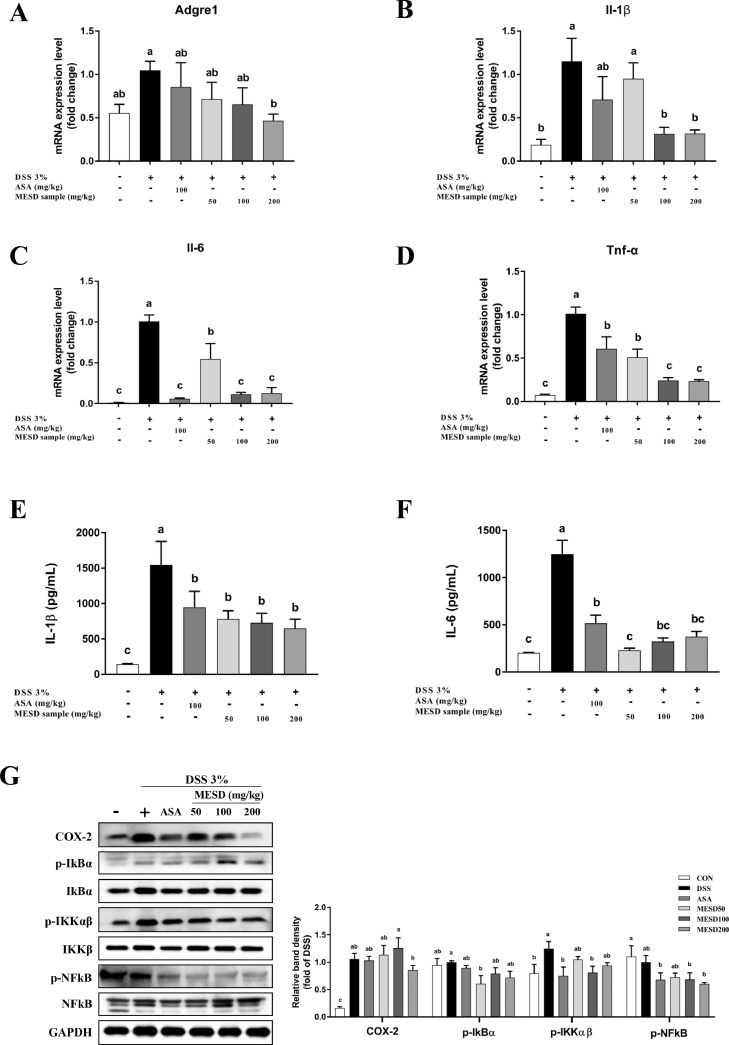
To determine whether MESD inhibited the expression of inflammatory markers in colon tissues inflamed by colitis, inflammatory markers in mouse colon tissues were identified using qRT-PCR (Fig. 3A-D). Compared with the CON group, the mRNA expression of inflammatory markers, such as adhesion G protein-coupled receptor E1 (Adgre1), Il-1β, Il-6, and Tnf-α, were induced in the DSS group and were significantly inhibited in the MESD treatment groups. In addition, the production of pro-inflammatory cytokines IL-1β and IL-6 were measured in colon tissue (Fig. 3E and 3F). Compared to the DSS group, the MESD treatment groups showed notably reduced levels of pro-inflammatory cytokines in the colon tissue. Afterward, in the colon tissue, COX-2 and the p-NFκB/NFκB, p-IκBα/IκBα, and p-IKKαβ/IKKβ kinase complex protein levels were detected (Fig. 3G). IKK protein levels were significantly elevated in the DSS group and decreased in the MESD group.
Fig 3.
Inflammatory markers gene and protein expression levels decreased treatment with MESD extract in mice with DSS-induced colitis.
mRNA levels of (A) adhesion G Protein-Coupled Receptor E1 (Adgre1) and (B) interleukin-1β (Il-1β), (C) interleukin-6 (Il-6) and (D) tumor necrosis factor- α (Tnf-α) in DSS-induced mice colon tissue. The expression of inflammatory cytokines, (E) interleukin-1β (IL-1β), and (F) interleukin-6 (IL-6), in colon tissue of DSS-induced mice, based on ELISA assay. (G) The protein expression levels of inflammation-related markers, including COX-2 and NFκB pathway in mice colon tissue of DSS-induced colitis. All data are presented as mean ± SEM, with significant differences at p <0.05.
