Abstract
Recent studies have shown that the health benefits of probiotics are not limited to those offered by living bacteria. It was reported that both live and killed cells of Propionibacterium freudenreichii MJ2 (MJ2) isolated from raw milk showed antiobesity activity in 3T3-L1 cells and high-fat diet-induced obese mice. This study was aimed at identifying the active component(s) responsible for the antiadipogenic activity of MJ2. Cell wall, surface protein, and cytoplasmic fractions of MJ2 were investigated for their inhibitory effects on adipogenesis in 3T3-L1 cells. Adipocytes treated with the surface protein fraction showed significantly lower lipid accumulation. Using the MASCOT algorithm following LC-MS/MS analysis, 131 surface proteins were identified and they were principally classified into three categories (network clusters related to ribosomes, carbon metabolism, and chaperones). Among them, chaperonin 60 (Cpn60) was selected as a potential candidate protein. Cpn60 inhibited lipid accumulation and adipogenesis during the early period of differentiation (days 0–2) and decreased expression of genes related to adipogenesis (Pparg and Cebpa) and lipogenesis (Fas and Scd1). The expression of Gata2/3, which suppresses adipogenesis, significantly increased in Cpn60-treated cells. Moreover, the nuclear translocation of C/EBPβ was inhibited by Cpn60 treatment. In conclusion, Cpn60, a surface protein in MJ2, shows antiadipogenic activity by reducing the expression of C/EBPβ through the upregulation of Gata2/3 expression followed by downregulation of Pparg and Cebpa expression.
Subject terms: Cell biology, Microbiology
Health concerns related to obesity are increasing worldwide because of the correlation between obesity and serious metabolic syndromes, such as type 2 diabetes, nonalcoholic fatty liver diseases, and insulin resistance1. Obesity, due to an imbalance in the generation and expenditure of energy, is caused by remodeling of the adipose tissue, which is mainly composed of mature adipocytes and various other cells, such as preadipocytes, mesenchymal stromal or stem cells, immune cells, and vascular epithelial cells2. Adipogenesis is the development of adipocytes from mesenchymal stem cells (MSCs). Hypertrophy and hyperplasia of adipocytes result in the development of obesity. Therefore, novel molecules or mechanisms that inhibit adipogenesis are required to prevent or treat obesity3. Adipogenesis from multipotent MSCs proceeds through the following stages: MSC is committed to the adipocyte lineage, mitotic clonal expansion (MCE), which replicates DNA and duplicates cells, and terminal differentiation stage in which major transcription factors related to adipogenesis, namely peroxisome proliferator activated receptor γ (PPARγ) and CCAAT/enhancer-binding protein α (C/EBPα) and genes related to lipogenesis, such as fatty acid synthase (FAS) and stearoyl-CoA desaturase (SCD1), are expressed4. These processes are regulated by a complex network of transcription factors and signaling pathways5. The master adipogenic regulators, PPARγ and C/EBPα, are induced to an epigenomic transition state to lead a transcriptional cascade during the early stage of cellular differentiation6.
The health-promoting effects of probiotics are no longer considered to be limited to live microorganisms. Postbiotics, whose consumption is beneficial, include components, such as metabolites (proteins, peptides, lipids, organic acids, and enzymes) produced by probiotics, bacterial components (cell surface proteins, lipoteichoic acids, teichoic acid, peptidoglycan, and polysaccharides), cell-free supernatants, and lysed bacteria7. The beneficial properties of postbiotics have led to various clinical applications, such as in the fields of immunomodulation and antitumor interventions, with advantages related to production technologies and safety vis-à-vis live probiotics8. Propionate, a short-chain fatty acid produced by Propionibacterium freudenreichii, induces apoptosis in human gastric cancer cells9. The surface proteins (SP) of P. freudenreichii show immunomodulatory effects by stimulating the expression of the anti-inflammatory cytokine, IL-1010. SlpB, a surface-layer protein B of P. freudenreichii, is involved in the adhesion of probiotic cells to human colon cells, which increases probiotic-host interactions11.
P. freudenreichii MJ2 (MJ2), isolated from raw milk, has been investigated for its beneficial physiological activities against obesity, osteoporosis, and rheumatoid arthritis12–14. However, the key components of MJ2 responsible for these beneficial effects have not been identified. Similarly, administration of live or heat-killed MJ2 reduced weight gain and insulin resistance in high-fat diet-induced obese mice12. Based on our previous study, we hypothesized that MJ2 may function as a postbiotic and may have roles beyond those as a probiotic bacterium. In this study, we identified an active component of MJ2 with antiadipogenic activity as a postbiotic and deciphered the mechanism underlying its inhibition of differentiation of 3T3-L1 preadipocytes into adipocytes.
Results
Effect of the various cell fractions from P. freudenreichii MJ2 on the viability of 3T3-L1 preadipocytes
3T3-L1 cells were treated with each MJ2 cell fraction from 1 × 108 cells/mL of MJ2 or heat-killed P. freudenreichii MJ2 (hkMJ2) for 24 h, and cell viability was measured using the 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT) assay. There was no significant decrease in the viability of cells treated with the various MJ2 fractions or hkMJ2 (Supplementary Fig. 1).
Surface proteins from MJ2 contribute to the inhibition of lipid accumulation during the differentiation of 3T3-L1
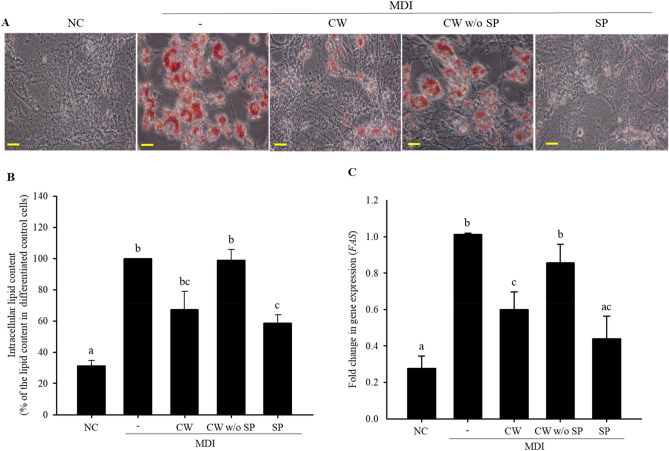
To investigate the effect of each MJ2 cell fraction on lipid accumulation, 3T3-L1 cells were treated with hkMJ2, or cell wall (CW) or cytoplasmic (Cyto) fractions during the induction of cell differentiation. Lipid accumulation decreased in hkMJ2- and CW-treated cells compared with that in differentiated control cells treated only with MDI (3-isobutyl-1-methylxanthine, dexamethasone, and insulin) (Supplementary Fig. 2), which indicates that the active component(s) that decrease lipid accumulation during adipocyte differentiation might be present in the CW. Thus, we measured lipid accumulation in cells treated with CW, CW without surface proteins (CW w/o SP), or SP fractions to specifically identify the active fraction. We found that the SP fraction of CW significantly inhibited lipid accumulation (Fig. 1A,B). In addition, the expression level of the lipogenesis factor, Fas, significantly decreased in CW- and SP-treated cells, but not in CW w/o SP-treated cells, compared with that in control cells that were treated only with MDI (Fig. 1C). These results suggest that the candidate active component(s) are present in the SP fraction.
Figure 1.
