Abstract
Six bacteriocinlike peptides (plantaricin A [PlnA], PlnE, PlnF, PlnJ, PlnK, and PlnN) produced by Lactobacillus plantarum C11 were detected by amino acid sequencing and mass spectrometry. Since purification to homogeneity was problematic, all six peptides were obtained by solid-phase peptide synthesis and were tested for bacteriocin activity. It was found that L. plantarum C11 produces two two-peptide bacteriocins (PlnEF and PlnJK); a strain-specific antagonistic activity was detected at nanomolar concentrations when PlnE and PlnF were combined and when PlnJ and PlnK were combined. Complementary peptides were at least 103 times more active when they were combined than when they were present individually, and optimal activity was obtained when the complementary peptides were present in approximately equal amounts. The interaction between complementary peptides was specific, since neither PlnE nor PlnF could complement PlnJ or PlnK, and none of these peptides could complement the peptides constituting the two-peptide bacteriocin lactococcin G. Interestingly, PlnA, which acts as an extracellular signal (pheromone) that triggers bacteriocin production, also possessed a strain-specific antagonistic activity. No bacteriocin activity could be detected for PlnN.
Ribosomally synthesized antimicrobial polypeptides, termed bacteriocins, are produced by various lactic acid bacteria (LAB). LAB bacteriocins are usually membrane-permeabilizing cationic peptides which seldom contain more than 60 amino acid residues (1, 14, 19, 21, 32). These peptide bacteriocins may be classified into two main groups; group I consists of posttranslationally modified bacteriocins, and group II consists of unmodified bacteriocins. The modified group I bacteriocins are called lantibiotics because they contain the thioether amino acid lanthionine, and often they contain other modified residues, such as methyllanthionine, dehydroalanine, dehydrobutyrine, and d-alanine (27, 29). The unmodified group II bacteriocins include the pediocinlike bacteriocins (all of which exhibit more than 25% sequence identity to pediocin PA-1), as well as several non-pediocin-like bacteriocins, such as lactococcin A and the two-peptide bacteriocin lactococcin G (13, 19, 21, 22).
Lactococcin G is classified as a two-peptide bacteriocin since it consists of two different peptides and antibacterial activity requires the complementary action of both peptides in approximately equal amounts (18, 22). The genes encoding the two peptides are next to each other in the same operon, together with the genes encoding the immunity protein (which protects the producer against lactococcin G) and the membrane-associated ABC transporter (which transfers lactococcin G across the membrane) (11a). Lactococcin G kills cells by permeabilizing the target cell membrane (18). When the two lactococcin G peptides interact with each other in the presence of membrane structures, they form amphiphilic α-helices that apparently are inserted into target cell membranes, which creates potassium-selective channels (11, 18). Since the isolation and characterization of lactococcin G, other two-peptide LAB bacteriocins that have been described include lactacin F (2), lactobin A (which exhibits sequence similarity to lactacin F) (5), and thermophilin 13 (17). The two genes encoding the lactacin F peptides are located in the same operon (2).
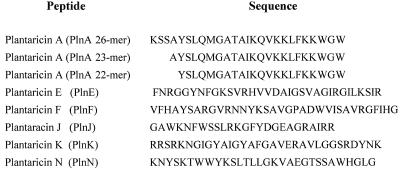
Plantaricin A (PlnA) is a short bacteriocinlike peptide produced by Lactobacillus plantarum C11. The plnA gene encodes the precursor of a 26-residue peptide (8). In addition to the 26-mer peptide, two N-terminally truncated shorter forms of PlnA (containing 22 and 23 residues) (Fig. 1) were isolated from L. plantarum C11 culture medium. Originally, it was thought that these two truncated peptides compose a two-peptide bacteriocin (23). Subsequent studies revealed, however, that all three variants of PlnA function as a peptide pheromone that induces transcription of pln genes (3, 6, 15). The bacteriocin activity originally attributed to the PlnA peptides was presumably caused at least in part by previously unidentified bacteriocins, probably two-peptide bacteriocins (see below).
FIG. 1.
Bacteriocinlike peptides encoded by pln genes. The peptides are characterized by their cationic character, their potential to form amphiphilic α-helices, and the fact that their genes encode precursors containing a typical double glycine leader peptide (7, 12). All of these peptides were detected in culture supernatants of L. plantarum C11 (see text for details).
