Abstract
Introduction
The Tousled-Like Kinases 1 and 2 (TLK1 and TLK2) are involved in many fundamental processes, including DNA replication, cell cycle checkpoint recovery and chromatin remodelling. Mutations in TLK2 were recently associated with “Mental Retardation Autosomal Dominant 57” (MRD57, OMIM 618050), a neurodevelopmental disorder characterized by a highly variable phenotype, including mild-to-moderate intellectual disability, behavioural abnormalities, facial dysmorphisms, microcephaly, epilepsy, and skeletal anomalies.
Methods
We re-evaluate whole exome sequencing and array-CGH data from a large cohort of patients affected by neurodevelopmental disorders. Using spatial proteomics (BioID) and single-cell gel electrophoresis, we investigated the proximity interaction landscape of TLK2 and analysed the effects of p.(Asp551Gly) and a previously reported missense variant (c.1850C>T; p.(Ser617Leu)) on TLK2 interactions, localization and activity.
Results
We identified three new unrelated MRD57 families. Two were sporadic and caused by a missense change (c.1652A>G; p.(Asp551Gly)) or a 39-kb deletion encompassing TLK2, and one was familial with three affected siblings who inherited a nonsense change from an affected mother (c.1423G>T; p.(Glu475Ter)). The clinical phenotypes were consistent with those of previously reported cases. The tested mutations strongly impaired TLK2 kinase activity. Proximal interactions between TLK2 and other factors implicated in neurological disorders, including CHD7, CHD8, BRD4, and NACC1, were identified. Finally, we demonstrated a more relaxed chromatin state in lymphoblastoid cells harbouring the p.(Asp551Gly) variant compared to control cells, conferring susceptibility to DNA damage.
Conclusion
Our study identified novel TLK2 pathogenic variants, confirming, and further expanding the MRD57 related phenotype. The molecular characterization of missense variants increases our knowledge about TLK2 function and provides new insights into its role in neurodevelopmental disorders.
Keywords: intellectual disability, TLK2, comet assay, chromatin condensation, BioID, proximal interactome, autism spectrum disorder, tousled like kinases, MRD57
INTRODUCTION
The Tousled-Like Kinases 1 and 2 (TLK1 and TLK2) are serine-threonine kinases involved in DNA replication and repair, transcription, cell cycle checkpoint recovery, chromatin maintenance and genomic stability[1–3]. Both kinases target the ASF1A and ASF1B histone H3/H4 chaperones and are regulated by DNA damage responsive checkpoint signalling[4]. Depletion of Tlk1 and Tlk2 in mice indicated that they are largely redundant, with the exception of an essential role for Tlk2 in placental development[5]. Null Tlk2 mice, generated with a conditional allele to bypass the placental defect, showed no gross developmental defects except for a slight growth delay compared to controls, suggesting that TLK1 can compensate for loss of TLK2 function after embryonic development. Further work in human cancer cells supports extensive redundancy at the cellular level. Co-depletion of TLK1 and TLK2, however, causes replication stress, DNA damage and altered chromatin maintenance, particularly affecting telomeres and other repetitive genome elements, while mild effects were observed with depletion of either proteins[6].
Mutations in TLK2 were associated with Mental Retardation Autosomal Dominant (MRD57, MIM: 618050) by Reijnders et al.[7], who described 40 cases from 38 unrelated families. MRD57 is clinically characterized by autism spectrum disorder (ASD), intellectual disability (ID), behavioural problems, growth delay and facial dysmorphism, including blepharophimosis, telecanthus, prominent nasal bridge, broad nasal tip, thin vermilion of the upper lip and upslanting palpebral fissures. Other features shared by a subset of cases are gastrointestinal problems, seizures, skeletal malformations and ocular problems.
In the majority of reported cases, the disease is likely due to TLK2 haploinsufficiency, as most of cases are heterozygous for loss-of-function (LoF) alleles, which is in line with the strong constraint against LoF variants in the gene (pLI = 1, GnomAD database). Reported missense variants cluster mainly in the C-terminal Protein Kinase Domain (PKD)[7–9], the core of TLK2 function. Four TLK2 variants localized in this region have previously been analysed, showing a strong reduction of kinase activity on ASF1A in vitro[10]. TLK2 activation requires dimerization through an N-terminal coiled coil motif, suggesting that inactive mutants could have a dominant negative effect[10]. Thus, based on the available data, the predominant pathogenic mechanism of TLK2 mutations appears to be a reduction in its overall activity. Of note, recent work suggested a possible autosomal recessive phenotype in a proband affected by severe neurodevelopmental disease with a homozygous missense variant (c.163A>G; p.(Lys55Glu))[8]. This variant is localized outside the TLK2 PKD and coiled-coil motifs, and may be hypomorphic, since the carrier parents are not affected.