Inhibitory effects of various cell fractions of Propionibacterium freudenreichii MJ2 on lipid accumulation in 3T3-L1 cells. 3T3-L1 cells were treated with different cell extracts: cell wall (CW), cell wall without surface proteins (CW w/o SP), and surface proteins (SP) from 1 × 108 P. freudenreichii MJ2 cells/mL. (A) Lipid droplets in differentiated cells were visualized using Oil Red O staining (× 400 magnification). Scale bars indicate 0.5 mm. (B) Lipid accumulation was quantified and compared with that in cells treated only with MDI. (C) Fold change in the gene expression of lipogenic transcription factor, Fas. The data indicate the mean ± SD of values from three independent experiments. The p values were determined using ANOVA and Tukey’s honest significant difference test. Different letters indicate statistically significant differences (p < 0.05). NC, negative control; MDI, 1 μM dexamethasone, 0.5 mM IBMX, and 5 μg/mL insulin.
Characterization of SP isolated from MJ2
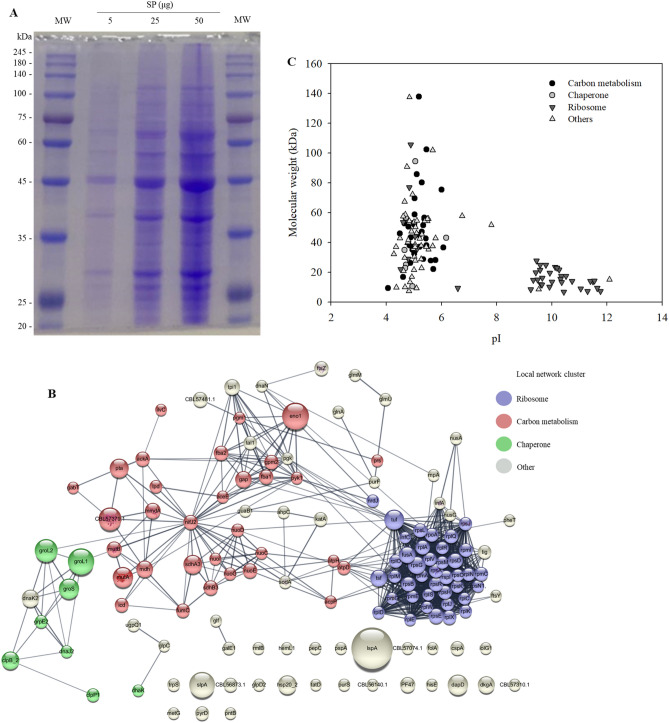
We analyzed the SP fraction using sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE; Fig. 2A), followed by peptide analysis using nano LC-MS/MS analysis. A total of 131 proteins were identified in the SP fraction of MJ2 (Supplementary Table 1). The molecular weight of these proteins was in the range of 7–138 kDa and the pI was in the range of 4.07–12.09 (Fig. 2B). The results of metabolomic pathway analysis, and Kyoto encyclopedia of genes and genomes (KEGG) classification and gene ontology (GO) analysis of proteins are provided in Supplementary Tables 2 and 3, respectively. The results showed the presence of several characteristic SPs, which were principally classified into three categories (network clusters related to ribosomes, carbon metabolism, and chaperones) (Fig. 2C).
Figure 2.
Characterization of surface proteins (SP) isolated from Propionibacterium freudenreichii MJ2 using 5 M guanidine hydrochloride. (A) SP (5‒50 μg) were separated by electrophoresis on 10% sodium dodecyl sulfate–polyacrylamide gel. The full-length SDS-PAGE image of SP is shown in Supplementary Fig. 3. (B) STRING analysis of interaction networks among SP. Proteins are presented as nodes connected by edges. The size of nodes represents the protein score analyzed using the MASCOT algorithm and the color of the node indicates the local network cluster defined using STRING. The interactions of proteins were assessed under a high confidence cutoff (0.7). Purple, red, and green nodes indicate the network clusters related to ribosome, carbon metabolism, and chaperone, respectively. The thickness of edges indicates the confidence level of protein interaction score. (C) Molecular weight and isoelectric point (pI) of the identified proteins.
Chaperonin 60, an MJ2 SP inhibits lipid accumulation in 3T3-L1 cells
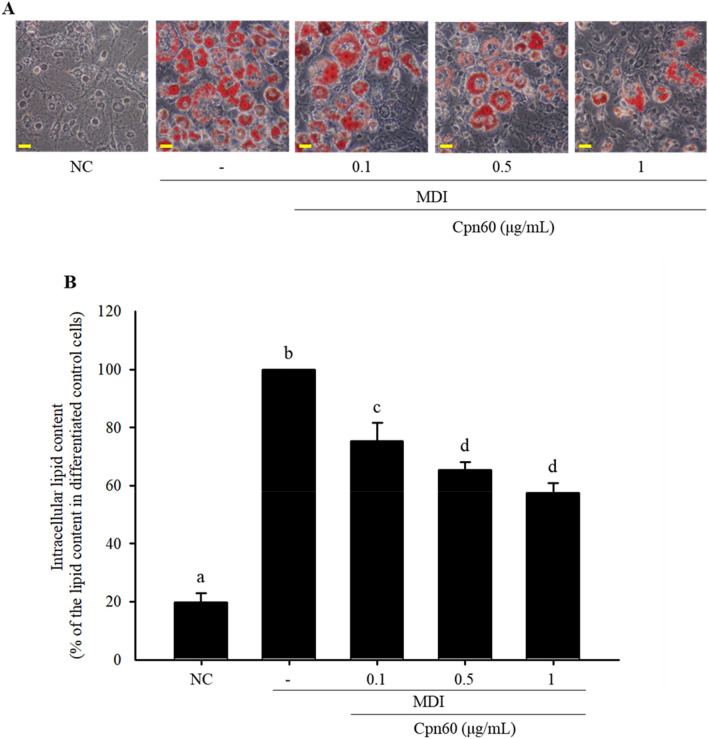
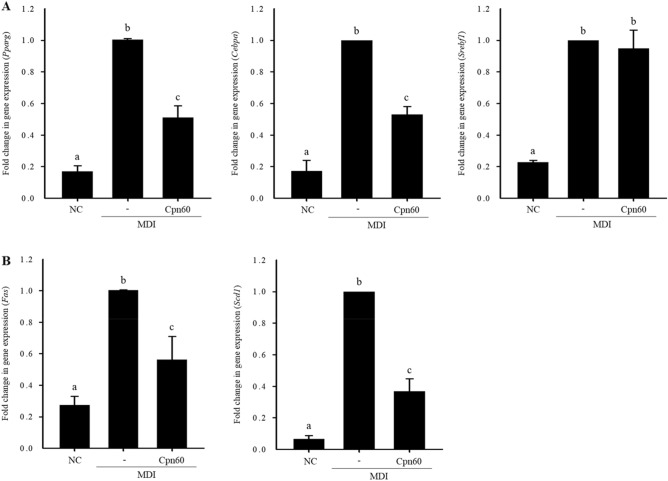
To select an active candidate for anti-adipogenesis, we fractionated the SP fraction by ion-exchange chromatography using a Q fast column, an anion exchange column, and size exclusive chromatography and investigated the effect of each fraction on lipid accumulation. The fraction strongly bound to Q fast column significantly decreased lipid accumulation, which means that the candidate protein(s) is acidic with relatively low pI (Supplementary Fig. 4). Next, size exclusion chromatography using Superdex 75 was carried out. The results showed that proteins with relatively small molecular weight (less than 20) did not affect lipid accumulation. Thus, we assumed that the active component(s) in SP might be an acidic protein with a molecular weight of more than 35 kDa. Therefore, chaperonin 60 (Cpn60) was selected as a potent active component among the proteins in the SP faction. We synthesized recombinant Cpn60 (Supplementary Fig. 5) and investigated its antiadipogenic effects. Accumulation of lipids in Cpn60-treated 3T3-L1 cells was significantly reduced in a dose-dependent manner compared with that in 3T3-L1 cells treated only with MDI (Fig. 3). The expression of genes related to adipogenesis (Pparg, Cebpa, and Srebf1) and lipogenesis (Fas and Scd1) was measured to confirm the effect of Cpn60 on the inhibition of adipogenesis. In 3T3-L1 cells treated with 1 μg/mL of Cpn60, the expression of Pparg, Cepba, Fas, and Scd1 was significantly decreased compared with that in control cells treated only with MDI (Fig. 4). However, the expression of Srepb1 did not differ between Cpn60-treated 3T3-L1 cells and control cells. These results showed that Cpn60 may be a potential protein component that exerts antiobesity effects by inhibiting adipogenesis.