3.4. MESD restores DSS-induced changes in gut microbial composition
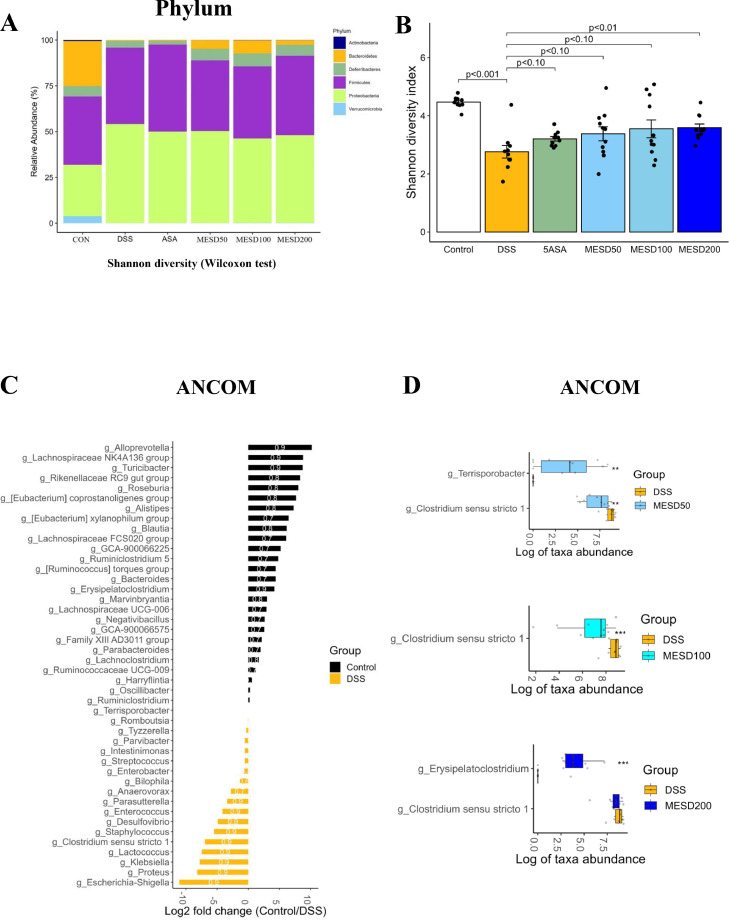
We evaluated the composition of the gut microbiota, which is known to influence the pathogenesis of colitis.39 The diversity of microbial composition and phylum and genus levels were analyzed to classify the bacterial taxa in each group. Six bacterial phyla were identified, including Actinobacteria, Bacteroidetes, Deferribacteres, Firmicutes, Proteobacteria, and Verrucomicrobia, at the phylum level (Fig. 4A). In the DSS group, the abundance of Bacteroidetes markedly reduced, whereas that of Proteobacteria increased. MESD restored the DSS-induced changes in the gut microbial composition (Fig. 4A). Alpha diversity analysis was performed to observe changes in the gut microbiota abundance and phylogenetic diversity. Alpha diversity was significantly reduced in the DSS group compared to the CON group, and gut microbiota abundance and phylogenetic diversity were restored in the MESD-treated group (Fig. 4B and Supplementary Fig. S1A-C). The abundance of genera that differed significantly among the DSS, CON, and MESD groups was assessed using ANCOM. Compared to the CON group, the abundances of Escherichia, Klebsiella, Clostridium sensu stricto 1, Desulfovibrio, and Enterococcus increased, and the abundances of Alloprevotella, Turicibacter, Roseburia, and Alistipes decreased in the DSS group (Fig. 4C). However, the MESD treatment significantly reduced the abundance of Clostridium sensu stricto 1 (Fig. 4D). PICRUSt2 was used to identify microbial functional pathways that were significantly altered by MESD treatment. Several MetaCyc pathways were differentially expressed between the DSS-and MESD treatment groups (Supplementary Fig. S2A-C). Interestingly, among these biochemical pathways, l-glutamate degradation VIII (to propanoate) was the most significantly increased in all MESD treatment groups compared to that in the DSS group.
Fig 4.
Effect of MESD on gut microbial composition in DSS-induced colitis.
(A) Average relative abundance of the gut microbiota at the phylum level. (B) Pairwise comparison of the alpha diversity of the gut microbiota. (C) Different abundant taxa between the Control and DSS groups. (D) Different abundant taxa between DSS group and groups with different concentrations of MESD. Microbiotas are determined by the Wilcoxon test and ANCOM. All data are presented as mean ± SEM, with significant differences at *p < 0.05, **p < 0.01, and ***p < 0.001 vs. DSS group.