During the screening of a large survey of patients with ID by array-CGH and whole exome sequencing (WES), we identified three new cases with TLK2-mutations. Two subjects were heterozygous for a de novo 39-kb deletion encompassing TLK2, and a de novo c.1652A>G; p.(Asp551Gly) missense change. The third case was familial, with a nonsense variant (c.1423G>T; p.(Glu475Ter)) occurring in three affected siblings and their affected mother. Here, we report their clinical description, confirming and expanding the disease phenotype. We also provide data documenting an altered chromatin state in patient-derived fibroblasts and lymphoblastoid cells (LCLs), consistent with a defect in the regulation of histone chaperones. Finally, we characterized the activity and proximal interactomes of the p.(Asp551Gly) variant, as well as another missense variant (p.(Ser617Leu)), reported in a several published studies[11–13]. Both variants exhibited impaired kinase activity and TLK2 proximal interactomes were enriched with proteins previously implicated in ID and ASD, suggesting connections to a larger chromatin maintenance network.
MATERIALS AND METHODS
Whole exome sequencing, prioritization, and variant calling
DNA was extracted from total blood using the ReliaPrep Blood gDNA Miniprep kit (Promega, Madison, WY, USA) following manufacturer’s protocol and quantified with a NanoDrop spectrophotometer (Thermo Fisher Scientifics, Waltham, MA, USA). Informed consent was obtained from participating families and the study protocol was approved by our internal Ethics Committee, according to the Declaration of Helsinki.
Array-CGH was performed using a 60K whole-genome oligonucleotide microarray (Agilent Technologies, Santa Clara, California, USA). Family 1 and 2 were enrolled in the Autism Sequencing Consortium (ASC) project and their gDNA samples were sequenced at the Broad Institute on Illumina HiSeq sequencers as previously described[11, 14]; variant calling was performed using targeted bioinformatic pipelines adapted for different pattern of inheritance. Identified variants were confirmed by Sanger sequencing using standard conditions and the primers in table S1. Additional information is provided in Supplementary materials and methods.
All variants are referred to GRCh37 annotation and to NM_001284333.2, in line with the previously published TLK2 structure[10]. For homogeneity with the clinical work from Reijnders et al.[7], we specified variants also in NM_006852.6 in Supplementary table S2.
In silico prediction of missense variants impact and splicing analysis
Variants were analysed with the VarSome tool[15] as a starting point for further analysis. This allowed evaluation of at least 15 in silico predictors simultaneously. Variants frequencies were evaluated using Genome Aggregation Database (GnomAD) Browser version 2.1.1. Impaired splicing was predicted using Human Splicing Finder (HSF) version 3.1[16] and experimentally verified as described in Supplementary materials and methods.
Cell cultures
Peripheral blood mononuclear cells (PBMCs) were isolated from whole blood using Histopaque®−1077 (Sigma-Aldrich) and subsequently immortalized with Epstein-Barr Virus (EBV) and cultured in RPMI medium (GIBCO, Thermo Fisher Scientific) supplemented with 10% Fetal Bovine Serum (FBS) (GIBCO), 1% Pen-Strep and 1% L-Glutamine. Primary fibroblasts were isolated from human skin biopsies after overnight incubation in Dulbecco’s modified Eagle’s medium (DMEM) (Sigma-Aldrich) supplemented with 10% FBS and 160 μg/ml collagenase. Fibroblasts were maintained in DMEM supplemented with 10% FBS, 1% Pen-Strep and 1 mM sodium pyruvate (Thermo Fisher Scientific) at 37°C, 5% CO2. AD-293 cells (Stratagene) were grown in DMEM supplemented with 10% FBS (Sigma-Aldrich), 50 U/mL penicillin and 50 μg/mL streptomycin (Thermo Fisher Scientific) at 37°C in a 5% CO2 incubator.
RNA isolation and quantitative real time PCR
Total RNA was extracted from fibroblasts and LCLs using the Direct-Zol RNA MiniPrep system (Zymo Research, Irvine, CA, USA) and complementary DNA (cDNA) was generated using the M-MLV Reverse Transcriptase kit (Invitrogen, Thermo Fisher Scientific). The expression level of TLK2 was measured using the Universal Probe Library system (Roche Diagnostics, Risch-Rotkreuz, Switzerland) with primers and probe in table S1 and HMBS and GAPDH as reference gene (assays numbers Hs00609297_m1 and Hs00266705_g1; Applied Biosystems, Thermo Fisher Scientific). Assays were carried out in triplicate on an ABI 7500 real-time PCR instrument using the ABI 2X TaqMan™ Universal PCR Master Mix II, according to the manufacturer’s protocol (Thermo Fisher Scientific). For each experiment, biological triplicates with at least two technical replicates were performed. Data were analysed with Prism-GraphPad Software performing unpaired t-test with Welch’s correction. p-values are indicated as follows: ns= P value > 0.05; *= P value ≤ 0.05; ** P value ≤ 0.01; ***= P value ≤ 0.001; ****= P value ≤ 0.0001.