Figure 3.
Inhibitory effect of chaperonin 60 (Cpn60) on lipid accumulation in 3T3-L1 adipocytes. 3T3-L1 cells were differentiated using MDI and treated with various concentrations of Cpn60. (A) Lipid droplets in differentiated cells were visualized using Oil Red O staining (× 400 magnification). Scale bars indicate 0.5 mm. (B) Intracellular lipid content was quantified and compared with that in cells treated only with MDI. The data indicate the mean ± SD of values from three independent experiments. The p values were determined using ANOVA and Tukey’s honest significant difference test. Different letters indicate statistically significant differences (p < 0.05). NC, negative control; MDI, 1 μM dexamethasone, 0.5 mM IBMX, and 5 μg/mL insulin.
Figure 4.
Inhibitory effect of chaperonin 60 (Cpn60) on the expression of genes related to adipogenesis and lipogenesis in 3T3-L1 adipocytes. 3T3-L1 cells were cultured with MDI or MDI + 1 μg/mL Cpn60 and mRNA expression of genes was analyzed using qPCR. Fold change in the expression of adipogenic transcription factor (Pparg, Cebpa, and Srebf1) (A) and lipogenic transcription factor (Fas and Scd1) genes (B). The data indicate the mean ± SD of values from three independent experiments. The p values were determined using ANOVA and Tukey’s honest significant difference test. Different letters indicate statistically significant differences (p < 0.05). NC negative control, MDI, 1 μM dexamethasone, 0.5 mM IBMX, and 5 μg/mL insulin.
Antiadipogenic effect of chaperonin 60 in 3T3-L1 cells during different periods of differentiation
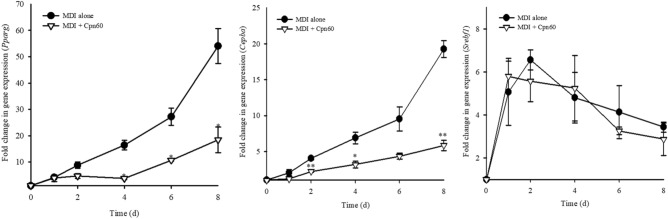
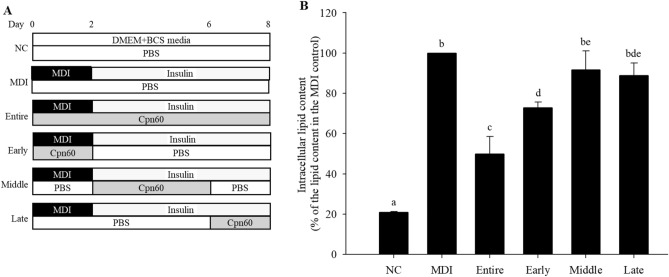
To determine the inhibitory effect of Cpn60 on adipogenesis during the different periods of differentiation, we measured the expression of Pparg, Cebpa, and Srebf1 every 2 days during the differentiation period (day 8). The expression of Pparg and Cebpa in Cpn60-treated cells decreased significantly starting day 4 and day 2, respectively, compared with that in control cells treated only with MDI. In contrast, the expression of Srebf1 did not differ between Cpn60-treated and control cells during the entire period (Fig. 5). These results are consistent with the gene expression results shown in Fig. 4. Adipogenesis master transcription factors, PPARγ and C/EBPα, act during the early period of cell differentiation, whereas SREBP1 is expressed relatively during the later stages of adipogenesis. We measured lipid accumulation in the different periods of differentiation by treating cells with Cpn60 during the early (day 0‒2), intermediate (day 2‒6), and late (day 6‒8) periods as well as during the entire differentiation period (entire) (Fig. 6A). The accumulation of lipid droplets in Cpn60-traeated cells was significantly decreased during the entire and early periods compared with that in the control cells treated only with MDI (Fig. 6B). The results suggest that Cpn60 inhibited adipogenesis by reducing the activity of the master transcription factors, especially PPARγ and C/EBPα, in early period of adipogenesis.
Figure 5.
Effect of chaperonin 60 (Cpn60) on the expression of genes related to adipogenesis (Pparg, Cebpa, and Srebf1) in 3T3-L1 adipocytes during different periods of differentiation. The cells were cultured with MDI or MDI + 1 μg/mL Cpn60 for 2, 4, 6, and 8 days and the expression of genes was analyzed using qPCR. The data indicate the mean ± SD of values from three independent experiments. The p values were determined using ANOVA and Tukey’s honest significant difference test. Differences were considered significant at p < 0.05 (*) and p < 0.01 (**). MDI, 1 μM dexamethasone, 0.5 mM IBMX, and 5 μg/mL insulin.
Figure 6.
Effect of chaperonin 60 (Cpn60) on lipid accumulation during different periods of treatment of 3T3-L1 cells with Cpn60. 3T3-L1 cells were differentiated by treatment with MDI or MDI + 1 μg/mL Cpn60 for different periods during differentiation (A). Lipid accumulation was quantified on day 8 by measuring lipid droplets stained with Oil Red O and comparing them with that in cells treated only with MDI (B). The data indicate the mean ± SD of values from three independent experiments. The p values were determined using ANOVA and Tukey’s honest significant difference test. Different letters indicate statistically significant differences (p < 0.05). NC, negative control; MDI, 1 μM dexamethasone, 0.5 mM IBMX, and 5 μg/mL insulin.
Chaperonin 60 elevates the gene expression of GATA-binding factor 2 and 3 (Gata2 and Gata3) and inhibits transcriptional activity of C/EPBβ
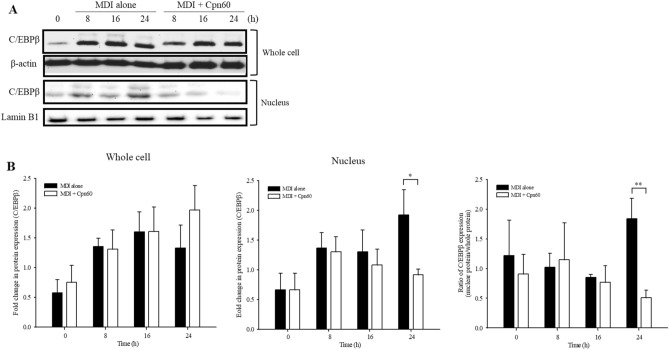
The expression and activity of PPARγ and C/EBPα, the major regulators of adipogenesis, are regulated through various signaling pathways2,6,15,16. We found that the expression of Pparg and Cebpa was regulated in the early period of adipogenesis upon treatment of 3T3-L1 cells with Cpn60. C/EBPβ is the major transcription factor that initiates adipogenesis induced by MDI and induces the expression of Pparg and Cebpa. Active C/EBPβ is translocated to the nucleus and induces the transcription of target genes17. To investigate the effect of Cpn60 on the nuclear translocation of C/EBPβ, the expression of C/EBPβ was measured during the early period of adipogenesis. The expression of C/EBPβ did not show any significant difference between control and Cpn60-treated cells during the initial 24 h (Fig. 7). However, the level of C/EBPβ in the nucleus was significantly decreased in Cpn60-treated cells at 24 h compared with that in control cells treated only with MDI.
