Abstract
Mutations in genes encoding nuclear pore proteins (NUPs) lead to the development of steroid-resistant nephrotic syndrome and focal segmental glomerulosclerosis (FSGS). However, the precise molecular mechanisms by which NUP dysfunction contributes to podocyte injury preceding FSGS remain unclear. The tightly regulated activity of Yes-associated protein (YAP) and WW-domain-containing transcription regulator 1 (TAZ), the transcriptional effectors of the Hippo pathway, is crucial for podocytes and the maintenance of the glomerular filter. In this study, we investigate the impact of NUPs on the regulation of YAP/TAZ nuclear import and activity in podocytes. In unbiased interactome studies using quantitative label-free mass spectrometry, we identify the FSGS disease gene products NUP107, NUP133, NUP205, and Exportin-5 (XPO5) as components of YAP and TAZ protein complexes in podocytes. Moreover, we demonstrate that NUP205 is essential for YAP/TAZ nuclear import. Consistently, both the nuclear interaction of YAP/TAZ with TEA domain transcription factor 1 and their transcriptional activity were dependent on NUP205 expression. Additionally, we elucidate a regulatory feedback mechanism whereby YAP activity is modulated in response to TAZ-mediated NUP205 expression. In conclusion, this study establishes a connection between the FSGS disease protein NUP205 and the activity of the transcriptional regulators and Hippo effectors YAP and TAZ and it proposes a potential pathological role of YAP/TAZ dysregulation in podocytes of patients with pathogenic NUP205 variants.
Keywords: podocyte, YAP, TAZ, FSGS, NUP205
Introduction
Focal segmental glomerulosclerosis (FSGS) is one of the main histopathological findings in proteinuric renal diseases that can ultimately lead to end-stage renal disease (1). FSGS is caused by dysfunction and loss of podocytes disrupting the glomerular filtration barrier. Podocytes are terminally differentiated cells whose primary and secondary processes cover the glomerular capillaries and are essential for the formation of the filtration barrier (2,3). Processes of adjacent podocytes form a specialized cell–cell contact at the filtration slits, bridged by a membrane-like junction called the slit diaphragm (4). Multiple causes can lead to foot process effacement and podocyte loss, including immunological and systemic processes and genetic variations (5). For hereditary FSGS, more than 50 mutations in various genes encoding for proteins of the slit diaphragm, cell membrane, cytoskeleton, and mitochondria have been identified to induce podocyte injury and the loss of integrity of the filtration barrier (6,7). Recently, mutations in specific genes of the nuclear pore complex and the related shuttling machinery have been added to the list of genes causing steroid-resistant nephrotic syndrome (SRNS), a disease complex that includes the histopathological finding of FSGS (8).
The nuclear pore complex (NPC) forms a channel at the nuclear membrane that allows nucleocytoplasmic transport in both directions (9,10). The NPC consists of 30 different nuclear pore proteins called nucleoporins (NUPs) (11). These NUPs interact in subclusters to form the major structural elements of the NPC: an inner, a nuclear, and a cytoplasmic ring; the nuclear basket; and cytoplasmic filaments (10). In families suffering from SRNS, mutations in genes encoding for components of the inner ring (NUP205, NUP93) and of the cytoplasmic and nuclear rings (NUP85, NUP107, NUP133, NUP160) have been identified (8,12). In addition, mutations in Exportin-5 (XPO5), one of the proteins involved in nuclear export acting in concert with nucleoporins, have been discovered (8,13). However, the exact mechanism of how the disruption of the nucleocytoplasmic shuttling affects podocyte homeostasis remains to be elucidated.
The Hippo signaling pathway and its two effector proteins, Yes-associated protein (YAP1, hereafter referred to as YAP) and WW-domain-containing transcription regulator 1 (WWTR1, hereafter referred to by the alternative name TAZ), are important regulators of many cellular processes, such as cell differentiation, proliferation, and apoptosis (14). YAP and TAZ shuttle between the cytoplasm and the nucleus in response to various inputs. In the nucleus, YAP and TAZ act as transcriptional coactivators and regulate the expression of downstream targets in synergy with other transcription factors like TEA domain family members (TEAD1–4) (15). In most healthy adult tissues, YAP and TAZ are usually maintained in an inactive state, i.e. phosphorylated and retained in the cytoplasm, only shuttling to the nucleus for regenerative or malignant growth (16). In podocytes, YAP and TAZ seem to have divergent roles. Inputs that regulate YAP and TAZ activity, like cell–cell adhesions, cell polarity, or mechanical forces, are highly relevant in podocyte biology. Nuclear YAP activity has been shown to promote cell survival and inhibit podocyte apoptosis in FSGS (17,18). Furthermore, mice lacking YAP or TAZ specifically in podocytes develop proteinuria and FSGS (19,20). Therefore, podocyte homeostasis relies on a well-balanced nuclear shuttling and activity of YAP and TAZ, a process whose regulation is so far not well understood (18, 21–26).
Here, we explore the interactome of YAP and TAZ in an in vitro mouse podocyte model. We identify several nuclear transport components that associate in protein complexes with both YAP and TAZ. Focusing on the FSGS gene NUP205, we describe its regulation of the nucleocytoplasmic shuttling of YAP and TAZ, relevant for their transcriptional activity. Finally, we elucidate a regulatory feedback mechanism whereby YAP localization and activity are influenced by TAZ-mediated NUP205 expression. These results unmask a molecular pathomechanism perspective underlying genetic FSGS.
Materials and Methods
Cells
Heat-sensitive mouse podocytes (hsMPs) were cultured in RPMI-1640 medium (Gibco, 61870) supplemented with 10% fetal bovine serum (FBS, Gibco, 10270), sodium pyruvate (Sigma, S8636), 20 mM HEPES (Sigma, H0887), and 0.25 μl/ml IFN-δ (PeproTech, 315-05). Podocytes were cultured on Collagen I-coated 10-cm primaria cell culture dishes (Corning, 353803), at low confluency and at 33°C, as previously described (27). The mouse podocyte cell line was obtained from Stuart Shankland (Seattle, WA). HEK293T cells were cultured in DMEM (Gibco, 31966) supplemented with 10% FBS at 37°C. For transfection experiments, cells were grown until 40% confluence and transfected with ON-TARGETplus SMARTpool siRNAs (Dharmacon) using Lipofectamine RNAiMAX Transfection Reagent (Invitrogen, 13778-150). ON-TARGETplus Non-targeting pool (Dharmacon) was used as a negative control, scrambled RNA. The sequences of all siRNAs are described in Table 1. All experiments were performed 48 h after transfection, unless mentioned otherwise. All cell lines were regularly tested for mycoplasma contamination by a regular kit. Cross-contamination with other cell lines was not observed and is currently not reported.
Table 1.
ON-TARGETplus SMARTpool siRNA sequences.