3.5. Effects of seven isolated compounds from MESD on the inflammatory mediators in RAW 264.7 cells
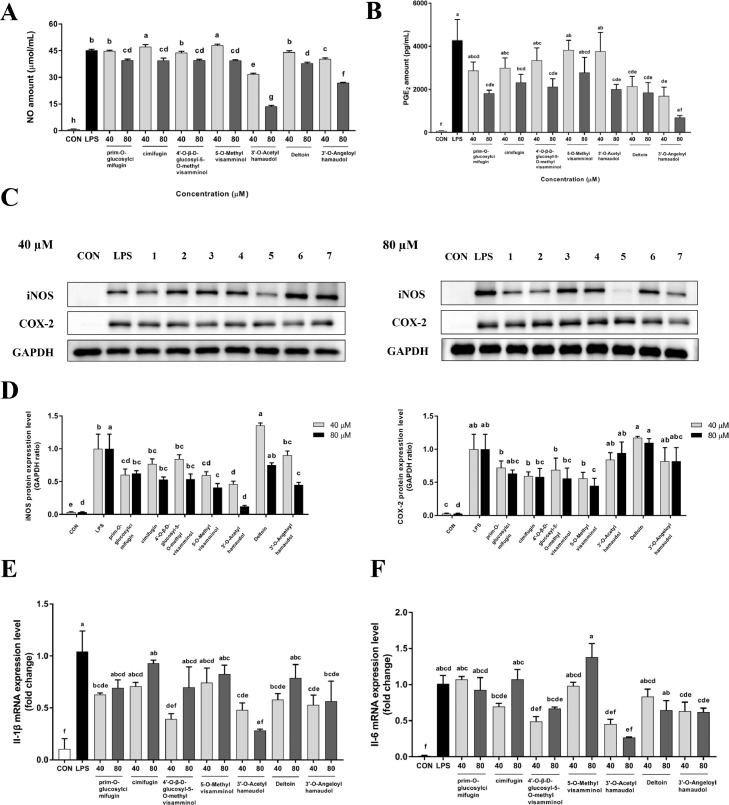
In this study, seven compounds were isolated from MESD, including six chromones and one coumarin compound (prim-O-glucosylcimifugin (1), cimifugin (2), 4′-O-β-D-glucosyl-5-O-methylvisamminol (3), 5-O-methylvisamminol (4), 3′-O-acetylhamaudol (5), deltoin (6), and 3′-O-angeloylhamaudol (7)) (Fig. 5). The effects of the isolated compounds on inflammation in vitro was evaluated to identify compounds with anti-inflammatory effects. All isolated compounds displayed a significant anti-inflammatory effect, as they inhibited NO and PGE2 levels in the cell culture medium (Fig. 6A and 6B) and the protein levels of iNOS and COX-2 in LPS-induced RAW 264.7 cells (Fig. 6C and 6D). The seven compounds also significantly inhibited the mRNA levels of the inflammatory cytokines Il-1β and Il-6 (Fig. 6E and 6F) in vitro.
Fig 5.
Structure of the identified and isolated compounds from MESD.
Chemical structure of the seven major compounds of MESD.
Fig 6.
Anti-inflammatory effects of the isolated compounds from MESD in LPS-induced RAW 264.7 cells.
(A, B) Nitric oxide (NO) and Prostaglandin E2 (PGE2) content in cell culture media in LPS-induced RAW 264.7 cells. (C, D) Protein expression and quantitative analysis of iNOS and COX-2 in RAW 264.7 cells treated with the isolated compounds (40 µM and 80 µM). (E, F) mRNA levels of interleukin-1β (Il-1β) and interleukin-6 (Il-6) in LPS-induced RAW 264.7 cells. All data are presented as mean ± SEM, with significant differences at p <0.05. (prim-O-glucosylcimifugin (1), cimifugin (2), 4′-O-β-d-glucosyl-5-O-methylvisamminol (3), 5-O-methylvisamminol (4), 3′-O-acetylhamaudol (5), deltoin (6), and 3′-O-angeloylhamaudol (7)).
4. Discussion
The present study revealed a significant anti-inflammatory effect of dried Saposhnikovia divaricata root and its bioactive components via the reduction of pro-inflammatory cytokines in vitro and in vivo. In this study, several findings supported the anti-inflammatory effects of MESD. In fact, the MESD decreased the levels of inflammatory markers in LPS-induced RAW 264.7 mouse macrophages. By administering MESD to a mouse model of DSS-induced IBD colitis, this compound was found to effectively inhibit pro-inflammatory cytokines in the infiltrated blood and colon tissues and suppress the mRNA expression of pro-inflammatory cytokines and crypt damage in the mouse colon. MESD induced significant changes in gut microbial diversity, composition, and functional pathways. Seven single compounds were isolated from MESD,21, 22, 23 and their anti-inflammatory activities were determined in vitro. Overall, MESD has potential effects on inflammatory markers and the gut microbial composition in a DSS-induced gut environment, as discussed below.
Inflammation is the first response of the immune system to pathogens, damaged cells, and toxic composition.40 However, inflammation mediated by abnormal immune activation can lead to many serious acute and chronic inflammatory diseases, such as systemic inflammatory response syndrome, acute lung injury, inflammatory bowel disease, rheumatoid arthritis, and atherosclerosis.41 During the inflammatory process, macrophages play an important role in the production of pro-inflammatory cytokines, with NO and PGE2 produced by iNOS and COX-2, respectively.42 The DSS-induced murine model is convenient for studying IBD due to its simplicity, easy control, and rapid disease development.16 Moreover, this colitis model has symptoms similar to those of UC patients, including weight loss and bloody diarrhea.43 A hallmark of IBD is diffuse inflammation, damage to crypt structures, and depletion of goblet cells in the mucosal region of the colon.44 The imbalance of inflammatory markers observed in IBD contributes to immune dysfunction and intervenes in tissue inflammation, organ damage,45 and bacterial disorders. We determined whether MESD inhibited the inflammatory factors characteristic of IBD in the blood and colon tissues. MESD ameliorated inflammatory cell infiltration and crypt injury in the colons of mice with DSS-induced colitis. Colonic goblet cells were also significantly damaged in DSS-treated mice but were restored following MESD treatment. DSS increased the levels of inflammatory markers, such as TNF-α, IL-1β, IL-6, iNOS, and COX-2, in the plasma and colon tissue, while MESD significantly reduced the levels of pro-inflammatory cytokines, such as TNF-α, IL-1β, and IL-6. Although the precise mechanism is still unclear, many references have suggested the NFκB pathway plays a central role in controlling the release of cytokines in patients with UC and contributes to inflammation and immune responses in the intestinal tract.46,47 Cytokines such as TNF-α and IL-1β act as NFκB inducers, and the degradation of IκB is an essential step for releasing and activating NFκB. A crucial regulatory step in this process is the signal-induced phosphorylation of IκB at specific amino-terminal serine residues, which is mediated by the IKK complex.48 NFκB/IKK is an attractive target for therapeutic intervention in inflammatory diseases.
