Abstract
High temperature and nitrate supported gene expression for sterigmatocystin biosynthesis in Aspergillus nidulans; ammonium did not. Homologous genes for aflatoxin biosynthesis in A. parasiticus showed the opposite transcript expression pattern, suggesting that the two mycotoxins are regulated differently. The aflR gene is postulated to require additional genetic elements to effect its own activation by the different culture conditions. A patulin polyketide synthase (PKS) gene was found to be regulated differently than the aflatoxin PKS. Thus, the biosyntheses of structurally similar compounds in these two fungi appear to be regulated very differently.
Aflatoxins (AF) are produced by Aspergillus flavus and A. parasiticus, while sterigmatocystin (ST) is produced by A. nidulans. AF and ST biosyntheses have similar multiple-step pathways except that the ST pathway lacks the last few biochemical steps (3, 12). These mycotoxins, commonly found as contaminants in corn, peanuts, and cottonseeds, are potent toxic and carcinogenic compounds. Neither ST nor AF and their pathway intermediates are essential for growth and development under laboratory conditions (7, 13). A number of highly conserved genes involved in AF and ST biosynthesis in A. parasiticus (12), A. flavus (12), and A. nidulans (3) have been characterized. All of these genes are tightly clustered in the three species, suggesting a common origin. These genes are expressed when primary growth slows or stops (7, 13). Expression of the AF biosynthetis genes in A. parasiticus is influenced by factors such as the carbon source and culture temperature (6, 7, 9). Less is known about the effects of similar environmental factors on ST biosynthesis genes. The environmental cue that leads to AF and ST biosynthesis is presumably manifested by activating aflR, which in turn activates transcription of the AF and ST genes (5, 11, 14). The nature of any genetic connection between various environmental stimuli and aflR activation has not been reported. In the present study we examined the effects of culture temperature and nitrogen source on the activation of ST and AF transcripts. While only a few transcripts were analyzed, they were assumed to reflect the activation of the entire ST and AF biosynthetic pathways. Our objective was to determine if ST and AF transcripts in A. nidulans and A. parasiticus are differentially transcribed.
Effects of culture temperature.
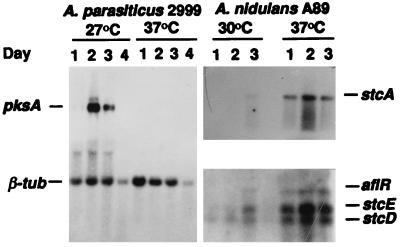
A. parasiticus NRRL2999 is a wild-type strain known to produce AF (1). A. nidulans FGSC A89 (biA1 argB2) is auxotrophic for biotin and arginine and produces ST (13). To test AF and ST production, we inoculated fresh conidia of the two strains onto the surface of 3 ml of supplemented minimal liquid medium (13) in 8-ml vials and incubated them in darkness. AF or ST was extracted and detected by thin-layer chromatography (7, 13). As reported previously (6, 7), no AF was detected when A. parasiticus was cultured at 37°C, but high levels (peak amount, >500 μg/vial) were produced at 27°C and only marginal amounts (peak amount, <20 μg/vial) were produced at 33°C. In A. nidulans, however, similar peak amounts of ST (30 to 45 μg/vial) were detected at all three temperatures, but the time of ST appearance varied: the 4th day peak was at 37°C, the 5th day peak was at 33°C, and the 6th day peak was at 30°C. At 27°C, ST was produced later and in smaller amounts (<5 μg/vial). Northern blot analysis revealed that the temperature effects on AF and ST production resulted from differential gene expression in the two species. The 6.8-kb transcript of the AF polyketide synthase (PKS) gene, pksA (4), also known as pksL1 (7), was expressed at 27°C but not at 37°C (Fig. 1). In A. nidulans (Fig. 1), however, three transcripts (7.0, 1.0, and 0.6 kb) for the ST biosynthetic genes stcA (pksA), stcE, and stcD (3) and another (1.6 kb) for the regulatory gene aflR were expressed earlier and more strongly at higher temperatures (33 and 37°C) than at lower temperatures (27 and 30°C).
FIG. 1.