PlnA induces transcription of genes organized in the following five operons: plnABCD, plnEFI, plnJKLR, plnMNOP, and plnGHSTUV. By analogy to functionally characterized genes from related species, it has been suggested that most of the pln genes are involved in bacteriocin production (7). plnA, plnE, plnF, plnJ, plnK, and plnN encode typical precursors of bacteriocinlike peptides (7) (Fig. 1), whereas it has been suggested that plnI, plnL, plnM, and plnP encode immunity proteins. Other genes often found in conjunction with bacteriocin production have also been detected in the pln operons; plnB, plnC, and plnD encode proteins involved in transducing the signal given by the PlnA pheromone (3, 8), and plnGH encodes the machinery necessary to secrete and process bacteriocin precursors (7).
Purification of PlnA, PlnE, PlnF, PlnJ, PlnK, and PlnN.
To purify LAB bacteriocins to apparent homogeneity, we developed a standard four-step method which includes (i) precipitation of bacteriocins from culture media with 40% (wt/vol) ammonium sulfate, (ii) binding of bacteriocins to a cation exchanger (SP-Sepharose) at pH 7 and elution with 1 M NaCl, (iii) binding of bacteriocins to a hydrophobic interaction column (Octyl-Sepharose) and elution with 70% (vol/vol) ethanol, and (iv) repeated reverse-phase chromatography (20, 22, 23, 30). However, purification of the various peptides from L. plantarum C11 culture medium by this standard and usually successful four-step method was problematic. N-terminal sequencing and mass spectrometry analysis showed that almost all peptide preparations contained different amounts of at least two of the plantaricin peptides. Fractions of PlnN and the PlnA 22- and 23-mer peptides that were approximately 80 to 90% pure could be obtained by the four-step method, as described previously (23). A slight change in the chromatographic conditions (3 M guanidine-HCl was added to the elution buffer in the ion-exchange and hydrophobic interaction chromatography steps) resulted in purification of PlnJ and PlnK to the extent (purity, approximately 80 to 90%) that sequence determination by peptide sequencing was possible for these peptides (results not shown). Peptide sequencing and mass spectrometry analysis (measured molecular masses were within 0.4 Da of the theoretical molecular masses) showed that the purified PlnJ, PlnK, PlnN, and PlnA 22- and 23-mer peptides were identical to the peptides predicted from the genes (Fig. 1).
Reasonably pure preparations of PlnE, PlnF, and the PlnA 26-mer peptide were not obtained by the four-step purification method. However, peptide sequencing and mass spectrometry analysis of various peptide mixtures obtained when we attempted to purify these three peptides did reveal their presence (the detected molecular masses were within 0.4 Da of the theoretical molecular masses). Thus, the data showed that L. plantarum C11 did secrete all three PlnA peptides, as well as the putative bacteriocins PlnE, PlnF, PlnJ, PlnK, and PlnN. Moreover, the data showed that none of the peptides contained posttranslationally modified amino acids.
Two new two-peptide bacteriocins: PlnEF and PlnJK.
To ensure that each plantaricin preparation used for further studies was completely devoid of other contaminating plantaricin peptides, preparations of the eight plantaricin peptides mentioned above (Fig. 1) were obtained by solid-phase synthesis. Synthesis was carried out with an Applied Biosystems model 430A peptide synthesizer by using the standard tert-butoxycarbonyl synthesis protocol of the manufacturer. The peptides were solubilized in 0.1% trifluoroacetic acid (TFA) so that the concentrations were about 5 mg/ml for PlnN and the PlnA peptides, 25 mg/ml for PlnJ, PlnK, and PlnE, and 50 mg/ml for PlnF. For purification, about 1 mg of synthesized peptide was applied to a C2-C18 reverse-phase PepRPC HR 5/5 column (Pharmacia Biotech) equilibrated with 0.1% TFA. The peptides were eluted from the column with a 20 to 40% linear gradient of 2-propanol containing 0.1% TFA and rechromatographed once or twice on the reverse-phase column as described above. The primary structure and purity of the peptides were verified by electrospray mass spectrometry analysis, protein sequencing, and capillary electrophoresis as previously described (10).