| Target gene | ON-TARGETplus SMARTpool siRNA | Target sequences | Catalog ID |
|---|---|---|---|
| Scrambled | UGGUUUACAUGUCGACUAA UGGUUUACAUGUUGUGUGA UGGUUUACAUGUUUUCUGA UGGUUUACAUGUUUUCCUA |
D-001810-10-20 | |
| Human NUP205 | J-010646-05 J-010646-06 J-010646-07 J-010646-08 |
GGAACGAGAUGAUAUGAUU GAGUAUAAGUCAGAAACUA CCACCUGGAUUUAGUCUGA CAACUGAUUAUUCGUGAUA |
L-010646-00-0005 |
| Mouse Yap1 | J-046247-09 J-046247-10 J-046247-11 J-046247-12 |
CCGAAAUCUUGGACGUGGA GAAUAAAGGAUGGCGUCUU UCUUAAAUCACAACGAUCA AAGGAGAGACUGCGGUUGA |
L-046247-01-0005 |
| Human YAP1 | J-012200-05 J-012200-06 J-012200-07 J-012200-08 |
GCACCUAUCACUCUCGAGA UGAGAACAAUGACGACCAA GGUCAGAGAUACUUCUUAA CCACCAAGCUAGAUAAAGA |
L-012200-00-0005 |
| Mouse Wwtr1 | J-041057-09 J-041057-10 J-041057-11 J-041057-12 |
CAAUUUAUGUCCACGUUAA CCAUUGAAAUAGAAACGCA GAGAUGACCUUCACGGCCA AUGUAUUGGCAGACGAGAA |
L-016083-00-0005 |
| Human WWTR1 | J-016083-05 J-016083-06 J-016083-07 J-016083-08 |
CCGCAGGGCUCAUGAGUAU GGACAAACACCCAUGAACA AGGAACAAACGUUGACUUA CCAAAUCUCGUGAUGAAUC |
L-041057-01-0005 |
Generation of stable cell lines
Using Transcription Activator-Like Effector Endonucleases (TALEN) technology, we generated stable hsMPs cell lines carrying a single copy integration of a transgenic construct expressing a flagged version of mouse Yap (3XFLAG.YAP), Taz (3XFLAG.TAZ), or Ruby (3XFLAG.RUBY) in the Rosa26 locus. Rosa26_TV_CMV-3xFL-mRuby-STOP_EF1a-Puro-T2A-copGFP plasmid was based on pDonor MCS Rosa26 (a gift from Charles Gersbach; addgene plasmid #37200) (28). Four point mutations were added in the sequence of the later plasmid: HindIII site 2253 killed (now AATCTT), KpnI site 657 killed (now GGTAGC), NotI site 821 killed (now GGGGCCGC), and NotI site 1492 killed (now GGGGCGGC). Mouse Yap or Taz (kindly provided by Michael Yaffe) was inserted after excising mRuby. The integration of the constructs in the mouse Rosa26 locus was possible by co-transfecting with TALEN-mROSA26 KKR and TALEN-mROSA26 ELD (a gift from Radislav Sedláček; addgene plasmid #60025 and #60026) (28,29). After transfection, stably transfected cells were selected with 1.5 μg/ml of puromycin (InvivoGen, ant-pr). Cells were maintained in the presence of puromycin and ready to be used. The correct expression of the tagged proteins was confirmed by immunoblotting (Supplementary Material, Fig. S1B).
Immunoprecipitation
For each replicate of the experiments, 2–4 × 10-cm dishes (60% confluency) were used. Cells were harvested with ice-cold phosphate buffered saline (PBS). The harvested cells were lysed in modified RIPA buffer [1% Igepal NP40, 50 mM Tris·HCl pH 7.5, 150 mM NaCl, 0.25% sodium deoxycholate, and complete protease inhibitors without EDTA (PIM; Roche)]. Cells were homogenized eight times with a 27-gauge needle and incubated on ice for 15 min. Afterward, the samples were sonicated with a Bioruptor (10 min, cycle 30/30 s) to degrade chromatin followed by centrifugation at 20 000xg for 30 min at 4°C and ultracentrifugation at 125 000xg for 30 min at 4°C. The samples for mass spectrometry (MS) analysis were incubated with Dynabeads protein G that were pre-coupled with the M2-Flag-antibody (Sigma, F1804) at 4°C overnight. Before adding of the antibody-coupled-beads, a small aliquot of each sample was preserved and diluted with 2x SDS-PAGE sample buffer for immunoblot analysis. The next day, the beads were washed five times and then resuspended in 50 μl of 5% SDS in PBS and boiled for 5 min. The magnetic beads were collected on a magnet. The eluate was further processed for MS analysis except a small aliquot that was used to validate the immunoprecipitation by immunoblotting. To prepare for MS analysis, DTT was added to a final concentration of 5 mM, vortexed, and incubated at 55°C for 30 min. After that, CAA was added to a final concentration of 40 mM, vortexed, and incubated in the dark for 30 min at room temperature. After centrifugation for 10 min at 20 000xg, the supernatant was stored at −20°C to be then further processed and measured by the CECAD Proteomics Facility, University of Cologne. For IgG or YAP-immunoprecipitation, whole-cell supernatants were incubated with 1 μg of rabbit IgG (Santa Cruz, sc-2027) or 0.2 μg of rabbit anti-YAP (Cell Signaling, 4912), overnight at 4°C. The next day, Protein A sepharose beads were added and incubated for 1 h at 4°C on an overhead shaker. Then, beads were washed five times and proteins eluted in 1x SDS-PAGE sample buffer for immunoblot analysis.
Mass spectrometry and data processing
Proteomics analysis was conducted at the CECAD Proteomics Facility, University of Cologne. All samples were analyzed on a Q-Exactive Plus (Thermo Scientific) mass spectrometer that was coupled to an EASY nLC 1000 or 1200 UPLC (Thermo Scientific). Samples were loaded onto an in-house packed analytical column (50 cm × 75 μm I.D., filled with 2.7 μm Poroshell EC120 C18, Agilent) that has been equilibrated in solvent A (0.1% formic acid in water), and peptides were separated at a constant flow rate of 250 nl/min using a 50-min gradient followed by a 10-min wash with 95% Solvent B (0.1% formic acid in 80% acetonitrile) for 10 min. The mass spectrometer was operated in the data-dependent acquisition mode. MS1 survey scans were acquired from 300 to 1750 m/z at a resolution of 70 000. The top 10 most abundant peptides were isolated within a 1.8-Th window and subjected to HCD fragmentation at a normalized collision energy of 27%. The AGC target was set to 5e5 charges, allowing a maximum injection time of 110 ms. Product ions were detected in the Orbitrap at a resolution of 35 000. Precursors were dynamically excluded for 10 s.
All mass spectrometric raw data were processed with MaxQuant (version 1.5.3.8) using default parameters. Briefly, MS2 spectra were searched against the Mouse reference (downloaded at 16.6.2017), including a list of common contaminants. False discovery rates on protein and PSM level were estimated by the target-decoy approach to 1% (Protein FDR) and 1% (PSM FDR), respectively. The minimal peptide length was set to seven amino acids, and carbamidomethylation at cysteine residues was considered as a fixed modification. Oxidation (M) and Acetyl (Protein N-term) were included as variable modifications. The match-between runs option was enabled within replicate groups. LFQ quantification was enabled using default settings. Student’s t-tests were calculated in Perseus (version 1.6.1.1) after removal of decoys and potential contaminants. Data were filtered for at least three out of five values in at least one condition. Remaining missing values were imputed with random values from the lower end of the intensity distribution using Perseus defaults.
Cellular fractions
For whole-cell lysates, hsMPs or HEK293T cells were harvested with ice-cold PBS and lysed in modified RIPA buffer. Cells were titrated eight times with a 27-gauge needle and incubated on ice for 15 min. After centrifugation at 20 000xg for 15 min at 4°C, the supernatant was mixed with 2x SDS-PAGE sample buffer and boiled for 5 min at 95°C.
Nuclear and cytoplasmic fractions were collected as previously described (30). Briefly, HEK293T cells were harvested on ice in 7 ml ice-cold PBS. One milliliter of the resultant cell suspension was collected for processing as a whole-cell lysate in SDS-PAGE sample buffer. The remaining cell suspension was pelleted at 48xg for 5 min at 4°C. The supernatant was discarded, and the cell pellet was resuspended in 150 μl of the hypotonic Cell Fraction Buffer [10 mM HEPES, 1.5 mM MgCl2, 10 mM KCl, pH 7.9 plus PIM (Roche)]. Cells were incubated on ice for 10 min and then disrupted 14 times using a 21-gauge needle. The mixture was gently centrifuged at 100xg for 30 min at 4°C to pellet the nuclear fraction. The supernatant was ultracentrifuged at 100 000xg for 30 min at 4°C. Eighty microliters of the resultant supernatant was transferred to a new 1.5 Eppendorf tube and mixed with 80 μl of 2 × SDS-PAGE sample buffer, boiled for 5 min at 95°C, and centrifuged for 1 min at 20 000xg to yield the cytosolic fraction. Meanwhile, the nuclear pellet was resuspended in 1 ml cold PBS and centrifuged at 9200xg for 10 min at 4°C. The supernatant was discarded, and the remaining pellet was washed once with PBS and centrifuged again for 10 min 9200xg at 4°C. The pellet was then resuspended in 100 μl of 2x SDS-PAGE sample buffer, boiled for 10 min at 95°C, and centrifuged for 5 min at 20°000xg to yield the nuclear fraction. Twenty microliters of the supernatant was analyzed by western blot.