Influence of culture temperature on expression of genes for AF and ST biosynthesis. Supplemented minimal medium was used with 3.7 g of NH4Cl per liter for A. parasiticus and 6 g of NaNO3 per liter for A. nidulans. Each of the above probes was from the relevant cloned gene in A. nidulans and A. parasiticus. The A. parasiticus probes were a 4.5-kb KpnI fragment for pksA and a cloned β-tub gene (6a) as an internal control. The A. nidulans probes were a 5.4-kb KpnI fragment for stcA (3, 13) and a 5.6-kb KpnI fragment for aflR, stcE, and stcD (3).
Effects of nitrogen source.
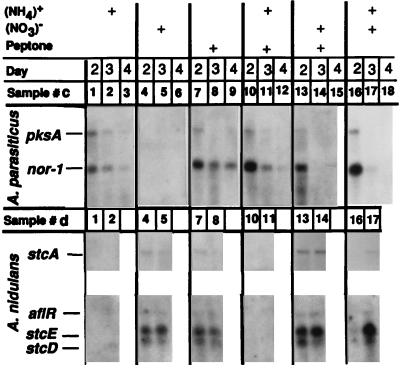
In A. parasiticus, nitrate represses synthesis of the AF intermediate, versicolorin, while ammonium supports it (8). In the present study, we observed that the effect was the opposite in A. nidulans; i.e., nitrate supported ST production (peak amount, 30 to 45 μg/vial) while ammonium did not. When ammonium was used as a sole nitrogen source for A. nidulans, the initial mycelial growth was slower and stopped after 2 days. No ST was detected (<1 μg/vial). Northern blot analysis showed that the effects of nitrogen sources on AF and ST production may result from their effects on AF and ST gene expression (Fig. 2). Three nitrogen sources (ammonium, nitrate, and peptone) were tested, individually or in combination, for their effects on transcript accumulation. In A. parasiticus, ammonium and peptone both supported AF gene expression, while nitrate had strong negative effects. The genes pksA and nor-1 (10) were expressed when strain NRRL2999 was grown in media with either ammonium, peptone, or both (Fig. 2, lanes c1 to c3, c7 to c9, and c10 to c12) but were not expressed when nitrate alone was used as the nitrogen source (Fig. 2, lanes c4 to c6. The expression pattern of these two genes changed in media where nitrate was combined with either peptone (Fig. 2, lanes c13 to c15) or ammonium (Fig. 2, lanes c16 to c18). In these media, the levels of transcripts were comparable to ammonium or peptone alone by day 2 but were significantly weaker or absent on days 3 and 4.
FIG. 2.
Influence of nitrogen sources in the culture medium on transcription of genes for AF and ST biosynthesis. The medium background was supplemented minimal medium. The sole nitrogen source was either 3.7 g of NH4Cl per liter for (NH4)+, 6 g of NaNO3 per liter for (NO3)−, or 20 g of peptone per liter. When used in combination, each was reduced to half strength. The media for A. nidulans were appropriately supplemented. The respective temperatures employed were 27°C for A. parasiticus and 33°C for A. nidulans. A PCR-amplified 0.8-kb fragment was used for nor-1 (10); other probes were the same as described for Fig. 1.
In contrast to A. parasiticus, nitrate supported ST gene expression in A. nidulans (Fig. 2). Four genes were tested: stcA, aflR, stcE, and stcD (3). The genes stcA and stcE in A. nidulans are homologous to the genes pksA and nor-1 in A. parasiticus. Consistent with higher yields of ST, the transcripts of the four genes in A. nidulans were present mostly at high levels when nitrate was used as a sole nitrogen source (Fig. 2, lanes d4 and d5) or with a combined nitrogen source (Fig. 2, lanes d13 and d14 and lanes d16 and d17). Also, in contrast to A. parasiticus, ammonium reduced ST production and ST gene expression in A. nidulans (Fig. 2). Transcripts of the four genes were not detectable whether ammonium was used alone (Fig. 2, lanes d1 and d2) or in combination with peptone (Fig. 2, lanes d10 and d11). Interestingly, when ammonium was combined with nitrate, the transcripts were barely detectable by day 2 but were readily detected by day 3 (Fig. 2, lanes d16 and d17).
The effects of nitrogen sources in the experiment described above were apparently not related to other compounds in the medium, because similar results were obtained when these nitrogen sources were tested in another chemically defined medium, GMS (1) (data not shown).