Single-cell gel electrophoresis
Samples were prepared according to the alkaline single-cell gel electrophoresis (SCGE) assay method, as previously described[17] and are briefly summarized in Supplementary material and methods, together with experimental details.
Site directed mutagenesis and in vitro kinase assays from cell lysates
In vitro kinase assays were performed as previously described[10] with minor modifications. Full methods provided in the Supplementary materials and methods and primers in table S3.
Western blotting
For affinity purification (AP), 40 μg of input protein and 20 μL of Strep-AP elution, with 6X SDS (0.2% Bromophenol blue and β-mercaptoethanol), were separated by SDS-PAGE and transferred to nitrocellulose membranes (0.2 or 0.45 μm pore, Amersham Protran; Sigma-Aldrich). For detection of Streptavidin, PVDF membranes (0.45 μm pore, Immobilon-P, Merck) were used. Membranes were blocked and antibodies prepared in 5% non-fat milk in PBS-T, with the exception of CHD7 and CHD8, where 5% BSA in PBS-T was used. Primary antibodies were detected with the appropriate secondary antibodies conjugated to Horseradish peroxidase (HRP) (table S4) and visualized by ECL-Plus (GE Healthcare).
Proximity-dependent biotin identification mass spectrometry (BioID-MS)
BioID-MS and BioID-Westerns were performed essentially as described in Silva et al. [18] with some modifications. Full methods are provided in the Supplementary materials and methods and data is available in the PRIDE database with accession number PXD019450 (for reviewer access: Username: reviewer30722@ebi.ac.uk Password: dGfoWR6c).
RESULTS
Identification of novel MRD57 patients
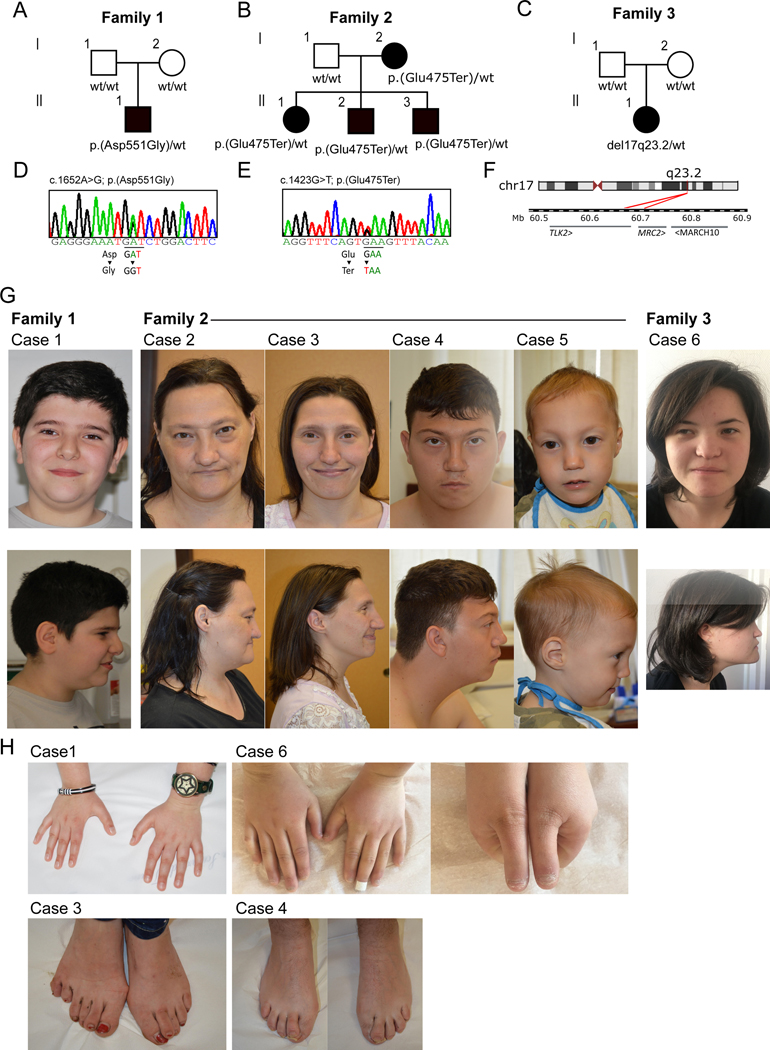
The identification of TLK2 as a risk gene for intellectual disability[7], prompted us to re-evaluate the genomic information available for a large cohort of patients affected by ASD and/or ID who had previously been analysed by WES. This included our in-house cohort (2,250 total samples, 736 affected), as well as a publicly available one from the Autism Sequencing Consortium (35,584 total samples, 11,986 affected)[11]. No further likely pathogenic variants or variant of unknown significance (VUS) in neurodevelopment-related genes[2] were identified. A search in the DECIPHER database[19] for novel TLK2 deletions was also performed. We found six cases in three independent families with detrimental variants in the TLK2 gene (figure 1A-F).