Figure 7.
Effects of chaperonin 60 (Cpn60) on the activation of C/EBPβ. 3T3-L1 cells were cultured with MDI or MDI + 1 μg/mL Cpn60 for 0, 8, 16, and 24 h and total and nuclear proteins were extracted. The relative protein levels of C/EBPβ were assessed using western blot analysis (A) and quantified densitometrically (B). The p values were determined using the Student’s t-test. Differences were considered significant at p < 0.05 (*) and p < 0.01 (**). The full-length images of the blots are shown in Supplementary Fig. 7. MDI, 1 μM dexamethasone, 0.5 mM IBMX, and 5 μg/mL insulin.
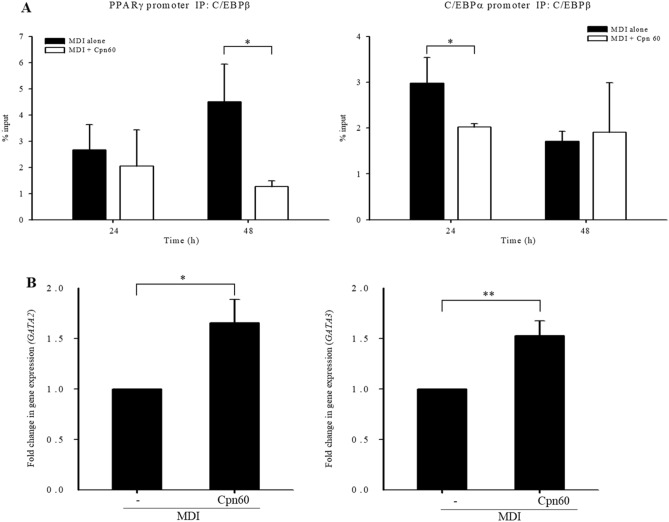
To evaluate the ability of C/EBPβ to bind to the promoter of major factors inducing adipogenesis, Pparg and Cebpa, we performed chromatin immunoprecipitation (ChIP) using extract of cells treated with MDI or Cpn60 for 24 or 48 h with an anti-C/EBPβ antibody. The percent input bound on the C/EBPα and PPARγ promoters was significantly decreased in cells treated with Cpn60 for 24 and 48 h, respectively (Fig. 8A).
Figure 8.
Binding of C/EBPβ on PPARγ or C/EBPα promoter and the expression of Gata2 and Gata3 investigated using chromatin immunoprecipitation (ChIP) assay. The cells were cultured with MDI or MDI + 1 μg/mL Cpn60 for 24 and 48 h and the binding of C/EBPβ on PPARγ or C/EBPα promoter was investigated using the ChIP assay (A). Fold change in the expression of Gata2 and Gata3 was analyzed using qPCR (B). The data indicate the mean ± SD of values from three independent experiments. The p values were determined using the Student’s t-test. Differences were considered significant at p < 0.05 (*) and p < 0.01 (**). MDI, 1 μM dexamethasone, 0.5 mM IBMX, and 5 μg/mL insulin.
GATA-binding factor (GATA) family is the transcription factor family comprising six members, among which GATA2 and GATA3 suppress adipocyte differentiation by binding to PPARγ, C/EBPα, and C/EBPβ and suppress their activity during adipogenesis18. The expression of GATA2 and GATA3 was significantly increased in Cpn60-treated cells compared with that in the control cells treated only with MDI (Fig. 8B). Cpn60 treatment resulted in reduced lipid accumulation in 3T3-L1 cells through the upregulation of Gata2/3, which inhibits the expression of Cebpa and Pparg and the translocation of C/EPBβ into the nucleus, the transcription factors that induce adipogenesis in the early stages of the differentiation period.
Discussion
The use of probiotics, comprising live bacteria, in health supplements and drugs is fraught with several problems in the manufacturing and distribution processes. Consumption of live bacteria is associated with concerns about side effects, such as systemic infections, including bacteremia, harmful metabolic activities stimulating the immune system, and genetic transfer19. Postbiotics exhibit various advantages, such as definite chemical structures, selection of safe dosages, long shelf life without denaturation, and bioactivity and health benefits similar to those of live probiotics20,21. Cell wall protein fractions extracted from Lactobacillus paracasei showed anticancer effects in human colon carcinoma Caco-2 cells by inducing apoptosis22. Lipoteichoic acid, located on the cell membrane of Bifidobacterium animalis subsp. lactis BPL1, reduced obesity biomarkers in Caenorhabditis elegans under hyperglycemic conditions via the IGF-1 pathway23. A novel protein, HM0539, secreted by Lactobacillus rhamnosus GG, protected the intestinal barrier by reinforcing mucin expression in Caco-2 cells and in neonatal rats infected with E. coli24. In this study, we found that the surface proteins of MJ2 inhibited lipid accumulation in 3T3-L1 cells, an effect similar to that seen for heat-killed intact MJ2, which indicates the potential of the surface proteins of MJ2 for use as postbiotics.
Moonlighting proteins are proteins produced from a single gene sequence that have more than one function in a cell or organism by sharing genes. Cpn60 is generally localized in the cytoplasm and plays a role in protein folding in many bacteria25. However, Cpn60 is a moonlighting protein that performs several other physiologically important biophysical and biochemical functions besides its main functions when localized to other subcellular locations. It is often located on the surface of bacteria26. In Lactococcus lactis, Cpn60, located on the cell surface, plays a role in binding to invertase from Saccharomyces cerevisiae27. Cpn60 of L. johnsonii La1, located on the cell surface, stimulates interleukin-8 through a CD14-dependent mechanism by binding to mucins and epithelial cells of the host28. In the present study, the surface proteins of MJ2 were sorted into four groups—chaperones, proteins involved in carbon metabolism, ribosomal proteins, and usually surface-located proteins. The chaperones and proteins involved in carbon metabolism identified in this study are known as moonlighting proteins that are highly conserved across phyla. Surface proteins isolated from MJ2 are generally located in parts other than the surface and may have functions different from their known functions. Identification of the active components of probiotics and prebiotics and understanding of the mechanisms underlying their beneficial effects are important to obtain maximum benefits with minimal side effects. Various cell fractions isolated from MJ2 were separated and surface fraction showed an inhibitory effect on lipid accumulation. Among the surface proteins, Cpn60, a well-known moonlighting protein, was selected as a potent active component based on its acidity, molecular weight, and moonlighting properties, however other proteins that satisfied the conditions could also be considered active components. Cpn60 was identified as a potentially active antiadipogenic SP of MJ2.
Heat shock protein 60 or chaperonin 60 (HSP60 or Cpn60) is highly conserved among species from bacteria to mammals, however, the sequences of Hsp60 do not show greater similarity between eukaryotic cells and Gram-positive bacteria like P. freudenreichii29. Mitochondrial HSP60 modulates energy metabolism in adipose tissue and reduced HSP60 level induces mild mitochondrial dysfunction that causes insufficient mitochondrial energy production resulted in the protection of diet-induced obesity in a high fat diet-induced obese male mice30. In this study, the addition of Cpn60 inhibited adipogenesis by regulating expression of adipogenesis-related transcription factors and we do not have direct evidences that the addition of Cpn60 decreases mitochondrial energy production, which induces antiobesity in a high fat-diet induced obese mice. Further studies are required to investigate whether Cpn60 isolated from the SP of P. freudenreichii MJ2 shows the antiobesity effect by the modulation of energy metabolism similar with HSP60 from C57BL/6N mice.