Normally, colonic microflora prevents the invasion of pathogenic organisms and contributes to host defense.49 However, dysbiosis of the gut microbiota can cause colonic inflammation and inflammatory bowel disease in humans and animal models.50 A healthy gut environment has four major bacterial phyla: Actinobacteria, Bacteroidetes, Firmicutes, and Proteobacteria.51 Gut dysbiosis in IBD and alterations in the normal composition and abundance of bacterial communities occur, including a decrease in the relative abundance of Bacteroidetes and Firmicutes, and an increase in Proteobacteria.52,53 DSS has been reported to increase the abundance of Proteobacteria, one of the most abundant phyla in patients with metabolic syndrome.54 In this study, DSS-induced colitis drove lowered abundance of Actinobacteria, Bacteroidetes, and Verrucomicrobia in the gut microbiota. In contrast, in the MESD treatment groups, especially the MESD 100 mg/kg group, gut microbial diversity and the abundance of Bacteroidetes increased, whereas the abundance of Proteobacteria in DSS-induced colitis mice decreased. At the genus level, the abundance of Escherichia, Klebsiella, and Clostridium sensu stricto 1, known as pathogens, was significantly higher in the DSS group than in the CON and MESD-treated groups. Clostridium sensu stricto 1, including Clostridium spp., is an opportunistic pathogen that plays a critical role in intestinal inflammation55 and is a recognized indicator of less healthy intestinal microbiota. These pathogens are possibly responsible for mild-to-severe diarrhea, serious infection, and mucosal necrosis.56 MESD significantly reduce the abundance of Clostridium sensu stricto 1 at the genus level, suggesting that MESD ameliorates dysbiosis. Microbial functional pathway analysis confirmed that the l-glutamate degradation VIII (to propanoate) metabolic pathway was significantly increased at all three concentrations in the MESD treatment groups. Thus, activation of this pathway promotes the production of the short-chain fatty acid propanoate, which affects intestinal development and the colonic barrier.57,58 Accordingly, MESD activates short-chain fatty acid-related pathways, leading to changes in gut microbial composition.
The therapeutic effects of Saposhnikovia divaricata roots on cold, headache, rheumatic disease, arthralgia, rubella, pruritus, and tetanus were first described in Shen Nong's Materia Medica written in 300 A.D. in China.10 Chromones and coumarins are bioactive components mainly found in the roots of S. divaraicata.9 Prior studies revealed that chromones isolated from S. divaraicata, including prim-O-glucosylcimifugin, cimifugin, 4′-O-β-D-glucosyl-5-O-methylvisamminol, 5-O-methylvisamminol, 3′-O-acetylhamaudol, 3′-O-angeloylhamaudol, and coumarin compounds such as deltoin, had anti-inflammatory properties.9,59 These compounds were also found to inhibit the mitogen-activated protein kinase (MAPK), phosphorylated-extracellular signal-regulated kinase, and c-Jun N-terminal kinase (JNK) pathways.10 To the best of our knowledge, this is the first study to reveal the anti-inflammatory activity of 3′-O-acetylhamaudol and 3′-O-angeloylhamaudol. 3′-O-acetylhamaudol has been reported to exhibit anti-tumor activity in colon cancer models through anti-angiogenesis and intestinal intraepithelial lymphocyte activation.60 In particular, 3′-O-acetylhamaudol modulates the immune system in the spleen and small intestine. Accordingly, 3′-O-acetylhamaudol can be hypothesized to modify the immune system of the colon, resulting in the protection of colon tissue destroyed by DSS-induced colitis. However, further studies on the mechanisms underlying their interaction are required. In this study, S. divaricata root extract and its isolated compounds showed inhibitory effects on NO and PGE2 production, inflammatory cytokines, and mediators in LPS-treated RAW 264.7 cells. In particular, deltoin and 3′-O-acetyhamaudol significantly decreased NO levels and the gene expression levels of Il-1β and Il-6 more effectively than the other compounds.