Immunoblotting
Proteins were separated by 8.5% sodium dodecyl sulfate (SDS) polyacrylamide gel electrophoresis and transferred onto a polyvinylidene fluoride membrane (Millipore, Immobilon-P, IPVH00010) using a semi-dry transfer unit (PEQLAB). After blocking in 5% bovine serum albumin, membranes were stained with 1:2000 of primary anti-beta-tubulin mouse monoclonal antibody (DSHB, E7), anti-fibrillarin rabbit monoclonal antibody (Abcam, ab166630), anti-NUP205 rabbit polyclonal antibody (Biomol, A303-935A), anti-YAP rabbit polyclonal antibody (Cell Signaling, 4912), anti-TAZ rabbit monoclonal antibody (Cell Signaling, 72 804), anti-TEAD1 mouse monoclonal antibody (BD Biosciences, 610 922), anti-beta-actin mouse monoclonal antibody (Cell Signaling, 3700 S), or anti-GAPDH rabbit monoclonal antibody (Cell Signaling, 5174), overnight at 4°C. Proteins were visualized using horseradish peroxidase-conjugated secondary antibody (1:30 000) and ECL detection. Images were acquired with a Fusion Solo S (Vilber Lourmat Germany GmbH, Eberhardzell, Germany). Densitometric analysis was performed using ImageJ/Fiji Software version 2.9.0/1.53t (NIH, Bethesda, MD) (31).
Immunocytochemistry
HEK293T cells were seeded onto 12-mm-coverslips and transfected using indicated siRNA. Sixteen hours or 48 h afterward, cells were rinsed with PBS and fixed with 4% formalin for 15 min. After washing with PBS, cells were permeabilized in 0.5% Triton X-100 in PBS, for 10 min. After blocking with 1% normal donkey serum (NDS, Biozol, END9010) and 1% bovine serum albumin (BSA, Thermo Fisher Scientific, 23 209) in PBS, cells were sequentially co-stained with 1:200 primary anti-NUP205 rabbit polyclonal antibody (Biomol, A303-935A), anti-YAP mouse monoclonal antibody (Santa Cruz, sc-376830), or anti-TAZ mouse monoclonal antibody (BD Biosciences, 560235). Afterward, the coverslips were mounted with Prolong gold with DAPI (Invitrogen) and subjected to immunofluorescence microscopy. Pictures were taken with a STELLARIS 5 LIAchroic inverse confocal microscope (Leica) and objective HC PL APO 63x/1.30 GLYC CORR CS2 was used for acquisition. Subsequent image processing and analysis was performed using ImageJ/Fiji Software version 2.9.0/1.53t (NIH, Bethesda, MD) (31). Nuclear and cytosolic expression levels were quantified by analyzing fluorescence intensities in the regions of interest (ROIs) and were plotted as proportions of nuclear versus cytoplasmic fluorescence.
qPCR analyses
RNA was isolated using TRIzol (Sigma, S8636) and the Direct-zol RNA MiniPrep (ZYMO research, R2052) according to the manufacturer’s instructions. In-column digestion with DNAse was performed for all samples to remove genomic DNA contamination. Quality and quantity of eluted RNA were checked using a NanoDrop spectrophotometer (PEQLAB). Complementary DNA (cDNA) was synthesized using the same amounts of RNA using the high-capacity cDNA reverse transcription (Thermo Fisher Scientific, 4368814). qPCR was performed using SYBR Green assays (Thermo Fisher Scientific, 4367659) and analyzed using a QuantStudio 12 K Flex cycler system. Primer sequences are listed in Table 2. Quantification of relative expression levels was performed using the 2−∆∆CT method as previously described (32), which involves normalization to a housekeeping gene. The gene Hprt was used as housekeeping gene. Afterward, expression was normalized to control condition, resulting in the final “relative mRNA expression” parameter. Primer pairs were tested for efficiency.
Table 2.
Primers used for qPCR.
| Name | Sequence forward | Sequence reverse |
|---|---|---|
| ANKRD1 | AGTAGAGGAACTGGTCACTGG | TGGGCTAGAAGTGTCTTCAGAT |
| CTGF | CTGCAGACTGGAGAAGCAGA | GATGCACTTTTTGCCCTTCTT |
| CYR61 | AGCCTCGCATCCTATACAACC | TTCTTTCACAAGGCGGCACTC |
| DIAPH3 | GCGGTATGCATTGTAGGGGA | CAGGAGATGTAACCAGGGCA |
| HPRT1 | TGACACTGGCAAAACAATGCA | GGTCCTTTTCACCAGCAAGCT |
| NUP205 | GATTTTAGAAGTGGGCTGGCT | CGTCTGACAAGAGCCTGTATGA |
| TAZ | CAGAACCACCCCACTCAGAAC | TCAGCGCATTGGGCATACT |
Live-cell imaging
hsMPs cells were seeded, with 5000 cells per well, into 96-well plates in triplicates. Twenty-four hours after seeding, cells were transfected with the indicated siRNAs. After 24 h, cells were treated either with 1 μg/ml Adriamycin (ADR, doxorubicin; Pharmacy of the University Hospital of Cologne, Charge number: AT0034) or 100 μM hydrogen peroxide (H2O2) (ROTH, 1A8Y.3), as indicated. Cell death was visualized by adding DiYO-1 (Biomol, ABD-17580). Immediately after treatment, the plates were analyzed every 2 h up to 24 h in the IncuCyte® S3 (Sartorius; 37°C and 5% CO2). Per well, three single images of the green channel with 300 ms exposure time and the phase-contrast channel were generated with ×20 objective for each time point. The analysis was done by training the machine for positive events within the included IncuCyte® Cell-by-Cell Analysis Software Module (Sartorius, #9600-0031).
Statistical analysis
Data are reported as mean ± SEM. Student’s t-test for unpaired samples was used for statistical analysis. P < 0.05 was accepted as significant difference.
Results
YAP and TAZ interact with the nuclear shuttling machinery in podocytes
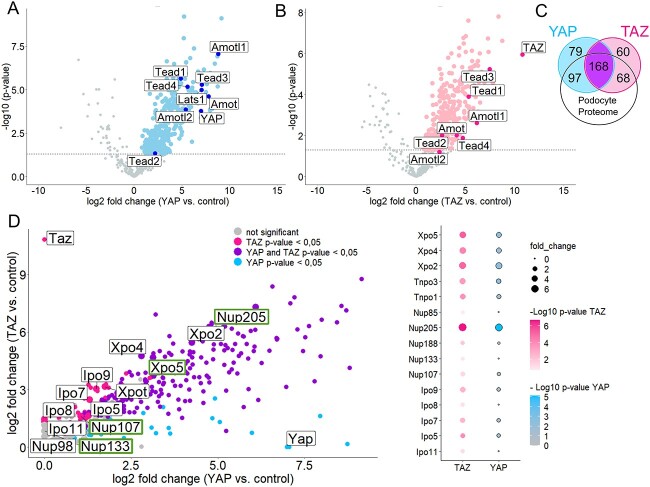
To unravel the importance of YAP and TAZ in podocytes, we aimed to identify novel podocyte-specific components of the YAP/TAZ protein complexes. Therefore, we used a single-copy integration system targeting the Rosa26 locus and generated podocyte cell lines stably expressing low levels of 3xFLAG.YAP, 3xFLAG.TAZ, or 3xFLAG.Ruby as negative control (Supplementary Material, Fig. S1A). Pulldown assays with Flag-antibody successfully immunoprecipitated FLAG-tagged Yap, Taz, or Ruby as confirmed by immunoblotting (Supplementary Material, Fig. S1B). Five biological replicates generated on different days were analyzed together by mass spectrometry to identify specific components of Yap and Taz protein complexes in podocytes. Principal component analysis (PCA) revealed that all samples within a group (i.e. RUBY, YAP, and TAZ) clustered together, whereas the groups were markedly separated (Supplementary Material, Fig. S1C). Further analysis led to the identification of a large number of proteins enriched in either the YAP or TAZ data set as compared with the control (Fig. 1A and B). As expected, Yap and Taz were among the most prominently enriched proteins in the 3XFLAG.YAP pull-down (Fig. 1A) and in the 3XFLAG.TAZ pull-down (Fig. 1B), respectively. Taken together, our data sets revealed 362 proteins significantly enriched in the Yap-samples and 314 proteins in the Taz-samples as components of YAP/TAZ protein complexes. Among these identified proteins, many well-established interactors of Yap and Taz were included, like proteins of the Tead family, different angiomotins, or large tumor suppressor kinase 1 (Lats1) verifying the accuracy of the data sets (Fig. 1A and B). Furthermore, comparison of our data set with a published proteome expression profile of mouse podocytes isolated directly from glomeruli confirmed the expression of the majority of the identified proteins from our study in podocytes in vivo, underlining the relevance of our data sets for podocyte biology (Fig. 1C) (33). There are many proteins in common to both complexes, which is well explained by the homology and common functions of YAP and TAZ. Strikingly, among the shared proteins were numerous proteins of the nuclear shuttling machinery, like importins (Ipo), exportins (Xpo), transportins (Tnpo), and nucleoporins (Nup) that form the NPC (Fig. 1D). Among those were NUP107, NUP133, NUP205, and XPO5. Variants in these corresponding genes have recently been identified to cause FSGS, making them even more interesting for podocyte biology (8,12,34). Specifically, Nup205 is the most abundant nuclear transport protein within the complex of both YAP and TAZ.
