SUMMARY
Nuclear pore complexes (NPCs) direct the nucleocytoplasmic transport of macromolecules. Here, we provide a composite multiscale structure of the yeast NPC, based on improved 3D density maps from cryogenic electron microscopy and AlphaFold2 models. Key features of the inner and outer rings were integrated into a comprehensive model. We resolved flexible connectors that tie together the core scaffold, along with equatorial transmembrane complexes and a lumenal ring that anchor this channel within the pore membrane. The organization of the nuclear double outer ring reveals an architecture that may be shared with ancestral NPCs. Additional connections between the core scaffold and the central transporter suggest that under certain conditions, a degree of local organization is present at the periphery of the transport machinery. These connectors may couple conformational changes in the scaffold to the central transporter to modulate transport. Collectively, this analysis provides insights into assembly, transport, and NPC evolution.
In brief
Akey et al. report a multiscale structure of the isolated yeast NPC that provides a framework to piece together the roles of nucleoporins in assembly and transport while providing insights into the evolution of critical sub-assemblies.
Graphical Abstract
INTRODUCTION
Nuclear pore complexes (NPCs) form a gateway between the cytoplasm and nucleus for the bidirectional transport of macro-molecules during cell growth, differentiation, and development. However, an understanding of NPC function and its transport mechanism remains incomplete, due in part to the lack of a high-resolution structure. Such a structure will provide insights into the hierarchical assembly of nucleoporins (Nups), conformational plasticity of the scaffold, and membrane association. This approach may also illuminate the central transport machinery.1–3 In addition, an improved structural model should deliver a framework for understanding the role of NPCs in aging, infectious disease, and cancer.4
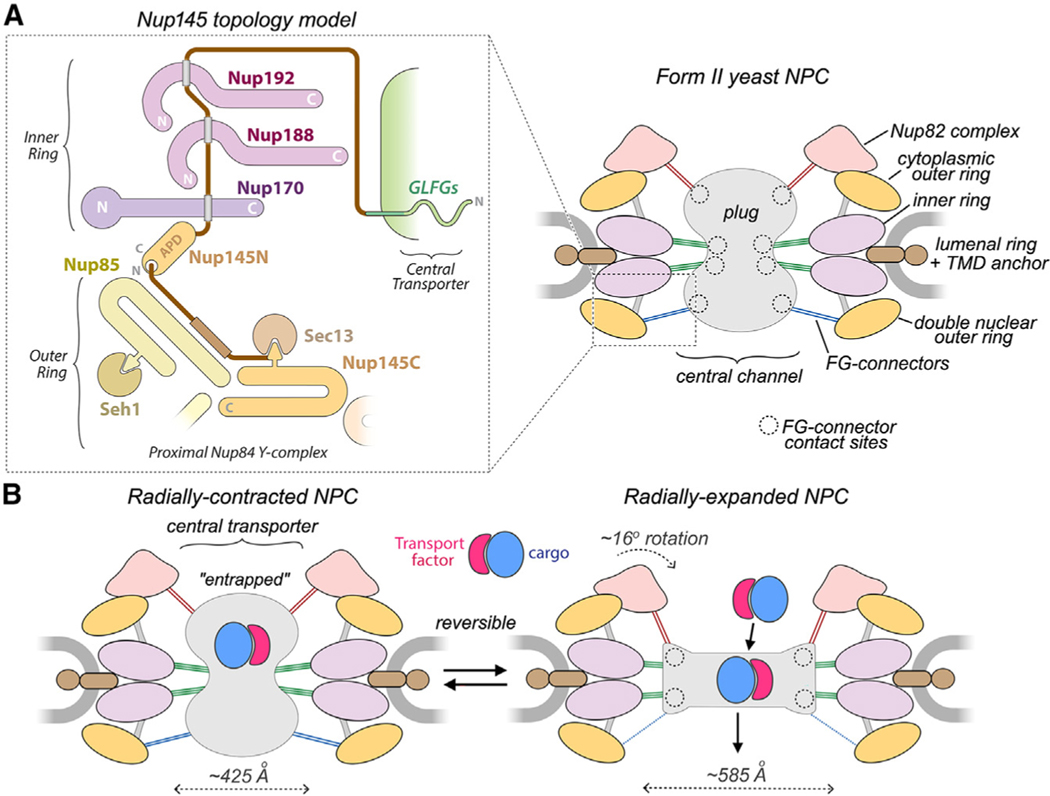
The canonical NPC from baker’s yeast has a mass of ~52 MDa, spanning a pore in the nuclear envelope (NE). This channel comprises ~30 distinct Nups (~550 in total) arranged as 8 spokes in the inner ring (IR), flanked by single cytoplasmic and nuclear outer rings comprising Y-shaped Nup84 complexes.5–7 However, multiple isoforms are present in yeast cells, including a subset with a double outer ring on the nuclear side.7 In addition, a basket functions as a nuclear RNA export platform8,9 and is roughly co-linear with Nup82 complexes on the cytoplasmic outer ring (CR); together, these form the core of an RNA remodeling and export machine.10 The yeast NPC is tethered to the NE by membrane anchors in the IR. Nup84 Y-complexes in the outer rings contribute additional membrane binding motifs (MBMs) and may interact with the transmembrane domain (TMD) of Ndc1.11 In addition, a pore membrane protein (Pom152) has an extended C-terminal domain (CTD) with tandem repeats of Ig-like domains12,13; 2 copies are anchored to each spoke, and 16 copies form a lumenal ring (LR) in the NE lumen.6,7 Flexible termini of ~10 different FG Nups with FXFG or GLFG repeats are anchored to the core scaffold and extend into the central channel,6,7 where they provide binding sites for transport factors and cargo complexes to facilitate transport—together, these form the central transporter.14–17
The NPC has been analyzed by cryogenic electron tomography (CET) and sub-volume averaging for human,18–21 frog,22 algal,23 and yeast complexes, including Saccharomyces cerevisiae5–7,24 and Schizosaccharomyces pombe.25 The core scaffold in these structures ranges from ~12 to 30 Å resolution, and molecular models have pinpointed the position and relative orientation of many Nups. Moreover, Artificial-intelligence-based structure prediction programs26,27 are able to leverage structures in the Protein Data Bank and per-residue correlations in sequence alignments to provide computed structure models (CSMs). Nup crystal structures, CSMs, and homology models can be used to model the NPC with electron density maps from CET or single-particle analysis, in concert with other data, such as chemical cross-links.6,18,24,28–32
Recent CET studies of the S. pombe NPC have shown that the core scaffold undergoes a large radial expansion of the IR with smaller displacements of the outer rings25 to upregulate trans-port. Similar expanded NPC conformations have been observed in other organisms.5,7,18,21,33 Current single-particle density maps of isolated yeast and vertebrate NPCs span a range from nanometer to near-atomic resolution for sub-assemblies of the core scaffold and have revealed a radially contracted NPC with a smaller diameter central channel.7,24,28–32,34 In this study, improved 3D density maps from the S. cerevisiae NPC were combined with flexible fitting of AlphaFold2 models26 to obtain a composite multiscale structure that provides insights into this remarkable channel.
RESULTS
Overview
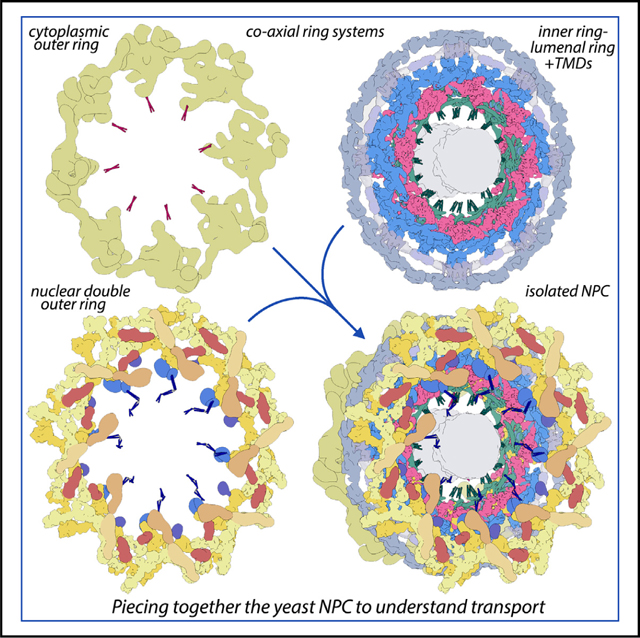
Yeast cells lack a nuclear lamina that allows intact NPCs to be released from the NE with mild detergent extraction of a cell cryolysate and purified with a single affinity step.6,7,35 However, the large size and flexibility of the NPC presents a formidable challenge to structure determination. Low-resolution 3D density maps can facilitate the integration of information from higher-resolution analysis of sub-assemblies and other sources.36,37 We adopted a multiscale approach in which individual ring systems were refined at 17–30 Å resolution to give a better representation of their connectivity within the NPC (Figures S1A and S1B, left-hand panels). This allowed the identification of additional spoke connectors, orphan Nup densities in the double outer ring, linker domains between the inner and outer double rings, and FG connectors to the central transporter.
We took advantage of the fact that yeast NPCs with nuclear double outer rings (NRs) (form II, 15%–20%) are more amenable to electron microscopy (EM) structural determination for certain sub-assemblies than canonical form I channels with single rings on both sides of the IR.7 Improved density maps for the spoke and double Y-complexes (Figure S2A; Table S1; STAR Methods) allowed us to increase the precision and extent of our previous NPC structure7 by incorporating Nup AlphaFold CSMs26 with molecular dynamics flexible fitting (MDFF).38 Focused 3D refinements also improved the clarity of the massive double outer ring and LR comprising Pom152 CTDs.12,13 Finally, we were able to visualize a more complete CR (Figure S2B; Table S1)7 with a rough Y shape for the Nup84 complex; however, residual blurring precluded the building of a model at this stage. Even so, we determined that the diameter of the CR is 15% smaller than its counterpart in situ5,7 (Figure S2C). In addition, density for the Nup82 complex, which resides on the CR, exhibits a rotation of ~16° when compared with the in situ density map5–7 (Figure S2C, inset). This suggests that the single outer ring and cytoplasmic export platform may alter their structure in concert with radial contraction of the IR.25
We then combined nine 3D density maps from the co-axial rings and sub-assemblies to create a composite multiscale structure of the form II yeast NPC (Figures S1A and S1B, right-hand panels). This structure covers a resolution range of 6.6–30 Å (excluding the central transporter). Next, 9 3D maps were segmented into 16 functional regions to give a more informative model (Figure 1, see key legend) and provide a framework for understanding NPC function.
Figure 1. A composite multiscale 3D structure of the isolated yeast NPC.
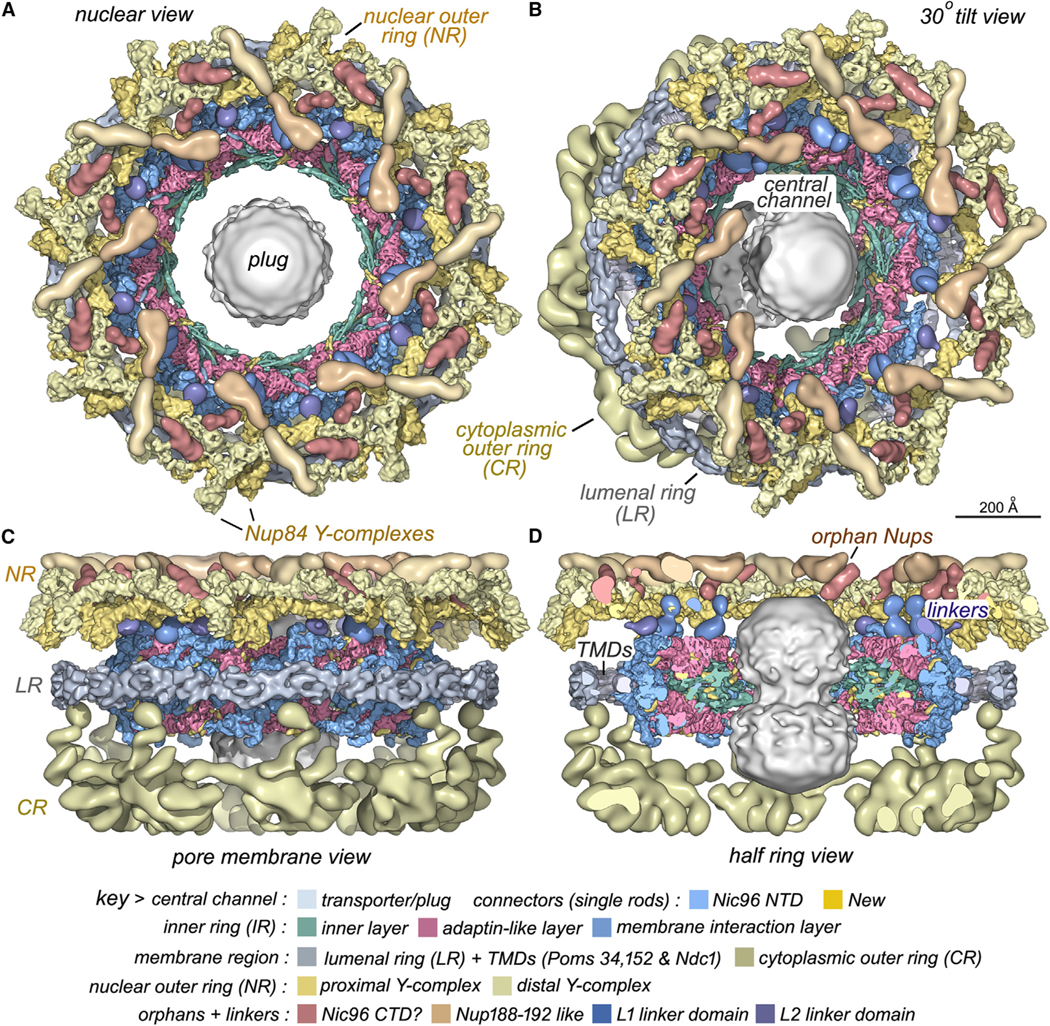
Nine 3D density maps were segmented into 16 regions to highlight functional sub-assemblies as indicated in the color-coded key. Labels: nuclear double outer ring (NR), inner ring (IR), and cytoplasmic outer ring (CR).
(A) A top view from the nuclear compartment reveals the double outer ring with associated orphan Nups and linker domains.
(B) The view in (A) has been rotated 30° counterclockwise about the vertical axis.
(C) A view from the pore membrane; the lipid-detergent micelle that encircles the inner ring is not shown.
(D) A cut-away view oriented as in (C) to reveal the co-axial rings, along with membrane anchor points (TMDs) and the hourglass-shaped central transporter/plug.
Structure and model of the IR
The intrinsic protomer of the IR, termed a spoke, comprises 28 Nups arranged with nearly perfect 2-fold symmetry in 3 layers that extend from the central channel to the pore membrane (Figure S3). There are 14 Nups in a half spoke; this includes an inner layer with two laterally offset Nsp1 heterotrimers (Nup49, Nup57, and Nsp1); a middle adaptin-like layer with Nup188 and its homolog Nup192; and a membrane-interacting layer containing two laterally offset copies of Nic96, a Nup53-Nup59 heterodimer, and single copies of Nup157 and Nup170.6,7,24 Each half spoke also contains 14 flexible connectors from Nic96 NTDs that interact with Nsp1 complexes, Nup188, and Nup192 to help tie together their respective layers.7,24 However, additional connectors and important features remain to be discovered.
To this end, we revisited a single-particle dataset and used better masking and non-uniform refinement in CryoSPARC39 to improve our 3D map of the IR protomer (STAR Methods). This provided a 3D density map with a global resolution of 6.6 Å and greater clarity for all three spoke layers (Figure S2A; Table S1). A 3D map of the IR was created from the spoke density map and segmented to highlight the cylindrical organization of the IR (Figures 2A–2D). AlphaFold2 CSMs26 and prior subunit models7 were combined to create a nearly complete structure of the IR (Figures 2E, 2F, and S4; Table S2, top; STAR Methods). Helices and some connecting loops were resolved, except in membrane proximal regions of Nup157, Nup170, and the outer copies of Nic96 CTDs, which are more flexible.
Figure 2. Inner ring structure and interactions between spokes.