Figure 1. Facial features and skeletal anomalies of individuals with TLK2 variants.
(A-C) Pedigrees of family 1, 2 and 3. Cases from family 1 and 3 carried respectively a heterozygous de novo missense variant (c.1652A>G; p.(Asp551Gly)) and a heterozygous de novo deletion encompassing TLK2 gene. Cases from family 2 shared a heterozygous premature stop variant (c.1423G>T; (p.Glu475Ter)) inherited from an affected mother. Analysis of maternal grandparent genotype were not possible, but familial clinical history did not suggest a possible MRD57-like phenotype for them. wt= wild type at variant position. (D-E) Sanger validation of variants identified in family 1 and 2. Validation was performed both on gDNA from affected cases and from their unaffected relatives. (F) 17q23.2 deletion (minimum size 39 kb, chr17–60683462-60722398) identified in case 6 from family 3. The 39 kb deletion encompassed TLK2 and MRC2 genes. (G) Frontal and lateral face photographs of our cases, showing overlapping facial dysmorphisms. Most frequently reported features were upward slanted palpebral fissures, broad nasal tip, thin lips, low hanging columella, prognathism, wide spaced eyes and downturned corners of the mouth. (H) Details of reported skeletal anomalies at the level of hands and feet. Upper left panel: tapered hands fingers from case 1; upper right panels: short hands with short distal phalanx from case 6; bottom left panel: postaxial polydactyly of left foot observed in case 3;bottom right panel: overriding second toe in case 4.
In family 1, the proband carried a de novo missense change c.1652A>G; p.(Asp551Gly) not reported in gnomAD and predicted to be damaging by multiple in silico predictors (table S5). This variant localizes in a highly Constrained Coding Region (CCR) (>93th percentile) within the PKD[20]. In family 2, three affected siblings carried a premature stop variant (c.1423G>T; p.(Glu475Ter)) inherited from their affected mother (case 2). This variant was not reported in gnomAD and was classified as pathogenic using the American College of Medical Genetics (ACMG) criteria[7, 8]. A sixth case (family 3) was identified in the DECIPHER database: a female carried a likely pathogenic (class 4)[7, 8] de novo del(17)(q23.2), whose minimum size was 39 kb (NC_000017.10:g.(60683462–60722398)del). The deletion encompassed TLK2, MRC2 and MARCH10 genes. Neither the MRC2 or MARCH10 genes are predicted to be haploinsufficient (GnomAD MRC2 pLi=0.67; MARCH10 pLi=0). MRC2 encodes the Mannose Receptor C Type 2 and has a role in the turnover of collagens in the cytomembrane and extracellular matrix[8]. Its upregulation has been linked to tumorigenic activity, and a role in breast cancer and hepatocellular carcinoma has been suggested[9, 10] but, to our knowledge, there is no supporting literature implicating MRC2 in neurodevelopmental disorders. The literature available for MARCH10 is limited but the gene is expressed almost exclusively in testis (https://gtexportal.org/home/gene/MARCH10) and was suggested to play a role in spermiogenesis[11]. Therefore, we think it is unlikely that haploinsufficiency in MRC2 or MARCH10 contribute to the patient’s phenotype.
Our patients (3 males and 3 females) ranged from 3 to 47 years of age, all of them were of Caucasian ethnicity. A broad range of behavioural disorders was present, including ASD (1/6), attention deficit hyperactivity disorder (ADHD) (4/6), anxiety (3/6), short attention span (2/6) and obsessive-compulsive behaviour (2/6). ID was reported for 4 patients in the borderline (IQ 70–85) or low (IQ≤70) range. Patient 5 was too young for formal assessment of a neurodevelopmental phenotype, but a global developmental delay was reported. No formal evaluation for intellectual disability was provided for case 6 but difficulties in memory and transcription were reported. All affected members from family 2 had microcephaly. Dysmorphic facial features were observed in all patients and included upslanting palpebral fissures, wide nose, low hanging columella, smooth philtrum, prognathism, pointed chin and hypertelorism (figure 1G). Minor skeletal anomalies of the hands and feet were reported for four patients (figure 1H). Interestingly, some of the features we observed in a portion of our patients had not been described in previously reported cases, expanding the clinical phenotype. Among them, there were neurodevelopmental (difficulties in reading, writing, memory and transcription and pavor nocturnus), dysmorphic (prognathism, bifid nasal tip, low hanging columella, absent ear lobe, low-implant auricle, synophrys and downturned corners of mouth) and skeletal anomalies (postaxial polydactyly of left foot, overriding second toe, tapering fingers, short hands with short distal phalanx). A summary of the clinical phenotypes is reported in table S6 and a description is provided in the Supplemental Data, while a comparison with the available literature is provided in table S7.