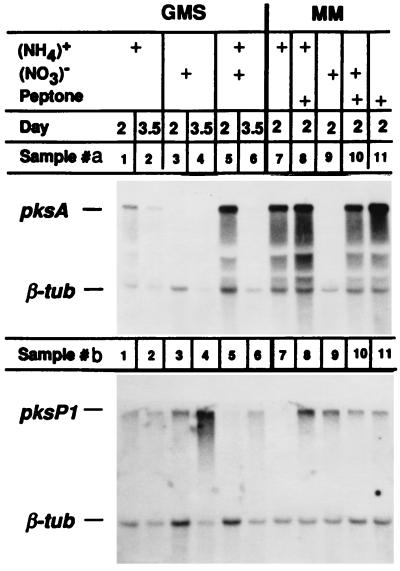
The nitrogen source also affected the expression of a second PKS gene in A. parasiticus (Fig. 3). In addition to the pksA gene (7), we have cloned another PKS gene, pksP1. The pksP1 gene is probably involved in the biosynthesis of patulin-related secondary compounds, since the putative PKSP1 polypeptide showed 56% identity at the amino acid sequence level and can be lined up throughout its entire length with a known patulin PKS (6-methylsalicylic acid synthase) in Penicillium patulun (2). To determine whether these two PKS genes are controlled by a common regulatory mechanism, we studied their expression in media with different nitrogen sources (Fig. 3). For pksA, Northern blot analysis of the poly(A)+ RNA was similar to that for the total RNA Northern blot in Fig. 2. Expression was favored by ammonium (Fig. 3, lanes a1, a2, and a7) and peptone (lane a11) but inhibited by nitrate (lanes a3, a4, and a9). For pksP1, however, nitrate had a positive effect, which is clearly seen by comparing lanes a3, a4, and a9 to lanes b3, b4, and b9 (Fig. 3). The transcript level of pksP1 in nitrate-supplemented media was higher than in ammonium-supplemented media. For example, samples b3 and b4 showed stronger hybridization than b1 and b2, and b9 was stronger than b7. These results suggest that the two PKS genes in A. parasiticus are controlled differently, although little can be said about the nature of pksP1 transcript regulation.
FIG. 3.
Influence of nitrogen sources in the culture media (GMS and minimal medium [MM]) on expression of the two PKS genes in A. parasiticus. Nitrogen sources were the same as described for Fig. 2. The culture temperature was 27°C. The A. parasiticus pksA probe was as described for Fig. 1. The probe for A. parasiticus pksP1 was a 2.6-kb fragment from the cloned pksP1 gene (6a).
The results presented show differential expression of the homologous AF and ST genes caused by both culture temperature (Fig. 1) and nitrate as a nitrogen source (Fig. 2). While ammonium showed similar results, the growth problems with A. nidulans when ammonium was used as a sole nitrogen source rendered these experiments less convincing. Nevertheless, the differential gene expression observed with temperature and nitrate was presumably manifested through the common regulatory gene aflR. There is persuasive evidence that aflR is a major ST and AF pathway regulator. It encodes a putative peptide with a zinc binuclear cluster DNA-binding domain (11, 14). When aflR was mutated (11) in A. flavus or knocked out in A. nidulans (14), expression of all of the AF and ST clustered genes was turned off or significantly reduced. When aflR was overexpressed, the nitrate inhibition of AF biosynthesis was relieved in A. parasiticus (5) and expression of the controlled pathway genes was activated in A. nidulans (5a). Forced expression of the A. flavus aflR gene in A. nidulans induced expression of genes in the ST cluster (14), suggesting that aflR is functionally conserved in the two species. Notwithstanding the important regulatory role of aflR, the differential transcription of the ST and AF pathways in response to similar environmental factors is difficult to explain if aflR is the sole regulatory gene. A testable hypothesis to explain differential transcription of ST and AF genes is that other genetic regulatory factors, located externally to the ST and AF gene clusters, regulate the activation of aflR. Presumably these external regulatory elements respond to the environmental cues. Analysis of mutations induced in a strain with an appropriate genetic background should lead to the identification of these additional regulatory elements.
Nucleotide sequence accession number.
The DNA sequence of pksP1 has been deposited in the GenBank database under accession no. U52151.
Acknowledgments
G.H.F. was supported by a training grant from the Department of Genetics, University of Wisconsin—Madison. This work was partially supported by the National Peanut Foundation and Best Foods.
We thank J. H. Yu for helpful discussions and T. Volk for comments on the manuscript.