A strain-specific antagonistic activity (bacteriocin activity) was detected at nanomolar concentrations when PlnE and PlnF were combined and when PlnJ and PlnK were combined (Table 1). The peptides were at least 103 times more active when they were combined (PlnE was combined with PlnF and PlnJ was combined with PlnK) than when they were tested individually, and optimal activity was obtained when complementary peptides were present in approximately equal amounts (results not shown). Thus, PlnEF and PlnJK clearly are two-peptide bacteriocin systems. This is consistent with the fact that the structural genes encoding the complementary peptides are located next to each other in the same operon (plnE is next to plnF, and plnJ is next to plnK) and thus are transcribed simultaneously in equal amounts (7).
TABLE 1.
MICs for plantaricin peptidesa
| Indicator strain | Peptide MIC (nM) of:
|
|||
|---|---|---|---|---|
| PlnJK | PlnEF | PlnA 22-mer | PlnA 26-mer | |
| Pediococcus pentosaceus Pac 1.0 | 50 | 5 | 100 | 100 |
| Lactobacillus plantarum 965 | 0.1 | 7 | 500 | 300 |
| Lactobacillus casei subsp. casei NCDO 161 | ≥400 | 20 | 250 | 250 |
| Lactobacillus casei NCDO 2713 | ≥400 | 150 | ≥6,000 | ≥6,000 |
| Lactobacillus sake NCDO 2714 | 50 | 4 | 250 | 60 |
| Lactobacillus viridescens NCDO 1655 | 5 | 2 | 50 | 20 |
| Pediococcus pentosaceus NCDO 990 | ≥400 | 30 | ≥6,000 | ≥6,000 |
| Carnobacterium piscicola UI49 | ≥400 | ≥400 | ≥6,000 | ≥6,000 |
| Lactobacillus plantarum UI50 | ≥400 | ≥400 | ≥6,000 | ≥6,000 |
| Lactobacillus sake 706 | ≥400 | 100 | ≥1,000 | ≥500 |
| Pediococcus acidilactici CH | ≥400 | 10 | NTb | ≥500 |
| Lactobacillus curvatus 89 | ≥400 | 5 | NT | ≥500 |
Bacteriocin activity was determined as described previously (22). The values are the concentrations of peptides that caused 50% growth inhibition of the indicator strains. The peptides making up PlnJK and PlnEF were mixed at a 1:1 ratio. PlnN was tested against all indicator strains, but no antagonistic activity was detected at any concentration (the highest concentration tested was approximately 1 μM). The PlnA 23-mer peptide was tested against L. plantarum 965 and exhibited antagonistic activity similar to that of the PlnA 22-mer peptide.
NT, not tested.
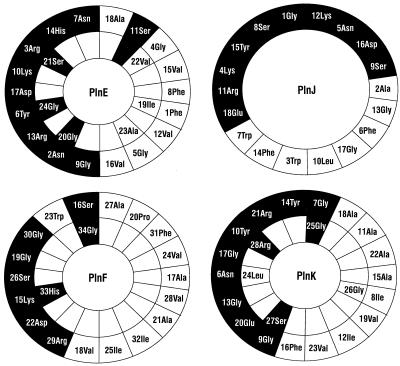
PlnE, PlnF, PlnJ, and PlnK all have regions (lengths, 18 to 24 residues) that become amphiphilic if they adopt an α-helical structure (Fig. 2). The peptides are in this respect similar to the α and β peptides that constitute the two-peptide bacteriocin lactococcin G (22) and to one of the peptides of the two-peptide bacteriocin lactacin F (2). The putative amphiphilic regions suggest that both PlnEF and PlnJK kill cells by permeabilizing cell membranes, as is the case for lactococcin G, lactacin F, and thermophilin 13 (2, 17, 18). The interaction between complementary peptides which presumably results in membrane permeabilization is specific, since neither PlnE nor PlnF could complement (act synergistically with) PlnJ or PlnK, and none of these peptides could complement the lactococcin G α or β peptides (results not shown).
FIG. 2.
Helical wheel representations of PlnE, PlnF, PlnJ, and PlnK. The black and white boxes indicate polar and nonpolar residues, respectively. Glycine is included in both black and white boxes as it is treated as being neutral with respect to polarity.
PlnN looks like a bacteriocin. First, the plnN gene encodes a precursor with a so-called double glycine leader peptide, which is very typical of bacteriocinlike peptides (7, 12). Second, the mature PlnN peptide has a putative amphiphilic α-helical region that is found in many bacteriocins. Even so, synthetic PlnN did not exhibit antagonistic activity toward the indicator strains tested (Table 1). Thus, the function of PlnN remains unknown.
The peptide pheromone PlnA has bacteriocinlike activity.
