Abstract
Objective
Endotracheal intubation (ETI) is critical to secure the airway in emergent situations. Although artificial intelligence algorithms are frequently used to analyze medical images, their application to evaluating intraoral structures based on images captured during emergent ETI remains limited. The aim of this study is to develop an artificial intelligence model for segmenting structures in the oral cavity using video laryngoscope (VL) images.
Methods
From 54 VL videos, clinicians manually labeled images that include motion blur, foggy vision, blood, mucus, and vomitus. Anatomical structures of interest included the tongue, epiglottis, vocal cord, and corniculate cartilage. EfficientNet-B5 with DeepLabv3+, EffecientNet-B5 with U-Net, and Configured Mask R-Convolution Neural Network (CNN) were used; EffecientNet-B5 was pretrained on ImageNet. Dice similarity coefficient (DSC) was used to measure the segmentation performance of the model. Accuracy, recall, specificity, and F1 score were used to evaluate the model's performance in targeting the structure from the value of the intersection over union between the ground truth and prediction mask.
Results
The DSC of tongue, epiglottis, vocal cord, and corniculate cartilage obtained from the EfficientNet-B5 with DeepLabv3+, EfficientNet-B5 with U-Net, and Configured Mask R-CNN model were 0.3351/0.7675/0.766/0.6539, 0.0/0.7581/0.7395/0.6906, and 0.1167/0.7677/0.7207/0.57, respectively. Furthermore, the processing speeds (frames per second) of the three models stood at 3, 24, and 32, respectively.
Conclusions
The algorithm developed in this study can assist medical providers performing ETI in emergent situations.
Keywords: Biomedical image processing, convolutional neural networks, deep learning, image segmentation, intubation
Introduction
Endotracheal intubation (ETI) is a key technique performed during emergencies, such as cardiopulmonary resuscitation, loss of consciousness, and respiratory failure, to secure the airway. 1 The purpose of ETI is to protect and secure the airway when mask ventilation is difficult or prolonged mechanical ventilation is necessary. 2 However, tracheal intubation in emergency situations has lower first-pass success rate than operating room environment with experienced anesthetists under elective conditions and repetitive efforts to secure the airway when the first intubation is unsuccessful increase the risk of aspiration, insufficient ventilation, and cause potential harm to the airway.1,3,4 In this context, the deployment of video laryngoscope (VL) can assist ETI by providing relatively good visualization of the larynx, improving the ETI success rate, and reducing unnecessary intubation, resulting in patient safety.5,6 In addition, VL could help teach emergency medicine residents or attendings about tracheal intubation and airway management and recording of the ETI can allow additional review and analyses.5,7
When dealing with laryngeal videos or images, artificial intelligence (AI) is a powerful tool applied for tasks such as classification, detection, and segmentation. For instance, the U-Net base model was used to automatically segment the glottis for the first step in a computer-aided diagnostic system using 928 images from 90 volunteers of routine examination (electronic laryngoscope for routine examination) using electronic laryngoscope.8,9 Similarly, Mask R-Convolution Neural Network (CNN) was used to segment the vocal folds and glottal region to provide clinicians with important medical information during diagnosis using manually segmented 536 images from 2 resection surgeries and 3045 images from various laryngeal videos and public videostroboscopy dataset.10,11 Additionally, a combined model of Transformer and CNN was used to segment the organ in the laryngoscope image using 822 images from 350 routine examination videos.12–14 Moreover, a study using a CNN-based classifier was used to classify laryngeal neoplasms to aid in diagnosis using 24,667 independent and clear consecutive laryngoscopy images from 9231 patients. 15 However, these models targeted normal anatomy measurement or disease detection, not for ETI use.
ETI is mainly performed in the intensive care unit, emergency department (ED), hospital ward, or operating room. Previous VL related AI studies used images from resection surgeries, images collected from YouTube, public VL datasets, or laryngoscope images during routine examinations in out-patient clinics in hospitals.9,11,16 The VL images acquired from predictable and stable conditions, such as elective operating rooms or out-patient clinics, are different from those acquired in unpredictable situations such as in the ED where various factors can contribute to physiologically challenging airways, including hemodynamic instability, vomiting, facial/cervical trauma, and the need for cervical immobilization.4,17 In contrast to previous studies, VL images obtained within the ED frequently contain foggy vision, motion blur, blood, vomitus, saliva, and mucus which make ETI more intricate task for clinicians.