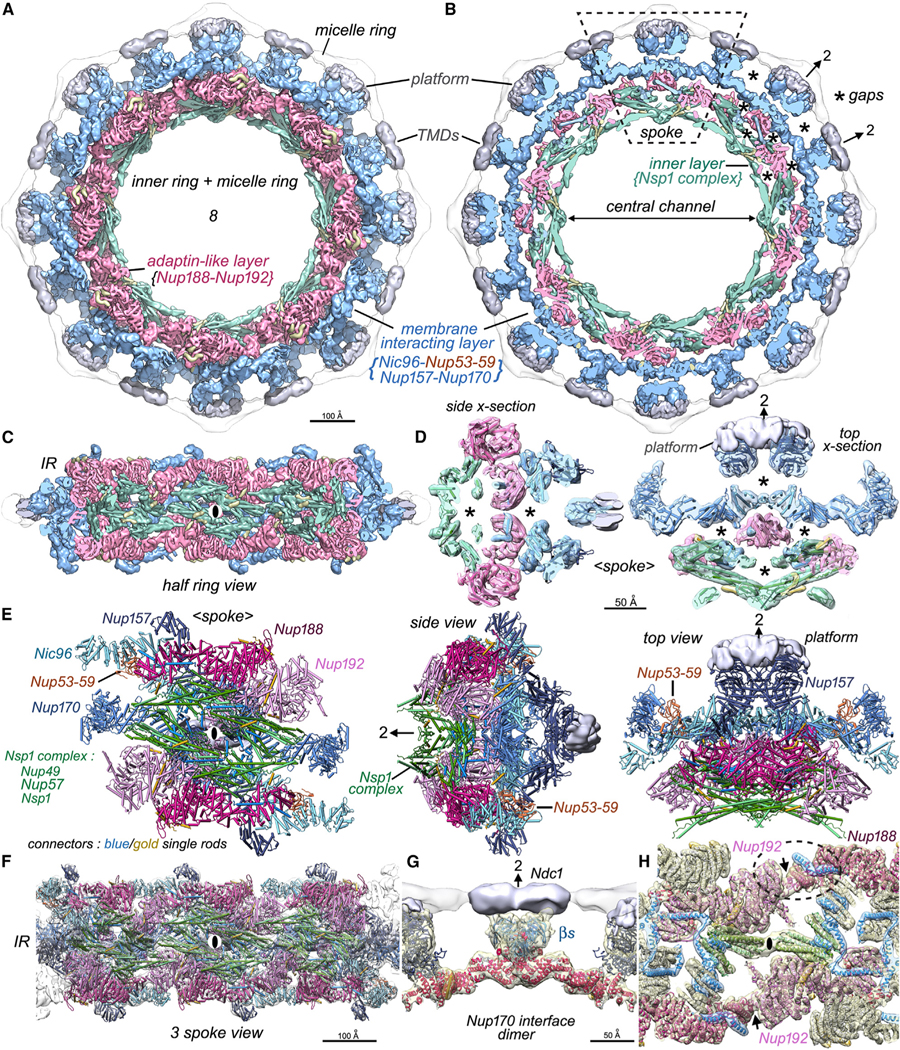
(A) The inner ring and associated membrane protein densities (silver) within the micelle ring are shown. The inner ring has been segmented into an inner layer (green), an intermediate adaptin-like layer (dark pink), and the membrane-interacting layer (light blue).
(B) The highly fenestrated architecture of the inner ring with gaps (asterisks) and membrane-spanning regions are revealed in a cross-section that also shows the central channel. A single spoke is outlined (dashed rhomboid).
(C) A “half ring” view of the inner ring is viewed from the pore membrane; the local 2-fold axis for the center spoke is indicated with a black ellipse.
(D) Central cross-sections through the spoke with molecular models depicted as rods are viewed from the side and top to show the open architecture with gaps and voids (asterisks). A local 2-fold axis in the plane is indicated with an arrow and “2”.
(E) A molecular model for Nups within the spoke is shown as “cylinders and planks.” Three views from left to right: a front view from the central channel, a side view, and a view from the cytoplasm. All views have platform density (silver) and single rod-like connectors in blue and gold.
(F) Molecular model for three spokes in the inner ring.
(G) Two Nup170 molecules form a major contact between adjacent spokes and may interact with an Ndc1 dimer at the local 2-fold axis with their β-propellers (βs).
(H) Three inter-spoke contacts are shown: a contact between adjacent Nsp1 complexes and a pair of symmetry-related contacts between the Nup192 NTD and Nup188 CTD tail that are indicated with black arrows. One Nup188-Nup192 contact is outlined with a dashed oval. Nic96 and new connectors are shown in blue and gold ribbons.
This analysis also revealed a platform-like region formed by Pom34 and Pom152, which interacts with Nup157 β-propellers at the pore membrane. A central cross-section shows the organization of the IR with large gaps both within and between the spokes (asterisks, Figures 2B and 2D). Functionally related Nups are arrayed in cylindrical layers that emphasize the evolutionary relationship of the NPC to membrane coating complexes.40–42 In addition, a lipid-detergent micelle (in white) encircles the membrane-interacting layer (shown in blue, Figures 2A and 2B). Nup157, Nup170, and Nic96 CTDs in the membrane-interacting layer, along with Nup84 Y-complexes in the outer rings, form a stable “coat” for the pore membrane.6,18,43,44 In addition, a pair of α helices extend from each copy of the innermost Nic96 CTD (aa 769–821) to support a Nup157 β-propeller at the membrane anchor site (not shown). Nups in the adaptin-like layer (dark pink) mediate the attachment of Nsp1 complexes (shown in green) to the membrane-interacting layer, while Nsp1 complexes with 4 copies per spoke encircle the central channel and contribute FG repeats from their N termini to the transport machinery. The open packing of Nups in the IR may facilitate rearrangements during radial expansion and transport.7,25
The resulting model of the IR also revealed four contact sites between adjacent spokes in the radially contracted conformation (Figures 2F–2H).7,24 The largest contact occurs between Nup170 β-propellers and their attendant α-helical solenoids; this feature forms part of a membrane anchor site on the local 2-fold axis (Figure 2G). Three additional contact sites have smaller buried surface areas. One contact at the midline of the IR is formed by the ends of adjacent, channel-facing 3-helical bundles from two Nsp1 complexes (Figure 2H; black ellipse). Two additional symmetry-related contacts involve Nup192-NTD helices that interact with the Nup188 tail domain (Figure 2H; arrows, dashed oval). Intriguingly, the Nup188 tail conformation may be stabilized by two Nic96 connector helices. The limited buried surface area for three of the inter-spoke contacts may be conducive to radial expansion of the IR during which, to a first approximation, each spoke behaves as a rigid body.7,25 Significant changes in the N-terminal regions of the two Nup170 molecules occur during radial expansion,7 and further rearrangements may allow the import of inner membrane proteins through a large gap between adjacent spokes in the expanded IR.45
Connectors in the IR
The resolution of our spoke map and completeness of the model allowed us to identify additional connectors in the IR that arise from the Nup145N/116/100 and Nup53/59 families (Figure 3A, in gold). These “new” connectors are aligned along a diagonal path when viewed from the central 8-fold axis and interact with Nups in all three spoke layers. In addition, Nic96 NTD connectors are better resolved (Figures 3A and 3B, in blue) and stabilize internal walls formed by α-helical bundles from Nsp1 complexes; they also connect Nup188 and Nup192 in the adaptin-like layer to Nic96-CTDs in the membrane-interacting layer.7 In brief, 16 new connectors were grouped with two previously identified connectors that do not arise from Nic96.7 Thus, 9 connectors, labeled C1–C9 and their symmetry mates (C1′–C9′) are shown within segmented 3D densities (Figures 3B and 3C, in gold; Table S3).
Figure 3. Connectors in the inner ring tie together Nup layers in the spoke.
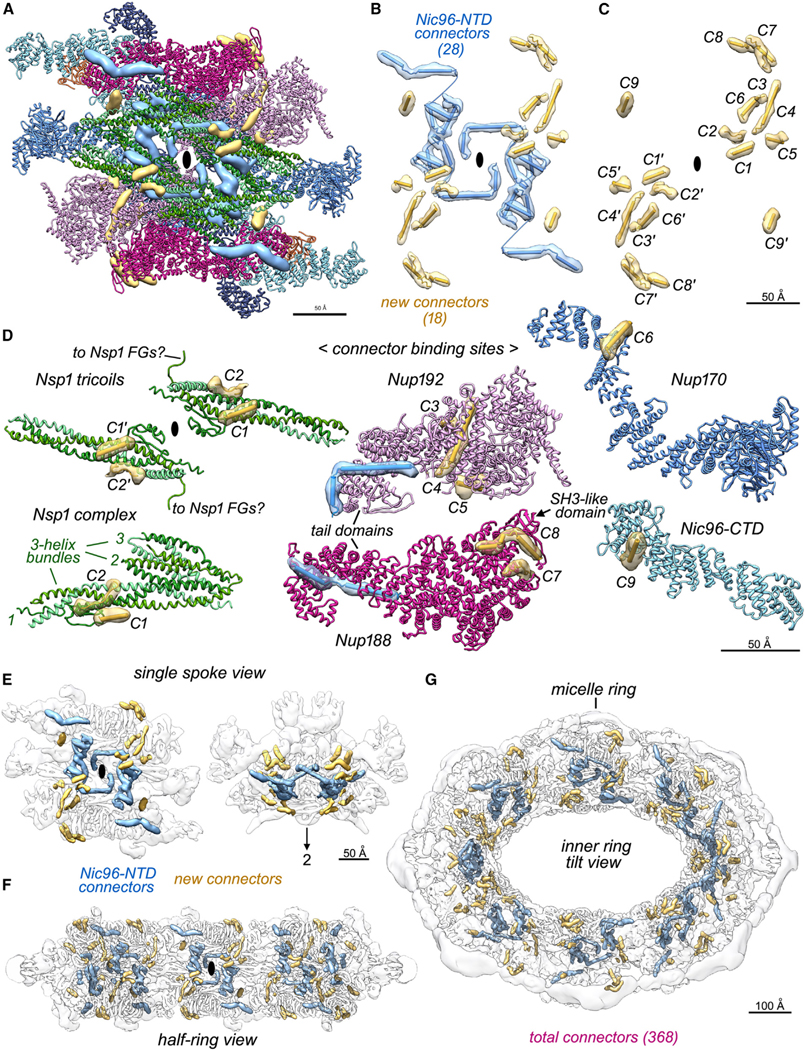
(A) A front view of the spoke with Nups displayed as ribbons and connectors as segmented and color-coded density maps: Nic96 NTD connectors (blue) and new connectors in gold.
(B) A front view is shown along the local 2-fold axis (black ellipse) with spoke connectors as cylinders and strands in their local density.
(C) Newly identified and grouped connectors labeled C1 to C9 (and their symmetry mates: C1′–C9′) start at the inner layer and extend into the membrane-interacting layer (Table S3).
(D) Interactions between new connectors (gold) and their Nup partners.
(E) Color-coded electron density for Nic96 NTD and new connectors are shown for a single transparent spoke (front and top views).
(F) All inner ring connectors are shown in a half ring view.
(G) Connectors within the inner ring are viewed in a 45° tilt view.
Crystal structures of Nup145N connectors with their cognate Nups from Chaetomium thermophilum19,46 allowed for the identification of similar S. cerevisiae connectors and their partners that include the following: the C3 connector with the Nup192-NTD (Figure 3D, Nup192 panel), the C6 connector with the Nup170 α-helical solenoid (Figure 3D, Nup170), and the C7 connector with Nup188 (Figure 3D, Nup188). In Chaetomium, these Nup145N connectors span ~119 residues and extend from the C-terminal autoclipping site toward the N terminus with sequential binding sites in Nup170, Nup188, and Nup192 (Table S3).19,46 We were not able to identify every new connector, but given their size and physical proximity, it is possible that connectors C3–C8 inclusive may arise from a single Nup145N molecule. Importantly, the new connector binding sites on the Nsp1 complex, Nup170, Nup188, and Nup192 (Figure 3D) are not laterally duplicated, as occurs for 2 copies of the Nic96 connectors in each half spoke (Figure 3B, blue rods).7 Hence, the remaining unidentified connector sites in the form II NPC may not originate from a second copy of Nup145N that is created by the cleavage of a Nup145 molecule in the distal Y-complex of each double outer ring protomer.47,48 Nup145N paralogs Nup116 and Nup100 are absent from Chaetomium49 and may contribute to some of these connectors in S. cerevisiae, as the distribution of new binding sites is symmetrical about the local 2-fold axis of the spoke. The C9 connector was modeled based on the crystal structure of a Chaetomium Nup53 α helix46 with its partner and interacts with the Nic96-CTD near the local 2-fold axis (Figure 3D, Nic96-CTD). Intriguingly, the C1 connector is present on the channel-facing surface of an Nsp1 complex (Figure 3D, left panel) and a short linker extends from Nsp1 toward FXFG repeats in the central channel. We also identified the C4 connector as a long α helix (Figure 3D, Nup192)19,46 that may arise from the Nup145N family (because of its proximity to C3) or the Nup53 family (Table S3). While four of the new con-nectors have been documented in crystal structures, speciesspecific differences may influence the position, size, and secondary structure of these features.
In total, we have identified 46 connectors per spoke with 368 within the IR (Figures 3E–3G). This analysis revealed an extensive network upon which the integrity of the NPC must depend. Connector binding to Nups may lock down hinge regions in α-helical solenoids to promote their interactions with Nup partners and drive assembly of the spoke and IR. In addition, the C7–C8 connectors are in close proximity to an SH3-like domain in Nup188 that contacts the nuclear outer ring in the radially expanded form I yeast NPC.5,7 We have shown that Nup145N, Nup116, Nup100, Nup59, and Nup53 may reversibly associate with the NPC, and readjustment of these Nup connectors may contribute to NPC flexibility,50 a proposal supported by recent structures.6,7,19 Importantly, these connectors may help to maintain the integrity of the IR during transport. A higher-resolution map is needed to place the connectors within their cognate Nups and to reveal additional connectors that may be present.19,46
Membrane anchor complexes and the LR
The primary sequences of Nup orthologs in the core scaffold are weakly conserved across species, while their structures and functions are generally conserved.51 However, membrane proteins in the inner and LRs are more divergent.52 We determined a 3D density map at ~10–12 Å resolution for the Nup157 membrane anchor site that involves Pom34 and Pom152 (Figures S5A and S5B; Table S1). Collectively, the IR Pom34-Pom152 TMDs with the LR and Ndc1 form an integrated superstructure that positions the NPC within the pore membrane, with the latter replaced by a micelle ring in this structure (Figures 4A and 4B). At a higher threshold, the 3D map of the anchor site contains 10 elongated densities arranged in two parallel rows that were modeled with 5 rods in each row (Figures 4C and 4D). These rods probably represent two Pom34 and three Pom152 TM α helices that are predicted by AlphaFold226 and would give 10 TM helices in the tetrameric anchor complex with 2 copies each of Pom34 and Pom152. This stoichiometry is in agreement with quantitative mass spectrometry.6 Interestingly, biochemical analyses suggest that Pom152 has one defined TM,53 raising the possibility that some of its membrane-interacting helices may be MBMs rather than full TM domains. Nevertheless, the Pom34 and Pom152 membranous hlices interact with Nup157 β-propellers, and all 10 helices may be necessary to form a stable membrane anchor site (Figures 4B and 4C).
Figure 4. Membrane anchor complexes for the inner ring.
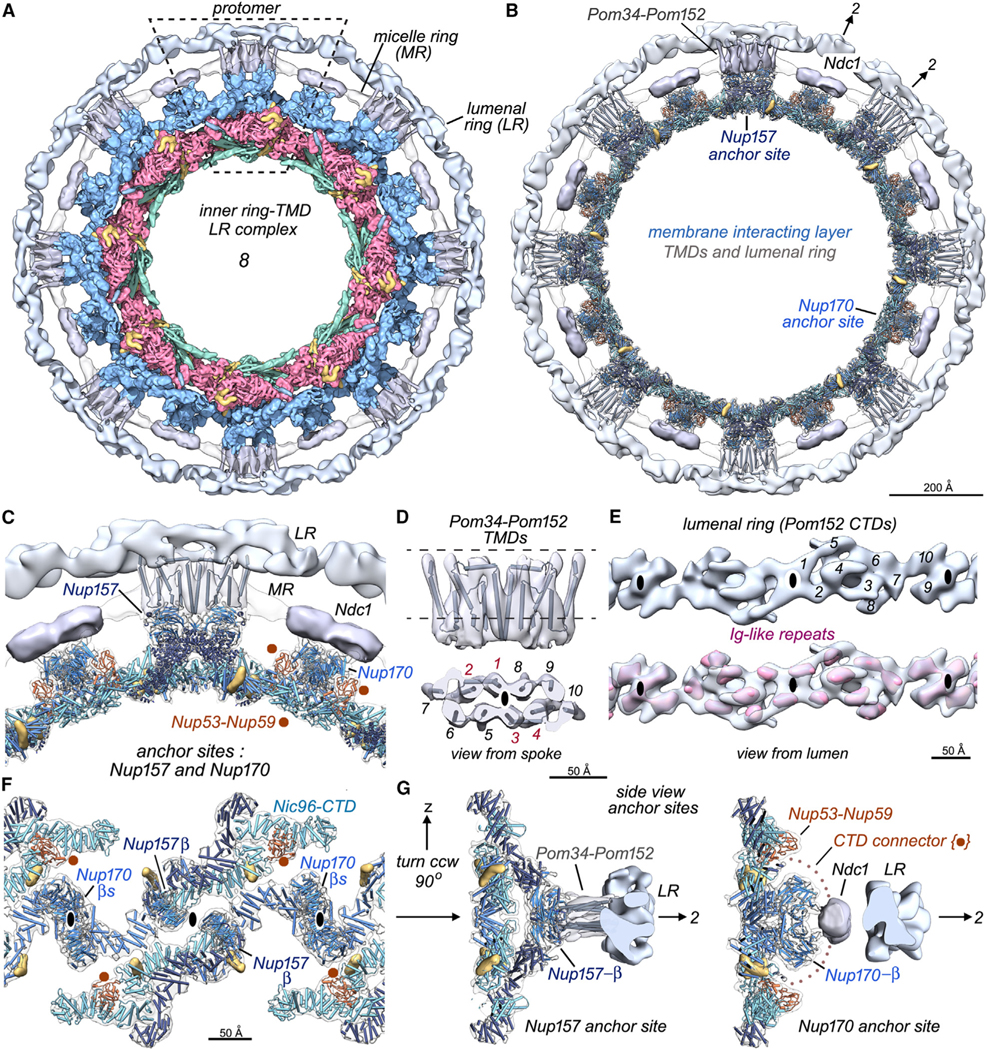
(A) An inner ring-TMD lumenal ring complex is viewed along the C8 axis with one protomer outlined (dashed rhomboid). The lumenal ring (LR) is light silver, TMDs for Pom34-Pom152 and Ndc1 are in gray, and the micelle ring is white.