TLK2 haploinsufficiency disrupts proper chromatin compaction
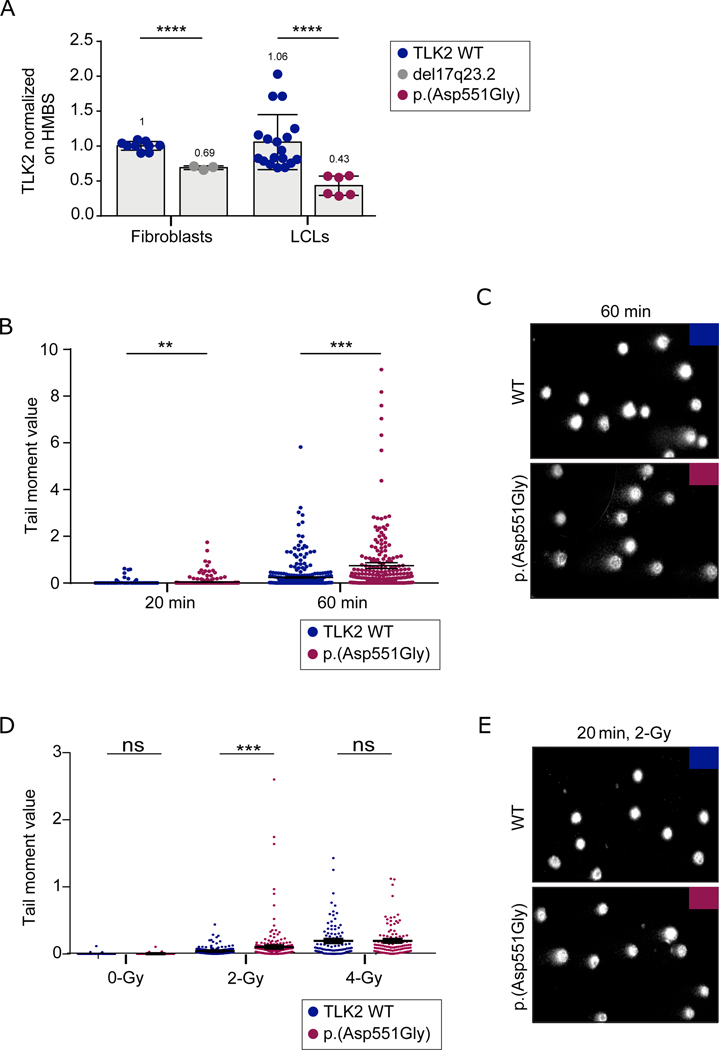
Using quantitative real time polymerase chain reaction (qRT-PCR), we analysed TLK2 mRNA expression in LCLs from case 1 and fibroblasts from case 6. We found an approximately 50% reduction in TLK2 mRNA expression in both cases (figure 2A and S1A). This was unexpected in case 1, who carried the c.1652A>G; p.(Asp551Gly) missense change. Variants encoding missense changes may induce unstable mRNA secondary structures leading to degradation of the altered allele[8, 9]. To verify if both alleles were expressed, we amplified and sequenced cDNA between exons 18 and 23. We found that both wild type and mutant allele were present (figure S1B), suggesting that the allele encoding the p.(Asp551Gly) variant was not degraded. Moreover, no differences were observed in band sizes from WT and p.(Asp551Gly) cDNAs (figure S1C), excluding that p.(Asp551Gly) led to the production of aberrantly spliced transcripts[21, 22]. Western Blot analysis on LCLs from case 1 compared to controls, uncovered a significant increase in TLK2 protein expression (figure S1D), motivating us to perform a deeper characterization of the impact of p.(Asp551Gly) on TLK2 activity.
Figure 2. TLK2-Asp551Gly affects chromatin density and confers susceptibility to DNA damage.