Figure 1.
Podocyte interactome shows interplay of YAP and TAZ with nuclear transport proteins. Volcano plots showing the interactors of YAP (A) and TAZ (B) in immortalized podocytes (hsMPs) overexpressing 3xFLAG.YAP or 3xFLAG.TAZ. Logarithmized ratios are plotted against the negative logarithmic P-value of the Student’s t-test. Data points with a P-value < 0.05 are printed bold (above dashed line). Known interactors are highlighted and labeled. (C) Venn diagram indicating numerical YAP and TAZ interactors overlapping with mouse podocyte proteome from Rinschen et al. (33). (d) Scatter plot combining the interactors of YAP and TAZ, with shared interactors (P-value for YAP and TAZ < 0.05) highlighted. Proteins of the nuclear-shuttling machinery are labeled and highlighted when related to FSGS. Dot plot showing proteins of nuclear shuttling machinery that are shared interactors by YAP and TAZ.
NUP205 regulates YAP and TAZ nuclear shuttling
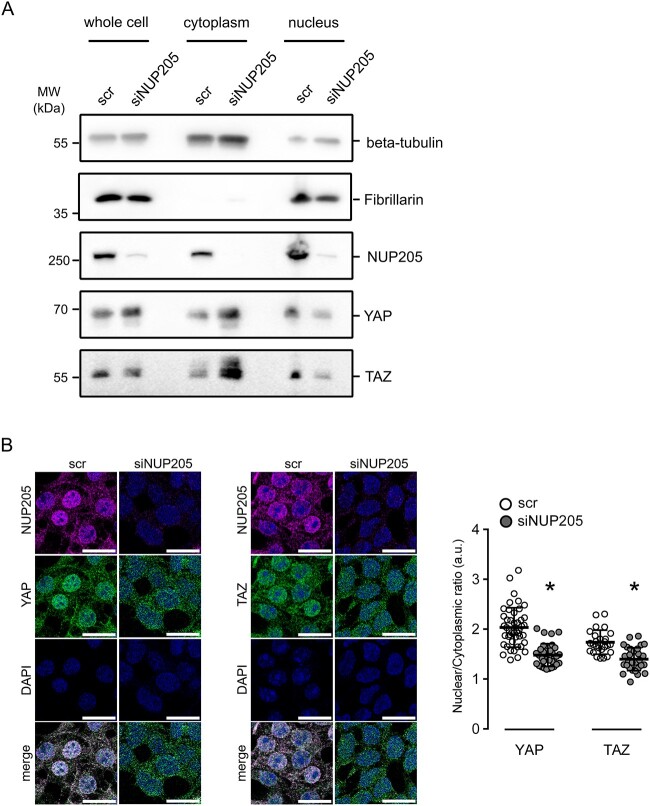
NUP205 is a protein from the inner ring subunit of the NPC, important for its assembly and function in active bidirectional transport of molecules between the nucleus and the cytoplasm (35,36). To investigate the influence of this specific nucleoporin on the nuclear shuttling of YAP and TAZ, we targeted endogenous NUP205 using small interfering RNA (siRNA) and analyzed changes in endogenous YAP or TAZ subcellular localization in HEK293T cells. Subcellular fractionation assays revealed that knockdown of NUP205 reduced the nuclear localization of YAP and TAZ (Fig. 2A). Meanwhile, the cytoplasmic expression levels of YAP and TAZ were increased, whereas total protein levels remained equal (Supplementary Material, Fig. S2A and B). The lack of constitutive nuclear shuttling of YAP and TAZ upon knockdown of NUP205 was further confirmed by assessing the shift in localization of endogenous proteins by immunofluorescence (Fig. 2B). From these results, it is evident that NUP205 is essential for the nuclear import of both YAP and TAZ. Furthermore, this role of NUP205 seems to be specific to the trafficking of YAP and TAZ since it did not have any impact on the subcellular localization of SMAD4 (SMAD family member 4), a protein that also shuttles between the nucleus and the cytoplasm (Supplementary Material, Fig. S3A). Notably, when NUP93, another structural component of the nuclear pore, was knocked down, it had no effect on the subcellular localization of YAP and TAZ (Supplementary Material, Fig. S3B). This underscores the specificity of NUP205 for the shuttling of YAP and TAZ.
Figure 2.
Downregulation of NUP205 compromises nuclear localization of YAP and TAZ. (A) Immunoblot showing the results of the cell fractionation assay for HEK293T cells transfected with siRNA NUP205 compared with control. The presence of the cytoplasmic protein beta-tubulin and the nuclear protein fibrillarin in the appropriate fraction was confirmed by western blotting with the corresponding antibodies. Downregulation of NUP205 decreases nuclear shuttling of YAP and TAZ, which stay confined in the cytoplasm. (B) Immunofluorescence staining of HEK293T cells transfected with siRNA NUP205 shows a decrease of nuclear YAP and TAZ compared with control. Co-staining for endogenous NUP205, endogenous YAP (left) or TAZ (right), and nuclear stain with DAPI. All scale bars = 20 μm. Summary of nuclear/cytoplasmatic fluorescence ratio quantification (n = 26–45 cells). Data are reported as mean ± SEM. Representative data from three independent experiments are shown. *Significant decrease of nuclear staining of YAP and TAZ (unpaired t-test; P < 0.05).
Lack of NUP205 leads to a decrease in YAP and TAZ-dependent transcriptional activity
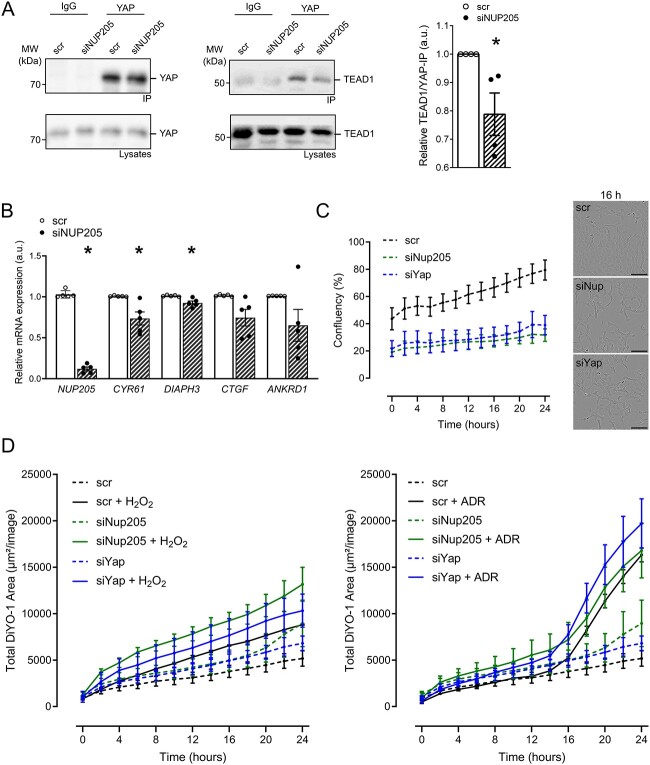
YAP and TAZ shuttle into the nucleus to function as co-transcription factors and initiate the transcription of various target genes (37–39). YAP/TAZ can contact the DNA only indirectly through transcription factor partners. One pivotal transcription factor they bind is TEAD1 (15). TEAD1 is localized mainly in the nucleus; therefore, YAP needs to be able to shuttle into the nucleus to interact with TEAD1. To determine whether NUP205 downregulation causes a reduced YAP/TEAD1 nuclear interaction, TEAD1 was co-immunoprecipitated with YAP after NUP205 knockdown and control cells. YAP pulled down TEAD1, and this co-immunoprecipitation was diminished upon NUP205 knockdown (Fig. 3A). In support of this, we found that the mRNA levels of the bona fide target genes of YAP and TAZ, CYR61 and DIAPH3, were reduced significantly (P = 0.027 and P = 0.018) and CTGF and ANKRD1 showed a similar tendency (P = 0.056 and P = 0.138, respectively), upon knockdown of NUP205 (Fig. 3B). Taken together, depletion of NUP205 led to a reduction of nuclear YAP and TAZ and, consequently, a reduced interaction with TEAD1 and inhibition of target genes expression, confirming that the transcriptional activity of YAP and TAZ is dependent on the proper expression of the nuclear pore component NUP205. The nuclear localization and activity of YAP and TAZ regulate the balance between proliferation, differentiation, and apoptosis (16). To explore the functional importance of the modulation of localization and transcriptional activity of YAP and TAZ by NUP205, we further investigated the effect of Nup205 downregulation on cell proliferation in podocytes using live-cell imaging. Over a time course of 24 h, the reduction of Nup205 compromised the proliferation of immortalized mouse podocytes, similar to the effects observed with the knockdown of Yap, as compared with the control conditions (Fig. 3C). We further analyzed the consequences of Nup205 and Yap knockdown on cell death under normal conditions and in response to oxidative stress in cultured podocytes. Under basal conditions, Nup205 downregulation resulted in an increase of dead cells, when compared with controls, similar to the effect of Yap downregulation, confirming the described antiapoptotic role of Yap in podocytes (Fig. 3D) (24). Application of oxidative stress either by H2O2 or ADR as a model mimicking induced FSGS resulted in augmented cell death and was even further enhanced in cells lacking Nup205 or Yap.