(B) A molecular model for the membrane-interacting layer is juxtaposed to Nup157 and Nup170 TMD anchor sites and the lumenal ring.
(C) A close up of the Nup157 and Nup170 anchor sites; Nup53-Nup59 heterodimers (brown) are in close proximity to Nup170 molecules at the interface between spokes.
(D) Orthogonal views are shown of the Pom34-Pom152 TMD map with gray cylinders to mark 10 α helices aligned in two parallel rows of 5. The protomer in this dimeric complex may correspond to an α-helical bundle formed by rods 1–2 and 5–7 (see text).
(E) Top: an improved 3D density map for the lumenal ring viewed from the NE lumen. Local 2-fold axes are indicated with black ellipses, and 10 densities are numbered in one repeat. Bottom: 10 Ig-like features (red) are present in the lumenal ring protomer.
(F) Anchor sites in the membrane-interacting layer as viewed from the pore membrane. These include Nup157 and Nup170 β-propellers at local 2-fold axes and likely contributions from Nup53-Nup59 heterodimers (see text).
(G) Cross-sections centered on anchor complexes for Nup157 (left) and Ndc1 (right) are viewed at right angles to the local 2-fold axes.
In a second approach, an integrative model of the TMD region was computed based on AlphaFold2 predictions for Pom34 and Pom152 (aa 1–249) monomers and other restraints (STAR Methods). The resulting ensemble satisfies 100% of the cross-links (Table S4) with a cross-correlation of 0.95 between the 3D density map and a model probability map. The integrative structure localizes Pom34 and Pom152 TMDs along with a non-TM helix of Pom34 that is aligned parallel to the membrane (Figures 4D, top panel and S5C–S5E). Integrative and manually built models identify a similar pattern of TMs; however, the third TM in the AlphaFold2 model of Pom152 was extended in the manually built model to better fit the cryogenic electron microscopy (cryo-EM) density map. In both models, Pom34 and Pom152 TMDs form a 5-helical bundle corresponding to rods 1–2 and 5–7, respectively (Figures 4D, bottom, S5B, and S5E) along with their symmetry copies (rods 3–4 and 8–10). Thus, the TM region of Pom152 may comprise three TM helices, rather than just the third helix as proposed previously,53 or alternatively, interactions with adjacent Nups during assembly may stabilize these two additional N-terminal TM helices. We note that rods in the Pom34-Pom152 TMDs are further apart than parallel TM helices in typical, solubilized membrane proteins; this may reflect the participation of pore membrane lipids54 or an atypical membrane interaction of TMDs.53 In addition, some Pom34-Pom152 TMDs extend ~20 Å beyond the pore membrane to contact the LR comprising Pom152 CTDs (Figures 4A–4C and 4G, left). Higher resolution will further clarify this membrane anchor complex.
We next used two different refinement strategies to obtain improved 3D maps of the LR (Figures S1 and S5F; STAR Methods). In previous work, the LR had a smooth, sinusoidal outline,6,7,13 whereas the ring in current maps is reticulated and nearly circular (Figures 4A, 4B, and 4E). We surmise that the reticulated appearance is due to small Ig-like domains that have been resolved in each repeat, while the more circular profile arises from extended Pom34- and Pom152-connecting rods that contact the LR at a higher radius than was visualized previously. We identified 10 Ig-like densities in the repeating unit based on their size and shape (Figure 4E, top; 1–10), and each Ig-like module is shown as an isosurface (in red, Figure 4E, bottom). Further work is needed to understand the packing and connectivity of Ig-like domains in the LR.
A second membrane anchor site is located on the local 2-fold axis between adjacent spokes and contains two Nup170 molecules (Figure 2G). We observed two high-density regions in the lipid-detergent micelle ring in close apposition to Nup170 with the correct volume to represent 12 TMs of an Ndc1 dimer, as predicted by AlphaFold255–57 (Figures 4A–4C). Thus, Ndc1 may be located at this site in agreement with biochemical data and quantitative mass spectrometry.6,58–60 However, the interface between spokes is flexible, and densities for Ndc1 CTDs were not resolved; these domains face the core scaffold and may become disordered during isolation, or they may be dynamic in vivo, in agreement with other studies.50,61 An overview of the anchor sites, as viewed from the pore membrane, shows Nup157 and Nup170 β-propellers on local 2-fold axes (Figures 4F and 4G) where they interact with TMDs from their membrane protein partners. We also identified folded RNA recognition motifs in the Nup53-Nup59 heterodimer (Figures 4C and 4F, brown dots). Nup53-Nup59 is located between adjacent Nic96 CTDs in each half spoke, and their flexible N and C termini contribute connectors to the IR. Moreover, Nup53-Nup59 heterodimers from adjacent spokes are in close proximity to their respective Nup170 molecules and could contribute C-terminal amphipathic helices to this membrane anchor site (Figures 2E, 4F, and 4G right).59,62–64 In addition, yeast Ndc1 also contains an amphipathic helix in its CTD that may interact with similar motifs in the Nup53-Nup59 heterodimer.11
In summary, 2 copies of Pom34-Pom152 form a tetrameric platform in the pore membrane that helps to anchor each spoke within the pore membrane through interactions with Nup157 (Figures 4A–4D, S5A, and S5B). A second, equatorial membrane anchor is located between adjacent spokes wherein Ndc1 interacts with Nup170 and perhaps Nup53-Nup59.6,59,60 Intriguingly, while there are some similarities in fold arrangement and usage between Pom152 and its vertebrate counterpart, Gp210,18,65 many aspects of the TM regions are less well conserved. These differences may be correlated with NE morphology as the distance between the inner and outer membranes is smaller in yeast cells when measured near the IR, ~250 versus ~320 Å in humans.7,18
Architecture of the NR
Double outer rings appear to be ubiquitous in vertebrate NPCs.22,66 Their presence in unicellular eukaryotes was recently confirmed in Chlamydomonas reinhardtii,23 S. pombe,25 and a subset of S. cerevisiae NPCs7; in these organisms, a double outer ring is located on the nuclear side, and a single outer ring is present on the cytoplasmic side. We reanalyzed our NPC dataset to create an improved density map for Y-complexes7,67–71 in the double outer ring (Figures S6–S8). We also identified orphan Nup densities and linker domains that connect to the IR, while confirming earlier data on orphans in this superstructure7 (Figure 1).
Refinements in CryoSPARC39 provided a 3D density map of the double Y-complex at ~9–10 Å resolution (Figures S6A and S6B; STAR Methods), which allowed for the placement of helical pairs and β-propellers from Nup AlphaFold CSMs26 in both the proximal and distal Y-complexes (Figures 5A and S7). A complete model for the junction between Nup120, Nup85, and Nup145C (e.g., the hub) was made for each Y-complex (Figures 5A and 5C, panels 1 and 2). We also identified an α helix that extends from the Nup145C N-terminal region after it forms a 4-stranded β sheet that is inserted into the Sec13 β-propeller72 (Figure S7D, Nup145C-Sec13 panels); this helix contacts the Nup85 arm in each Y-complex (Figure 5C, panels 1 and 2). Notably, 7 of 8 β-propellers in each protomer were docked in well-fitting density, including propellers from Nup120, the proximal copy of Nup133, and 2 copies each of Seh1 and Sec13 (Figures 5A, 5B, and S7A–S7C).
Figure 5. Conformation of Y-complexes and characterization of orphan Nup densities and linker domains in the double outer ring.
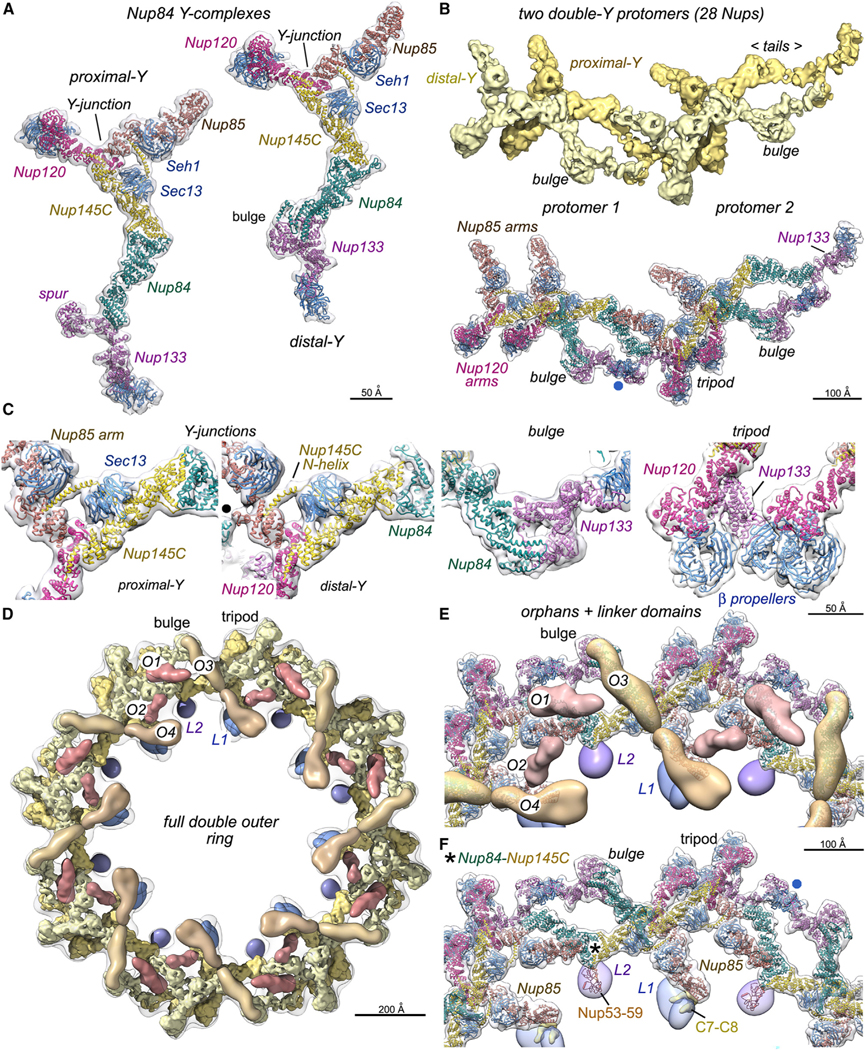
(A) Left: ribbon model for the proximal Nup84 Y-complex oriented vertically. Right: model of the distal Y-complex with the vertical offset that is present in the double ring protomer.
(B) Top: 3D density map with four Nup84 complexes in two protomers of the double outer ring. Bottom: molecular model of two adjacent protomers with notable features labeled. A non-canonical interaction of the distal Nup133 β-propeller with the spur of the proximal Nup133 is marked (blue dot).
(C) Four panels with close-up views of notable regions in Nup84 Y-complexes, including the Y-junction/hub, a non-canonical bulge in the distal Y-complex, and a tripod of membrane-interacting β-propellers. The black dot in panel 2 marks a lateral contact between Nup85 and Nup84 in an adjacent protomer.
(D) Overview of a full double outer ring with putative Nic96 CTDs (O1 and O2; dark brown), possible Nup188–192 orphans (O3 and O4; medium brown), linker domains (L1, dark blue; L2, purple), proximal and distal Y-complexes (gold and light yellow).
(E) A zoomed-in view of the full double outer ring with models of the double Y-protomers and orphans.
(F) A similar view to (E) without orphan Nups; semi-transparent L1 and L2 linker domains reveal their respective footprints on Nup188 in the spoke (above the C7– C8 connectors) and the Nup53–Nup59 heterodimer. A contact site for L2 on the proximal Y-complex is indicated (asterisk) along with a non-canonical interaction of the distal Nup133 β-propeller (blue dot).
A model was also built for a distinctive, torus-like bulge formed by the CTDs of Nup84 and Nup1337; this bulge creates a shorter contour length for the distal Y-complex (Figures 5A and 5C, panel 3). A tripod is formed by β-propellers from two Nup120 molecules and the proximal copy of Nup133, which may position their MBMs (also known as ALPS motifs73,74) to interact with the NE (Figures 5B, bottom, 5C, panel 4, and S9A, left).7 In addition, the distal Nup133 β-propeller interacts with the C-terminal spur domain of Nup133 from the proximal Y-complex (Figures 5A, 5B, and 5F; blue dot) to help stabilize the double ring. Conversely, β-propellers from Xenopus Nup160, Elys, and Nup133 form a tripod, and two adjacent tripods in each protomer create an extended membrane binding site in the NR (Figure S9A, right).29,32 These distinctive tripod arrangements may reflect differences in NE topology, while accommodating the flat orientation of the double outer ring on the yeast NE and the greater tilt of its vertebrate counterpart. In yeast, altered conformations for Nup85, Nup84, and Nup133 are present in the two Y-complexes due to the local geometry of the double outer ring (Figure S7D). Long loops extend from β-propellers to make contacts with adjacent Nups and the NE. Thus, Sec13 and Seh1 form stable hubs that act like gripping hands with extended finger-like loops to help maintain both their position and the integrity of their local neighborhood (Figures 5C, panels 1 and 2 and S7D). This key structural role may explain why these proteins are also utilized in other homologous yet functionally unrelated coating complexes.42
We next investigated how the massive double outer ring is positioned on the nuclear-facing surface of the IR. A subset of NPCs with double outer rings was identified by 3D classification (~20%), and refinements with C8 symmetry provided a density map at ~23 Å resolution that captures many of its salient features (Figures S1, left, S6C, and S8A; Table S1). A 3D map from the double ring of Nup84 Y-complexes fits into the density map of the full ring, thereby allowing the tripod of β-propellers and the torus-like bulge in the distal Y-complex to be identified (Figures 5D and S8).7 At this stage, orphan Nup densities were identified (Figures 5D and S8B; Table S5, orphan 1 [O1]–O3),7 including O3 that runs along the top surface of the distal Y-complex; this orphan may stabilize the torus-like bulge to promote double ring assembly. We extended our orphan catalog by docking a low-pass filtered density map of Nup188 into a large density that faces into the central channel (Figures 5D and S8D, O4). O4 interacts with the Nup85 arm of the proximal Y-complex and may position a linking domain that connects to the IR. We could not discern whether the larger orphans (O3 and O4) arise from Nup188 or its paralog Nup1927; however, Nup205 (a Nup192 ortholog) is present in vertebrate double outer rings.18–20,28–30,32,34 In addition, two elongated orphans (O1 and O2) are the correct size for Nic96 CTDs; each of these features is in close proximity to a cognate Nup188–192 orphan with which they may interact through their NTDs (Figures 5D and 5E, O1/O3 and O2/O4).7,24
When taken together, the data suggest that two Nic96 CTD/Nup188–192 pairs are present in each protomer of the yeast double outer ring. Intriguingly, the Xenopus NR has a single Nup93-Nup205 pair in each protomer,29,32 while the cytoplasmic double outer ring has two pairs per protomer.28–30,34 Moreover, the predicted Nup cognate pairs are located within a double Y-complex protomer in S. cerevisiae, while the single pair in Xenopus spans two protomers of the NR.29,32 Densities for these additional Nups have not been found in single outer rings of the form I NPC in S. cerevisiae.5,7 This suggests a possible role for the additional structural Nups, which may impart more rigidity to the double outer rings.