Based on our findings, Saposhnikovia divaricata exerts anti-inflammatory activity by inhibiting pro-inflammatory cytokines in LPS-induced RAW 264.7 cells. MESD displayed high IL-1β, IL-6, and NO inhibitory activities, indicating that the bioactive compounds present in the extract can reduce inflammation. MESD significantly lowered the activity of inflammatory cytokines and alleviated DSS-induced colitis symptoms. Oral administration of MESD reduced colitis-induced histological changes in the colon tissue, decreased pro-inflammatory cytokine levels in the serum and colon tissue, and improved the diversity of the gut microbial composition. However, the use of MESD in the treatment of IBD requires further improvement and in-depth research.
Our findings suggest that MESD may be a potential anti-inflammatory treatment for IBD, as it inhibits inflammation at the cellular and organism levels and regulates the gut microbiota. The results of this study suggest that MESD may ameliorate DSS-induced colitis in mice through the synergistic effects of various active compounds. Since effective treatment of IBD is urgently needed, the development of novel therapeutics is crucial, and it may be appropriate to identify and analyze natural compounds with anti-inflammatory properties and the capacity to regulate gut microbiota. Nevertheless, the anti-inflammatory effects of MESD in a DSS-induced colitis model have been validated; however, limitations remain. Considering the toxicological limitations of organic solvents,61 water and ethanol are preferred over methanol for solvent extraction. However, methanol has the advantages of relatively higher extraction yields than other solvents and the ability to dissolve a wide range of polar bioactive compounds, such as phenolics, flavonoids, and alkaloids. This solvent could also evaporate at 40 °C.62,63 Further mechanistic studies are needed to identify the bioactive compounds that drive the IBD-relieving effects of MESD. In addition, the mechanisms of action of a single compound and additional therapeutic targets have not been fully characterized because of the limitations of the study methodology. The correlation between gut microbiota and MESD remains to be elucidated. Considering the limitations of the present study, future research should focus on resolving or correcting these issues.
CRediT authorship contribution statement
Saruul Erdenebileg: Conceptualization, Methodology, Formal analysis, Investigation, Writing – original draft, Visualization. Yang-Ju Son: Methodology, Formal analysis, Writing – review & editing, Project administration. Myungsuk Kim: Formal analysis, Data curation, Writing – review & editing. Sarangerel Oidovsambuu: . Kwang Hyun Cha: Formal analysis, Data curation. Jaeyoung Kwon: Formal analysis. Da Seul Jung: Formal analysis. Chu Won Nho: Writing – review & editing, Supervision, Project administration, Funding acquisition.
Acknowledgments
Conflict of interests
The authors declare no conflict of interest.
Funding
This research was supported by the Intramural Project of the Korea Institute of Science and Technology [grant number 2Z06670] and the Korea-Mongolia Cooperation Project of the National Research Foundation of Korea (NRF; Grant number 2008-00592).
Ethical statement
All animal experiments were approved by the International Animal Care and Use Committee of the Korea Institute of Science and Technology (Approval NO: KIST-2019-07003).
Data availability
Data will be made available upon reasonable request to the authors.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.imr.2023.100998.
Appendix. Supplementary materials
Fig S1. Effect of MESD on the alpha-diversity of the gut microbiota in DSS-induced colitis. Fig S2. Comparison of the MetaCyc pathways between the DSS and MESD groups.
References
- 1.Yeshi K., Ruscher R., Hunter L., Daly N.L., Loukas A., Wangchuk P. Revisiting inflammatory bowel disease: pathology, treatments, challenges and emerging therapeutics including drug leads from natural products. J Clin Med. 2020;9(5) doi: 10.3390/jcm9051273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bevins C.L., Salzman N.H. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat Rev Microbiol. 2011;9(5):356–368. doi: 10.1038/nrmicro2546. [DOI] [PubMed] [Google Scholar]
- 3.Kaplan G.G. The global burden of IBD: from 2015 to 2025. Nat Rev Gastroenterol Hepatol. 2015;12(12):720–727. doi: 10.1038/nrgastro.2015.150. [DOI] [PubMed] [Google Scholar]
- 4.Alatab S., Sepanlou S.G., Ikuta K., Vahedi H., Bisignano C., Safiri S., et al. The global, regional, and national burden of inflammatory bowel disease in 195 countries and territories, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol Hepatol. 2020;5(1):17–30. doi: 10.1016/S2468-1253(19)30333-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ben Ghezala I., Charkaoui M., Michiels C., Bardou M., Luu M. Small molecule drugs in inflammatory bowel diseases. Pharmaceuticals (Basel) 2021;14(7) doi: 10.3390/ph14070637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernstein C.N., Eliakim A., Fedail S., Fried M., Gearry R., Goh K.L., et al. World gastroenterology organisation global guidelines inflammatory bowel disease: update August 2015. J Clin Gastroenterol. 2016;50(10):803–818. doi: 10.1097/MCG.0000000000000660. [DOI] [PubMed] [Google Scholar]