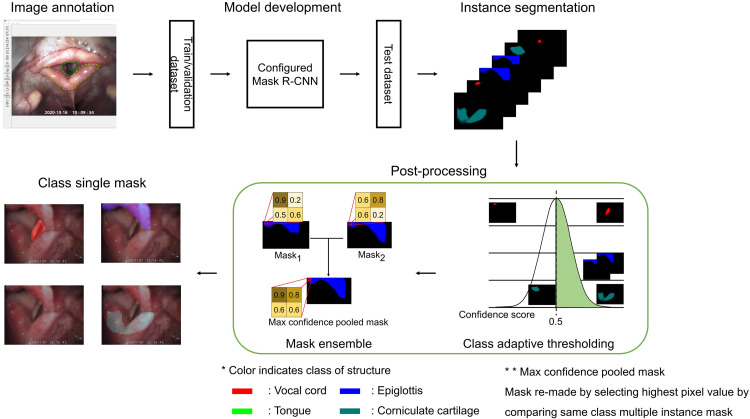
The primary objective of this study is to develop an AI algorithm that can provide real-time assistance to clinicians conducting ETI procedures in emergent situations. In this study, we developed AI algorithms to segment the tongue, epiglottis, vocal cord (VC), and corniculate cartilage (CC) using VL images acquired from the ED while performing emergent ETI. Semantic segmentation models, DeepLabv3+ and U-Net and an instance segmentation model, Mask R-CNN, were used. The blueprint of the study is illustrated in Figure 1.
Figure 1.
Flowchart of the study. The color in the mask indicates the class of the structure. Vocal cord in red, epiglottis in blue, corniculate cartilage in jade, and tongue in green. The max confidence pooled mask is the mask remade by selecting the highest pixel value by comparing multiple instance masks of the same class.
Methods
This study is an experimental retrospective study developing AI algorithms using VL images.
Dataset
Intubation videos in clinical emergencies were collected between October 2020 and January 2021 at the ED of Seoul National University Bundang Hospital. This is a tertiary teaching hospital with an annual ED volume of approximately 80,000 patients and 600 ETI cases in Gyeonggi Province, South Korea. The inclusion criteria were the videos from the patients who are equal to or greater than 18 years old. The videos were excluded if patients have intraoral or laryngeal medical diseases such as neoplasm, epiglottis, or trauma.
VL videos were recorded with a resolution of 640 × 480 pixels at 30 frames per second (FPS) in AVI format using the GlideScope Go (Verathon, US), and 54 cases were collected.
This study was approved by the Institutional Review Board of Seoul National University Bundang Hospital (number B-2112-725-102). The study protocol adhered to the ethical guidelines of the 1975 Declaration of Helsinki and its subsequent revisions. The requirement for informed consent was waived by the Institutional Review Board of Seoul National University Bundang Hospital.
Images were extracted from the videos using the VirtualDub program. From the recorded videos, the range to extract the images was manually set from the beginning of intubation to the end of intubation and then extracted at two FPS. A total of 8973 images were extracted. Images with foggy vision, motion blur, blood, vomitus, saliva, and mucus were included in this study to reflect unpredictable situation images with ETI (Figure 2). Three clinicians manually excluded non-informative images and images captured outside of intraoral cavity. The structures from the remaining images were labeled using the Labelme program, and 4956 images were labeled. The anatomical structures including tongue, epiglottis, VC, and CC were labeled as landmarks for ETI. These structures were chosen according to three medical experts’ consensus. Medical experts agreed that these structures are observed in the process of ETI in time sequential with clinical importance. In cases of disagreement with labeling, consensus among the three clinicians was derived through discussion.
Figure 2.
Images used in this study. The image sample with foggy vision, motion blur, blood, vomitus, saliva, and mucus are in (a) and image samples without them are in (b).