At this point, we identified two orphan densities that form links between the double outer ring and the IR. The first is a bilobed density that contacts the Nup85 arm of the proximal Y-complex and sits beneath the putative Nup188–192 orphan (O4), which faces the central channel (Figures 5D–5F, S8B, and S8E–S8G; linker domain 1 [L1]). The orphan Nup(s) in this bilobed density may contact Nup85 in the proximal Y-complex and is located directly above a Nup145N connector binding site in the spoke (C7 and possibly C8, L1; Figure 5F) that is in close proximity to an SH3-like domain in Nup188 (Figures 3D and S8G; Table S5; Nup188). Intriguingly, this SH3-like site is also involved in a link between the single outer ring and the nuclear face of the IR in the form I yeast NPC.5 The position of the L1 domain suggests that it might contain a monomer or dimer of the Nup145N autoproteolytic domain (APD)6,75,76 and additional Nups may be present. In addition, the L1 density is near a long a helix that extends from the N terminus of Nup145C and contacts the proximal copy of Nup85. A second linker density interacts with the Nup145C-Nup84 interface in the tail of the proximal Y-complex and contacts the Nup53-Nup59 heterodimer in the spoke (Figures 5D–5F, S8B, and S8E–S8G; L2). Importantly, the yeast NPC does not appear to use Nup157 or Nup170 to form links between the inner and outer rings, as occurs with the Nup155 homolog in vertebrates.66 This may account in part for the smaller vertical spacing between the inner and outer rings in the yeast NPC relative to vertebrates (Figure S8G).
Our model of the NR suggests that IR components may be recruited to help stabilize the yeast superstructure.7 However, these IR components are positioned in a different manner relative to those in vertebrates,18–20,28–30,32,34 underscoring the modular nature of the NPC in which different architectures can be constructed from similar Nup building blocks.51 NRs have been found in all branches of the eukaryotic family tree that have been studied; this arrangement may be an ancestral form that has subsequently evolved by addition or subtraction in different lineages.
Cylindrical arrays of FG connectors in the central channel
FG-repeat-containing Nups are anchored to the core scaffold and extend flexible termini with transport-factor binding sites (FXFG/GLFG repeats) into the central channel.15,77,78 In previous work,6,7 we documented FG-repeat connectors from Nsp1 complexes that are located above and below the mid-plane of the IR. However, additional features of the central channel and FG connectors remain to be characterized. To test this idea, ~5,000 form II yeast NPCs with NRs were refined with C8 symmetry and appropriate cylindrical masks. Importantly, a subset with better-resolved, CRs was identified by 3D classification (~1,400 particles). Although the resolution is low (20–30 Å), these density maps revealed connections to the central transporter that emanate from the outer rings on both sides of the core scaffold. Potential FG connectors were modeled with blue and red cylinders in their respective 3D maps (Figures 6A and 6B). Altogether, FG connectors extend from the CR, the mid-plane of the IR, and the NR (Figures 6C–6F).6,7 Each bridge-like density spans a gap of ~50–70 Å to make contact with sites on the channel periphery (Figures 6C–6F, S9B, and S9C, dotted circles) and may contain two or three FG connectors to provide the necessary stiffness to cross the local gap.
Figure 6. Bridging FG connectors between the core scaffold and central transporter.
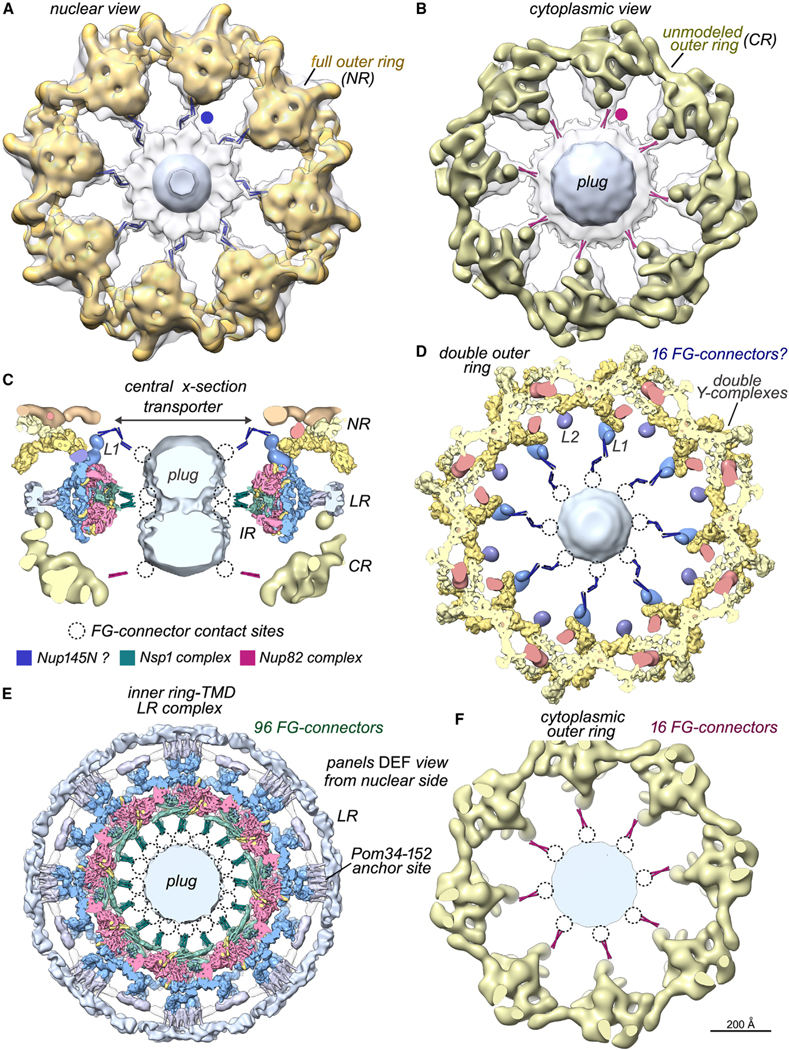
(A) A map of the full nuclear double outer ring (gold, NR) is docked within a density map from the entire NPC (transparent gray). A thick slab shows connectors from the double outer ring to the central transporter/plug (blue dot), which were modeled with blue rods for visualization.
(B) A map of the single cytoplasmic outer ring (bronze, CR) is docked into a map of the entire NPC. This view reveals FG connectors (red dot and rods) appropriately positioned to originate from the Nup82 complex associated with the single outer ring.
(C) Modeled FG connectors are present at three levels along the central channel in this central cross-section; labels: inner ring (IR), lumenal ring (LR), and linker domain 1 (L1). Possible contact sites on the plug (dotted circles) and a color key for FG connectors are shown.
(D) A thick section of the double outer ring viewed along the central C8 axis with modeled FG-connector bundles to the central transporter. (D)–(F) are viewed from the nuclear side.
(E) Thick section of the inner ring-TMD lumenal ring complex with modeled FG connectors from Nsp1 complexes.6,7
(F) Thick section of the cytoplasmic outer ring with modeled FG connectors.
On the nuclear side, connections to the central transporter originate from the bilobed L1 density attached to the Nup85 arm of the proximal Y-complex; this feature also forms a linker domain to the IR (Figures 5F and S8G). One component of this orphan density may correspond to a Nup145N-APD monomer or dimer, and in this scenario, bridges to the central transporter would represent the N terminus of Nup145N, which contributes about 200 residues with GLFG repeats to the transport channel (blue rods, Figures 6A, 6C, and 6D). On the cytoplasmic side, FG connections from the Nup82 complex were resolved previously with CET7 for the in situ NPC. Hence, cytoplasmic bridges in the isolated NPC may originate from Nsp1 molecules in dimeric Nup82 complexes that bind to each Nup84 Y-complex in the single outer ring (red rods, Figures 6B, 6C, and 6F).10,79 These density features may also contain FG connectors from Nup159 and Nup116, as the latter’s autoproteolytic-like domain associates with the Nup82 complex.6,10,80 In total, ~48 bridging densities have been identified that link the scaffold with the central transporter, which may be formed by connectors from ~128 FG Nups (Figures 6D–6F and S9C; Table S6).6,7 These data suggest that FG connectors may confer a limited degree of organization on the central transporter with FG repeats arrayed in discrete bundles at the channel periphery, which then spread into a more flexuous configuration near the central axis.
DISCUSSION
Implications of a multiscale NPC structure
In this study, we created a composite, multiscale structure of the form II yeast NPC that attains α-helical resolution in the IR and 9–10 Å for Nup84 Y-complexes in the double outer ring (Figure 1). The accuracy and completeness of Nups in the IR and double outer ring were increased with Nup AlphaFold CSMs,26 MDFF,38 and integrative structure modeling.81,82 How do these observations impact our understanding of the functional architecture of NPCs? Vertebrate pore complexes contain an IR framed by two double outer rings,66 while the major isoform in S. cerevisiae (form I) has single outer rings on both cytoplasmic and nuclear faces5–7 and is less massive (~52 versus ~109 MDa).6,7,18 Even so, both of these NPC isoforms have a quasisymmetric architecture that reflects the environment of the pore membrane. However, adaptations that confer a sidedness to the outer rings include the presence of structural Nups that are found in the spoke, along with the cytoplasmic Nup82 complex, cytoplasmic filaments (in vertebrates), and the nuclear basket. In addition, the form II yeast NPC displays a larger asymmetry due to the presence of a double NR.7 Regions of the core scaffold that interact with the NE show significant divergence across species, while the diameter of the central channel and the ability to undergo radial expansion are generally conserved.5,7,18,21,25
An emergent theme is the versatility of α-helical solenoid Nups that reside within the spoke and outer rings of yeast and vertebrate NPCs. This is exemplified by the conformational flexibility of Nups 84, 85, and 133 in Y-complexes of the yeast double outer ring44,69–71,74 and extends to multiple copies of laterally offset Nups that are present in each half spoke of yeast and vertebrate NPCs.6,7,18,31,46 In addition, vertebrate Nup93/Nup205 and their yeast orthologs (Nic96 and Nup188–192) may be present in both the spoke and double outer rings,7,18–20,28–30,32,34 while form I yeast NPCs do not contain Nic96 and Nup188–192 densities in the single outer rings.5,7
The ubiquitous presence of double NRs, along with Nups that are building blocks of the spokes, would suggest that this super-structure evolved at an early stage and was present in the last common eukaryotic ancestor.83 Single outer rings in C. reinhardtii, S. pombe, and S. cerevisiae5–7,23,25 might be logical intermediates in the evolution of double outer rings or could have arisen as the result of secondary losses, as may occur in the incomplete or more flexible CRs of S. pombe NPCs.25,84 In S. cerevisiae, the presence of a single outer ring on the cytoplasmic side may have provided the impetus for dimerization of the Nup82 complex to maintain a functional stoichiometry that is in line with double outer rings from other NPCs. In either case, the presence of single outer rings in unicellular organisms may create a more dynamic scaffold that is responsive to changes in transport cargo levels and the nuclear environment. This also suggests that the NR in single-celled organisms may be associated with an important function such as basket anchoring. Indeed, the basket appears to be dynamically assembled on NPCs through an RNA-dependent mechanism.85 It is thus reasonable to hypothesize that assembly of the NR and basket might be co-ordinated for specific RNA processing roles.
Membrane anchor sites may act as transducers for mechanical processes associated with the NE.25 Remarkably, membrane anchor sites show a large structural variation between yeast and vertebrate NPCs. Thus, vertebrate Nup155 forms links between the inner and outer rings that provide additional interactions with the NE through MBMs in their β-propellers.86,87 In yeast, two distinctive membrane anchor complexes are present at local 2-fold axes in the equatorial plane of the IR.6,7 One anchor complex contains a tetramer with 2 copies each of Pom34 and Pom152 that may interact with the MBMs of Nup157 β-propellers. However, there are no clear Pom orthologs in vertebrates, although a distant evolutionary relationship may exist between yeast Pom152 and vertebrate Gp210, as both contain numerous tandem Ig-like modules.12,13,18,65 Extended regions of Ig-like repeats in Pom152 and Gp210 both form LRs that interconnect adjacent spokes in their respective NPCs, yet these rings have radically different subunit copy numbers and structures.6,7,18,65 The larger LR in vertebrates may be necessary to restrict the dynamic expansion of the more massive NPC or may have other functions.65
A second membrane anchor complex in yeast NPCs contains Ndc1 and Nup170.59,78 As shown herein, Ndc1 and Pom34-Pom152 interact with β-propellers in Nup170 and Nup157, respectively.6 Ndc1 monomers in vertebrate NPCs are positioned at a greater distance from the local 2-fold axis and interact with the β-propeller of Aladdin, rather than Nup155 (a Nup170 ortholog).18 Thus, membrane anchor complexes have diverged in yeast and vertebrate NPCs. However, a recurring motif involves the use of a Nup β-propeller to contact the membrane protein, and this may extend to newly discovered interactions between Nup84 Y-complexes and yeast Ndc1.11
The Nup53-Nup59 heterodimer is illustrative of the divergence in multitasking NPC modules. Nup53 and Nup59 are paralogs with each subunit having extended N- and C-terminal tails that provide spoke connectors and membrane anchoring motifs.59,62–64 The Nup53-Nup59 heterodimer has been located between the inner and outer CTDs of Nic96, faces the NE, and may contribute amphipathic C-terminal helices to the Nup170-Ndc1 membrane anchor complex.6,7 However, the equivalent Nup35 dimer was not resolved in the Xenopus IR,31 while human Nup35 dimers have been positioned between adjacent spokes, near the NE in a lower-resolution 3D map. In this case, Nup35 dimers may provide connectors between adjacent spokes to stabilize the vertebrate IR.18,19
Overall, significant alterations have occurred in yeast and vertebrate NPC architecture that may reflect changes in the NE, while features that lie at the heart of nucleocytoplasmic transport are more strongly conserved. Moreover, Nup composition and the organization of critical sub-assemblies may reflect the optimization of physical properties that govern interactions with the NE, which in turn direct the scaffolds’ response to mechanical stress.25,88 Finally, large-scale structural variations in NPCs may provide functional specialization in different nuclear regions7 and could be present in other organisms.
Role of connectors and the topology of Nup145
An unusual feature of the NPC is the presence of flexible connectors in the N and/or C termini of Nups that in yeast include Nic96, Nup53-Nup59, and the Nup145N family (Nup145N, Nup116, and Nup100).6,7,89 These connectors form links between the three major layers in the spoke and have additional functions.6,7,18,19 In previous work, we showed that 7 connectors in the Nic96 NTD (28 in total) can be divided into two groups. One group binds to pairs of 3-helical bundles from Nsp1 complexes (Nsp1, Nup49, and Nup57)46,90,91 to stabilize diagonal walls within the spoke.7 These diagonal walls connect channel-facing Nsp1 3-helical bundles, which anchor N-terminal FG repeats within the central channel, to Nup188 and Nup192 in the adaptin-like layer. A second group tethers Nic96-CTDs in the membrane-interacting layer to C-terminal tails of Nup188 and Nup192 in the adaptin-like layer.7,46,49,89
We have now identified 16 additional connectors in each spoke, which were grouped with two previously unassigned connectors.7 We used extant crystal structures of Nup-connector complexes from Chaetomium thermophilum19,46 to identify 8 connectors in each S. cerevisiae spoke; 6 connectors arise from the Nup145N family, and 2 connectors arise from Nup53 or Nup59.46 Thus, Nup145N-connector binding sites have been identified on Nup170, Nup188, and Nup192 along with a Nup53-Nup59 site on the innermost copy of Nic96 CTD. These connectors tie together Nups within the adaptin-like and membrane-interacting layers6,19,89; higher-resolution data will be required to complete the 3D connector puzzle. Overall, the spoke connectors (368 in total) may confer structural plasticity to the IR by providing a large set of inter-Nup linkages that stabilizes the highly fenestrated spokes when they are subjected to potentially disruptive forces during transport. Connectors may also stabilize productive Nup conformations for spoke assembly and promote interactions required to assemble the IR. Some connectors form dynamic interactions within the scaffold even though they tie together much of the NPC50; hence, a regulated reversal of their associations with other Nups may underlie key elements of mitotic NPC disassembly in organisms with an open mitosis.7
Nup145 provides connectors that bind to specific sites in the spoke and undergoes a well-documented internal self-cleavage during its biogenesis.47 Autoproteolytic cleavage into Nup145N and Nup145C is mediated by a central APD, and the resulting proteins play important roles; however, the overall topology of Nup145 remains elusive. We now propose an initial model for Nup145 in the proximal Y-complex of the double outer ring and the IR (Figure 7A). Our data suggest that regions of Nup145C and Nup145N near the cleavage site may remain in close proximity at the proximal Nup84 Y-complex. Thus, Nup145C forms an α-helical domain in the stem of the Y-complex and contributes a β sheet that is inserted into the Sec13 β-propeller, and this cylindrical domain binds to the Nup145C near the Y-junction.72,92 A long α helix extends from the inserted β sheet toward the underside of the Nup85-Seh1 arm. We identified an orphan density that links the Nup85 arm and Nup188 in the IR; this linker domain may contain at least one copy of the Nup145N APD. In addition, three confirmed Nup145N-connector binding sites have been identified in the adaptin-like and membrane-interacting layers of the half spoke.6,19,46,89
Figure 7. Nup145 topology diagram and interplay between the core scaffold and central transporter/plug.