- 7.Hazel K., O'Connor A. Emerging treatments for inflammatory bowel disease. Ther Adv Chronic Dis. 2020;11 doi: 10.1177/2040622319899297. 2040622319899297-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Triantafyllidi A., Xanthos T., Papalois A., Triantafillidis J.K. Herbal and plant therapy in patients with inflammatory bowel disease. Ann Gastroenterol. 2015;28(2):210–220. [PMC free article] [PubMed] [Google Scholar]
- 9.Kreiner J., Pang E., Lenon G.B., Yang A.W.H. Saposhnikoviae divaricata: a phytochemical, pharmacological, and pharmacokinetic review. Chin J Nat Med. 2017;15(4):255–264. doi: 10.1016/S1875-5364(17)30042-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang M., Wang C.-C., Wang W.-L., Xu J.-P., Wang J., Zhang C.-H., et al. Saposhnikovia divaricata—an ethnopharmacological. Phytochem Pharmacol Rev Chin J Integr Med. 2020;26(11):873–880. doi: 10.1007/s11655-020-3091-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tai J., Cheung S. Anti-proliferative and antioxidant activities of Saposhnikovia divaricata. Oncol Rep. 2007;18(1):227–234. [PubMed] [Google Scholar]
- 12.Jia P., Zhang Y., Zhang Q., Sun Y., Yang H., Shi H., et al. Metabolism studies on prim-O-glucosylcimifugin and cimifugin in human liver microsomes by ultra-performance liquid chromatography quadrupole time-of-flight mass spectrometry. Biomed Chromatogr: BMC. 2016;30(9):1498–1505. doi: 10.1002/bmc.3711. [DOI] [PubMed] [Google Scholar]
- 13.Kim S.H., Jong H.S., Yoon M.H., Oh S.H., Jung K.T. Antinociceptive effect of intrathecal sec-O-glucosylhamaudol on the formalin-induced pain in rats. Korean J Pain. 2017;30(2):98–103. doi: 10.3344/kjp.2017.30.2.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Y., Zhang T., Chen C., Xu Z., Liu C. Transcriptomics explores the potential of flavonoid in non-medicinal parts of Saposhnikovia divaricata (Turcz.) Schischk. Front Plant Sci. 2023;14 doi: 10.3389/fpls.2023.1067920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu W., Zhang Z., Lu Y., Li M., Li J., Tao W. Traditional Chinese medicine Tongxie Yaofang treating irritable bowel syndrome with diarrhea and type 2 diabetes mellitus in rats with liver-depression and spleen-deficiency: a preliminary study. Front Nutr. 2022;9 doi: 10.3389/fnut.2022.968930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chassaing B., Aitken J.D., Malleshappa M., Vijay-Kumar M. Dextran sulfate sodium (DSS)-induced colitis in mice. Curr Protoc Immunol. 2014;104 doi: 10.1002/0471142735.im1525s104. 15.25.1-15.25.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carroll K.C., Hobden J.A., Miller S., Morse S.A., Mietzner T.A., Detrick B., et al. McGraw-Hill Education; New York, NY: 2019. Normal Human Microbiota. Jawetz, Melnick, & Adelberg's Medical Microbiology, 27e. [Google Scholar]
- 18.Xu P., Lv T., Dong S., Cui Z., Luo X., Jia B., et al. Association between intestinal microbiome and inflammatory bowel disease: insights from bibliometric analysis. Comput Struct Biotechnol J. 2022;20:1716–1725. doi: 10.1016/j.csbj.2022.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leonardi I., Gerstgrasser A., Schmidt T.S.B., Nicholls F., Tewes B., Greinwald R., et al. Preventive Trichuris suis ova (TSO) treatment protects immunocompetent rabbits from DSS colitis but may be detrimental under conditions of immunosuppression. Sci Rep. 2017;7(1):16500. doi: 10.1038/s41598-017-16287-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manichanh C., Borruel N., Casellas F., Guarner F. The gut microbiota in IBD. Nat Rev Gastroenterol Hepatol. 2012;9(10):599–608. doi: 10.1038/nrgastro.2012.152. [DOI] [PubMed] [Google Scholar]
- 21.Kang J., Sun J.H., Zhou L., Ye M., Han J., Wang B.R., et al. Characterization of compounds from the roots of Saposhnikovia divaricata by high-performance liquid chromatography coupled with electrospray ionization tandem mass spectrometry. Rapid communications in mass spectrometry. RCM. 2008;22(12):1899–1911. doi: 10.1002/rcm.3559. [DOI] [PubMed] [Google Scholar]
- 22.Li Y.-Y., Wang X.-X., Zhao L., Zhang H., Lv L., Zhou G-c, et al. High-performance liquid chromatography–electrospray ionization time-of-flight mass spectrometry analysis of radix saposhnikoviae for metabolomic research. J Chromatogr Sci. 2012;51(2):99–106. doi: 10.1093/chromsci/bms111. [DOI] [PubMed] [Google Scholar]
- 23.Wang S., Qian Y., Sun M., Jia L., Hu Y., Li X., et al. Holistic quality evaluation of Saposhnikoviae Radix (Saposhnikovia divaricata) by reversed-phase ultra-high performance liquid chromatography and hydrophilic interaction chromatography coupled with ion mobility quadrupole time-of-flight mass spectrometry-based untargeted metabolomics. Arab J Chem. 2020;13(12):8835–8847. [Google Scholar]