Labeled data from the Labelme program were saved in JSON format. These were converted into a single gray scale PNG file for each dataset. The dataset was randomly split into a train set (32 cases, 2888 images), validation set (11 cases, 1177 images), and test set (11 cases, 891 images).
Environment
The study was performed on Ubuntu 20.04.3 LTS, AMD Ryzen 9 5900X, Nvidia RTX 3090 24GB, and 48GB of RAM using Python 3.7 and pytorch 1.7.1.
Segmentation
To segment the tongue, epiglottis, VC, and CC from the collected data, DeepLabv3+, U-Net and Mask R-CNN were used. DeepLabv3+ and U-Net were trained using the segmentation models pytorch library and Mask R-CNN was trained using torchvision.8,10,18 EfficientNet-B5, pretrained with ImageNet, was used for the encoder of DeepLabv3+ and U-Net. 19 For the previous two models, the batch size was set to 8, and the Adam optimizer with a learning rate of 1e-4 was used. The dice similarity coefficient (DSC) loss was used to train the model. 20 The model was evaluated using the validation set for each epoch and stored at the lowest validation loss. The learning rate decreased by 1/10 when the validation loss did not change during 30 epochs. Early stopping was applied if the validation loss was not updated for 50 epochs. The input RGB images were scaled between 0 and 1.
For the Mask R-CNN model, the batch size was set to 16, the Adam optimizer was used, and the learning rate was set to 1e-4. The model was trained using the sum of the region proposal network (RPN) and region of interest (ROI) losses, which were embedded in the torchvision Mask R-CNN code. 11 The ResNet-50-Feature Pyramid Network backbone, pretrained on COCO, was used. The input RGB images were scaled and normalized. The model was saved using the DSC loss, and the prediction mask to calculate the DSC loss was reconstructed.
As a result of the model's prediction, one or more prediction masks could exist for each class. Validation loss was calculated after going through the process of converting several masks that could exist for each class into one mask for each class. In this process, after multiplying the pixel value of each mask by the class score of the mask, the generated masks were made into one mask by comparing each pixel and replacing it with the pixel with the largest value. The Mask R-CNN trained using this method is called the Configured Mask R-CNN, and this method is presented in Figure 3. Then DSC loss was then calculated to evaluate the model and save the model when it had the lowest validation loss. The same learning rate decrease method was used for all models.
Figure 3.
Method used to train the Configured Mask R-CNN model. Prediction masks (Mask1, Mask2) were multiplied with corresponding confidence score (class confidence score). Each pixel in the masks were compared and the max value of each pixel was pooled to make max confidence pooled mask. The dice similarity coefficient loss was calculated using max confidence pooled mask and ground truth mask. The values in the figure do not depict the real value of the image.
Post-processing
The softmax activation function was used for the semantic segmentation models because the sigmoid activation function cannot clearly define the class to which the pixel belongs. Conversely, Mask R-CNN, which is an instance segmentation model, uses a sigmoid for generating masks, as the class of the mask is determined by the result of the classification part in the model. 10 The RPN module of the Mask R-CNN suggests several locations where objects could exist. A mask was obtained in each proposed area, and the class of the mask was determined by the class obtained from the model. If multiple areas were proposed for a class, the class would receive multiple masks. Therefore, a refinement process was required, as shown in Figure 4. The refinement process first starts with dropping out prediction masks that have a low-class prediction score; in this study the threshold was set to 0.5. Subsequently, each pixel in the prediction masks was compared, leaving only the pixel with the highest value for each class.
Figure 4.
Post-processing step used for the Configured Mask R-CNN. The color in the mask indicates the class of the structure. Vocal cord in red, epiglottis in blue, corniculate cartilage in jade, and tongue in green. Mask ensemble was used to make multiple instance mask into a single mask.