In the intestine, proteins regardless of origin usually break down into small peptides and amino acids by mainly microbial proteases and are absorbed cross the intestinal tract. In case of intact proteins, they bind to receptors on the surface of the membrane of intestinal epithelial cells and are translocated into the cytoplasm by endocytic mechanism31. In addition, lysosome-rich enterocytes, specialized intestinal cells, internalize proteins via receptor-mediated endocytosis followed by uptake proteins into the intestinal tract by endocytic machinery32. Although it is not clear the mechanism of absorption of probiotic-derived secreted proteins in the intestinal tract, Cpn60 from a probiotic bacterium might be absorbed by endocytic mechanism. In this study, we found the inhibitory effects of Cpn60 on adipogenesis in vitro. Thus, further studies need to confirm the effects of Cpn60 on anti-adipogenesis in vivo study and investigate its absorption mechanism in the intestine.
Preadipocytes, including 3T3-L1 preadipocytes, are differentiated into adipocytes through MCE, which is an essential step in differentiation during the early stage of adipogenesis and is followed by induction of the expression of key regulators of adipogenesis, such as PPARγ and C/EBPα. Differentiated adipocytes produce lipid droplets in the cytosol and express mature adipocyte markers, such as adipokines, adipose triglyceride lipase, and perilipin33. C/EBPs, comprised of six members, are key factors that regulate cellular differentiation. Among them, C/EBPβ is expressed immediately after the induction of adipogenesis by an adipogenic cocktail (MDI). The expressed C/EBPβ gains DNA binding ability upon phosphorylation by a mitogen-activated protein kinase (MAPK) and then induces expression of transcription factors, PPARγ and C/EBPα, which are critical factors in adipogenesis. Thus, interrupting the processes that occur in the early stages of adipogenesis contributes to the prevention or treatment of obesity. In this study, the translocation of C/EBPβ in preadipocytes was inhibited by Cpn60 treatment, which subsequently decreased the expression of Pparg and C/EBPβ, demonstrating the potential of Cpn60 as a therapeutic molecule for obesity.
GATA2/3 forms a complex with C/EBPα and C/EBPβ, which plays a critical role in the ability of GATA to suppress adipocyte differentiation18. C/EBPβ is the major transcription factor that initiates adipogenesis induced by MDI and induces the expression of Pparg and Cebpa. Cpn60 inhibits the activation of C/EBPβ in the nucleus by upregulating the expression of GATA2/3 and subsequently downregulating the expression of PPARγ and C/EBPα, which eventually inhibits adipogenesis and decreases lipid accumulation in 3T3-L1 cells. However, in our further study, it should be investigated whether inhibition of GATA2/3 restores the Cpn60-induced reduction of the DNA binding capacity of C/EBPβ and Pparg and Cebpa expression. In addition, further studies need to investigate whether inhibition of Cpn60 decreases its anti-adipogenesis activity.
In conclusion, Cpn60 reduced lipid accumulation in 3T3-L1 cells by upregulating the expression of GATA2/3, which might hinder the translocation of C/EPBβ and result in the inhibition of the expression of Cebpa and Pparg, the transcription factors necessary for the induction of adipogenesis in the early stages of differentiation.
Materials and methods
Materials
Dulbecco’s modified Eagle’s medium (DMEM), bovine calf serum (BCS), penicillin/streptomycin (P/S), and trypsin-EDTA used for cultivation of cells were purchased from HyClone (Logan, UT, USA). DNaseI, RNase, 3-isobutyl-1-methylxanthine (IBMX), dexamethasone, and insulin were obtained from Sigma-Aldrich (St. Louis, MO, USA). MTT was purchased from Amresco (Solon, OH, USA).
Bacterial strains and culture conditions
P. freudenreichii MJ2 strain (KCCM12272P) (MJ2)14 was cultured in Reinforced clostridial medium (RCM) (Oxoid, Hampshire, United Kingdom) at 30 °C under anaerobic conditions for 48 h until the stationary phase was reached. Thereafter, the cells were collected by centrifugation at 8000×g for 10 min at 4 °C and then washed twice with 20 mM Tris-HCl (pH 8). Washed cells were used to obtain cell fractions or were heated at 100 °C for 30 min to get hkMJ2. HkMJ2 showed no growth.
Preparation of bacterial cell fractions
The CW and Cyto fractions of MJ2 cells were extracted as described previously34–38. In brief, MJ2 cells were washed, resuspended in 1/10 volume of 20 mM Tris-HCl (pH 8), and incubated at − 80 °C for 2 h. The frozen cell solution was subjected to freeze–thaw cycles to weaken the cell wall. The cells were harvested by centrifugation at 8000×g for 10 min at 4 °C and the cell pellet was resuspended in 20 mM Tris-HCl (pH 8). The cells were disrupted by bead beating with sterile glass beads (diameter, 100 μm) and vortexing for 1 min for 15 cycles, and were left on ice for 1 min. The lysate was centrifuged at 1000×g for 10 min at 4 °C, and the supernatant, thus obtained, was centrifuged at 16,000×g for 10 min at 4 °C to remove unbroken cells. The supernatant was further centrifuged at 40,000×g for 10 min at 4 °C. The pellet containing the crude extract of the CW was treated with 50 μg/mL DNaseI and 100 μg/mL RNase A for 4 h at 37 °C and then centrifuged at 40,000×g for 10 min at 4 °C. The pellet containing CW was collected and the supernatant was ultracentrifuged at 110,000×g for 1 h at 4 °C using an ultracentrifuge (Optima L-100 K, Beckman Coulter, Brea, CA, USA) to fractionate the pellet containing the crude extract of the membrane and the supernatant containing the crude extract of cytoplasm (Cyto). The cell fractionation scheme is shown in Supplementary Fig. 6.
Preparation of SP from MJ2
The extraction of SP from MJ2 was carried out using two different methods as described previously10,39. In brief, MJ2 cells were incubated at 30 °C under anaerobic conditions for 48 h and subsequently collected by centrifugation at 8000×g for 10 min at 4 °C. The collected cells were resuspended in 5 M GuHCl (DAEJUNG, Seoul, Korea) to a final OD600 of 20 and incubated for 15 min at room temperature with gentle shaking. The suspension was centrifuged at 40,000×g for 10 min at 4 °C to collect SP. The supernatant was dialyzed against 20 mM Tris–HCl buffer using a dialysis cassette (MWCO 10 K, Slide-A-Lyzer™, Thermo Fisher Scientific, Waltham, MA, USA) for 24 h at 4 °C. The CW fraction without SP (CW w/o SP) was washed twice with 20 mM Tris-HCl and extracted as described above (Supplementary Fig. 6).
3T3-L1 cell culture and adipocyte differentiation
3T3-L1 preadipocytes were purchased from the American Type Culture Collection (ATCC CL-173). The cells were cultured at 37 °C in a humidified 5% CO2 environment in DMEM with 10% BCS and 1% P/S. The culture medium was changed every two days. For two days, the cells were maintained at 100% confluence. Thereafter, the culture medium was replaced with the MDI medium (DMEM containing 10% FBS, 1% P/S, and MDI) for inducing adipocyte differentiation. The day on which the culture medium was replaced with the MDI medium was considered day 0. On day 2, the medium was replaced with insulin medium (DMEM containing 5 μg/mL insulin), which was refreshed every two days until day 8.