- 24.Bolyen E., Rideout J.R., Dillon M.R., Bokulich N.A., Abnet C.C., Al-Ghalith G.A., et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019;37(8):852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Callahan B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J., Holmes S.P. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13(7):581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Price M.N., Dehal P.S., Arkin A.P. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5(3):e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bokulich N.A., Kaehler B.D., Rideout J.R., Dillon M., Bolyen E., Knight R., et al. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2′s q2-feature-classifier plugin. Microbiome. 2018;6(1):90. doi: 10.1186/s40168-018-0470-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yilmaz P., Parfrey L.W., Yarza P., Gerken J., Pruesse E., Quast C., et al. The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res. 2014;42(D1):D643–DD48. doi: 10.1093/nar/gkt1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McMurdie P.J., Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8(4):e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Douglas G.M., Maffei V.J., Zaneveld J.R., Yurgel S.N., Brown J.R., Taylor C.M., et al. PICRUSt2 for prediction of metagenome functions. Nat Biotechnol. 2020;38(6):685–688. doi: 10.1038/s41587-020-0548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barbera P., Kozlov A.M., Czech L., Morel B., Darriba D., Flouri T., et al. EPA-ng: massively parallel evolutionary placement of genetic sequences. Syst Biol. 2019;68(2):365–369. doi: 10.1093/sysbio/syy054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Czech L., Stamatakis A. Scalable methods for analyzing and visualizing phylogenetic placement of metagenomic samples. PLoS One. 2019;14(5) doi: 10.1371/journal.pone.0217050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caspi R., Billington R., Ferrer L., Foerster H., Fulcher C.A., Keseler I.M., et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2016;44(D1):D471–D480. doi: 10.1093/nar/gkv1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oksanen J., Blanchet F.G., Friendly M., Kindt R., Legendre P., McGlinn D., et al. Vegan: community ecology package. R package version 2.5-2. https://CRANR-projectorg/package=vegan 2018.
- 36.McArdle B.H., Anderson M.J. Fitting multivariate models to community data - a comment on distance-based redundancy analysis. Ecology. 2001;82(1):290–297. [Google Scholar]
- 37.Kaul A., Mandal S., Davidov O., Peddada S.D. Analysis of microbiome data in the presence of excess zeros. Front Microbiol. 2017;8:2114. doi: 10.3389/fmicb.2017.02114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cappello M., Randazzo C., Bravatà I., Licata A., Peralta S., Craxì A., et al. Liver function test abnormalities in patients with inflammatory bowel diseases: a hospital-based survey. Clin Med Insights Gastroenterol. 2014;7:25–31. doi: 10.4137/CGast.S13125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nell S., Suerbaum S., Josenhans C. The impact of the microbiota on the pathogenesis of IBD: lessons from mouse infection models. Nat Rev Microbiol. 2010;8(8):564–577. doi: 10.1038/nrmicro2403. [DOI] [PubMed] [Google Scholar]
- 40.Medzhitov R. Inflammation 2010: new adventures of an old flame. Cell. 2010;140(6):771–776. doi: 10.1016/j.cell.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 41.Zhu Z., Gu Y., Zhao Y., Song Y., Li J., Tu P. GYF-17, a chloride substituted 2-(2-phenethyl)-chromone, suppresses LPS-induced inflammatory mediator production in RAW264.7 cells by inhibiting STAT1/3 and ERK1/2 signaling pathways. Int Immunopharmacol. 2016;35:185–192. doi: 10.1016/j.intimp.2016.03.044. [DOI] [PubMed] [Google Scholar]
- 42.Posadas I., Terencio M.C., Guillén I., Ferrándiz M.L., Coloma J., Payá M., et al. Co-regulation between cyclo-oxygenase-2 and inducible nitric oxide synthase expression in the time-course of murine inflammation. Naunyn Schmied Arch Pharmacol. 2000;361(1):98–106. doi: 10.1007/s002109900150. [DOI] [PubMed] [Google Scholar]
- 43.Zhang Y., Wu Z., Liu J., Zheng Z., Li Q., Wang H., et al. Identification of the core active structure of a Dendrobium officinale polysaccharide and its protective effect against dextran sulfate sodium-induced colitis via alleviating gut microbiota dysbiosis. Food Res Int (Ottawa, Ont) 2020;137 doi: 10.1016/j.foodres.2020.109641. [DOI] [PubMed] [Google Scholar]
- 44.Wang K., Li Y-f, Lv Q., Li X-m, Dai Y., Wei Z-f. Bergenin, Acting as an agonist of PPARγ, ameliorates experimental colitis in mice through improving expression of SIRT1, and therefore inhibiting NF-κB-mediated macrophage activation. 2018;8. [DOI] [PMC free article] [PubMed]