Statistical analysis
To evaluate the performance of the model, a DSC metric was used for assessing the model.20–22 The DSC measures the similarity between the ground truth mask and the prediction mask. The method used to evaluate the detection model was used to intuitively determine if the prediction mask properly segmented the structure. If the intersection over union (IoU) of the prediction mask and the ground truth mask is 0.5 or more, the prediction mask was considered correct (true positive).23–26 False positive was defined when the prediction mask and ground truth mask did not overlap or the IoU was lower than 0.5. False negative was defined when there was a ground truth mask but no prediction mask. True negative was defined as the presence of neither a prediction mask nor a ground truth mask. The method to divide true positive, false positive, false negative, and true negative is shown in Figure 5. A confusion matrix was generated to evaluate the model in terms of accuracy, recall, specificity, and F1 score,
| (1) |
| (2) |
| (3) |
| (4) |
Figure 5.
Flow of how the method used in detection was applied in this study. The purple mask on the left is the ground truth mask and the yellow mask is the prediction mask. The mask below diamond shape written IoU ≥ 0.5 inside indicates ground truth mask in purple, prediction mask in yellow and intersection area in sky blue. The colored mask on the right represents the class of each structure, the vocal cord is red, epiglottis is blue, the corniculate cartilage is jade, and the tongue is green. How each section is divided is shown through masks. IoU: intersection over union.
and they were calculated according to equations (1), (2), (3), and (4), respectively.
Results
DSC was evaluated separately for each structure. The threshold of the prediction mask was set to 0.5 for all the models. As shown in Table 1, EfficientNet-B5 with DeepLabv3+ showed the highest values for tongue (0.3351) and VC (0.766), EfficientNet-B5 with U-Net showed the highest value for CC (0.6906), and Configured Mask R-CNN showed the highest value for epiglottis (0.7677). The FPS of the models were measured when the models were inferencing as follows: 3 FPS, 24 FPS, and 32 FPS in EfficientNet-B5 with DeepLabv3+, EfficientNet-B5 with U-Net, and Configured Mask R-CNN, respectively.
Table 1.
DSC result of the model for each structure and its FPS.
| Model | Metric | Tongue | Epiglottis | Vocal cord | Corniculate cartilage | FPS |
|---|---|---|---|---|---|---|
| EfficientNet-B5 with DeepLabv3+ | DSC | 0.3351 | 0.7675 | 0.766 | 0.6539 | 3 |
| EfficientNet-B5 with U-Net | DSC | 0.0 | 0.7581 | 0.7395 | 0.6906 | 24 |
| Configured Mask R-CNN | DSC | 0.1167 | 0.7677 | 0.7207 | 0.57 | 32 |
DSC: dice similarity coefficient; FPS: frames per second.
The accuracy, recall, specificity, and F1 score obtained for each structure with the different models are summarized in Tables 2, 3, and 4. In terms of the tongue, the EfficientNet-B5 with DeepLabv3+ model showcased the highest accuracy (0.8743), recall (0.4252), and F1 score (0.4909). On the other hand, the EfficientNet-B5 with U-Net model yielded the highest specificity (1.0). For the epiglottis, the Configured Mask R-CNN model took the lead in accuracy (0.862) and specificity (0.7517), whereas the EfficientNet-B5 with DeepLabv3+ model excelled in recall (0.9794), and F1 score (0.9012). Concerning the VC, the EfficientNet-B5 with DeepLabv3+ model stood out with the highest accuracy (0.8283), recall (0.9219), and F1 score (0.8852). Conversely, the Configured Mask R-CNN model exhibited the highest specificity (0.8281). Finally, in the case of the CC, the EfficientNet-B5 with U-Net model demonstrated a remarkable performance with respect to accuracy (0.7755), recall (0.9231), and F1 score (0.8494), whereas the Configured Mask R-CNN model exhibited notable specificity (0.7571).
Table 2.
Performance metrics to assess the efficacy of EfficientNet-B5 with DeepLabv3+ in targeting different structures.
| Model | Metric | Tongue | Epiglottis | Vocal cord | Corniculate cartilage |
|---|---|---|---|---|---|
| EfficientNet-B5 with DeepLabv3+ | Accuracy | 0.8743 | 0.8597 | 0.8283 | 0.7553 |
| Recall | 0.4252 | 0.9794 | 0.9219 | 0.8867 | |
| Specificity | 0.949 | 0.6343 | 0.5896 | 0.4845 | |
| F1 score | 0.4909 | 0.9012 | 0.8852 | 0.83 |
Table 3.