Assessment of cell viability
The cell viability was measured by MTT assay to determine the effect of various MJ2 cell fractions on the viability 3T3-L1 preadipocytes. We assessed the same concentration of cell extract from 1 × 108 hkMJ2 cells/mL, which was previously shown to inhibit lipid accumulation in 3T3-L1 cells without cytotoxicity12. 3T3-L1 preadipocytes (5 × 104 cells/mL) were seeded in a 96-well plate containing DMEM supplemented with 10% BCS and cultured at 37 °C in a humidified 5% CO2 atmosphere for 24 h. The medium was then replaced with DMEM without 10% BCS and treated with various crude cell fractions prepared from 1 × 108 MJ2 cells/mL (CW, Cyto, and SP) or from 1 × 108 hkMJ2 cells/mL. After incubation for 24 h, the medium was discarded and MTT reagent (0.125 mg/mL) was added into the wells. The plate was incubated at 37 °C for 1 h in the dark. The supernatant was completely removed, and 200 μL of dimethyl sulfoxide was added to each well. After 30 min, absorbance was measured at 540 nm using a microplate reader (SpectraMax 340PC, Molecular Devices, Sunnyvale, CA, USA). Relative cell viability (%) compared to that of the negative control was calculated.
Oil red O staining
Oil Red O staining was performed on day 8 when preadipocytes were differentiated into adipocytes more than 80%40. Briefly, cells were washed with PBS twice and fixed with 10% formalin for 1 h at room temperature. The fixed cells were washed twice with deionized water, incubated with isopropanol (60%) for 5 min, and soaked in Oil Red O working solution for 30 min at room temperature. After staining, the cells were washed four times with deionized water and allowed to dry completely. Isopropanol (100%) was added to each well to extract the stain; the solution from each well was transferred to a new microplate, and its absorbance was measured at 500 nm for quantification of the Oil Red O stain. The relative lipid accumulation (%) in cells subjected to each treatment was calculated by comparing with the absorbance of the cells treated only with MDI (differentiation control).
Quantitative real-time polymerase chain reaction (qPCR)
Total RNA was extracted with TRIzol reagent (Thermo Fisher Scientific) following the manufacturer’s instructions. The concentration of RNA was measured using a NanoDrop spectrophotometer (ND-1000 spectrophotometer, Thermo Fisher Scientific). After quantification, RNA was reverse transcribed into cDNA using a RevertAid First Stand cDNA Synthesis Kit (Thermo Fisher Scientific). cDNA was amplified using qPCR with a KAPA SYBR FAST qPCR Kit (KAPA Biosystems, Woburn, MA, USA) on a QuantStudio 6 Flex system (Life Technologies, Carlsbad, CA, USA). The primers used in this study were purchased from Bioneer (Daejeon, Korea). The sequences of the primers are Pparg (Forward 5′-AGA CAT CAG CGC CTA CAT CG-3′, Reverse 5′-GCT CCC GGG TAG TCA AAG TC-3′), Cebpa (Forward 5′- TGG ACA AGA ACA GCA ACG AG-3′, Reverse 5′-TCA CTG GTC AAC TCC AGC AC-3′), Srebf1 (Forward 5′-AAC CAG AAG CTC AAG CAG GA-3′, Reverse 5′-TTT CAT GCC CTC CAT AGA CA-3′), Scd1 (Forward 5′-TGA CTA TCA TCA TGC CGG CC-3′, Reverse 5′-CTT TGA CAG CCG GGT GTT TG-3′), Gata2 (Forward 5′-CGA CCA CAC TTG TTG CAC AG-3′, Reverse 5′-GGG TAA ACA GAC AGA GGC CC-3′), Gata3 (Forward 5′-GCT ACG GTG CAG AGG TAT CC-3′, Reverse 5′-GAG GGT AAA CGG ACA GAG GC-3′), and β-actin (Forward 5′-GAC ATG GAG AAG ATC TGG CA-3′, Reverse 5′-GGT CTT TAC GGA TGT CAA CG-3′). The reaction was preheated to 95 °C, and then subjected to 40 cycles of 95 °C for 15 s, 60 °C for 15 s, and 72 °C for 30 s. β-actin was used as the reference gene. The normalized mRNA expression level was indicated as 2−ΔΔCt (the expression level of β-actin control was set to 1)41.
LC-MS/MS for peptides analysis
Nano-LC-MS/MS analysis was performed using an Easy n-LC system (Thermo Fisher Scientific) and an LTQ Orbitrap XL mass spectrometer (Thermo Fisher Scientific) equipped with a nano-electrospray source. Samples were separated on a C18 nanopore column (150 mm × 0.1 mm, 3 μm pore size; Agilent, Santa Clara, CA, USA). Mobile phase A for LC separation comprised of 0.1% formic acid and 3% acetonitrile in deionized water, and mobile phase B was 0.1% formic acid in acetonitrile. The linear solvent gradient profile used was as follows: 0% mobile phase B for 40 min; 40–60% mobile phase B for 4 min; 95% mobile phase B for 4 min; 100% mobile phase A for 6 min. The solvent flow rate was maintained at 1.5 μL/min. Mass spectra were acquired using data-dependent acquisition with a full mass scan (350–1200 m/z), followed by 10 MS/MS scans. For MS1 full scans, the Orbitrap resolution was 15,000, and the automatic gain control (AGC) was 2 × 105. For MS/MS in the LTQ, AGC was 1 × 104. The MASCOT algorithm (Matrix Science, Boston, MA, USA) was used to identify peptide sequences present in the UniProt protein sequence database. The database search criteria were as follows: taxonomy: Propionibacterium freudenreichii; fixed modification: carbamidomethylated at cysteine residues; variable modification: oxidized at methionine residues; maximum allowed missed cleavage: 2; MS tolerance: 10 ppm; MS/MS tolerance: 0.8 Da. The peptides were filtered using a significance threshold of p < 0.05. The 131 identified proteins in the SP fraction were analyzed using the STRING database version 11, which indicates networks of interactions among proteins. The interactions of proteins were assessed under a high-confidence cutoff (0.7), which indicated the minimum required interaction score. Protein networks were visualized using Cytoscape version 3.7.1.
Proteins expression and purification
Based on the results of the inhibition of lipid accumulation in 3T3L1 cells treated with the fractions obtained from ion exchange and size exclusive chromatography (Supplementary Fig. 4), Cpn60 was selected as the most likely active substance inhibiting lipid accumulation. To investigate whether Cpn60 has antiobesity properties, recombinant Cpn60 was generated by cloning the MJ2 Cpn60 into the pBT7-N-His plasmid vector (Bioneer) and transforming Escherichia coli cells with the vector construct. The transformed E. coli was grown at 37 °C in LB broth. Recombinant Cpn60 was purified using an Automated Protein Production System (ExiProgen, Bioneer) and its purity was confirmed by electrophoresing on a 10% SDS-polyacrylamide gel (Supplementary Fig. 5).