REFERENCES
- 1.Abodollahi A, Buchanan R L. Regulation of aflatoxin biosynthesis: characterization of glucose as an apparent inducer of aflatoxin production. J Food Sci. 1981;46:143–146. [Google Scholar]
- 2.Beck J, Ripka S, Siegner A, Schiltz E, Schweizer E. The multifunctional 6-methylsalicylic acid synthase gene of Penicillium patulun: its gene structure relative to that of other polyketide synthases. Eur J Biochem. 1990;192:487–498. doi: 10.1111/j.1432-1033.1990.tb19252.x. [DOI] [PubMed] [Google Scholar]
- 3.Brown D W, Yu J, Kellar H, Fernandes M, Nesbitt C, Keller N P, Adams T H, Leonard T J. Twenty-five coregulated transcripts define a secondary metabolite gene cluster in Aspergillus nidulans. Proc Natl Acad Sci USA. 1996;93:1418–1422. doi: 10.1073/pnas.93.4.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang P-K, Cary J W, Yu J, Bhatnagar D, Cleveland T E. The Aspergillus parasiticus polyketide synthase gene pksA, a homolog of Aspergillus nidulans wA, is required for aflatoxin B1 biosynthesis. Mol Gen Genet. 1995;248:270–277. doi: 10.1007/BF02191593. [DOI] [PubMed] [Google Scholar]
- 5.Chang P-K, Ehrlich K C, Yu J, Bhatnagar D, Cleveland T J. Increased expression of Aspergillus parasiticus aflR, encoding a sequence-specific DNA binding protein, relieves nitrate inhibition of aflatoxin biosynthesis. Appl Environ Microbiol. 1995;61:2372–2377. doi: 10.1128/aem.61.6.2372-2377.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5a.Feng, G. H. Unpublished results.
- 6.Feng G H, Chu F S, Leonard T J. Molecular cloning of genes related to aflatoxin biosynthesis by differential screening. Appl Environ Microbiol. 1992;58:455–460. doi: 10.1128/aem.58.2.455-460.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6a.Feng, G. H., and T. J. Leonard. Unpublished results.
- 7.Feng G H, Leonard T J. Characterization of the polyketide synthase gene (pksL1) required for aflatoxin biosynthesis in Aspergillus parasiticus. J Bacteriol. 1995;177:6246–6254. doi: 10.1128/jb.177.21.6246-6254.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Niehaus W G, Jiang W. Nitrate induces enzymes of the mannitol cycle and suppresses versicolorin synthesis in Aspergillus parasiticus. Mycopathologia. 1989;107:131–137. doi: 10.1007/BF00707550. [DOI] [PubMed] [Google Scholar]
- 9.Skory C D, Chang P K, Cary J, Linz J E. Isolation and characterization of a gene from Aspergillus parasiticus associated with the conversion of versicolorin A to sterigmatocystin in aflatoxin biosynthesis. Appl Environ Microbiol. 1992;58:3527–3537. doi: 10.1128/aem.58.11.3527-3537.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trail F, Chang P-K, Cary J, Linz J E. Structural and functional analysis of the nor-1 gene involved in the biosynthesis of aflatoxins by Aspergillus parasiticus. Appl Environ Microbiol. 1994;60:4078–4085. doi: 10.1128/aem.60.11.4078-4085.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woloshuk C P, Foutz K R, Brewer J F, Bhatnagar D, Cleveland T E, Payne G A. Molecular characterization of aflR, a regulatory locus for aflatoxin biosynthesis. Appl Environ Microbiol. 1994;60:2408–2414. doi: 10.1128/aem.60.7.2408-2414.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu J, Chang P-K, Cary J W, Wright M, Bhatnagar D, Cleveland T, Payne G A, Linz J E. Comparative mapping of aflatoxin pathway gene clusters in Aspergillus parasiticus and Aspergillus flavus. Appl Environ Microbiol. 1995;61:2365–2371. doi: 10.1128/aem.61.6.2365-2371.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu J H, Leonard T J. Sterigmatocystin biosynthesis in Aspergillus nidulans requires a type I polyketide synthase. J Bacteriol. 1995;177:4792–4800. doi: 10.1128/jb.177.16.4792-4800.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu J H, Butchko R A E, Fernandes M, Keller N P, Leonard T L, Adams T H. Conservation of structure and function of the aflatoxin regulatory gene aflR from Aspergillus nidulans and A. flavus. Curr Genet. 1996;29:549–555. doi: 10.1007/BF02426959. [DOI] [PubMed] [Google Scholar]