Figure 3.
YAP and TAZ activity is decreased upon NUP205 downregulation. (A) Co-immunoprecipitation of endogenous YAP shows a decrease in interaction with TEAD1 in HEK293T cells transfected with siRNA NUP205 compared with control. IgG is used as negative control for the IP. Quantification of relative TEAD1 pulldown normalized to TEAD1 lysates strength and further normalization to strength of YAP-immunoprecipitation (n = 4). (B) qPCR shows decrease of relative mRNA expression of YAP/TAZ target genes in HEK293T cells transfected with siRNA NUP205 compared with control (n = 5). *Significant decrease by siRNA NUP205 (unpaired t-test; P < 0.05). (C) Proliferation rate of cultured podocytes by live-cell imaging over the period of 24 h. Knockdown of Nup205 or Yap inhibited the proliferation activity as compared with control cells. Representative images for cell confluency after 16 h. (D) Cell death assessed by live-cell imaging over the period of 24 h. Dead cells marked with DiYO-1. Basal cell death (untreated cells; dashed lines depicted in both left and right panels) and after treatment with either 100 μM H2O2 (lines, left panel) or 1 μg/ml ADR (lines, right panel). Lack of Nup205 or Yap resulted in decreased resistance to oxidative stress, when compared with scrambled controls. Live-cell images were captured every 2 h (n = 3–4 plates). Data are reported as mean ± SEM.
Downregulation of TAZ impairs NUP205-dependent nuclear shuttling of YAP
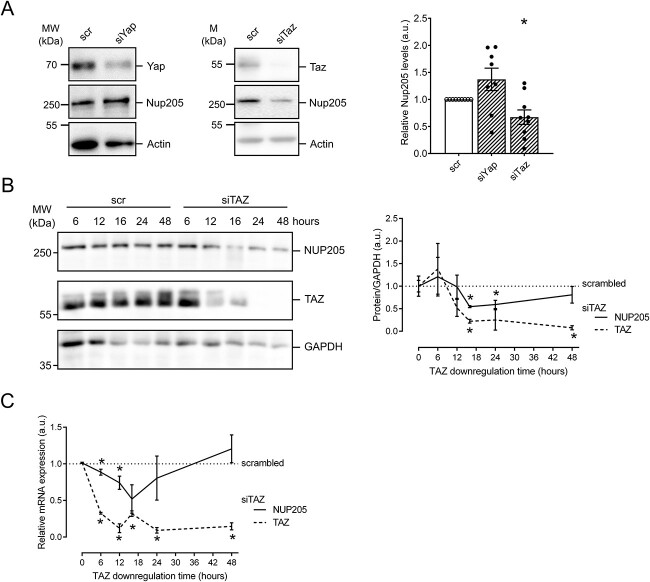
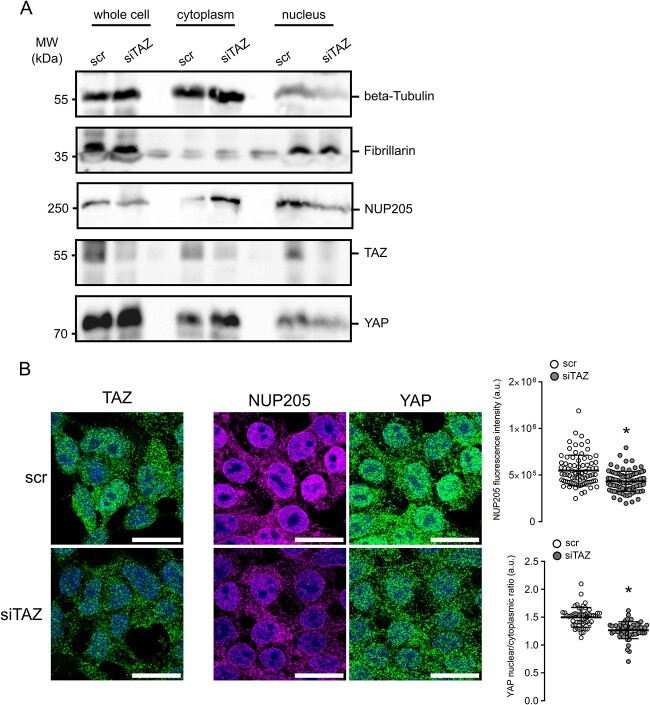
Protein complex partners can mutually affect expression levels, stability, and function. Therefore, we further analyzed the effect of YAP or TAZ knockdown on NUP205. In cultured podocytes, knockdown of Taz, but not Yap, resulted in a significant reduction of Nup205 expression after 48 h (Fig. 4A). To characterize the time course of this NUP205 reduction, we examined different instances of TAZ knockdown in HEK293T cells. Firstly, immunoblotting revealed the chronological sequence of TAZ depletion upon transfection with TAZ siRNA (Fig. 4B). While after 6 h of knockdown, TAZ protein levels were still equivalent to control, a strong reduction could be observed from 12 h on. A total depletion of TAZ was obtained after 24 h. The decrease of NUP205 expression levels followed TAZ downregulation with a stronger transient reduction after 16 h. Further qPCR analyses in HEK293T cells indicated that NUP205 serves as a transcriptional target gene of TAZ as evidenced by a transient decrease in relative mRNA expression of NUP205 following TAZ downregulation. Intriguingly, after 24 h, NUP205 mRNA levels showed an increase, suggesting the activation of a compensatory mechanism (Fig. 4C). To examine whether the downregulation of TAZ indirectly affected YAP subcellular localization through NUP205 downregulation, we again performed subcellular fractionation assays. We selected a period of 16 h for the TAZ knockdown because it resulted in the most efficient reduction of NUP205 expression. After 16 h of siRNA, not only TAZ but also NUP205 showed a reduction in protein expression in the whole-cell samples compared with control (Fig. 5A). Strikingly, NUP205 also showed a shift in subcellular localization from the nuclear to the cytoplasmatic fractions. YAP whole-cell expression levels were unaffected by the knockdown of TAZ, but it was retained in the cytoplasm, showing an increased expression in the cytoplasmic fraction while being reduced in the nucleus. This confirmed that TAZ was indispensable for proper NUP205 expression, which, in turn, affected the nuclear shuttling of YAP. To validate this effect, we examined the same conditions by immunofluorescence staining (Fig. 5B). Once again, downregulation of TAZ affected NUP205 expression levels, quantified by whole-cell fluorescence intensity. Notably, cells showing a reduced NUP205 expression also displayed a reduced nuclear expression of YAP, confirmed by assessing the nuclear/cytoplasmatic ratio. Taken together, the present results confirm the two-step regulation of YAP localization by TAZ through NUP205, the NPC protein essential for the shuttling of the Hippo pathway effector proteins.