(A) Left: proposed Nup145 topology with a region of the double outer ring shown as an outset in the dashed box from the right-hand panel. Nup145N binding sites are indicated by gray rectangles. Right: schematic model of the form II yeast NPC with FG connectors to the plug/central transporter and their contact sites (dashed circles).
(B) Radial expansion of the NPC scaffold may trigger functional changes in the central transporter. FG connectors appear to be maintained in the transition; connectors are color-coded (Figure 6C) based on location and possible Nup anchor. Left: transport factors with cargo may be entrapped within the FG mesh of the plug-like transporter. Right: reorganization of the core scaffold and FG mesh promotes translocation of transport factor-cargo complexes.
As diagrammed in Figure 7A, this suggests a topology for Nup145N in which a reverse C to N tracing starts at the APD, traverses at least 3 connector binding sites in the membrane-interacting and adaptin-like layers, and then loops back to form an FG connector at the outer ring that delivers GLFG repeats (~200 residues) to the central transporter. This model will be refined in higher-resolution studies and raises questions about the role of possible connectors from the second copy of Nup145 that contributes a Nup145C molecule to the distal Y-complex. Moreover, the newly identified binding sites are present in both halves of the spoke, which suggests that a similar path may apply to Nup145N connectors or flexible regions in its paralogs (Nup100 and Nup116) that associate with the CR. Hence, Nup145N and its paralogs may provide flexible and dynamic links between the inner and outer rings of the yeast NPC.
The NPC scaffold creates a central transport assembly
The core scaffold creates a passageway across the NE and provides anchor sites for FG-containing Nups that extend transport-factor binding sites into the central channel. These natively disordered FG-repeat regions may be flexible and locally disordered within the central channel.93,94 However, the cylindrical symmetry of FG-Nup binding sites could create annular regions of similar FG repeats that, depending upon their chemical characteristics, may transiently interact with one another in the high local concentrations within the transport channel.15 Moreover, ring-like features have been observed in frozen-hydrated NPCs,95,96 and a plug-like feature has been observed in 3D maps (this work).6,7,16,96,97 In addition, surface imaging of fixed and metal-coated nuclei revealed central granules along with connecting filaments to the scaffold on the cytoplasmic side.98 A full complement of FG repeats would account for ~9.6 MDa in the central channel (Table S6),6 and these polypeptides may occupy a larger volume than folded proteins of comparable molecular weight. Transport factors and cargo are also present in the channel. This is based on imaging of micro-injected nucleoplasmin-gold conjugates after isolation of Xenopus germinal vesicles,99 mass spectrometry of yeast NPCs,6 and cryo-EM of 60S pre-ribosomal large subunits caught in transit.100 Thus, an amalgam of FG repeats, transport factors, and cargo complexes may create a blurred plug-like feature in isolated NPCs that is, in effect, cylindrically averaged during refinements.101
We have documented connections that extend from the midplane of the IR to the central transporter in both isolated and in situ yeast NPCs.6,7 FG connectors may also originate from two Nsp1 molecules in Nup82 complex dimers on the CR6,7 and from Nup145N on the double nuclear outer ring (Figure 7A, right). In total, FG connectors in 48 bridging densities may account for ~128 FG Nups and could provide ~77% of the expected FG repeats (by mass) in the central channel (Table S6). Collectively, the data are consistent with the hypothesis that some local order may exist where connectors extend from the core scaffold into the central channel. This idea seems inconsistent with prior views of FG repeats in the central transporter as a homogeneous phase,102,103 but it may be consistent with segregated pathways at different regions of the central channel for different kinds of transport.6,15,104,105
Recent analysis of S. pombe NPCs under conditions of osmotic shock or energy depletion showed that transport is downregulated when the scaffold undergoes a radial contraction.25 When mechanical forces are applied to the nucleus, the core scaffold has been shown to dilate,18,25,106 an effect that is associated with a weakening of the permeability barrier and an increase in facilitated transport.88 This data and prior work suggest that the NPC may function like a mechano-sensitive channel that responds to changes in the NE (Figure 7B).107 Against this background, transport models must account for the ~2-fold volume increase of the central channel during radial expansion of the IR and a smaller expansion of the outer rings (this work).7,25 Moreover, the plug-like feature in the central channel may undergo a remarkable change from an hourglass-shaped density in isolated NPCs (this work)6,7,16 to a flattened disk-like feature in human pore complexes imaged in situ (Figure S9C).21
We suggest that the plug-like feature, and the FG repeats in particular, may act as a gate to impede transport in NPCs with a radially contracted conformation (Figure 7B, left). Transport factors bind FG repeats with a relatively low intrinsic affinity (1–10 mM) that may be enhanced by the high local concentrations of these repeats in the central channel.93,108,109 Hence, a balance must be struck between FG-repeat density and transport-factor affinity to allow facilitated diffusion driven by binding and unbinding events. Radial expansion of the core scaffold may reorganize the FG repeats by lowering their packing density in the central channel to enhance transport (Figure 7B, right).18,25,88 A cylindrical perimeter of quasi-ordered FG domains attached to connectors (Figures 7A and 7B, righthand panels) could guide this process by forcing transport complexes toward the channel center99 while blocking non-productive interactions with the IR.
In this model, FG repeats have two functions: they may act as a barrier that entraps transport factors and cargo complexes in radially contracted NPCs, while reorganization of the FG matrix during expansion “unplugs” the central channel to facilitate a higher rate of transport. However, the link between the conformation of the core scaffold and the distribution of FG repeats in the central channel may be modulated by physical properties of the cargo complex100 and lateral tension in the NE.7,25 Thus, additional levels of complexity and adaptability remain to be investigated in this mechano-sensitive translocation channel.
Limitations of the study
Protomers in the co-axial rings of the NPC are dynamic and sensitive to isolation conditions; thus, the ability to determine 3D density maps with side-chain resolution is generally limited. In addition, Nups within the spoke and double outer ring are subjected to variations in local geometry that are not built into AlphaFold2 models. Our analysis indicates that X-ray crystal structures, Nup AlphaFold CSMs, and electron density maps may provide accurate models for Nups in the NPC when combined with flexible fitting. Caveats concerning the resolution and functional interpretation of various co-axial rings and sub-assemblies within the NPC are given in the results and discussion. Moving forward, experimental challenges in achieving near-atomic resolution over the entire ensemble will require a coordinated approach that is aided by improved 3D density maps110 and will build on the composite 3D structure reported herein.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by Christopher Akey (cakey@bu.edu).
Materials availability
Yeast strain W303 [MATa ade2–1 ura3–1 his3–11,15 trp1–1 leu2–3,112 can1–100 MLP1-PPX-ProteinA::HIS5] and plasmids generated for this study will be distributed without restriction upon request.
Data and code availability
Cryo-EM 3D density maps and related models have been deposited in the EMDB and RCSB, respectively. Integrative modeling data have been deposited in the PDB-Dev.
All original code has been deposited at Zenodo and is publicly available as of the date of publication. DOIs are listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Deposited data | ||
|
| ||
| Chemical Cross-linking with Mass Spectrometry readout datasets | Akey et al.7 | Zenodo (https://zenodo.org/) Accession#: 5721514 |
| Files and input data, scripts and results for integrative modeling of the Pom34-Pom152 tetramer | This paper | Zenodo (https://zenodo.org/) DOI-10.5281 Accession#: 8226857 |
| Integrative models for Pom152 rings | This paper | PDBDEV_00000213 |
| Full nuclear double outer ring of the yeast NPC | This paper | EMD-41114 |
| Double ring of Nup84 Y-complexes from the yeast NPC | This paper | EMD-41285 |
| Lumenal ring of the isolated yeast NPC | This paper | EMD-41116 |
| Pom34-152 membrane attachment site in the yeast NPC and preliminary model | This paper | EMD-41117 PDB: 8T9L |
| Inner ring spoke of the yeast NPC | This paper | EMD-41300 |
| Cytoplasmic outer ring of the yeast NPC | This paper | EMD-41119 |
| Central transporter/plug in 3D map of the yeast NPC | This paper | EMD-41120 |
| Connectors from the double outer ring of the yeast NPC | This paper | EMD-41121 |
| Connectors from the cytoplasmic outer ring of the yeast NPC | This paper | EMD-41122 |
| Composite multi-scale 3D structure of the yeast NPC | This paper | EMD-41123 |
| Molecular model of the inner ring protomer (spoke) | This paper, Akey et al.7; Jumper et al.26 | PDB: 8TIE PDB: 7N85 Alphafold CSMs |
| Molecular model of the double Nup84 Y-complex protomer | This paper, Akey et al.7; Jumper et al.26 | PDB: 8TJ5 PDB: 7N84 Alphafold CSMs |
| Crystal structures of domains of the Nup84 Y-complex | Stuwe et al.91; Brohawn and Schwartz92; Brohawn et al.111 | PDB: 3EWE, 3JRO PDB: 4XMM |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| S. cerevisiae strain W303 MATa ade2–1 ura3–1 his3–11,15 trp1–1 leu2–3,112 can1–100 MLP1-PPX-ProteinA::HIS5 | Kim et al.6 | N/A |
|
| ||
| Software and algorithms | ||
|
| ||
| UCSF Chimera, version 1.13 | Pettersen et al.112 | https://www.cgl.ucsf.edu/chimera/ |
| RELION 3.0.7 | Nakane and Scheres113 | https://github.com/3dem/relion |
| MotionCor2 v1.2.3 | Zheng et al.114 | https://msg.ucsf.edu/software |
| GCTF v1.18 | Zhang115 | https://www2.mrc-lmb.cam.ac.uk/research/locally-developed-software/zhang-software/ |
| Gautomatch v0.55 | N/A | https://www2.mrc-lmb.cam.ac.uk/research/locally-developed-software/zhang-software/ |
| CryoSPARC 3.3.2 | Punjani et al.39 | https://guide.cryosparc.com/setup-configuration-and-management/how-to-download-install-and-configure |
| Coot 0.8.9.2 | Casanal et al.116 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| NAMD/MDFF Molecular dynamics flexible fitting | Trabuco et al.38 |
https://www.ks.uiuc.edu/Development/Download/download.cgi?PackageName=NAMD
https://github.com/ryanmcgreevy/mdff |
| EMAN2 | Tang et al.117 | https://blake.bcm.edu/emanwiki/EMAN2/Install |
| IMP, version 2.8 | Russel et al.82 | https://integrativemodeling.org/ |
|
| ||
| Other | ||
|
| ||
| Quantifoil holy carbon grids | Quantifoil Micro Tools | R2/2 400 mesh copper |
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
The S. cerevisiae strain W303 (see key resources table) was grown and processed as described to provide detergent-solubilized, isolated NPCs.7 Sample preparation, grid freezing, data collection, and movie processing were described previously.7
METHOD DETAILS
Statistics for the current analysis of yeast NPC rings and sub-asemblies are summarized in Tables S1A and S1B. We extracted the inner ring and double Y-complex from NPC images and reprocessed them with focused refinements and more efficient masks in RELION 3.0113 and CryoSPARC.39,118 At the same time, we took a step back to produce lower resolution 3D maps of sub-assemblies including the complete nuclear double outer ring with orphan Nup densities and linker domains, the cytoplasmic single outer ring, the lumenal ring, and a pore membrane anchor site on the spoke. Connecting bridges between FG repeat Nups in the core scaffold and the central channel were also identified. All computational steps were done with scripts due to the complexity of analyzing NPC subassemblies at different bin sizes (pixel sizes). Half maps and gold standard Fourier shell curves (FSCs; not shown) for the low resolution structures have been deposited in the EMDB. Improved molecular models for the inner ring and double Y-complexes were created with the aide of Nup AlphaFold CSMs26 and crystal structures (Table S2). Sub-assembly 3D maps and molecular models were combined to create a multiscale model that provides additional insights into the structure and function of this large channel.
Single-particle processing of yeast NPCs
Inner ring spoke and membrane anchor site
The spoke with 28 Nups was refined as follows. A cleaned and pre-aligned, single particle dataset for the yeast NPC (bin2, 2.66 Å/pixel, 26K) was aligned with C8 symmetry to a reference volume containing a D8 enforced 3D map of the inner ring with a suitable mask, to insure that the local 2-fold axis of a spoke within the inner ring was aligned along the +Y axis while maintaining the overall polarity of the NPC. The particle stack file and star file from the alignment was symmetry expanded to give both the requisite image stack file and star file (relion_particle_symmetry_expand) with alignment parameters for each protomer (208K). SEGGER119 (in Chimera112) and masking strategies were then used with RELION to create a volume containing the outer rings and the central transporter by subtracting the inner and lumenal rings (relion_image_handler). Projections of this volume were then subtracted from NPC images in the C8 expanded stack file (relion_project) to create an image stack file containing the inner and lumenal rings, along with residual noise that accrues from imperfect subtractions due to flexibility and other factors.
A focused multi-body refinement was run with a 2x spoke-lumenal ring volume coupled with on-the-fly subtraction of the complimentary 6x spoke-lumenal ring volume during each alignment cycle; the 2x-spoke-lumenal ring mask and reference volume were centered on the local 2-fold axis aligned on the +Y-axis. At this stage, a star file and extracted 2x-spoke-lumenal ring stack file were then created (relion_flex_analyze_mpi) and particles refined with C1 symmetry. Then a 1.1x spoke stack file was created by subtraction with the appropriate 3D volume (relion_project). A final C1 refinement was done in RELION 3.0.7 using a tight mask for the spoke that included the membrane anchor sites adjacent to Nup157 and Nup170; since the spoke has nearly perfect 2-fold symmetry, we used an initial C2 symmetrized 3D reference volume and final half-maps were post-processed with either C1 symmetry or with the local C2 symmetry enforced before post-processing (FSC0.143 = 7.6Å resolution). Appropriate files for the spoke were exported to CryoSPARC for local non-uniform refinement with C1 symmetry; final half maps were then post-processed in RELION 3.0.7 after C2 symmetrization (Table S1; FSC0.143 = 6.6 Å resolution) and a local resolution map was computed in RELION (local resolution sampling = 50 Å, Bfactor = −600). The C2 enforced map was used in subsequent work and new features were cross-checked in a C1 map.
The data processing increased both the clarity and resolution of the spoke with significant improvements in the membrane interacting layer, including the presence of a partly resolved platform at the membrane contact site for Nup157 β-propellers and clear density for Nup53-Nup59 heterodimers located between adjacent Nic96 CTDs. At the current resolution, 3D classification did not reveal a significant subset of particles that should be discarded, which may reflect the quality of the initial cleaning. We then combined 8 copies of the 1.1x spoke 3D map to create a map for the inner ring; importantly, refinement masks did not truncate inter-spoke contacts in the inner ring.
Focused refinement of the Nup157 membrane anchor site
To further investigate a platform-like feature located on the central 2-fold axis of the spoke that intersects the lipid-detergent micelle, we used a focused mask that included density adjacent to Nup157 and most of the membrane interacting layer. This mask was used to create a reference volume from our best spoke 3D map and to carry out a subsequent refinement in RELION with C1 symmetry that used particle stack and star files from the final spoke C1 refinement in RELION. Half maps were again C2 symmetrized before post-processing and local resolution calculation in RELION (local resolution sampling = 100Å; Table S1). Thus, the final density map was in good alignment with the spoke 3D map; the TMD region was trimmed of extraneous density by mask multiplication and used for modeling and figures.
Double Y-complexes
A particle stack file which contained the best ordered particle subset from our previous multi-body analysis of the Nup84 double Y-complex (bin3, pixel size 3.98 Å; ~14% of total NPC protomers7), was refined in RELION and CryoSPARC. The mask excluded orphan densities and was centered roughly on the overlap region of the two Nup84 complexes, which forms a double-V and we extended the mask to either side to effectively cover a complete protomer in the double outer ring (Table S1). This density map from 3D refinements in RELION was used to create an improved mask by zoning in Chimera112 around a docked model for the two Nup84 Y-complexes (7N84.pdb). This mask and the 3D reference with aligned star file were imported into CryoSPARC and processed by local, non-uniform refinement. CryoSPARC half maps were post-processed in RELION including the calculation of a locally filtered map which indicated a resolution of ~9–10 Å, consistent with features observed during model building (Table S1). A complete C8 ring of double Y-complexes was recreated in Chimera.112
Complete nuclear double outer ring
Complete double outer rings and flexible single rings on the nuclear side remained in NPC images (bin3, pixel size 3.99 Å) after subtracting density for all other ring assemblies and the central transporter. Focused 3D classification with C8 symmetry and a mask was then used to identify particles with a well ordered double ring. Subsequent 3D refinements with improved masks that included relevant orphan and linker domain density regions, along with a small region of the inner ring adjacent to Nup188, provided a representative map with C8 symmetry which was post processed (~23 Å resolution; Table S1). In a final step, adjacent inner ring density that remained in the 3D map was subtracted from the full double outer ring.