(A) TLK2 mRNA levels in fibroblasts from case 6 and in lymphoblastoid cell line derived from case 1. TLK2 expression was significantly reduced both in LCL carrying p.(Asp551Gly) variant and in fibroblasts carrying the 17q23.2 deletion. All experiments were performed at least in triplicate. UPL probe #72 and primers indicated in Supplementary material and methods were used; HMBS mRNA expression was used as reference. Statistical analysis was performed using t-test with Welch’s correction; ****= P value ≤ 0.0001. Numbers at the top of the bars indicated mean values. (B) SCGE assays highlighted significant differences in chromatin condensation between LCLs carrying the p.(Asp551Gly) amino acid change and control cells after 20 minutes of electrophoresis run time (**p < 0.05; two-tailed unpaired Student’s t test with Welch’s correction), that became more evident after 60 minutes of electrophoresis run time (***p 0.0001; two-tailed unpaired Student’s t-test with Welch’s correction). DNA migration was quantified as Tail moment values, which is defines as the product between the tail length and the percentage of DNA in the tail. For each point, at least 100 cells were analysed. Values are represented as mean ± SEM of three independent experiments. (C) Representative images of nucleoids derived from control LCLs and LCLs from affected subject 1 referred to experiment shown in figure 2B. (D) Single and double strand breaks were induced by γ-ray irradiation (2-Gy or 4-Gy). Tail moment values specify the amount of γ-ray-induced DNA damage measured immediately after the treatment. The mutant LCLs showed a higher vulnerability to 2-Gy γ-ray irradiation (***p 0.0006; two-tailed unpaired Student’s t-test with Welch’s correction ). Following 4-Gy treatment, no differences were observed between control and mutant cells, which is likely explained by the observation that the overall damage, especially double strand breaks, prevails on the condensation state of chromatin at high doses of γ-ray irradiation. For each point, at least 100 cells were analysed and four independent experiments were performed. (E) Representative images of nucleoids derived from control LCLs and LCLs from affected subject 1 referred to experiment shown in figure 2D.
Based on the role of TLK2 in controlling chromatin remodelling[2], we investigated possible changes in chromatin compaction, performing the single-cell gel electrophoresis (SCGE) assay with LCLs derived from case 1. Relatively short electrophoresis time was already sufficient to unmask slight differences between control and mutant cells. Longer run times allowed DNA loops to stretch under the electric field and revealed a significantly more relaxed state of nucleoids in LCLs from the affected subject compared to control cells (figure 2B-C). Differences were quantified as “tail moment” values, which are defined as the product of the tail length and the percentage of DNA in the tail. LCLs harbouring the p.(Asp551Gly) variant also exhibited a higher sensitivity to γ-ray irradiation, documenting increased susceptibility to DNA damage (i.e., single and double strand breaks), which is in line with a more relaxed state of chromatin (figures 2D, E). Of note, a defective heterochromatin state was also observed in fibroblasts derived from subject 6 carrying the 17q23.2 deletion encompassing TLK2 (figure S2), suggesting a similar effect between the 17q23.2 deletion and the p.(Asp551Gly) variant. Overall, these findings demonstrated that TLK2 haploinsufficiency disrupts proper chromatin organization and confers susceptibility to DNA damage.
TLK2 missense mutations alter the activity and subcellular localization of the protein