Performance metrics to assess the efficacy of EfficientNet-B5 with U-Net in targeting different structures.
| Model | Metric | Tongue | Epiglottis | Vocal cord | Corniculate cartilage |
|---|---|---|---|---|---|
| EfficientNet-B5 with U-Net | Accuracy | 0.8328 | 0.8519 | 0.7991 | 0.7755 |
| Recall | 0.0 | 0.963 | 0.9057 | 0.9231 | |
| Specificity | 1.0 | 0.6563 | 0.5333 | 0.4536 | |
| F1 score | 0.0 | 0.8923 | 0.8655 | 0.8494 |
Table 4.
Performance metrics to assess the efficacy of Configured Mask R-CNN in targeting different structures.
| Model | Metric | Tongue | Epiglottis | Vocal cord | Corniculate cartilage |
|---|---|---|---|---|---|
| Configured Mask R-CNN | Accuracy | 0.7946 | 0.862 | 0.807 | 0.7048 |
| Recall | 0.1929 | 0.9174 | 0.8011 | 0.6887 | |
| Specificity | 0.9068 | 0.7517 | 0.8281 | 0.7571 | |
| F1 score | 0.2278 | 0.8984 | 0.8669 | 0.781 |
Discussion
In this study, we used VL images acquired at the ED of patients with emergent ETI. This is the first AI algorithm to segment anatomical structures related to ETI in emergency situations. We obtained DSC of 0.1167, 0.7677, 0.7207, and 0.57 in Configured Mask R-CNN for tongue, epiglottis, VC, and CC, respectively.
In AI algorithms used for detection, predicted bounding box and ground truth bounding box were used to calculate IoU, and the predicted bounding box from the model was considered correct if the IoU is equal to, or greater than 0.5.23–26 Unlike the detection algorithms, segmentation algorithms use a polygonal mask instead of a bounding box. The evaluation method used in detection was used to evaluate whether the model correctly detected the class by IoU, which was calculated using a bounding box. However, the IoU in this study was not calculated by a bounding box that could include unnecessary areas, as boxes are square shaped. When calculated using its mask, unnecessary areas are not used, resulting in a tight and accurate calculation. Therefore, when IoU was calculated using the masks, the structure was segmented enough to be clearly distinguished.
The inclusion of the Mask R-CNN model alongside semantic segmentation methods stemmed from related works that showed Mask R-CNN model had better recall and precisions or lesser false negatives.27,28 In this work for epiglottis, VC, and CC, the specificity for the semantic segmentation models (EfficientNet-B5 with DeepLabv3+ and EfficientNet-B5 with U-Net) was lower than that of the Configured Mask R-CNN. The low specificity of the two models indicates that there were many false positives. This is because the two models determined the existence of a structure using a mask obtained through softmax. The Configured Mask R-CNN first identifies the location of a structure through a RPN; therefore, if a ROI area is not obtained in the corresponding process, there is little false positive because there is no prediction mask for the structure. Therefore, the Configured Mask R-CNN had a lower recall than the other two models. The Configured Mask R-CNN shows high specificity for epiglottis, VC, and CC; however, the ROI in the RPN is not sufficiently obtained because it shows low recall. If the performance of the RPN is increased, low recall can be overcome.
The FPS of EfficientNet-B5 with DeepLabv3+, EfficientNet-B5 with U-Net, and Configured Mask R-CNN models were 3, 24, and 32 FPS, respectively. The VL video used in this study was 30 FPS. For real-time use, the inference FPS of the model should be close to or higher than FPS compared to what it should be applied to because FPS decreases during video processing in real time. As the Configured Mask R-CNN used in this study performed DSC for the epiglottis and VC of 0.7677 and 0.7207, respectively, the Configured Mask R-CNN showed an advantage over the other two models for real-time applications with the highest FPS mentioned to be rapid and fast experiment only adding minor computational cost than Faster R-CNN.29–31 The application of the developed algorithm to VL devices can be considered in future studies.