In a previous study (6) PlnA did not exhibit any antagonistic activity against the standard indicator strain L. plantarum 965. This study was refined and extended by testing the effect of very pure PlnA peptides at high concentrations on the growth of various indicator strains. Table 1 shows that the PlnA peptides do exhibit strain-specific antagonistic activity, although the peptides are clearly less potent than PlnEF, PlnJK, and bacteriocins in general with most of the indicator strains tested (including L. plantarum 965). As is the case for PlnEF, PlnJK, and the lactococcin G α and β peptides, the PlnA peptides may form an amphiphilic α-helix which may interact with target cell membranes. Thus, membrane perturbation could be the means by which PlnA acts antagonistically. All combinations of the PlnA 22-, 23-, and 26-mer peptides and all combinations of these peptides with PlnEF or PlnJK resulted in an additive (not a synergistic) antimicrobial effect (results not shown). Thus, PlnA should not be classified as a two-peptide bacteriocin as originally suggested (23). Rather, PlnA may (by definition) be considered a group II one-peptide bacteriocin, although it is a less potent antagonist than PlnEF and PlnJK and the biological significance of its antagonistic activity remains uncertain. Interestingly, it has been shown that in some other LAB transcription of the genes necessary for bacteriocin production is induced by the bacteriocin itself (e.g., nisin and carnobacteriocin B2) (16, 24, 26, 28). PlnA may be an (evolutionary) intermediate between these inducing factors with full bacteriocin activity and the very short inducing factors with no bacteriocin activity, such as the 19-mer peptide which induces the production of the bacteriocin sakacin P (3, 9). PlnA induces transcription of several genes with unknown functions (7), and it would be interesting to know whether one of these genes encodes an immunity protein for PlnA. The absence of an immunity protein would strongly suggest that the primary biological function of PlnA is as a peptide pheromone.
One might speculate that the truncated PlnA 22- and 23-mer peptides result from an alternate export and processing system that does not depend on the dedicated bacteriocin transport and processing machinery (encoded by plnGH) and thus does not depend on the expression of bacteriocin-related genes. This would permit slow accumulation of the 22- and 23-mer peptides in the absence of expression of the ABC transporter gene. Upon reaching the threshold concentration for induction, the truncated peptides would then induce expression of all genes involved in bacteriocin production, resulting in production and secretion of the intact PlnA 26-mer peptide, as well as PlnEF, PlnJK, and PlnN.
The present data provide a striking example of the gradually emerging general notion that many LAB known to produce bacteriocins actually produce several of these peptides. In addition to the “classic” example of the lactococcins produced by several Lactococcus lactis strains (31), several new examples have recently been described (3–5, 25). These observations are of great importance from an applied point of view, since they show that a complete analysis of the bacteriocin activity of an LAB may require a thorough search for more than one peptide.
Acknowledgments
We thank P. Hans Middelhoven for help with some of the experiments.
This work was supported by a grant from the Norwegian Research Council.
REFERENCES
- 1.Abee T. Pore-forming bacteriocins of gram-positive bacteria and self-protection mechanisms of producer organisms. FEMS Microbiol Lett. 1995;129:1–10. doi: 10.1016/0378-1097(95)00137-T. [DOI] [PubMed] [Google Scholar]
- 2.Allison G E, Fremaux C, Klaenhammer T R. Expansion of bacteriocin activity and host range upon complementation of two peptides encoded within the lactacin F operon. J Bacteriol. 1994;176:2235–2241. doi: 10.1128/jb.176.8.2235-2241.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brurberg M B, Nes I F, Eijsink V G H. Pheromone-induced production of antimicrobial peptides in Lactobacillus. Mol Microbiol. 1997;26:347–360. doi: 10.1046/j.1365-2958.1997.5821951.x. [DOI] [PubMed] [Google Scholar]