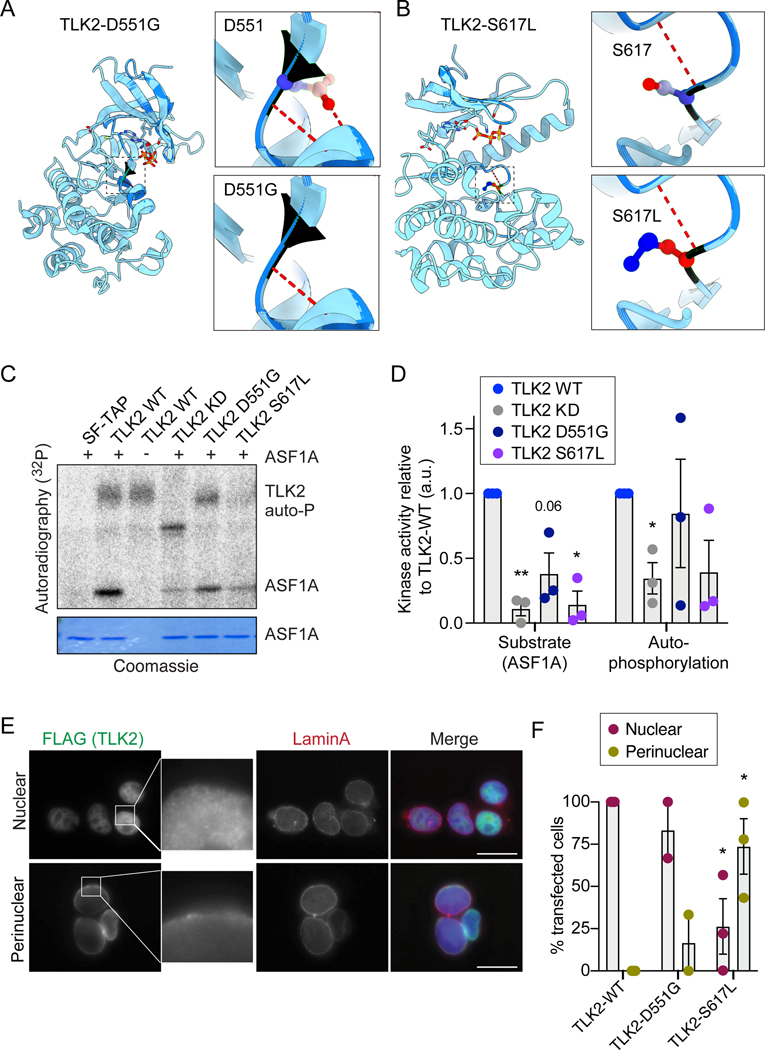
All of the TLK2 missense mutations in the PKD from MRD57 analysed to date exhibited decreased kinase activity, including H493R, H518R, R720A and D629N[10](Figure S4B). Given that the high conservation of the mutated residues in the PKD suggested reduced kinase activity, we examined the potential structural impact of the missense p.(Asp551Gly) (hereafter indicated as D551G) variant identified in our survey, as well as the p.(Ser617Leu) (hereafter indicated as S617L) variant, found in a patient with ASD already described in whole exome sequencing works[11–13]. The patient was also reported in the ASC exome analysis browser, a freely accessible portal containing de novo variants identified in more than six thousand affected probands enrolled in ASC WES projects (https://asc.broadinstitute.org/). Residue S617 is located one residue after the Asp-PheGly (DGF) motif[23, 24], that together with the activation and catalytic loop, constitute the kinase core[10]. We previously identified S617 as a TLK2 auto-phosphorylation site and demonstrated that alterations of this site are able to significantly enhance (S617A) or reduce (S617D) TLK2 kinase activity[10]. Therefore, it remained an open question how S617L, associated with autism spectrum disorder, would influence the kinase activity of TLK2.
We modelled both of the mutations using the crystal structure of the TLK2 PKD[10]. TLK2-D551G was predicted to weaken hydrogen bonds with the subsequent helix and S617L introduces a hydrophobic residue in place of the auto-phosphorylation site in the activation loop (figure 3A-B).
Figure 3. TLK2 autism mutations alter the activity and subcellular localization of TLK2.
(A-B) Modelling of the D551G and S617L missense mutations on the crystal structure of the TLK2 PKD. Hydrogen bonds are shown in red dashed lines. (C) Representative in vitro kinase assays with Strep-purified TLK2-WT, TLK2-KD (kinase dead; D592V) and indicated missense variants. Autophosphorylation and substrate (ASF1A) phosphorylation is shown. Coomassie is shown as loading control for ASF1A. (D) Quantification of n=3 independent kinase assays. Individual results (circles) are shown for each assay on purified ASF1A substrate or affinity purified TLK2 autophosphorylation relative to corresponding TLK2-WT and bars depict mean with SEM. (E) Representative immunofluorescence microscopy of overexpressed TLK2 in AD-293 cells is shown, indicating the 2 main localization patterns observed. The nuclear localization image corresponds to TLK2-WT, while the perinuclear localization to TLK2-S617L. Scale bar = 20 μM. (F) Quantification of TLK2 localization patterns for WT and indicated missense variants. Ten random fields were scored in 2 (D551G) or 3 (WT and S617L) biological replicates. Bars depict mean with SEM. Statistical significance was determined using an unpaired t test with Welch’s correction (****P<0.0001, ***P<0.001, **P<0.01, *P<0.05).
To determine if these mutations affected kinase activity, we analysed TLK2 activity using in vitro kinase assays. Ectopically expressed, Strep-FLAG tagged TLK2, TLK2-KD (kinase dead, D592V), TLK2-D551G and TLK2-S617L were affinity purified from AD-293 cells using an N-terminal Strep tag and incubated with purified substrate (ASF1A) for kinase assays. Both mutations led to a notable reduction in substrate modification (figures 3C-D). Quantification of multiple experiments demonstrated that TLK2-S617L severely impaired kinase activity, comparable to the TLK2-KD protein, while TLK2-D551G was more mildly impaired and showed slightly higher autophosphorylation levels than TLK2-WT in some experiments (figure 3D).