- 45.Lauwerys B., FJAiem Houssiau. Involvement of cytokines in the pathogenesis of systemic lupus erythematosus. Biology (Basel) 2003;520:237–251. doi: 10.1007/978-1-4615-0171-8_14. [DOI] [PubMed] [Google Scholar]
- 46.Hegazy S.K., El-Bedewy M.M. Effect of probiotics on pro-inflammatory cytokines and NF-kappaB activation in ulcerative colitis. World J Gastroenterol. 2010;16(33):4145–4151. doi: 10.3748/wjg.v16.i33.4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu P.D., Zhao Y.H. Targeting NF-κB pathway for treating ulcerative colitis: comprehensive regulatory characteristics of Chinese medicines. Chin Med. 2020;15:15. doi: 10.1186/s13020-020-0296-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li Q., Verma I.M. NF-κB regulation in the immune system. Nat Rev Immunol. 2002;2(10):725–734. doi: 10.1038/nri910. [DOI] [PubMed] [Google Scholar]
- 49.Macfarlane G.T., Cummings J.H. Probiotics and prebiotics: can regulating the activities of intestinal bacteria benefit health? BMJ (Clinical research ed) 1999;318(7189):999–1003. doi: 10.1136/bmj.318.7189.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mueller C., Macpherson A.J. Layers of mutualism with commensal bacteria protect us from intestinal inflammation. Gut. 2006;55(2):276–284. doi: 10.1136/gut.2004.054098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tanaka Y., Matsuzawa H., Tamaki H., Tagawa M., Toyama T., Kamagata Y., et al. Isolation of novel bacteria including rarely cultivated phyla, acidobacteria and verrucomicrobia, from the roots of emergent plants by simple culturing method. Microbes Environ. 2017;32(3):288–292. doi: 10.1264/jsme2.ME17027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kanwal S., Joseph T.P., Aliya S., Song S., Saleem M.Z., Nisar M.A., et al. Attenuation of DSS induced colitis by Dictyophora indusiata polysaccharide (DIP) via modulation of gut microbiota and inflammatory related signaling pathways. J Funct Foods. 2020;64 [Google Scholar]
- 53.Morgan X.C., Tickle T.L., Sokol H., Gevers D., Devaney K.L., Ward D.V., et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13(9):R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rizzatti G., Lopetuso L.R., Gibiino G., Binda C., Gasbarrini A. Proteobacteria: a common factor in human diseases. Biomed Res Int. 2017;2017 doi: 10.1155/2017/9351507. 9351507-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hu C., Niu X., Chen S., Wen J., Bao M., Mohyuddin S.G., et al. A comprehensive analysis of the colonic flora diversity, short chain fatty acid metabolism. Transcr, Biochem Indexes Heat-Stressed Pigs. 2021:12. doi: 10.3389/fimmu.2021.717723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lakshminarayanan B., Harris H.M.B., Coakley M., O'Sullivan Ó., Stanton C., Pruteanu M., et al. Prevalence and characterization of Clostridium perfringens from the faecal microbiota of elderly Irish subjects. J Med Microbiol. 2013;62(Pt 3):457–466. doi: 10.1099/jmm.0.052258-0. [DOI] [PubMed] [Google Scholar]
- 57.Buckel W. Unusual enzymes involved in five pathways of glutamate fermentation. Appl Microbiol Biotechnol. 2001;57(3):263–273. doi: 10.1007/s002530100773. [DOI] [PubMed] [Google Scholar]
- 58.Zhang Y., Chen H., Zhu W., Yu K. Cecal infusion of sodium propionate promotes intestinal development and jejunal barrier function in growing pigs. Animals (Basel) 2019;9(6):284. doi: 10.3390/ani9060284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen N., Wu Q., Chi G., Soromou L.W., Hou J., Deng Y., et al. Prime-O-glucosylcimifugin attenuates lipopolysaccharide-induced acute lung injury in mice. Int Immunopharmacol. 2013;16(2):139–147. doi: 10.1016/j.intimp.2013.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kimura Y., Sumiyoshi M., Baba K. Anti-tumor actions of major component 3′-O-acetylhamaudol of Angelica japonica roots through dual actions, anti-angiogenesis and intestinal intraepithelial lymphocyte activation. Cancer Lett. 2008;265(1):84–97. doi: 10.1016/j.canlet.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 61.Ikeda M. Public health problems of organic solvents. Toxicol Lett. 1992;64(65):191–201. doi: 10.1016/0378-4274(92)90189-q. [DOI] [PubMed] [Google Scholar]
- 62.Truong D.-H., Nguyen D.H., Ta N.T.A., Bui A.V., Do T.H., Nguyen H.C. Evaluation of the use of different solvents for phytochemical constituents, antioxidants, and in vitroanti-inflammatory activities of Severinia buxifolia. J Food Qual. 2019;2019 [Google Scholar]
- 63.Uddin N., Hasan M.R., Hasan M.M., Hossain M.M., Alam M.R., Hasan M.R., et al. Assessment of toxic effects of the methanol extract of Citrus macroptera Montr. Fruit via biochemical and hematological evaluation in female Sprague-Dawley rats. PLoS One. 2014;9(11) doi: 10.1371/journal.pone.0111101. e111101-e01. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1. Effect of MESD on the alpha-diversity of the gut microbiota in DSS-induced colitis. Fig S2. Comparison of the MetaCyc pathways between the DSS and MESD groups.
Data Availability Statement
Data will be made available upon reasonable request to the authors.