Western blot analysis
Whole protein was extracted from 5 × 106 3T3-L1 cells using the PRO-PREP™ protein extraction solution (iNtRON Biotechnology, Seongnam, Korea) according to the manufacturer’s instructions. Nuclear and cytoplasmic proteins were extracted using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Fisher Scientific), according to the manufacturer’s instructions. Protein concentration was determined using Bradford assay with bovine serum albumin as a standard. Each protein sample (10 μg) was separated by electrophoresis on a 10% SDS-polyacrylamide gel. The proteins were transferred onto a polyvinylidene difluoride membrane (0.45 μm, Millipore, Bedford, MA, USA); the membrane was blocked with 6% skimmed milk in Tris-buffered saline with 0.05% Tween 20 (TBST) and incubated with primary antibodies overnight at 4 °C. β-actin antibody (1:10,000 dilution, GTX109639, GeneTex, Irvine, CA, USA) and lamin B1 antibody (1:1000 dilution, GTX103292, GeneTex) were used as endogenous controls for whole and nuclear proteins, respectively. Anti-C/EBPβ (1:1000 dilution, sc-7962; SANTA CRUZ Biotechnology, Dallas, TX, USA) was used as the primary antibody. Membranes were washed with TBST and incubated with secondary antibodies for 1 h at room temperature. Goat anti-rabbit IgG horseradish peroxidase (HRP)-conjugated antibody (1:2000 dilution, GTX213110-01, GeneTex) was used as the secondary antibody for anti-β-actin and anti-lamin B1. A goat anti-mouse IgG antibody (HRP) (1:2000 dilution, GTX213111-01, GeneTex) was used as a secondary antibody for anti-C/EBPβ. The membranes were washed with TBST and the bands were detected with a SuperSignal West Femto Maximum Sensitivity Substrate Kit (Thermo Fisher Scientific) using a FluorChem E system (ProteinSimple, San Jose, CA, USA). Densitometric analysis was performed using the ImageJ software (Softomic, Barcelona, Spain).
ChIP assay
ChIP assay was performed using a ChIP assay kit (Sigma-Aldrich), according to the manufacturer’s instructions. Briefly, 3T3-L1 preadipocytes were differentiated into adipocytes and treated with 1 μg/mL of Cpn60 for 24 or 48 h. The cells were crosslinked using 1% formaldehyde for 10 min at room temperature, and then formaldehyde was quenched with 2.5 M glycine. The crosslinked cells were washed with ice-cold PBS twice and then harvested by centrifugation at 800×g at 4 °C for 5 min. The cell pellet was resuspended in a cell lysis buffer and incubated for 15 min on ice. The lysed cells were centrifuged and the pellet was resuspended in the nuclear lysis buffer. The cell lysate was reacted with 1 unit of micrococcal nuclease (Sigma-Aldrich) with 0.1 M CaCl2 at 28 °C to fragment DNA (0.5‒1 kb) and then centrifuged at 10,000×g for 10 min at 4 °C to remove insoluble materials. The lysates were diluted with ChIP dilution buffer and 2% of the diluted lysates were removed as input and stored at − 20 °C until immunoprecipitation. The chromatin-protein complexes were immunoprecipitated with 5 μg of anti-C/EBPβ antibody (H-7) (sc-7962, SANTA CRUZ Biotechnology) and 20 μL of protein G magnetic beads at 4 °C for overnight. After immunoprecipitation, protein G-antibody complexes were washed with low-salt immune complex wash buffer, high-salt immune complex wash buffer, LiCl buffer, and TE buffer for 3 min each, and then the protein G–antibody complexes and input were reversed crosslinked by incubating with ChIP elution buffer and proteinase K at 62 °C for 2 h. After reverse crosslinking, DNA was purified using spin-column extraction. Purified DNA was amplified using qPCR on a 7500 Fast Real-Time PCR System (Applied Biosystems). The sequences of primer synthesized by Bioneer: PPARγ promoter (Forward 5′-TTC AGA TGT GTG ATT AGG AG-3′, Reverse 5′-AGA CTT GGT ACA TTA CAA GG-3′) and C/EPBα promoter (Forward 5′-TCC CTA GTG TTG GCT GGA AG-3′, Reverse 5′-CAG TAG GAT GGT GCC TGC TG-3′). The resulting Ct values from the ChIP analysis were normalized using the percent input method (%input = 100 × 2Adjusted input—Ct (IP)).
Statistical analysis
Statistical Package for the Social Science (SPSS) analysis program (ver. 25) was used for all statistical analyses. The statistical analysis was performed using one-way analysis of variance (ANOVA) and Tukey’s honest significant difference (HSD) method as a post hoc test to confirm significant differences between the groups in case the experiments involved various groups. Single comparisons were performed to evaluate the significance between the negative control and treatment groups using the Student’s t-test. Data are presented as mean ± standard deviation (SD). Statistical significance was set at p < 0.05.
Supplementary Information
Author contributions
Conceptualization, M.A. and Y.H.L.; experiments, M.A. and Y.H.L.; validation, M.A. and Y.H.L.; visualization, M.A. and Y.H.L.; original draft preparation, M.A.; review and editing of the manuscript, Y.H.L.; supervision, Y.H.L.; project administration and funding acquisition, Y.H.L. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Korea University grant (K2222081).
Data availability
All generated data have been included in the published manuscript. For other information, contact with the corresponding author (Young-Hee Lim. yhlim@korea.ac.kr).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-023-46436-x.
References
- 1.Sarma S, Sockalingam S, Dash S. Obesity as a multisystem disease: Trends in obesity rates and obesity-related complications. Diabetes Obes. Metab. 2021;23:3–16. doi: 10.1111/dom.14290. [DOI] [PubMed] [Google Scholar]
- 2.Ambele MA, Dhanraj P, Giles R, Pepper MS. Adipogenesis: A complex interplay of multiple molecular determinants and pathways. Int. J. Mol. Sci. 2020;21:4283. doi: 10.3390/ijms21124283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haider N, Larose L. Harnessing adipogenesis to prevent obesity. Adipocyte. 2019;8:98–104. doi: 10.1080/21623945.2019.1583037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahmad B, Serpell CJ, Fong IL, Wong EH. Molecular mechanisms of adipogenesis: The anti-adipogenic role of AMP-activated protein kinase. Front. Mol. Biosci. 2020;7:76. doi: 10.3389/fmolb.2020.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosen ED, MacDougald OA. Adipocyte differentiation from the inside out. Nat. Rev. Mol. Cell Biol. 2006;7:885–896. doi: 10.1038/nrm2066. [DOI] [PubMed] [Google Scholar]
- 6.Cristancho AG, Lazar MA. Forming functional fat: A growing understanding of adipocyte differentiation. Nat. Rev. Mol. Cell Biol. 2011;12:722–734. doi: 10.1038/nrm3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsilingiri K, et al. Probiotic and postbiotic activity in health and disease: Comparison on a novel polarised ex-vivo organ culture model. Gut. 2012;61:1007–1015. doi: 10.1136/gutjnl-2011-300971. [DOI] [PubMed] [Google Scholar]
- 8.Żółkiewicz J, Marzec A, Ruszczyński M, Feleszko W. Postbiotics—a step beyond pre-and probiotics. Nutrients. 2020;12:2189. doi: 10.3390/nu12082189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cousin FJ, Jouan-Lanhouet S, Dimanche-Boitrel M-T, Corcos L, Jan G. Milk fermented by Propionibacterium freudenreichii induces apoptosis of HGT-1 human gastric cancer cells. PLoS ONE. 2012;7:e31892. doi: 10.1371/journal.pone.0031892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Le Maréchal C, et al. Surface proteins of Propionibacterium freudenreichii are involved in its anti-inflammatory properties. J. Proteomics. 2015;113:447–461. doi: 10.1016/j.jprot.2014.07.018. [DOI] [PubMed] [Google Scholar]