- 4.Casaus P, Nilsen T, Cintas L M, Nes I F, Hernández P E, Holo H. Enterocin B, a new bacteriocin from Enterococcus faecium T136 which can act synergistically with enterocin A. Microbiology. 1997;143:2287–2294. doi: 10.1099/00221287-143-7-2287. [DOI] [PubMed] [Google Scholar]
- 5.Contreras B G L, De Vuyst L, Devreese B, Busanyova K, Raymaeckers J, Bosman F, Sablon E, Vandamme E J. Isolation, purification, and amino acid sequence of lactobin A, one of the two bacteriocins produced by Lactobacillus amylovorus LMG P-13139. Appl Environ Microbiol. 1997;63:13–20. doi: 10.1128/aem.63.1.13-20.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diep D B, Håvarstein L S, Nes I F. A bacteriocin-like peptide induces bacteriocin synthesis in L. plantarum C11. Mol Microbiol. 1995;18:631–639. doi: 10.1111/j.1365-2958.1995.mmi_18040631.x. [DOI] [PubMed] [Google Scholar]
- 7.Diep D B, Håvarstein L S, Nes I F. Characterization of the locus responsible for the bacteriocin production in Lactobacillus plantarum C11. J Bacteriol. 1996;178:4472–4483. doi: 10.1128/jb.178.15.4472-4483.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diep D B, Håvarstein L S, Nissen-Meyer J, Nes I F. The gene encoding plantaricin A, a bacteriocin from Lactobacillus plantarum C11, is located on the same transcriptional unit as an agr-like regulatory system. Appl Environ Microbiol. 1994;60:160–166. doi: 10.1128/aem.60.1.160-166.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eijsink V G H, Brurberg M B, Middelhoven P H, Nes I F. Induction of bacteriocin production in Lactobacillus sake by a secreted peptide. J Bacteriol. 1996;178:2232–2237. doi: 10.1128/jb.178.8.2232-2237.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fimland G, Blingsmo O, Sletten K, Jung G, Nes I F, Nissen-Meyer J. New biologically active hybrid bacteriocins constructed by combining regions from various pediocin-like bacteriocins: the C-terminal region is important for determining specificity. Appl Environ Microbiol. 1996;62:3313–3318. doi: 10.1128/aem.62.9.3313-3318.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hauge H H, Nissen-Meyer J, Nes I F, Eijsink V G H. Amphiphilic α-helices are important structural motifs in the α and β peptides that constitute the bacteriocin lactococcin G. Eur J Biochem. 1998;251:565–572. doi: 10.1046/j.1432-1327.1998.2510565.x. [DOI] [PubMed] [Google Scholar]
- 11a.Håvarstein, L. S. Unpublished data.
- 12.Håvarstein L S, Diep D B, Nes I F. A family of ABC transporters carry out proteolytic processing of their substrates concomitant with export. Mol Microbiol. 1995;16:229–240. doi: 10.1111/j.1365-2958.1995.tb02295.x. [DOI] [PubMed] [Google Scholar]
- 13.Holo H, Nilssen Ø, Nes I F. Lactococcin A, a new bacteriocin from Lactococcus lactis subsp. cremoris: isolation and characterization of the protein and its gene. J Bacteriol. 1991;173:3879–3887. doi: 10.1128/jb.173.12.3879-3887.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klaenhammer T R. Genetics of bacteriocins produced by lactic acid bacteria. FEMS Microbiol Rev. 1993;12:39–86. doi: 10.1111/j.1574-6976.1993.tb00012.x. [DOI] [PubMed] [Google Scholar]
- 15.Kleerebezem M, Quadri L E N, Kuipers O P, De Vos W M. Quorum sensing by peptide pheromones and two-component signal-transduction systems in Gram-positive bacteria. Mol Microbiol. 1997;24:895–904. doi: 10.1046/j.1365-2958.1997.4251782.x. [DOI] [PubMed] [Google Scholar]
- 16.Kuipers O P, Beerthuyzen M M, de Ruyter P G G A, Luesink E J, de Vos W M. Autoregulation of nisin biosynthesis in Lactococcus lactis by signal transduction. J Biol Chem. 1995;270:27299–27304. doi: 10.1074/jbc.270.45.27299. [DOI] [PubMed] [Google Scholar]