Figure 4.
Lack of TAZ disturbs NUP205 expression. (A) Immunoblot showing whole-cell expression levels of endogenous NUP205 in hsMPs transfected with siRNA YAP or TAZ compared with control for 48 h. densitometry analysis of NUP205 expression normalized to actin (n = 8–9). Data are reported as mean ± SEM. *Significant decrease by siRNA TAZ (unpaired t-test; P < 0.05). (B) Immunoblot showing endogenous expression levels of NUP205, TAZ, and GAPDH, in HEK293T cells transfected with siRNA TAZ compared with control, after 6, 12, 16, and 24 h. Representative blot for n = 3. Quantification of relative TAZ (dashed) and NUP205 (line) whole-cell expression normalized to GAPDH (n = 3). (C) qPCR shows decrease of relative mRNA expression of TAZ (dashed) and NUP205 (line) in HEK293T cells transfected with siRNA TAZ compared with control (n = 3). Representative data from three independent experiments are shown. Data are reported as mean ± SEM. *Significant decrease by siRNA TAZ (unpaired t-test; P < 0.05).
Figure 5.
Lack of TAZ alters YAP localization. (A) Immunoblot showing downregulation of NUP205 and the nuclear-to-cytoplasmatic shift of YAP upon downregulation of TAZ after 16 h in HEK293T cell fractionation assay. Representative blot for n = 3. (B) Immunofluorescence staining of HEK293T cells transfected with siRNA TAZ for 16 h compared with control. TAZ downregulation on the left. Co-staining for endogenous NUP205, endogenous YAP, and nuclear stain with DAPI. Representative data from three independent experiments are shown. All scale bars = 20 μm. Summary of correlated total cell fluorescence of NUP205 (n = 81–95 cells) and summary of YAP nuclear/cytoplasmatic ratio quantification (n = 53–57 cells). Data are reported as mean ± SEM. *Significant decrease by siRNA TAZ (unpaired t-test; P < 0.05).
Discussion
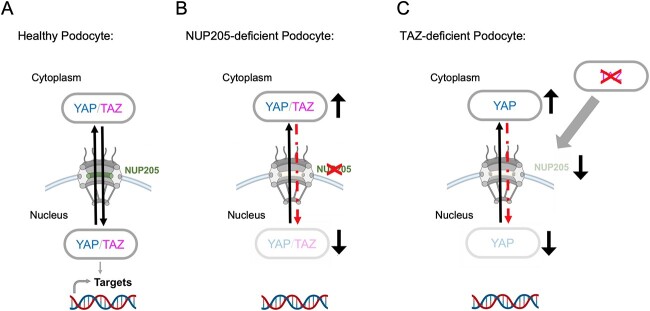
Mutations in genes encoding for components of the NPC have been identified to cause FSGS, whereas the exact role of the nuclear-shuttling machinery in podocytes remains elusive (8,12,34,40). NPCs regulate the transport of molecules shuttling from the cytoplasm to the nucleus, but have also been shown to be involved in transcriptional regulation (41). By exploring our podocyte interactome data of YAP and TAZ, we identify novel associations of NUPs and nuclear transport receptors (NTRs), like importins and exportins, with YAP and TAZ. This is captivating because YAP and TAZ activity or phosphorylation depends on smooth nucleocytoplasmic shuttling (14). So far, not much is known on this direct regulation of the nuclear translocation of YAP and TAZ, besides a dependency on NUP37 in hepatocellular carcinoma cells and oocytes, and a reported regulation of the nuclear pore size followed by the entry of YAP by mechanical forces (42–44). Further, Ipo7, which was also identified in our podocyte interactome data, has been shown to regulate the import of YAP in retinal pigment epithelium (RPE) cells (45). Among the newly identified nuclear-shuttling components of the YAP/TAZ complexes in our data are various proteins, mutations of which are known to cause FSGS, like XPO5, NUP107 and NUP133, and NUP205 (8,12). Of those, NUP205 showed the most robust enrichment in both protein complexes. Four different human mutations of NUP205 have been described so far to cause FSGS (6,8,46,47). One of the first studies that identified mutations in NUP205 or NUP93 to cause FSGS also described a role of NUP93 in the resistance to oxidative stress and the nuclear shuttling of SMAD (8). The consequences of NUP205 mutations on nuclear shuttling in podocytes, despite a reduced interaction with NUP93, had not been studied. Transport of proteins through the NPC is regulated by NUPs that are rich in phenylalanine-glycine repeats (FG-NUPs). FG-NUPs form liquid–liquid phase separates at the inner channel of the pore and impair free diffusion of macromolecules (48). They interact with NTRs to allow them and their cargo proteins to translocate rapidly in an energy-dependent manner (49). Theoretically, YAP and TAZ should, based on their size (55 kDa), be imported into the nucleus assisted by NTRs. However, YAP and TAZ lack a classic nuclear localization signal that is thought to be crucial for nuclear translocation coupled to NTRs (50). Interestingly, NUP205 can transport macromolecules independently of NTRs. NUP205 has been reported to interact directly with the arginine-rich motif (ARM) of adenovirus for its nuclear import (51). Moreover, Nup192 (NUP205 homolog in yeast) has been shown to carry structural and functional similarities with NTRs and can thus translocate proteins through intact NPCs in an NTR-independent way by facilitated diffusion (52). We postulate that NUP205 forms a protein complex with YAP and TAZ, allowing their nuclear import through translocation (Fig. 5B) of NUP205 within the pore as a dominant shuttling process in healthy podocytes (Fig. 6A).
Figure 6.
NUP205 is critical for nuclear shuttling of YAP in healthy podocytes. (A) In a healthy podocyte, NUP205 is located at the nuclear membrane as part of the nuclear pore complex where YAP and TAZ can shuttle between the cytoplasm and the nucleus. (B) In the diseased podocyte lacking proper NUP205 expression, the nuclear translocation of YAP and TAZ is disrupted. YAP and TAZ are “trapped” in the cytoplasm resulting in a reduced nuclear transcriptional activity. (C) TAZ expression is crucial for NUP205 expression in podocytes. Reduced TAZ expression leads to reduced NUP205 expression, which then results in a shift of YAP localization. Created with BioRender.com.
For podocyte homeostasis, nuclear localization and activity of YAP have been reported to be essential for protecting the podocyte from apoptosis (17,24). The importance of balanced YAP activity in podocytes is further supported by studies showing an initial increase of YAP activity followed by a decrease in rodent FSGS disease models (18,21,26). This temporary upregulation before the onset of disease suggests a transient protective role of YAP in the podocyte. Furthermore, increased activity of the Hippo Signaling cascade, that leads to an increased phosphorylation and cytosolic retention of YAP, consequently decreased its activity, led to disruption of the cytoskeletal integrity and podocyte morphology (53). Overall, YAP and TAZ seem to have a protective, antiapoptotic role, and their nuclear localization is pivotal for the healthy podocyte. Here, we show that NUP205 is not only required for the nuclear import of YAP and TAZ but is also crucial for their transcriptional activity in cooperation with TEAD1, affecting proliferation, protection from cell death and resistance to oxidative stress. Our results present a potential pathomechanism of genetic FSGS: when genetic mutations of NUP205 lead to a dysregulation of the indispensable nuclear shuttling and activity of YAP and TAZ, it results in loss of protective properties thus, podocytes injury (Fig. 6B).
Our identification of the association of NUPs with YAP and TAZ in podocytes led us to also investigate if YAP and TAZ influence the regulation of the nuclear import and export in podocytes. Recently, it was shown that YAP regulates Ipo7, an NTR, by monopolizing its nuclear import in response of mechanical cues in RPE cells (45). To our knowledge, our data describes for the first time the regulation of NUPs by components of the Hippo signaling pathway. We demonstrate that NUP205 expression is dependent on TAZ in podocytes. Moreover, our findings reveal an indirect decrease of YAP nuclear translocation by TAZ, suggesting a negative feedback regulation (Fig. 6C).