The DSC of the tongue was relatively low in all models compared with the other structures in the oral cavity. This is because the anatomical structure of the tongue differs between dorsal and ventral views. Furthermore, the presence of foreign substances, such as mucosa, blood, or vomitus, and light reflection of VL on the tongue during ETI cause variance in the ground truth mask. Although different characteristics are observed in the same structure, they are all labeled as having the same structure (Figure 6). In addition, the amount of data on the tongue is significantly smaller than that on other structures because the tongue is pushed aside and hidden from the VL screen for ETI. Future studies require subdividing labeling of the tongue, such as the dorsal and ventral side of tongue as different anatomical structures.
Figure 6.
Sample images showing structural differences between the dorsal and ventral view of the tongue. The examples include diverse variations such as vomitus, saliva, blood, and light reflection on the tongue. The tongue is shown in light green.
Previous studies were performed on datasets collected from resection surgeries, examinations, YouTube, and public videostroboscopy datasets. From the data, previous studies have attempted to segment or detect the VC to provide medical information for clinical diagnoses and reduce the risk of complications, which are important for clinical practice. 11 When treating patients or situations in stable conditions with controlled variables, previous studies could fulfill their goals. However, in the ED, various situations such as a barely visible view due to foggy vision, vomitus, and moving vocal cords with chest compression make it difficult to perform emergent ETI. To address this difficulty, we used VL images for ETI in the ED. The raw data images, which are different from previous research, are the main strength of this study. For further research, additional methods to process noisy images can be used to improve performance.
Notably, the mean and standard deviation for the train, validation, and test set are 90.25 ± 104.916, 107.0 ± 66.957, and 81.0 ± 33.723, respectively. While variations exist in the number of images across cases, the primary objective of this study remains the development of an AI algorithm capable of segmenting oral structures from ETI videos in emergency situations. Consequently, the model was developed based on images extracted from videos at a rate of two FPS, resulting in varying image counts per case. To mitigate this disparity and potential bias, forthcoming studies will involve the collection of more data.
This study has several limitations. First, the model evaluation was not conducted on the complete pool of extracted frames, a decision necessitated by the time-intensive nature of frame-by-frame labeling. Addressing this aspect of unused frames warrants consideration in future research. Second, the patients’ diverse conditions during data acquisition encompass factors such as the presence of foreign substances in the oral cavity, degrees of bleeding, and motion blur. Future explorations might harness Generative Adversarial Networks (GANs) to generate images with reduced motion blur, enabling the model's segmentation performance even on images afflicted by this phenomenon. 32 Third, as the acquired data came from real-world clinical situations, the number of structures present in each case and their distribution across different sets varied. Consequently, cross-validation was not suitable because of the variation in clinical situations. Further research can explore the feasibility of creating sub-groups for each clinical scenario, considering the potential influence of clinical changes or situations on the acquired data. Fourth, the algorithm developed in this study has not been validated using an external dataset. The next research study can be conducted by applying this model to an externally acquired dataset from different ED settings.
Conclusion
Herein, we developed an AI algorithm to segment anatomical structures related to emergent ETI. EfficientNet-B5 with DeepLabv3+, EfficientNet-B5 with U-Net and Configured Mask R-CNN were used for segmentation. The Configured Mask R-CNN achieved 32 FPS in inference time with the highest specificity shown for VC, epiglottis, and CC.
Our future endeavors are aimed at pursuing rigorous testing of the model on medical manikins and animal subjects to assess its clinical utility in varied scenarios. Additionally, we hope VL devices with AI algorithms for real-time segmentation will help clinicians perform ETI in unexpected emergent situations.
Footnotes
Contributorship: SJC: contributed to the model development, data analysis, and original draft writing as co-first authors. DKK: contributed to the data collection, data labeling, research funding, and original draft writing as co-first authors. BSK: contributed to the model development and data analysis as second author. MC: contributed to the model development and draft editing. JJ: contributed to the data collection protocol development and data labeling. YHJ: contributed to the data collection, protocol development and research process supervision. KJS: contributed to the research process supervision and research funding. YJK: supervised the study design, data collection, study protocol development, data labeling, and draft editing as co-corresponding authors. SK: supervised the model development, data analysis, and draft editing as co-corresponding authors.