- 17.Marciset O, Jeronimus-Stratingh M C, Mollet B, Poolman B. Thermophilin 13, a nontypical antilisterial poration complex bacteriocin, that functions without a receptor. J Biol Chem. 1997;272:14277–14284. doi: 10.1074/jbc.272.22.14277. [DOI] [PubMed] [Google Scholar]
- 18.Moll G, Ubbink-Kok T, Hauge H H, Nissen-Meyer J, Nes I F, Konings W N, Driessen A. Lactococcin G is a potassium ion-conducting, two-component bacteriocin. J Bacteriol. 1996;178:600–605. doi: 10.1128/jb.178.3.600-605.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nes I F, Diep D B, Håvarstein L S, Brurberg M B, Eijsink V, Holo H. Biosynthesis of bacteriocins in lactic acid bacteria. Antonie Leeuwenhoek. 1996;70:113–128. doi: 10.1007/BF00395929. [DOI] [PubMed] [Google Scholar]
- 20.Nieto Lozano J C, Nissen-Meyer J, Sletten K, Peláz C, Nes I F. Purification and amino acid sequence of a bacteriocin produced by Pediococcus acidilactici. J Gen Microbiol. 1992;138:1985–1990. doi: 10.1099/00221287-138-9-1985. [DOI] [PubMed] [Google Scholar]
- 21.Nissen-Meyer, J., H. H. Hauge, G. Fimland, V. G. H. Eijsink, and I. F. Nes. Ribosomally synthesized antimicrobial peptides produced by lactic acid bacteria: their function, structure, biogenesis, and their mechanism of action. Recent Res. Dev. Microbiol., in press. [PubMed]
- 22.Nissen-Meyer J, Holo H, Håvarstein S L, Sletten K, Nes I F. A novel lactococcal bacteriocin whose activity depends on the complementary action of two peptides. J Bacteriol. 1992;174:5686–5692. doi: 10.1128/jb.174.17.5686-5692.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nissen-Meyer J, Granly Larsen A, Sletten K, Daeschel M, Nes I F. Purification and characterization of plantaricin A, a Lactobacillus plantarum bacteriocin whose activity depends on the action of two peptides. J Gen Microbiol. 1993;139:1973–1978. doi: 10.1099/00221287-139-9-1973. [DOI] [PubMed] [Google Scholar]
- 24.Quadri L E N, Yan L Z, Stiles M E, Vederas J C. Effect of amino acid substitutions on the activity of carnobacteriocin B2. J Biol Chem. 1997;272:3384–3388. doi: 10.1074/jbc.272.6.3384. [DOI] [PubMed] [Google Scholar]
- 25.Quadri L E N, Sailer M, Roy K L, Vederas J C, Stiles M E. Chemical and genetic characterization of bacteriocins produced by Carnobacterium piscicola LV17B. J Biol Chem. 1994;269:12204–12211. [PubMed] [Google Scholar]
- 26.Ra S R, Qiao M, Immonen T, Pujana I, Saris P E J. Genes responsible for nisin synthesis, regulation and immunity form a regulon of two operons and are induced by nisin in Lactococcus lactis N8. Microbiology. 1996;142:1281–1288. doi: 10.1099/13500872-142-5-1281. [DOI] [PubMed] [Google Scholar]
- 27.Sahl H-G, Jack R W, Bierbaum G. Biosynthesis and biological activities of lantibiotics with unique post-translational modifications. Eur J Biochem. 1995;230:827–853. doi: 10.1111/j.1432-1033.1995.tb20627.x. [DOI] [PubMed] [Google Scholar]
- 28.Saucier L, Paradkar A S, Frost L S, Jensen S E, Stiles M E. Transcriptional analysis and regulation of carnobacteriocin production in Carnobacterium piscicola LV17. Gene. 1997;188:271–277. doi: 10.1016/s0378-1119(96)00822-0. [DOI] [PubMed] [Google Scholar]
- 29.Skaugen M, Nissen-Meyer J, Jung G, Stevanovic S, Sletten K, Mørtvedt Abildgaard C I, Nes I F. In vivo conversion of l-serine to d-alanine in a ribosomally synthesized polypeptide. J Biol Chem. 1994;269:27183–27185. [PubMed] [Google Scholar]
- 30.Tichaczek P S, Nissen-Meyer J, Nes I F, Vogel R F, Hammes W P. Characterization of the bacteriocins curvacin A from Lactobacillus curvatus LTH1174 and sakacin P from L. sake LTH673. Syst Appl Microbiol. 1992;15:460–468. [Google Scholar]
- 31.Van Belkum M J, Hayema B J, Jeeninga R E, Kok J, Venema G. Organization and nucleotide sequence of two lactococcal bacteriocin operons. Appl Environ Microbiol. 1991;57:492–498. doi: 10.1128/aem.57.2.492-498.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Venema K, Venema G, Kok J. Lactococcal bacteriocins: mode of action and immunity. Trends Microbiol. 1995;3:299–304. doi: 10.1016/s0966-842x(00)88958-1. [DOI] [PubMed] [Google Scholar]