In conclusion, our findings highlight the important role of YAP/TAZ signaling in podocyte homeostasis by connecting the shuttling of YAP and TAZ to a nucleoporin, known to cause FSGS. Our work identifies a novel interdependency between transcription factors and NPCs that highlights new specific therapeutic targets for FSGS like stabilizing nuclear YAP activity in podocytes.
Supplementary Material
Acknowledgements
We would like to thank Angelika Köser for expert technical assistance. We acknowledge the help of the CECAD imaging facility and the CECAD proteomics facility. In addition, we would like to express our gratitude to all members of our laboratory for helpful discussions and support.
Conflict of Interest statement. None declared.
Contributor Information
Lioba Ester, Department II of Internal Medicine and Center for Molecular Medicine Cologne, University of Cologne, Faculty of Medicine and University Hospital Cologne, Kerpener Str. 62, 50937 Cologne, Germany; Cologne Excellence Cluster on Cellular Stress Responses in Aging-Associated Diseases (CECAD), University of Cologne, Faculty of Medicine and University Hospital Cologne, Joseph-Stelzmann-Str. 26, 50931 Cologne, Germany.
Inês Cabrita, Department II of Internal Medicine and Center for Molecular Medicine Cologne, University of Cologne, Faculty of Medicine and University Hospital Cologne, Kerpener Str. 62, 50937 Cologne, Germany; Cologne Excellence Cluster on Cellular Stress Responses in Aging-Associated Diseases (CECAD), University of Cologne, Faculty of Medicine and University Hospital Cologne, Joseph-Stelzmann-Str. 26, 50931 Cologne, Germany.
Michel Ventzke, Department II of Internal Medicine and Center for Molecular Medicine Cologne, University of Cologne, Faculty of Medicine and University Hospital Cologne, Kerpener Str. 62, 50937 Cologne, Germany; Cologne Excellence Cluster on Cellular Stress Responses in Aging-Associated Diseases (CECAD), University of Cologne, Faculty of Medicine and University Hospital Cologne, Joseph-Stelzmann-Str. 26, 50931 Cologne, Germany.
Emilia Kieckhöfer, Department II of Internal Medicine and Center for Molecular Medicine Cologne, University of Cologne, Faculty of Medicine and University Hospital Cologne, Kerpener Str. 62, 50937 Cologne, Germany; Cologne Excellence Cluster on Cellular Stress Responses in Aging-Associated Diseases (CECAD), University of Cologne, Faculty of Medicine and University Hospital Cologne, Joseph-Stelzmann-Str. 26, 50931 Cologne, Germany.
Marita Christodoulou, Department II of Internal Medicine and Center for Molecular Medicine Cologne, University of Cologne, Faculty of Medicine and University Hospital Cologne, Kerpener Str. 62, 50937 Cologne, Germany; Cologne Excellence Cluster on Cellular Stress Responses in Aging-Associated Diseases (CECAD), University of Cologne, Faculty of Medicine and University Hospital Cologne, Joseph-Stelzmann-Str. 26, 50931 Cologne, Germany.
Amrei M Mandel, Department II of Internal Medicine and Center for Molecular Medicine Cologne, University of Cologne, Faculty of Medicine and University Hospital Cologne, Kerpener Str. 62, 50937 Cologne, Germany; Cologne Excellence Cluster on Cellular Stress Responses in Aging-Associated Diseases (CECAD), University of Cologne, Faculty of Medicine and University Hospital Cologne, Joseph-Stelzmann-Str. 26, 50931 Cologne, Germany.
Paul Diefenhardt, Department II of Internal Medicine and Center for Molecular Medicine Cologne, University of Cologne, Faculty of Medicine and University Hospital Cologne, Kerpener Str. 62, 50937 Cologne, Germany; Cologne Excellence Cluster on Cellular Stress Responses in Aging-Associated Diseases (CECAD), University of Cologne, Faculty of Medicine and University Hospital Cologne, Joseph-Stelzmann-Str. 26, 50931 Cologne, Germany.
Francesca Fabretti, Department II of Internal Medicine and Center for Molecular Medicine Cologne, University of Cologne, Faculty of Medicine and University Hospital Cologne, Kerpener Str. 62, 50937 Cologne, Germany; Cologne Excellence Cluster on Cellular Stress Responses in Aging-Associated Diseases (CECAD), University of Cologne, Faculty of Medicine and University Hospital Cologne, Joseph-Stelzmann-Str. 26, 50931 Cologne, Germany.
Thomas Benzing, Department II of Internal Medicine and Center for Molecular Medicine Cologne, University of Cologne, Faculty of Medicine and University Hospital Cologne, Kerpener Str. 62, 50937 Cologne, Germany; Cologne Excellence Cluster on Cellular Stress Responses in Aging-Associated Diseases (CECAD), University of Cologne, Faculty of Medicine and University Hospital Cologne, Joseph-Stelzmann-Str. 26, 50931 Cologne, Germany.
Sandra Habbig, Department of Pediatrics, University of Cologne, Faculty of Medicine and University Hospital Cologne, Kerpener Str. 62, 50937 Cologne, Germany.
Bernhard Schermer, Department II of Internal Medicine and Center for Molecular Medicine Cologne, University of Cologne, Faculty of Medicine and University Hospital Cologne, Kerpener Str. 62, 50937 Cologne, Germany; Cologne Excellence Cluster on Cellular Stress Responses in Aging-Associated Diseases (CECAD), University of Cologne, Faculty of Medicine and University Hospital Cologne, Joseph-Stelzmann-Str. 26, 50931 Cologne, Germany.
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeX change Consortium via the PRIDE partner repository with the data set identifier PXD040448 (54).
Data sharing statement
All data are available from the authors upon request.
Funding
This work was supported by the German Research Foundation (DFG) in the framework of the clinical research unit 329 (KFO 329; RP7 to S.H./B.S and RP1 to T.B.). L.E. and M.V. was supported by the Koeln Fortune program/Faculty of Medicine, University of Cologne.
Author contributions
L.E., I.C., F.F., B.S., S.H., and T.B. designed the study; L.E, I.C., E.K., M.V., and M.C. carried out experiments; L.E., I.C., A.M.M., P.D. and E.K. analyzed the data; L.E. made the figures; L.E. and I.C. drafted the paper; all authors approved the final version of the manuscript.