Cytoplasmic single outer ring
In previous work, there was significant but circumferentially disconnected and blurred density for the cytoplasmic outer ring in a 3D map of detergent-extracted and isolated NPCs. To reconstruct this ring, we used a subset of NPC images that contained well preserved double outer rings based on the rationale that these particles may be more robust. Thus, we refined this particle subset at bin3 (pixel size = 3.99Å; ~5K NPCs) with C8 symmetry, which revealed weak connectivity for the cytoplasmic outer ring. We then used appropriate masks and volumes to subtract all other NPC components from the aligned stack file images, leaving blurred density associated with the cytoplasmic outer ring. These particle images were refined with C8 symmetry (Table S1; Map 1) and then subjected to multi-body refinements with 2.5X and 5.5X protomer volumes as described for the spoke-lumenal ring processing (Table S1; Map 2). A combination of protomer flexibility and low particle numbers prevented us from refining the cytoplasmic outer ring to a resolution where a reliable molecular model could be built; this task must await a much larger dataset.
Lumenal ring
The lumenal ring has been refractory to analysis in the isolated NPC. We undertook two complementary approaches to improve the density for this flexible feature. First, we used an aligned 2x-spoke-lumenal ring stack file at bin3 for a 3D classification with skip_align to identify and eliminate roughly half of the particles based on weak lumenal ring density. We then transferred the combined class star file to a bin4 environment and started with an aligned 1.5X-spoke-lumenal ring stack file (bin4, 180 box), which by selective masking contained only a small region of the membrane interacting layer in the spoke, including Nup157 and portions of Nup170. A C1 refinement, with C2 post processing created the next 3D reference which was more tightly masked to remove residual spoke density and part of the micelle ring density. After another round of refinement, a 3D classification with skip_align reduced the particle number (85322) and a C1 alignment with C2 post-processing produced the final map (Table S1), which was then expanded to a full lumenal ring and the remaining micelle ring density trimmed by multiplication with a radial mask.
To corroborate our map of the lumenal ring, we started with an NPC image stack at bin4 (5.32 Å/pixel in 500 × 500 boxes) based on the strategy of first isolating the full ring by subtracting all other components. Thus, we used a D8 symmetry enforced, 3D map of the inner ring to align the full NPC images, followed by alignment with a 3D reference and mask based on computationally reassembled lumenal ring with D8 symmetry from our previous refinements that was blurred by low pass filtering to 30Å. After 3D classification with skip_align, we obtained an NPC subset (21372) with reasonable density for the lumenal ring. At this point, we subtracted all other NPC components to create a lumenal ring stack file and carried out D8 symmetry expansion (relion_particle_symmetry_expand) to generate an expanded star file. This star file was used to create a D8 stack file (341952 protomers) commensurate with the full symmetry of this ring, in 360 × 360 boxes (relion_image_handler). The extracted ring images were subjected to a multi-body protocol to create 2X lumenal ring protomers (as described for the spoke-lumenal ring), centered on the central local 2-fold axis of the spoke, and were further refined with C1 symmetry and post-refined with C2 enforced symmetry for the half maps. The final 2X protomer map was then used to recreate a full D8 lumenal ring in Chimera.112 During this process, we retained density from the pore membrane and from the tips of the spokes, which was subsequently removed by mask multiplication (relion_image_handler).
Connecting bridges to the central transporter
The ability to create improved 3D density maps for double outer rings and a single outer ring suggested that a subset of isolated yeast We first re-extracted the ~5K NPC particles with well ordered double outer rings at bin4 (5.32 Å/pixel, box size = 500 × 500 pixels) and created an appropriate star file. This data set was aligned with C8 symmetry with a mask focused on the inner ring and produced a 3D map after post-processing that revealed connecting bridges from the outer rings on both nuclear and cytoplasmic surfaces of the NPC (Table S1; Map 1). Cytoplasmic connections from the scaffold to the central channel have been resolved previously.7 We focused on the nuclear side. In particular, we started with the aligned 5.4K subset of the full NPC extracted at bin3 (3.99 Å/pixel, box size = 414 × 414) and did a skip_align 3D classification with a mask that contained the nuclear double outer ring and density for the nuclear half of the central transporter. This gave a single class (~1.4K) with clear density bridging between the nuclear double outer ring and the central transporter. This subset was subjected to a 3D refinement with C8 symmetry using a reference volume and mask that focused on the inner and lumenal rings. The resulting map was post-processed in RELION (Table S1; Map 2).
Modeling NPC sub-assemblies
Inner ring
We started with an extant model (PDB: 7N85) docked into a spoke density map (EMD-24232).7 We retained chain models for four Nsp1 heterotrimers, two Nup53-Nup59 heterodimers and four sets of Nic96-NTD connectors (Table S2, top half). AlphaFold2 CSMs for the Nic96-CTD and Nups 157, 170, 188 and 192 were trimmed to remove unstructured regions and then rigid body docked in Chimera112 (fit_in_map) and aligned to the spoke model from 7N85.pdb with MatchMaker. Initial manual rebuilding involved breaking some Nups into smaller domains guided by the density map to accommodate local flexibility, repositioning of certain ɑ-helices followed by rebuilding loops in Coot. Cycles of MDFF and manual rebuilding provided a molecular model with few missing loops; final model refinement steps used an improved, 3D density map of the spoke from a non-uniform refinement in CryoSPARC. The overall completeness of the hybrid AlphaFold model allowed the identification of 16 additional orphan densities (8 per half spoke) (Table S3); a careful trace back of computational steps involved in creating the extracted spoke 3D density map indicated that a mixing of top and bottom halves of the spoke did not occur. Orphan densities were of similar intensity in the two half spokes of a C1 map and their apparent occupancy was similar to adjacent Nups in the maps. This allowed the ad hoc enforcement of the local 2-fold symmetry at the post-processing step.
Nup84 double Y-complex
A comprehensive model was created in a stepwise process for two Nup84 Y-complexes in the C8 protomer of the nuclear double outer ring. Initially, an extant 3D density map of the double outer ring (EMD-24231)7 and the associated model (7N84.pdb) were used to guide the piecewise assembly of an AlphaFold2 model, which also utilized X-ray crystal structures as guides (Table S2, bottom half). In this approach, AlphaFold2 CSMs for 7 yeast Nups in the Nup84 complex were pruned where necessary to remove unmodeled termini. Crystal structures for conjoined Nup pairs were used to create models for Nup85/Seh1 and Nup145C/Sec13 with AlphaFold2 CSMs and MatchMaker in Chimera.112 In each case, a 4-stranded β-sheet (known as a blade) is donated by the ɑ-helical solenoids of Nup145C and Nup85 to a Nup partner with 6 blades to create seven blade β-propellers for Sec1372 and Seh1, respectively.111 A complete model of the Y-junction (hub) between Nups 120, 85 and 145C was created in a similar fashion using a crystal structure template (4XMM.pdb).91 All AlphaFold models were then superimposed on 7N84.pdb in Chimera112 and docked into the electron density map with MDFF and manual refinement, while maintaining Nup interfaces at the Y-intersection and between Nup145C and Nup84 in the stem.
In the next step, an improved 3D density map of the double Y-complex was obtained from CryoSPARC which allowed an accurate rigid body docking and subsequent refinement, because helix pair motifs and some single ɑ-helices were resolved at ~9–10Å resolution. In addition, β-propellers with large loops between β-strands were well fit. An improved model of the torus-like bulge in the distal Y-complex that is formed by C-terminal domains of Nup84 and Nup133 was built manually in Coot along with a large contact between the β-propeller of the distal Nup133 and spur domain of the proximal Nup133. The full model of the double Y-complex was refined in steps with MDFF and manual rebuilding in Coot. During refinements, the density map was carefully trimmed to remove orphan densities in the double outer ring that could erroneously draw protein features away from their local density maximum. In addition, we extended the density map and model to ~2 protomers, centered on a single central protomer, to provide contact regions for the flexible fitting. The inherent flexibility of ɑ-helical solenoid Nups and differences in local contacts and conformation arise from altered geometries for the two Y-complexes in the double ring; these features necessitated a large number of manual rebuilding steps to achieve the best fit to the electron density.
Pom34-Pom152 membrane anchor site
We took a stepwise approach to create an initial molecular model of the membrane anchor site located on the spoke 2-fold axis containing Pom34 and Pom152. First, we uploaded an AlphaFold2 model for Pom34 (AF_AFQ12445F1), which was trimmed to contain two TM helices with a short, lumenal helix between the TM helices (aa46–153). 2 copies of this model were manually docked in 4 central TM density rods, related by the local 2-fold axis with the correct sidedness. Second, full-length Pom152 was modeled as a dimer with AlphaFold2 and this produced a TM helix bundle in which each Pom152 monomer has a 3-helix bundle (TMH1-TMH3) followed by a fourth short helix located just above the pore membrane in the NE lumen. The Pom152 monomer was trimmed to residues aa105–214 and we fused helices 3 and 4 to create a longer TM3 that extends beyond the pore membrane as indicated by the density map; 2 copies of this Pom152 3-helix TMD were manually fit into the remaining density with the correct polarity to fill the remaining 6 rods. Extraneous density that is visible at a lower threshold was removed by a generous mask to create figures to showcase the 10 highest density rod-like features. Higher resolution wiill be needed to precisely dock the ɑ-helices from the AlphaFold2 CSMs in this membrane anchor complex.
Integrative modeling of the Pom34-Pom152 transmembrane domains
A structural model of the Pom34-Pom152 transmembrane domains (TMDs) was computed by integrative modeling,6,37,81,82 based on data summarized in Table S4. Relying on the AlphaFold2 predictions of Pom34 and Pom152 (aa1–249), we represented each predicted helix as a rigid body using a 1-residue-per-bead representation; all other regions were represented as flexible strings of beads (Table S4). The spatial restraints included: cross-link restraints corresponding to upper distance bounds based on a total of 15 DSS cross-links6; an EM density restraint corresponding to a cross-correlation between the Gaussian Mixture Model (GMM) representation of the Pom34-Pom152 subunits and the GMM representation of the cryo-EM density maps for the transmembrane region (i.e., 3D density maps; Table S4)120; transmembrane domain localization restraints that localize the predicted transmembrane domains (Pom3462–131, Pom34133–153, Pom34111–131, Pom152149–169, and Pom152173–193) within the pore membrane6; Pom152 peri-nuclear localization restraints applied to the CTD region of the Pom152 fragment (Pom152134–249); pore-side localization restraints to regions facing the pore (Pom341–43 and Pom1521–104); and excluded volume and sequence connectivity restraints were applied to all components.6
Structural models were computed using Replica Exchange Gibbs sampling, based on the Metropolis Monte Carlo (MC) algorithm.44,121 Each MC step consisted of a series of random transformations (i.e., rotations and/or translations) of the flexible beads and rigid bodies (Table S4). We improved the efficiency of structural sampling by explicitly considering the C2-symmetry of the inner ring. Analysis and validation of the structural models of the Pom34-Pom152 TMD region followed the previously published five steps122 (Table S4). The integrative structure modeling protocol (i.e., stages 2, 3, and 4) was scripted using the Python Modeling Interface (PMI) package and our open-source Integrative Modeling Platform (IMP) package,82 version 2.8 (https://integrativemodeling.org). Input data, scripts, and results are available at https://github.com/integrativemodeling/NPC_TMD. The model and data are also deposited in the PDB-dev archive.123
Figures
All Figure panels with density maps and models were created with Chimera,112 and Adobe Photoshop and Illustrator.
QUANTIFICATION AND STATISTICAL ANALYSIS
All quantitative and statistical analysis were done as part of the image processing and modeling pipeline within software packages listed in the main text, STAR Methods, and key resources table.
Supplementary Material
Highlights.
A composite multiscale 3D structure of the isolated yeast NPC has been determined
Double nuclear outer rings represent an ancient feature of NPCs
Membrane anchor sites on the inner ring use a nucleoporin β-propeller/TMD partner motif
Radial expansion of the NPC scaffold unplugs the central channel to promote transport
ACKNOWLEDGMENTS
We thank Drs. Kangkang Song and Chen Xu for advice and help in data collection at the University of Massachusetts Medical School Cryo-EM facility and Dr. J.C. Gumbart for helpful discussions on MDFF. We also thank members of the Chait and Rout laboratories and Drs. D. Singh, S.J. Ludtke, and E. Villa for technical and intellectual support. This work was supported by NIH R01 GM45377 (to C.W.A.), NIH P41 GM109824 (to B.T.C., M.P.R., and A.S.), NIH R01 GM083960 (to A.S.), NIH R01 GM112108 and NIH GM117212 (to M.P.R.), and NSF-1818129 and Spanish Ministerio de Ciencia e Innovacion PID2020-116404GB-I00 (to J.F.-M.).
APPENDIX
INCLUSION AND DIVERSITY
We support inclusive, diverse, and equitable conduct of research.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.molcel.2023.08.025.