Data availability: The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Ethical approval: This study was approved by the Institutional Review Board of Seoul National University Bundang Hospital (number: B-2112-725-102). The requirement for informed consent was waived by the Institutional Review Board of Seoul National University Bundang Hospital.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the National Research Foundation of Korea (grant number 2021R1C1C101035213) and AI Institute at Seoul National University.
Guarantor: SK and YJK.
ORCID iD: Sungwan Kim https://orcid.org/0000-0002-9318-849X
References
- 1.Peters J, Van Wageningen B, Hendriks I, et al. First-pass intubation success rate during rapid sequence induction of prehospital anaesthesia by physicians versus paramedics. Eur J Emerg Med 2015; 22: 391–394. [DOI] [PubMed] [Google Scholar]
- 2.Hurford WE. Techniques for endotracheal intubation. Int Anesthesiol Clin 2000; 38: 1–28. [DOI] [PubMed] [Google Scholar]
- 3.Matava C, Pankiv E, Raisbeck S, et al. A convolutional neural network for real time classification, identification, and labelling of vocal cord and tracheal using laryngoscopy and bronchoscopy video. J Med Syst 2020; 44: 44. [DOI] [PubMed] [Google Scholar]
- 4.Biro P, Hofmann P, Gage D, et al. Automated tracheal intubation in an airway manikin using a robotic endoscope: a proof of concept study. Anaesthesia 2020; 75: 881–886. [DOI] [PubMed] [Google Scholar]
- 5.Levitan RM, Heitz JW, Sweeney M, et al. The complexities of tracheal intubation with direct laryngoscopy and alternative intubation devices. Ann Emerg Med 2011; 57: 240–247. [DOI] [PubMed] [Google Scholar]
- 6.Paolini J-B, Donati F, Drolet P. Review article: video-laryngoscopy: another tool for difficult intubation or a new paradigm in airway management? Can J Anesth 2013; 60: 184–191. [DOI] [PubMed] [Google Scholar]
- 7.Carlson JN, Das S, De la Torre F, et al. A novel artificial intelligence system for endotracheal intubation. Prehosp Emerg Care 2016; 20: 667–671. [DOI] [PubMed] [Google Scholar]
- 8.Ronneberger O, Fischer P, Brox T. U-net: convolutional networks for biomedical image segmentation. In: Medical Image Computing and Computer-Assisted Intervention (MICCAI) 2015: 18th International Conference, Munich, Germany, October 5-9, 2015, Proceedings, Part III 18, 2015, pp. 234–241. Springer.
- 9.Ding H, Cen Q, Si X, et al. Automatic glottis segmentation for laryngeal endoscopic images based on U-Net. Biomed Signal Process Control 2022; 71: 103116. [Google Scholar]
- 10.He K, Gkioxari G, Dollár P, et al. Mask r-cnn. In: Proceedings of the IEEE international conference on computer vision, 2017, pp. 2961–2969.
- 11.Chin C-L, Chang C-L, Liu Y-C, et al. Automatic segmentation and indicators measurement of the vocal folds and glottal in laryngeal endoscopy images using mask R-CNN. Biomed Eng Appl Basis Commun 2021; 33: 2150027. [Google Scholar]
- 12.Vaswani A, Shazeer N, Parmar N, et al. Attention is all you need. Adv Neural Inf Process Syst 2017: 30: 5998–6008. [Google Scholar]
- 13.Dosovitskiy A, Beyer L, Kolesnikov A, et al. An image is worth 16x16 words: transformers for image recognition at scale. arXiv preprint arXiv 2020: 201011929.