References
- 1. Hogg R, Middleton J, Vehaskari VM. Focal segmental glomerulosclerosis--epidemiology aspects in children and adults. Pediatr Nephrol 2007;22:183–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Benzing T, Salant D. Insights into glomerular filtration and albuminuria. N Eng. J Med 2021;384:1437–46. [DOI] [PubMed] [Google Scholar]
- 3. Brinkkoetter PT, Ising C, Benzing T. The role of the podocyte in albumin filtration. Nat Rev Nephrol 2013;9:328–36. [DOI] [PubMed] [Google Scholar]
- 4. Assady S, Benzing T, Kretzler M. et al. Glomerular podocytes in kidney health and disease. Lancet 2019;393:856–8. [DOI] [PubMed] [Google Scholar]
- 5. Kopp JB, Anders HJ, Susztak K. et al. Podocytopathies. Nat Rev Dis Primers 2020;6:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Warejko JK, Tan W, Daga A. et al. Whole exome sequencing of patients with steroid-resistant nephrotic syndrome. Clin J Am Soc Nephrol 2018;13:53–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li AS, Ingham JF, Lennon R. Genetic disorders of the glomerular filtration barrier. Clin J Am Soc Nephrol 2020;15:1818–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Braun DA, Sadowski CE, Kohl S. et al. Mutations in nuclear pore genes NUP93, NUP205 and XPO5 cause steroid-resistant nephrotic syndrome. Nat Genet 2016;48:457–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Stewart M. Function of the nuclear transport machinery in maintaining the distinctive compositions of the nucleus and cytoplasm. Int J Mol Sci 2022;23:2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Beck M, Hurt E. The nuclear pore complex: understanding its function through structural insight. Nat Rev Mol Cell Biol 2017;18:73–89. [DOI] [PubMed] [Google Scholar]
- 11. Matsuda A, Mofrad MRK. On the nuclear pore complex and its emerging role in cellular mechanotransduction. APL Bioeng 2022;6:011504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Braun DA, Lovric S, Schapiro D. et al. Mutations in multiple components of the nuclear pore complex cause nephrotic syndrome. J Clin Invest 2018;128:4313–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hussain HMJ, Cai Y, Weng Q. et al. Mutation in XPO5 causes adult-onset autosomal dominant familial focal segmental glomerulosclerosis. Hum Genomics 2022;16:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Manning SA, Kroeger B, Harvey KF. The regulation of Yorkie, YAP and TAZ: new insights into the hippo pathway. Development 2020;147:dev179069. [DOI] [PubMed] [Google Scholar]
- 15. Zhao B, Ye X, Yu J. et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev 2008;22:1962–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Moya IM, Halder G. Hippo-YAP/TAZ signalling in organ regeneration and regenerative medicine. Nat Rev Mol Cell Biol 2019;20:211–26. [DOI] [PubMed] [Google Scholar]
- 17. Campbell KN, Wong JS, Gupta R. et al. Yes-associated protein (YAP) promotes cell survival by inhibiting proapoptotic dendrin signaling. J Biol Chem 2013;288:17057–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhuang Q, Li F, Liu J. et al. Nuclear exclusion of YAP exacerbates podocyte apoptosis and disease progression in Adriamycin-induced focal segmental glomerulosclerosis. Lab Investig 2021;101:258–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schwartzman M, Reginensi A, Wong JS. et al. Podocyte-specific deletion of yes-associated protein causes FSGS and progressive renal failure. J Am Soc Nephrol 2016;27:216–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen J, Wang X, He Q. et al. TAZ is important for maintenance of the integrity of podocytes. Am J Physiol Renal Physiol 2022;322:F419–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rinschen MM, Grahammer F, Hoppe AK. et al. YAP-mediated mechanotransduction determines the podocyte's response to damage. Sci Signal 2017;10:eaaf8165. [DOI] [PubMed] [Google Scholar]
- 22. Keyvani Chahi A, Martin CE, Jones N. Nephrin suppresses hippo signaling through the adaptor proteins Nck and WTIP. J Biol Chem 2016;291:12799–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kann M, Ettou S, Jung YL. et al. Genome-wide analysis of Wilms' tumor 1-controlled gene expression in podocytes reveals key regulatory mechanisms. J Am Soc Nephrol 2015;26:2097–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bonse J, Wennmann DO, Kremerskothen J. et al. Nuclear YAP localization as a key regulator of podocyte function. Cell Death Dis 2018;9:850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wennmann DO, Vollenbroker B, Eckart AK. et al. The hippo pathway is controlled by angiotensin II signaling and its reactivation induces apoptosis in podocytes. Cell Death Dis 2014;5:e1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chung JJ, Goldstein L, Chen YJ. et al. Single-cell transcriptome profiling of the kidney glomerulus identifies key cell types and reactions to injury. J Am Soc Nephrol 2020;31:2341–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Griffin SV, Hiromura K, Pippin J. et al. Cyclin-dependent kinase 5 is a regulator of podocyte differentiation, proliferation, and morphology. Am J Pathol 2004;165:1175–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Perez-Pinera P, Ousterout DG, Brown MT. et al. Gene targeting to the ROSA26 locus directed by engineered zinc finger nucleases. Nucleic Acids Res 2012;40:3741–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kasparek P, Krausova M, Haneckova R. et al. Efficient gene targeting of the Rosa26 locus in mouse zygotes using TALE nucleases. FEBS Lett 2014;588:3982–88. [DOI] [PubMed] [Google Scholar]
- 30. Borgal L, Habbig S, Hatzold J. et al. The ciliary protein nephrocystin-4 translocates the canonical Wnt regulator Jade-1 to the nucleus to negatively regulate β-catenin signaling. J Biol Chem 2012;287:25370–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schindelin J, Arganda-Carreras I, Frise E. et al. Fiji: an open-source platform for biological-image analysis. Nat Methods 2012;9:676–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta C(T)) method. Methods 2001;25:402–8. [DOI] [PubMed] [Google Scholar]
- 33. Rinschen MM, Godel M, Grahammer F. et al. A multi-layered quantitative in vivo expression atlas of the podocyte unravels kidney disease candidate genes. Cell Rep 2018;23:2495–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rogg M, Maier JI, Ehle M. et al. NUP133 controls nuclear pore assembly, transcriptome composition, and cytoskeleton regulation in podocytes. Cell 2022;11: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Grandi P, Dang T, Pane N. et al. Nup93, a vertebrate homologue of yeast Nic96p, forms a complex with a novel 205-kDa protein and is required for correct nuclear pore assembly. Mol Biol Cell 1997;8:2017–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chen X, Xu L. Specific nucleoporin requirement for Smad nuclear translocation. Mol Cell Biol 2010;30:4022–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Calvo F, Ege N, Grande-Garcia A. et al. Mechanotransduction and YAP-dependent matrix remodelling is required for the generation and maintenance of cancer-associated fibroblasts. Nat Cell Biol 2013;15:637–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dupont S, Morsut L, Aragona M. et al. Role of YAP/TAZ in mechanotransduction. Nature 2011;474:179–83. [DOI] [PubMed] [Google Scholar]
- 39. Yuen HF, McCrudden CM, Huang YH. et al. TAZ expression as a prognostic indicator in colorectal cancer. PLoS One 2013;8:e54211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rosti RO, Sotak BN, Bielas SL. et al. Homozygous mutation in NUP107 leads to microcephaly with steroid-resistant nephrotic condition similar to Galloway-Mowat syndrome. J Med Genet 2017;54:399–403. [DOI] [PubMed] [Google Scholar]
- 41. D'Angelo MA. Nuclear pore complexes as hubs for gene regulation. Nucleus 2018;9:142–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Luo X, Liu Y, Feng W. et al. NUP37, a positive regulator of YAP/TEAD signaling, promotes the progression of hepatocellular carcinoma. Oncotarget 2017;8:98004–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Guo Q, Liu Q, Wang N. et al. The function of nucleoporin 37 on mouse oocyte maturation and preimplantation embryo development. J Assist Reprod Genet 2022;39:107–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Elosegui-Artola A, Andreu I, Beedle AEM. et al. Force triggers YAP nuclear entry by regulating transport across nuclear pores. Cell 2017;171:1397–1410.e1314. [DOI] [PubMed] [Google Scholar]
- 45. Garcia-Garcia M, Sanchez-Perales S, Jarabo P. et al. Mechanical control of nuclear import by Importin-7 is regulated by its dominant cargo YAP. Nat Commun 2022;13:1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Najafi M, Riedhammer KM, Rad A. et al. High detection rate for disease-causing variants in a cohort of 30 Iranian pediatric steroid resistant nephrotic syndrome cases. Front Pediatr 2022;10:974840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rao J, Liu X, Mao J. et al. Genetic spectrum of renal disease for 1001 Chinese children based on a multicenter registration system. Clin Genet 2019;96:402–10. [DOI] [PubMed] [Google Scholar]
- 48. Celetti G, Paci G, Caria J. et al. The liquid state of FG-nucleoporins mimics permeability barrier properties of nuclear pore complexes. J Cell Biol 2020;219:e201907157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Timney BL, Raveh B, Mironska R. et al. Simple rules for passive diffusion through the nuclear pore complex. J Cell Biol 2016;215:57–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang S, Lu Y, Yin MX. et al. Importin alpha1 mediates Yorkie nuclear import via an N-terminal non-canonical nuclear localization signal. J Biol Chem 2016;291:7926–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lu Y, Kucharski TJ, Gamache I. et al. Interaction of adenovirus type 5 E4orf4 with the nuclear pore subunit Nup205 is required for proper viral gene expression. J Virol 2014;88:13249–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Andersen KR, Onischenko E, Tang JH. et al. Scaffold nucleoporins Nup188 and Nup192 share structural and functional properties with nuclear transport receptors. eLife 2013;2:e00745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Meliambro K, Wong JS, Ray J. et al. The Hippo pathway regulator KIBRA promotes podocyte injury by inhibiting YAP signaling and disrupting actin cytoskeletal dynamics. J Biol Chem 2017;292:21137–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Perez-Riverol Y, Bai J, Bandla C. et al. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res 2022;50:D543–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeX change Consortium via the PRIDE partner repository with the data set identifier PXD040448 (54).