REFERENCES
- 1.Hoogenboom BW, Hough LE, Lemke EA, Lim RYH, Onck PR, and Zilman A. (2021). Physics of the nuclear pore complex: theory, modeling and experiment. Phys. Rep 921, 1–53. 10.1016/j.physrep.2021.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dultz E, Wojtynek M, Medalia O, and Onischenko E. (2022). The nuclear pore complex: birth, life, and death of a cellular behemoth. Cells 11, 1456. 10.3390/cells11091456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Petrovic S, Mobbs GW, Bley CJ, Nie S, Patke A, and Hoelz A. (2022). Structure and function of the nuclear pore complex. Cold Spring Harb. Perspect. Biol 14, a041264. 10.1101/cshperspect.a041264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sakuma S, and D’Angelo MA (2017). The roles of the nuclear pore complex in cellular dysfunction, aging and disease. Semin. Cell Dev. Biol 68, 72–84. 10.1016/j.semcdb.2017.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allegretti M, Zimmerli CE, Rantos V, Wilfling F, Ronchi P, Fung HKH, Lee CW, Hagen W, Turoňová B, Karius K, et al. (2020). In-cell architecture of the nuclear pore and snapshots of its turnover. Nature 586, 796–800. 10.1038/s41586-020-2670-5. [DOI] [PubMed] [Google Scholar]
- 6.Kim SJ, Fernandez-Martinez J, Nudelman I, Shi Y, Zhang W, Raveh B, Herricks T, Slaughter BD, Hogan JA, Upla P, et al. (2018). Integrative structure and functional anatomy of a nuclear pore complex. Nature 555, 475–482. 10.1038/nature26003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akey CW, Singh D, Ouch C, Echeverria I, Nudelman I, Varberg JM, Yu Z, Fang F, Shi Y, Wang J, et al. (2022). Comprehensive structure and functional adaptations of the yeast nuclear pore complex. Cell 185, 361–378.e25. 10.1016/j.cell.2021.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cibulka J, Bisaccia F, Radisavljevic K, Gudino Carrillo RM, and Köhler A. (2022). Assembly principle of a membrane-anchored nuclear pore basket scaffold. Sci. Adv 8, eabl6863. 10.1126/sciadv.abl6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vial A, Costa L, Dosset P, Rosso P, Boutières G, Faklaris O, Haschke H, Milhiet PE, and Doucet CM (2023). Structure and mechanics of the human nuclear pore complex basket using correlative AFM-fluorescence superresolution microscopy. Nanoscale 15, 5756–5770. 10.1039/d2nr06034e. [DOI] [PubMed] [Google Scholar]
- 10.Fernandez-Martinez J, Kim SJ, Shi Y, Upla P, Pellarin R, Gagnon M, Chemmama IE, Wang J, Nudelman I, Zhang W, et al. (2016). Structure and function of the nuclear pore complex cytoplasmic mRNA export platform. Cell 167, 1215–1228.e25. 10.1016/j.cell.2016.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amm I, Weberruss M, Hellwig A, Schwarz J, Tatarek-Nossol M, Lüchtenborg C, Kallas M, Bruügger B, Hurt E, and Antonin W. (2022). New motifs in Ndc1 mediating interaction with the Nup84 complex and nuclear membranes. 10.1101/2022.07.31.501833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hao Q, Zhang B, Yuan K, Shi H, and Blobel G. (2018). Electron microscopy of Chaetomium pom152 shows the assembly of ten-bead string. Cell Discov. 4, 56. 10.1038/s41421-018-0057-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Upla P, Kim SJ, Sampathkumar P, Dutta K, Cahill SM, Chemmama IE, Williams R, Bonanno JB, Rice WJ, Stokes DL, et al. (2017). Molecular architecture of the major membrane ring component of the nuclear pore complex. Structure 25, 434–445. 10.1016/j.str.2017.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Akey CW (2010). The NPC-transporter, a ghost in the machine. Structure 18, 1230–1232. 10.1016/j.str.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamada J, Phillips JL, Patel S, Goldfien G, Calestagne-Morelli A, Huang H, Reza R, Acheson J, Krishnan VV, Newsam S, et al. (2010). A bimodal distribution of two distinct categories of intrinsically disordered structures with separate functions in FG nucleoporins. Mol. Cell. Proteomics 9, 2205–2224. 10.1074/mcp.M000035-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Akey CW, and Radermacher M. (1993). Architecture of the Xenopus nuclear pore complex revealed by three-dimensional cryo-electron microscopy. J. Cell Biol 122, 1–19. 10.1083/jcb.122.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kalita J, Kapinos LE, Zheng T, Rencurel C, Zilman A, and Lim RYH (2022). Karyopherin enrichment and compensation fortifies the nuclear pore complex against nucleocytoplasmic leakage. J. Cell Biol 221, e202108107. 10.1083/jcb.202108107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mosalaganti S, Obarska-Kosinska A, Siggel M, Taniguchi R, Turoňová B, Zimmerli CE, Buczak K, Schmidt FH, Margiotta E, Mackmull MT, et al. (2022). AI-based structure prediction empowers integrative structural analysis of human nuclear pores. Science 376, eabm9506. 10.1126/science.abm9506. [DOI] [PubMed] [Google Scholar]
- 19.Petrovic S, Samanta D, Perriches T, Bley CJ, Thierbach K, Brown B, Nie S, Mobbs GW, Stevens TA, Liu X, et al. (2022). Architecture of the linker-scaffold in the nuclear pore. Science 376, eabm9798. 10.1126/science.abm9798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bley CJ, Nie S, Mobbs GW, Petrovic S, Gres AT, Liu X, Mukherjee S, Harvey S, Huber FM, Lin DH, et al. (2022). Architecture of the cytoplasmic face of the nuclear pore. Science 376, eabm9129. 10.1126/science.abm9129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schuller AP, Wojtynek M, Mankus D, Tatli M, Kronenberg-Tenga R, Regmi SG, Dip PV, Lytton-Jean AKR, Brignole EJ, Dasso M, et al. (2021). The cellular environment shapes the nuclear pore complex architecture. Nature 598, 667–671. 10.1038/s41586-021-03985-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eibauer M, Pellanda M, Turgay Y, Dubrovsky A, Wild A, and Medalia O. (2015). Structure and gating of the nuclear pore complex. Nat. Commun 6, 7532. 10.1038/ncomms8532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mosalaganti S, Kosinski J, Albert S, Schaffer M, Strenkert D, Salomé PA, Merchant SS, Plitzko JM, Baumeister W, Engel BD, and Beck M. (2018). In situ architecture of the algal nuclear pore complex. Nat. Commun 9, 2361. 10.1038/s41467-018-04739-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Z, Chen S, Zhao L, Huang G, Pi X, Sun S, Wang P, and Sui SF (2022). Near-atomic structure of the inner ring of the Saccharomyces cerevisiae nuclear pore complex. Cell Res. 32, 437–450. 10.1038/s41422-022-00632-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zimmerli CE, Allegretti M, Rantos V, Goetz SK, Obarska-Kosinska A, Zagoriy I, Halavatyi A, Hummer G, Mahamid J, Kosinski J, and Beck M. (2021). Nuclear pores dilate and constrict in cellulo. Science 374, eabd9776. 10.1126/science.abd9776. [DOI] [PubMed] [Google Scholar]
- 26.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žı´dek A, Potapenko A, et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baek M, DiMaio F, Anishchenko I, Dauparas J, Ovchinnikov S, Lee GR, Wang J, Cong Q, Kinch LN, Schaeffer RD, et al. (2021). Accurate prediction of protein structures and interactions using a three-track neural network. Science 373, 871–876. 10.1126/science.abj8754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fontana P, Dong Y, Pi X, Tong AB, Hecksel CW, Wang L, Fu TM, Bustamante C, and Wu H. (2022). Structure of cytoplasmic ring of nuclear pore complex by integrative cryo-EM and AlphaFold. Science 376, eabm9326. 10.1126/science.abm9326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tai L, Zhu Y, Ren H, Huang X, Zhang C, and Sun F. (2022). 8 A structure of the outer rings of the Xenopus laevis nuclear pore complex obtained by cryo-EM and AI. Protein Cell 13, 760–777. 10.1007/s13238-021-00895-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu X, Huang G, Zeng C, Zhan X, Liang K, Xu Q, Zhao Y, Wang P, Wang Q, Zhou Q, et al. (2022). Structure of the cytoplasmic ring of the Xenopus laevis nuclear pore complex. Science 376, eabl8280. 10.1126/science.abl8280. [DOI] [PubMed] [Google Scholar]
- 31.Huang G, Zhan X, Zeng C, Liang K, Zhu X, Zhao Y, Wang P, Wang Q, Zhou Q, Tao Q, et al. (2022). Cryo-EM structure of the inner ring from the Xenopus laevis nuclear pore complex. Cell Res. 32, 451–460. 10.1038/s41422-022-00633-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang G, Zhan X, Zeng C, Zhu X, Liang K, Zhao Y, Wang P, Wang Q, Zhou Q, Tao Q, et al. (2022). Cryo-EM structure of the nuclear ring from Xenopus laevis nuclear pore complex. Cell Res. 32, 349–358. 10.1038/s41422-021-00610-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mahamid J, Pfeffer S, Schaffer M, Villa E, Danev R, Cuellar LK, Förster F, Hyman AA, Plitzko JM, and Baumeister W. (2016). Visualizing the molecular sociology at the HeLa cell nuclear periphery. Science 351, 969–972. 10.1126/science.aad8857. [DOI] [PubMed] [Google Scholar]
- 34.Huang G, Zhang Y, Zhu X, Zeng C, Wang Q, Zhou Q, Tao Q, Liu M, Lei J, Yan C, and Shi Y. (2020). Structure of the cytoplasmic ring of the Xenopus laevis nuclear pore complex by cryo-electron microscopy single particle analysis. Cell Res. 30, 520–531. 10.1038/s41422-020-0319-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nudelman I, Fernandez-Martinez J, and Rout MP (2022). Affinity isolation of endogenous Saccharomyces cerevisiae nuclear pore complexes. Methods Mol. Biol 2502, 3–34. 10.1007/978-1-0716-2337-4_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Belnap DM, Kumar A, Folk JT, Smith TJ, and Baker TS (1999). Low-resolution density maps from atomic models: how stepping “back” can be a step “forward”. J. Struct. Biol 125, 166–175. 10.1006/jsbi.1999.4093. [DOI] [PubMed] [Google Scholar]
- 37.Rout MP, and Sali A. (2019). Principles for integrative structural biology studies. Cell 177, 1384–1403. 10.1016/j.cell.2019.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trabuco LG, Villa E, Mitra K, Frank J, and Schulten K. (2008). Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure 16, 673–683. 10.1016/j.str.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Punjani A, Zhang H, and Fleet DJ (2020). Non-uniform refinement: adaptive regularization improves single-particle cryo-EM reconstruction. Nat. Methods 17, 1214–1221. 10.1038/s41592-020-00990-8. [DOI] [PubMed] [Google Scholar]
- 40.Devos D, Dokudovskaya S, Alber F, Williams R, Chait BT, Sali A, and Rout MP (2004). Components of coated vesicles and nuclear pore complexes share a common molecular architecture. PLoS Biol. 2, e380. 10.1371/journal.pbio.0020380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Field MC, and Rout MP (2019). Pore timing: the evolutionary origins of the nucleus and nuclear pore complex. F1000Res 8, 369. 10.12688/f1000research.16402.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Field MC, and Rout MP (2022). Coatomer in the universe of cellular complexity. Mol. Biol. Cell 33, pe8. 10.1091/mbc.E19-01-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kosinski J, Mosalaganti S, von Appen A, Teimer R, DiGuilio AL, Wan W, Bui KH, Hagen WJ, Briggs JA, Glavy JS, et al. (2016). Molecular architecture of the inner ring scaffold of the human nuclear pore complex. Science 352, 363–365. 10.1126/science.aaf0643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shi Y, Fernandez-Martinez J, Tjioe E, Pellarin R, Kim SJ, Williams R, Schneidman-Duhovny D, Sali A, Rout MP, and Chait BT (2014). Structural characterization by cross-linking reveals the detailed architecture of a coatomer-related heptameric module from the nuclear pore complex. Mol. Cell. Proteomics 13, 2927–2943. 10.1074/mcp.M114.041673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meinema AC, Laba JK, Hapsari RA, Otten R, Mulder FA, Kralt A, van den Bogaart G, Lusk CP, Poolman B, and Veenhoff LM (2011). Long unfolded linkers facilitate membrane protein import through the nuclear pore complex. Science 333, 90–93. 10.1126/science.1205741. [DOI] [PubMed] [Google Scholar]
- 46.Lin DH, Stuwe T, Schilbach S, Rundlet EJ, Perriches T, Mobbs G, Fan Y, Thierbach K, Huber FM, Collins LN, et al. (2016). Architecture of the symmetric core of the nuclear pore. Science 352, aaf1015. 10.1126/science.aaf1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Teixeira MT, Fabre E, and Dujon B. (1999). Self-catalyzed cleavage of the yeast nucleoporin Nup145p precursor. J. Biol. Chem 274, 32439–32444. 10.1074/jbc.274.45.32439. [DOI] [PubMed] [Google Scholar]
- 48.Rosenblum JS, and Blobel G. (1999). Autoproteolysis in nucleoporin biogenesis. Proc. Natl. Acad. Sci. USA 96, 11370–11375. 10.1073/pnas.96.20.11370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Amlacher S, Sarges P, Flemming D, van Noort V, Kunze R, Devos DP, Arumugam M, Bork P, and Hurt E. (2011). Insight into structure and assembly of the nuclear pore complex by utilizing the genome of a eukaryotic thermophile. Cell 146, 277–289. 10.1016/j.cell.2011.06.039. [DOI] [PubMed] [Google Scholar]
- 50.Hakhverdyan Z, Molloy KR, Keegan S, Herricks T, Lepore DM, Munson M, Subbotin RI, Fenyö D, Aitchison JD, Fernandez-Martinez J, et al. (2021). Dissecting the structural dynamics of the nuclear pore complex. Mol. Cell 81, 153–165.e7. 10.1016/j.molcel.2020.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fernandez-Martinez J, and Rout MP (2021). One ring to rule them all? Structural and functional diversity in the nuclear pore complex. Trends Biochem. Sci 46, 595–607. 10.1016/j.tibs.2021.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Neumann N, Lundin D, and Poole AM (2010). Comparative genomic evidence for a complete nuclear pore complex in the last eukaryotic common ancestor. PLoS One 5, e13241. 10.1371/journal.pone.0013241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tcheperegine SE, Marelli M, and Wozniak RW (1999). Topology and functional domains of the yeast pore membrane protein Pom152p. J. Biol. Chem 274, 5252–5258. 10.1074/jbc.274.8.5252. [DOI] [PubMed] [Google Scholar]
- 54.Gonen T, Cheng Y, Sliz P, Hiroaki Y, Fujiyoshi Y, Harrison SC, and Walz T. (2005). Lipid-protein interactions in double-layered two-dimensional AQP0 crystals. Nature 438, 633–638. 10.1038/nature04321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Winey M, Hoyt MA, Chan C, Goetsch L, Botstein D, and Byers B. (1993). NDC1: a nuclear periphery component required for yeast spindle pole body duplication. J. Cell Biol 122, 743–751. 10.1083/jcb.122.4.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stavru F, Hülsmann BB, Spang A, Hartmann E, Cordes VC, and Görlich D. (2006). NDC1: a crucial membrane-integral nucleoporin of metazoan nuclear pore complexes. J. Cell Biol 173, 509–519. 10.1083/jcb.200601001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mansfeld J, Güttinger S, Hawryluk-Gara LA, Panté N, Mall M, Galy V, Haselmann U, Mühlhäusser P, Wozniak RW, Mattaj IW, et al. (2006). The conserved transmembrane nucleoporin NDC1 is required for nuclear pore complex assembly in vertebrate cells. Mol. Cell 22, 93–103. 10.1016/j.molcel.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 58.Chial HJ, Rout MP, Giddings TH, and Winey M. (1998). Saccharomyces cerevisiae Ndc1p is a shared component of nuclear pore complexes and spindle pole bodies. J. Cell Biol 143, 1789–1800. 10.1083/jcb.143.7.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Onischenko E, Stanton LH, Madrid AS, Kieselbach T, and Weis K. (2009). Role of the Ndc1 interaction network in yeast nuclear pore complex assembly and maintenance. J. Cell Biol 185, 475–491. 10.1083/jcb.200810030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Eisenhardt N, Redolfi J, and Antonin W. (2014). Interaction of Nup53 with Ndc1 and Nup155 is required for nuclear pore complex assembly. J. Cell Sci 127, 908–921. 10.1242/jcs.141739. [DOI] [PubMed] [Google Scholar]
- 61.Onischenko E, Noor E, Fischer JS, Gillet L, Wojtynek M, Vallotton P, and Weis K. (2020). Maturation kinetics of a multiprotein complex revealed by metabolic labeling. Cell 183, 1785–1800.e26. 10.1016/j.cell.2020.11.001. [DOI] [PubMed] [Google Scholar]
- 62.Marelli M, Lusk CP, Chan H, Aitchison JD, and Wozniak RW (2001). A link between the synthesis of nucleoporins and the biogenesis of the nuclear envelope. J. Cell Biol 153, 709–724. 10.1083/jcb.153.4.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vollmer B, Schooley A, Sachdev R, Eisenhardt N, Schneider AM, Sieverding C, Madlung J, Gerken U, Macek B, and Antonin W. (2012). Dimerization and direct membrane interaction of Nup53 contribute to nuclear pore complex assembly. EMBO J. 31, 4072–4084. 10.1038/emboj.2012.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Patel SS, and Rexach MF (2008). Discovering novel interactions at the nuclear pore complex using bead halo: a rapid method for detecting molecular interactions of high and low affinity at equilibrium. Mol. Cell. Proteomics 7, 121–131. 10.1074/mcp.M700407-MCP200. [DOI] [PubMed] [Google Scholar]
- 65.Zhang Y, Li S, Zeng C, Huang G, Zhu X, Wang Q, Wang K, Zhou Q, Yan C, Zhang W, et al. (2020). Molecular architecture of the luminal ring of the Xenopus laevis nuclear pore complex. Cell Res. 30, 532–540. 10.1038/s41422-020-0320-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bui KH, von Appen A, DiGuilio AL, Ori A, Sparks L, Mackmull MT, Bock T, Hagen W, Andrés-Pons A, Glavy JS, and Beck M. (2013). Integrated structural analysis of the human nuclear pore complex scaffold. Cell 155, 1233–1243. 10.1016/j.cell.2013.10.055. [DOI] [PubMed] [Google Scholar]