- 14.Pan X, Bai W, Ma M, et al. RANT: a cascade reverse attention segmentation framework with hybrid transformer for laryngeal endoscope images. Biomed Signal Process Control 2022; 78: 103890. [Google Scholar]
- 15.Ren J, Jing X, Wang J, et al. Automatic recognition of laryngoscopic images using a deep-learning technique. Laryngoscope 2020; 130: E686–E693. [DOI] [PubMed] [Google Scholar]
- 16.Laves M-H, Bicker J, Kahrs LA, et al. A dataset of laryngeal endoscopic images with comparative study on convolution neural network-based semantic segmentation. Int J Comput Assist Radiol Surg 2019; 14: 483–492. [DOI] [PubMed] [Google Scholar]
- 17.Vasconcelos Pereira A, Simões AV, Rego L, et al. New technologies in airway management: a review. Medicine (Baltimore) 2022; 101: e32084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen L-C, Zhu Y, Papandreou G, et al. Encoder-decoder with atrous separable convolution for semantic image segmentation. In: Proceedings of the European conference on computer vision (ECCV), 2018, pp. 801–818.
- 19.Tan M, Le Q. Efficientnet: rethinking model scaling for convolutional neural networks. In: International conference on machine learning. PMLR, 2019, pp. 6105–6114.
- 20.Luo G, Yang Q, Chen T, et al. An optimized two-stage cascaded deep neural network for adrenal segmentation on CT images. Comput Biol Med 2021; 136: 104749. [DOI] [PubMed] [Google Scholar]
- 21.Chen J, Lu Y, Yu Q, et al. Transunet: transformers make strong encoders for medical image segmentation. arXiv preprint arXiv:210204306 2021.
- 22.Uhm K-H, Jung S-W, Choi MH, et al. Deep learning for end-to-end kidney cancer diagnosis on multi-phase abdominal computed tomography. NPJ Precis Oncol 2021; 5: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Azam MA, Sampieri C, Ioppi A, et al. Deep learning applied to white light and narrow band imaging videolaryngoscopy: toward real-time laryngeal cancer detection. Laryngoscope 2022; 132: 1798–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nogueira-Rodríguez A, Domínguez-Carbajales R, Campos-Tato F, et al. Real-time polyp detection model using convolutional neural networks. Neural Comput Appl 2022; 34: 10375–10396. [Google Scholar]
- 25.Li Y. Detecting lesion bounding ellipses with gaussian proposal networks. In: Machine Learning in Medical Imaging: 10th International Workshop, MLMI 2019, Held in Conjunction with MICCAI 2019, Shenzhen, China, October 13, 2019, Proceedings 10, 2019, pp. 337–344. Springer.
- 26.Zlocha M, Dou Q, Glocker B. Improving RetinaNet for CT lesion detection with dense masks from weak RECIST labels. In: Medical Image Computing and Computer Assisted Intervention—MICCAI 2019: 22nd International Conference, Shenzhen, China, October 13–17, 2019, Proceedings, Part VI 22. Springer, 2019, pp. 402–410.
- 27.Vuola AO, Akram SU, Kannala J. Mask-RCNN and U-Net ensembled for nuclei segmentation. In: 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019) 8-11 April 2019 2019, pp.208–212.
- 28.Vyshnav MT, Sowmya V, Gopalakrishnan EA, et al. Deep learning based approach for multiple myeloma detection. In: 2020 11th International Conference on Computing, Communication and Networking Technologies (ICCCNT), July 1–3, 2020, pp. 1–7.
- 29.Jin J, Zhang Q, Dong B, et al. Automatic detection of early gastric cancer in endoscopy based on Mask region-based convolutional neural networks (Mask R-CNN)(with video). Front Oncol 2022; 12: 927868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alfonso-Francia G, Pedraza-Ortega JC, Badillo-Fernández M, et al. Performance evaluation of different object detection models for the segmentation of optical cups and discs. Diagnostics 2022; 12: 3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alzahrani N, Al-Baity HH. Object recognition system for the visually impaired: a deep learning approach using Arabic annotation. Electronics 2023; 12: 541. [Google Scholar]
- 32.Goodfellow I, Pouget-Abadie J, Mirza M, et al. Generative adversarial networks. Commun ACM 2020; 63: 139–144. [Google Scholar]