- 11.do Carmo FL, et al. Propionibacterium freudenreichii surface protein SlpB is involved in adhesion to intestinal HT-29 cells. Front. Microbiol. 2017;8:1033. doi: 10.3389/fmicb.2017.01033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.An M, Park Y-H, Lim Y-H. Antiobesity and antidiabetic effects of the dairy bacterium Propionibacterium freudenreichii MJ2 in high-fat diet-induced obese mice by modulating lipid metabolism. Sci. Rep. 2021;11:2481. doi: 10.1038/s41598-021-82282-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yeom J, Ma S, Lim Y-H. Probiotic Propionibacterium freudenreichii MJ2 enhances osteoblast differentiation and mineralization by increasing the OPG/RANKL ratio. Microorganisms. 2021;9:673. doi: 10.3390/microorganisms9040673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yeom J, Yim DJ, Ma S, Lim Y-H. Propionibacterium freudenreichii inhibits RANKL-induced osteoclast differentiation and ameliorates rheumatoid arthritis in collagen-induced arthritis mice. Microorganisms. 2021;10:48. doi: 10.3390/microorganisms10010048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang E, Kim CY. Natural products and obesity: A focus on the regulation of mitotic clonal expansion during adipogenesis. Molecules. 2019;24:1157. doi: 10.3390/molecules24061157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farmer SR. Transcriptional control of adipocyte formation. Cell Metab. 2006;4:263–273. doi: 10.1016/j.cmet.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsukada J, Yoshida Y, Kominato Y, Auron PE. The CCAAT/enhancer (C/EBP) family of basic-leucine zipper (bZIP) transcription factors is a multifaceted highly-regulated system for gene regulation. Cytokine. 2011;54:6–19. doi: 10.1016/j.cyto.2010.12.019. [DOI] [PubMed] [Google Scholar]
- 18.Tong Q, Tsai J, Tan G, Dalgin GK, Hotamisligil GKS. Interaction between GATA and the C/EBP family of transcription factors is critical in GATA-mediated suppression of adipocyte differentiation. Mol. Cell. Biol. 2005;25:706–715. doi: 10.1128/MCB.25.2.706-715.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doron S, Snydman DR. Risk and safety of probiotics. Clin. Infect. Dis. 2015;60:S129–S134. doi: 10.1093/cid/civ085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aguilar-Toalá J, et al. Postbiotics: An evolving term within the functional foods field. Trends Food Sci. Technol. 2018;75:105–114. doi: 10.1016/j.tifs.2018.03.009. [DOI] [Google Scholar]
- 21.Sabahi, S. et al. Postbiotics as the new frontier in food and pharmaceutical research. Crit. Rev. Food Sci. Nutr.63, 8375−8402 (2023). [DOI] [PubMed]
- 22.Nozari S, et al. Potential anticancer effects of cell wall protein fractions from Lactobacillus paracasei on human intestinal Caco-2 cell line. Lett. Appl. Microbiol. 2019;69:148–154. doi: 10.1111/lam.13198. [DOI] [PubMed] [Google Scholar]
- 23.Balaguer F, et al. Lipoteichoic acid from Bifidobacterium animalis subsp. lactis BPL1: A novel postbiotic that reduces fat deposition via IGF-1 pathway. Microb. Biotechnol. 2022;15:805–816. doi: 10.1111/1751-7915.13769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao J, et al. A novel postbiotic from Lactobacillus rhamnosus GG with a beneficial effect on intestinal barrier function. Front. Microbiol. 2019;10:477. doi: 10.3389/fmicb.2019.00477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zeilstra-Ryalls J, Fayet O, Georgopoulos C. The universally conserved GroE (Hsp60) chaperonins. Annu. Rev. Microbiol. 1991;45:301–325. doi: 10.1146/annurev.mi.45.100191.001505. [DOI] [PubMed] [Google Scholar]
- 26.Jeffery CJ. Protein moonlighting: What is it, and why is it important? Philos. Trans. R. Soc. B. Biol. Sci. 2018;373:20160523. doi: 10.1098/rstb.2016.0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katakura Y, Sano R, Hashimoto T, Ninomiya K, Shioya S. Lactic acid bacteria display on the cell surface cytosolic proteins that recognize yeast mannan. Appl. Microbiol. Biotechnol. 2010;86:319–326. doi: 10.1007/s00253-009-2295-y. [DOI] [PubMed] [Google Scholar]
- 28.Bergonzelli GE, et al. GroEL of Lactobacillus johnsonii La1 (NCC 533) is cell surface associated: Potential role in interactions with the host and the gastric pathogen Helicobacter pylori. Infect. Immun. 2006;74:425–434. doi: 10.1128/IAI.74.1.425-434.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gupta RS. Evolution of the chaperonin families (Hsp60, Hsp10 and Tcp-1) of proteins and the origin of eukaryotic cells. Mol. Microbiol. 1995;15:1–11. doi: 10.1111/j.1365-2958.1995.tb02216.x. [DOI] [PubMed] [Google Scholar]
- 30.Hauffe R, et al. HSP60 reduction protects against diet-induced obesity by modulating energy metabolism in adipose tissue. Mol. Metab. 2021;53:101276. doi: 10.1016/j.molmet.2021.101276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gardner ML. Gastrointestinal absorption of intact proteins. Ann. Rev. Nutr. 1988;8:329–350. doi: 10.1146/annurev.nu.08.070188.001553. [DOI] [PubMed] [Google Scholar]
- 32.Park J, et al. Lysosome-rich enterocytes mediate protein absorption in the vertebrate gut. Dev. Cell. 2019;51:7–20.e26. doi: 10.1016/j.devcel.2019.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ghaben AL, Scherer PE. Adipogenesis and metabolic health. Nat. Rev. Mol. Cell Biol. 2019;20:242–258. doi: 10.1038/s41580-018-0093-z. [DOI] [PubMed] [Google Scholar]
- 34.Aderiye B, Oluwole O. How to obtain the organelles of prokaryotic and microbial eukaryotic cells. J. Cell Anim. Biol. 2014;8:95–109. doi: 10.5897/JCAB2014.0413. [DOI] [Google Scholar]
- 35.Gabay J, Horwitz MA. Isolation and characterization of the cytoplasmic and outer membranes of the Legionnaires’ disease bacterium (Legionella pneumophila) The J. Exp. Med. 1985;161:409–422. doi: 10.1084/jem.161.2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heumann D, Barras C, Severin A, Glauser M, Tomasz A. Gram-positive cell walls stimulate synthesis of tumor necrosis factor alpha and interleukin-6 by human monocytes. Infect. Immun. 1994;62:2715–2721. doi: 10.1128/iai.62.7.2715-2721.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Osborn M, Gander J, Parisi E, Carson J. Mechanism of assembly of the outer membrane of Salmonella typhimurium: Isolation and characterization of cytoplasmic and outer membrane. J. Biol. Chem. 1972;247:3962–3972. doi: 10.1016/S0021-9258(19)45127-2. [DOI] [PubMed] [Google Scholar]
- 38.Xu X, et al. Purified cell wall from the probiotic bacterium Lactobacillus gasseriactivates systemic inflammation and at higher doses, produces lethality in a rat model. Crit. Care. 2014;18:R140. doi: 10.1186/cc13966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson B, Selle K, O’Flaherty S, Goh YJ, Klaenhammer T. Identification of extracellular surface-layer associated proteins in Lactobacillus acidophilus NCFM. Microbiology. 2013;159:2269. doi: 10.1099/mic.0.070755-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramirez-Zacarias J, Castro-Munozledo F, Kuri-Harcuch W. Quantitation of adipose conversion and triglycerides by staining intracytoplasmic lipids with Oil red O. Histochemistry. 1992;97:493–497. doi: 10.1007/BF00316069. [DOI] [PubMed] [Google Scholar]
- 41.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All generated data have been included in the published manuscript. For other information, contact with the corresponding author (Young-Hee Lim. yhlim@korea.ac.kr).