- 67.Siniossoglou S, Lutzmann M, Santos-Rosa H, Leonard K, Mueller S, Aebi U, and Hurt E. (2000). Structure and assembly of the Nup84p complex. J. Cell Biol 149, 41–54. 10.1083/jcb.149.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lutzmann M, Kunze R, Buerer A, Aebi U, and Hurt E. (2002). Modular self-assembly of a Y-shaped multiprotein complex from seven nucleoporins. EMBO J. 21, 387–397. 10.1093/emboj/21.3.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fernandez-Martinez J, Phillips J, Sekedat MD, Diaz-Avalos R, Velazquez-Muriel J, Franke JD, Williams R, Stokes DL, Chait BT, Sali A, and Rout MP (2012). Structure-function mapping of a heptameric module in the nuclear pore complex. J. Cell Biol 196, 419–434. 10.1083/jcb.201109008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kampmann M, and Blobel G. (2009). Three-dimensional structure and flexibility of a membrane-coating module of the nuclear pore complex. Nat. Struct. Mol. Biol 16, 782–788. 10.1038/nsmb.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kelley K, Knockenhauer KE, Kabachinski G, and Schwartz TU (2015). Atomic structure of the Y complex of the nuclear pore. Nat. Struct. Mol. Biol 22, 425–431. 10.1038/nsmb.2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hsia KC, Stavropoulos P, Blobel G, and Hoelz A. (2007). Architecture of a coat for the nuclear pore membrane. Cell 131, 1313–1326. 10.1016/j.cell.2007.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Drin G, Casella JF, Gautier R, Boehmer T, Schwartz TU, and Antonny B. (2007). A general amphipathic alpha-helical motif for sensing membrane curvature. Nat. Struct. Mol. Biol 14, 138–146. 10.1038/nsmb1194. [DOI] [PubMed] [Google Scholar]
- 74.Kim SJ, Fernandez-Martinez J, Sampathkumar P, Martel A, Matsui T, Tsuruta H, Weiss TM, Shi Y, Markina-Inarrairaegui A, Bonanno JB, et al. (2014). Integrative structure-function mapping of the nucleoporin Nup133 suggests a conserved mechanism for membrane anchoring of the nuclear pore complex. Mol. Cell. Proteomics 13, 2911–2926. 10.1074/mcp.M114.040915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sampathkumar P, Ozyurt SA, Do J, Bain KT, Dickey M, Rodgers LA, Gheyi T, Sali A, Kim SJ, Phillips J, et al. (2010). Structures of the autoproteolytic domain from the Saccharomyces cerevisiae nuclear pore complex component, Nup145. Proteins 78, 1992–1998. 10.1002/prot.22707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hodel AE, Hodel MR, Griffis ER, Hennig KA, Ratner GA, Xu S, and Powers MA (2002). The three-dimensional structure of the autoproteolytic, nuclear pore-targeting domain of the human nucleoporin Nup98. Mol. Cell 10, 347–358. 10.1016/s1097-2765(02)00589-0. [DOI] [PubMed] [Google Scholar]
- 77.Terry LJ, and Wente SR (2009). Flexible gates: dynamic topologies and functions for FG nucleoporins in nucleocytoplasmic transport. Eukaryot. Cell 8, 1814–1827. 10.1128/EC.00225-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. (2007). The molecular architecture of the nuclear pore complex. Nature 450, 695–701. 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- 79.Gaik M, Flemming D, von Appen A, Kastritis P, Mücke N, Fischer J, Stelter P, Ori A, Bui KH, Baßler J, et al. (2015). Structural basis for assembly and function of the Nup82 complex in the nuclear pore scaffold. J. Cell Biol 208, 283–297. 10.1083/jcb.201411003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yoshida K, Seo HS, Debler EW, Blobel G, and Hoelz A. (2011). Structural and functional analysis of an essential nucleoporin heterotrimer on the cytoplasmic face of the nuclear pore complex. Proc. Natl. Acad. Sci. USA 108, 16571–16576. 10.1073/pnas.1112846108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. (2007). Determining the architectures of macromolecular assemblies. Nature 450, 683–694. 10.1038/nature06404. [DOI] [PubMed] [Google Scholar]
- 82.Russel D, Lasker K, Webb B, Velázquez-Muriel J, Tjioe E, Schneidman-Duhovny D, Peterson B, and Sali A. (2012). Putting the pieces together: integrative modeling platform software for structure determination of macromolecular assemblies. PLoS Biol. 10, e1001244. 10.1371/journal.pbio.1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Makarov AA, Padilla-Mejia NE, and Field MC (2021). Evolution and diversification of the nuclear pore complex. Biochem. Soc. Trans 49, 1601–1619. 10.1042/BST20200570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Asakawa H, Kojidani T, Yang HJ, Ohtsuki C, Osakada H, Matsuda A, Iwamoto M, Chikashige Y, Nagao K, Obuse C, et al. (2019). Asymmetrical localization of Nup107–160 subcomplex components within the nuclear pore complex in fission yeast. PLoS Genet. 15, e1008061. 10.1371/journal.pgen.1008061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bensidoun P, Reiter T, Montpetit B, Zenklusen D, and Oeffinger M. (2022). Nuclear mRNA metabolism drives selective basket assembly on a subset of nuclear pore complexes in budding yeast. Mol. Cell 82, 3856–3871.e6. 10.1016/j.molcel.2022.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.De Magistris P, Tatarek-Nossol M, Dewor M, and Antonin W. (2018). A self-inhibitory interaction within Nup155 and membrane binding are required for nuclear pore complex formation. J. Cell Sci 131, jcs208538. 10.1242/jcs.208538. [DOI] [PubMed] [Google Scholar]
- 87.von Appen A, Kosinski J, Sparks L, Ori A, DiGuilio AL, Vollmer B, Mackmull MT, Banterle N, Parca L, Kastritis P, et al. (2015). In situ structural analysis of the human nuclear pore complex. Nature 526, 140–143. 10.1038/nature15381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Andreu I, Granero-Moya I, Chahare NR, Clein K, Molina-Jordán M, Beedle AEM, Elosegui-Artola A, Abenza JF, Rossetti L, Trepat X, et al. (2022). Mechanical force application to the nucleus regulates nucleocytoplasmic transport. Nat. Cell Biol 24, 896–905. 10.1038/s41556-022-00927-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fischer J, Teimer R, Amlacher S, Kunze R, and Hurt E. (2015). Linker Nups connect the nuclear pore complex inner ring with the outer ring and transport channel. Nat. Struct. Mol. Biol 22, 774–781. 10.1038/nsmb.3084. [DOI] [PubMed] [Google Scholar]
- 90.Chug H, Trakhanov S, Hülsmann BB, Pleiner T, and Görlich D. (2015). Crystal structure of the metazoan Nup62*Nup58*Nup54 nucleoporin complex. Science 350, 106–110. 10.1126/science.aac7420. [DOI] [PubMed] [Google Scholar]
- 91.Stuwe T, Bley CJ, Thierbach K, Petrovic S, Schilbach S, Mayo DJ, Perriches T, Rundlet EJ, Jeon YE, Collins LN, et al. (2015). Architecture of the fungal nuclear pore inner ring complex. Science 350, 56–64. 10.1126/science.aac9176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Brohawn SG, and Schwartz TU (2009). Molecular architecture of the Nup84-Nup145C-Sec13 edge element in the nuclear pore complex lattice. Nat. Struct. Mol. Biol 16, 1173–1177. 10.1038/nsmb.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hough LE, Dutta K, Sparks S, Temel DB, Kamal A, Tetenbaum-Novatt J, Rout MP, and Cowburn D. (2015). The molecular mechanism of nuclear transport revealed by atomic-scale measurements. eLife 4, e10027. 10.7554/eLife.10027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lim RY, Huang NP, Köser J, Deng J, Lau KH, Schwarz-Herion K, Fahrenkrog B, and Aebi U. (2006). Flexible phenylalanine-glycine nucleoporins as entropic barriers to nucleocytoplasmic transport. Proc. Natl. Acad. Sci. USA 103, 9512–9517. 10.1073/pnas.0603521103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Akey CW (1990). Visualization of transport-related configurations of the nuclear pore transporter. Biophys. J 58, 341–355. 10.1016/S0006-3495(90)82381-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yang Q, Rout MP, and Akey CW (1998). Three-dimensional architecture of the isolated yeast nuclear pore complex: functional and evolutionary implications. Mol. Cell 1, 223–234. 10.1016/s1097-2765(00)80023-4. [DOI] [PubMed] [Google Scholar]
- 97.Unwin PN, and Milligan RA (1982). A large particle associated with the perimeter of the nuclear pore complex. J. Cell Biol 93, 63–75. 10.1083/jcb.93.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kiseleva E, Allen TD, Rutherford S, Bucci M, Wente SR, and Goldberg MW (2004). Yeast nuclear pore complexes have a cytoplasmic ring and internal filaments. J. Struct. Biol 145, 272–288. 10.1016/j.jsb.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 99.Akey CW, and Goldfarb DS (1989). Protein import through the nuclear pore complex is a multistep process. J. Cell Biol 109, 971–982. 10.1083/jcb.109.3.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li Z, Chen S, Zhao L, Huang G, Xu H, Yang X, Wang P, Gao N, and Sui SF (2023). Nuclear export of pre-60S particles through the nuclear pore complex. Nature 618, 411–418. 10.1038/s41586-023-06128-y. [DOI] [PubMed] [Google Scholar]
- 101.Lim RY, Huang B, and Kapinos LE (2015). How to operate a nuclear pore complex by Kap-centric control. Nucleus 6, 366–372. 10.1080/19491034.2015.1090061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Frey S, and Görlich D. (2007). A saturated FG-repeat hydrogel can reproduce the permeability properties of nuclear pore complexes. Cell 130, 512–523. 10.1016/j.cell.2007.06.024. [DOI] [PubMed] [Google Scholar]
- 103.Frey S, Richter RP, and Görlich D. (2006). FG-rich repeats of nuclear pore proteins form a three-dimensional meshwork with hydrogel-like properties. Science 314, 815–817. 10.1126/science.1132516. [DOI] [PubMed] [Google Scholar]
- 104.Fiserova J, Richards SA, Wente SR, and Goldberg MW (2010). Facilitated transport and diffusion take distinct spatial routes through the nuclear pore complex. J. Cell Sci 123, 2773–2780. 10.1242/jcs.070730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yang W. (2013). Distinct, but not completely separate spatial transport routes in the nuclear pore complex. Nucleus 4, 166–175. 10.4161/nucl.24874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Elosegui-Artola A, Andreu I, Beedle AEM, Lezamiz A, Uroz M, Kosmalska AJ, Oria R, Kechagia JZ, Rico-Lastres P, Le Roux AL, et al. (2017). Force triggers YAP nuclear entry by regulating transport across nuclear pores. Cell 171, 1397–1410.e14. 10.1016/j.cell.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 107.Andreu I, Granero-Moya I, Garcia-Manyes S, and Roca-Cusachs P. (2022). Understanding the role of mechanics in nucleocytoplasmic transport. APL Bioeng. 6, 020901. 10.1063/5.0076034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bayliss R, Littlewood T, and Stewart M. (2000). Structural basis for the interaction between FxFG nucleoporin repeats and importin-beta in nuclear trafficking. Cell 102, 99–108. 10.1016/s0092-8674(00)00014-3. [DOI] [PubMed] [Google Scholar]
- 109.Kapinos LE, Schoch RL, Wagner RS, Schleicher KD, and Lim RY (2014). Karyopherin-centric control of nuclear pores based on molecular occupancy and kinetic analysis of multivalent binding with FG nucleoporins. Biophys. J 106, 1751–1762. 10.1016/j.bpj.2014.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Terwilliger TC, Poon BK, Afonine PV, Schlicksup CJ, Croll TI, Millán C, Richardson JS, Read RJ, and Adams PD (2022). Improved AlphaFold modeling with implicit experimental information. Nat. Methods 19, 1376–1382. 10.1038/s41592-022-01645-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Brohawn SG, Leksa NC, Spear ED, Rajashankar KR, and Schwartz TU (2008). Structural evidence for common ancestry of the nuclear pore complex and vesicle coats. Science 322, 1369–1373. 10.1126/science.1165886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera – A visualization system for exploratory research and analysis. J. Comput. Chem 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- 113.Nakane T, and Scheres SHW (2021). Multi-body refinement of cryo-EM images in RELION. Methods Mol. Biol 2215, 145–160. 10.1007/978-1-0716-0966-8_7. [DOI] [PubMed] [Google Scholar]
- 114.Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, and Agard DA (2017). MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhang K. (2016). Gctf: real-time CTF determination and correction. J. Struct. Biol 193, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Casañal A, Lohkamp B, and Emsley P. (2020). Current developments in coot for macromolecular model building of Electron Cryo-microscopy and Crystallographic Data. Protein Sci. 29, 1069–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, and Ludtke SJ (2007). EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol 157, 38–46. [DOI] [PubMed] [Google Scholar]
- 118.Punjani A, Rubinstein JL, Fleet DJ, and Brubaker MA (2017). cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296. 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 119.Pintilie GD, Zhang J, Goddard TD, Chiu W, and Gossard DC (2010). Quantitative analysis of cryo-EM density map segmentation by watershed and scale-space filtering, and fitting of structures by alignment to regions. J. Struct. Biol 170, 427–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bonomi M, Hanot S, Greenberg CH, Sali A, Nilges M, Vendruscolo M, and Pellarin R. (2019). Bayesian weighing of electron cryo-microscopy data for integrative structural modeling. Structure 27, 175–188.e6. 10.1016/j.str.2018.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Swendsen RH, and Wang JS (1986). Replica Monte Carlo simulation of spin glasses. Phys. Rev. Lett 57, 2607–2609. 10.1103/PhysRevLett.57.2607. [DOI] [PubMed] [Google Scholar]
- 122.Saltzberg DJ, Viswanath S, Echeverria I, Chemmama IE, Webb B, and Sali A. (2021). Using Integrative Modeling Platform to compute, validate, and archive a model of a protein complex structure. Protein Sci. 30, 250–261. 10.1002/pro.3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Burley SK, Kurisu G, Markley JL, Nakamura H, Velankar S, Berman HM, Sali A, Schwede T, and Trewhella J. (2017). PDBdev: a prototype system for depositing integrative/hybrid structural models. Structure 25, 1317–1318. 10.1016/j.str.2017.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Cryo-EM 3D density maps and related models have been deposited in the EMDB and RCSB, respectively. Integrative modeling data have been deposited in the PDB-Dev.
All original code has been deposited at Zenodo and is publicly available as of the date of publication. DOIs are listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Deposited data | ||
|
| ||
| Chemical Cross-linking with Mass Spectrometry readout datasets | Akey et al.7 | Zenodo (https://zenodo.org/) Accession#: 5721514 |
| Files and input data, scripts and results for integrative modeling of the Pom34-Pom152 tetramer | This paper | Zenodo (https://zenodo.org/) DOI-10.5281 Accession#: 8226857 |
| Integrative models for Pom152 rings | This paper | PDBDEV_00000213 |
| Full nuclear double outer ring of the yeast NPC | This paper | EMD-41114 |
| Double ring of Nup84 Y-complexes from the yeast NPC | This paper | EMD-41285 |
| Lumenal ring of the isolated yeast NPC | This paper | EMD-41116 |
| Pom34-152 membrane attachment site in the yeast NPC and preliminary model | This paper | EMD-41117 PDB: 8T9L |
| Inner ring spoke of the yeast NPC | This paper | EMD-41300 |
| Cytoplasmic outer ring of the yeast NPC | This paper | EMD-41119 |
| Central transporter/plug in 3D map of the yeast NPC | This paper | EMD-41120 |
| Connectors from the double outer ring of the yeast NPC | This paper | EMD-41121 |
| Connectors from the cytoplasmic outer ring of the yeast NPC | This paper | EMD-41122 |
| Composite multi-scale 3D structure of the yeast NPC | This paper | EMD-41123 |
| Molecular model of the inner ring protomer (spoke) | This paper, Akey et al.7; Jumper et al.26 | PDB: 8TIE PDB: 7N85 Alphafold CSMs |
| Molecular model of the double Nup84 Y-complex protomer | This paper, Akey et al.7; Jumper et al.26 | PDB: 8TJ5 PDB: 7N84 Alphafold CSMs |
| Crystal structures of domains of the Nup84 Y-complex | Stuwe et al.91; Brohawn and Schwartz92; Brohawn et al.111 | PDB: 3EWE, 3JRO PDB: 4XMM |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| S. cerevisiae strain W303 MATa ade2–1 ura3–1 his3–11,15 trp1–1 leu2–3,112 can1–100 MLP1-PPX-ProteinA::HIS5 | Kim et al.6 | N/A |
|
| ||
| Software and algorithms | ||
|
| ||
| UCSF Chimera, version 1.13 | Pettersen et al.112 | https://www.cgl.ucsf.edu/chimera/ |
| RELION 3.0.7 | Nakane and Scheres113 | https://github.com/3dem/relion |
| MotionCor2 v1.2.3 | Zheng et al.114 | https://msg.ucsf.edu/software |
| GCTF v1.18 | Zhang115 | https://www2.mrc-lmb.cam.ac.uk/research/locally-developed-software/zhang-software/ |
| Gautomatch v0.55 | N/A | https://www2.mrc-lmb.cam.ac.uk/research/locally-developed-software/zhang-software/ |
| CryoSPARC 3.3.2 | Punjani et al.39 | https://guide.cryosparc.com/setup-configuration-and-management/how-to-download-install-and-configure |
| Coot 0.8.9.2 | Casanal et al.116 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| NAMD/MDFF Molecular dynamics flexible fitting | Trabuco et al.38 |
https://www.ks.uiuc.edu/Development/Download/download.cgi?PackageName=NAMD
https://github.com/ryanmcgreevy/mdff |
| EMAN2 | Tang et al.117 | https://blake.bcm.edu/emanwiki/EMAN2/Install |
| IMP, version 2.8 | Russel et al.82 | https://integrativemodeling.org/ |
|
| ||
| Other | ||
|
| ||
| Quantifoil holy carbon grids | Quantifoil Micro Tools | R2/2 400 mesh copper |