In previous work, we noted that loss of the coiled-coil domains of TLK2 led to perinuclear accumulation[10]. To determine if the TLK2-D551G and TLK2-S617L missense mutations altered TLK2 localization, we transfected FLAG-tagged mutants in AD-293 cells and performed immunofluorescence (IF) microscopy. Lamin A was used as an inner nuclear membrane marker and nuclear DNA was stained with DAPI. TLK2-WT showed diffuse nuclear localization in transfected cells, as observed previously (figures 3E, S3A). In contrast, TLK2-D551G and TLK2-S617L showed perinuclear localization to different extents (figures 3E-F, S2A). This was particularly prominent for the TLK2-S617L mutant, where 75% of transfected cells showed a perinuclear localization of TLK2 (figures 3E-F).
The proximal interactome of TLK2 is altered by missense mutations
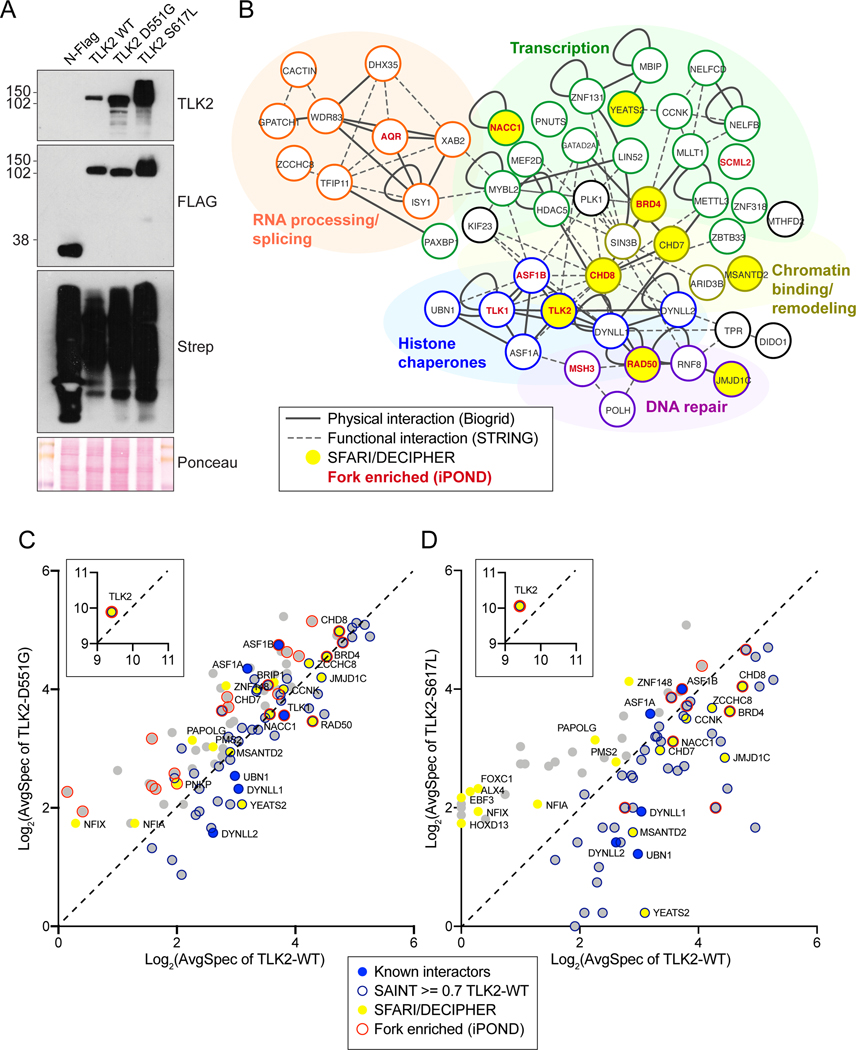
TLK2 is involved in many biological processes and few clear substrates aside from ASF1A and ASF1B have been well characterized[2]. As kinases often bind to substrates with low affinity, we previously used an unbiased proximity biotinylation assay coupled to mass spectrometry (BioID-MS), as it does not require high-affinity interactions that can withstand purification procedures[5, 25]. We used this approach to further characterize the cellular environment of TLK2 and determine if the missense mutations influenced its interactome.
BirA-tagged TLK2-WT, TLK2-D551G and TLK2-S617L were expressed in AD-293 cells by transient transfection (figure 4A). Network clustering of results from wild type TLK2 (TLK2-WT) identified the known TLK2 substrates ASF1A, ASF1B and TLK1, as well as the DYNLL1/2 (LC8) proteins that we previously validated[5, 10]. The proximal interactome grouped into five functional clusters consistent with the known functions of TLK2, including RNA processing and splicing, transcriptional regulation, chromatin binding or remodelling, DNA repair and histone chaperones (figure 4B and table S8). We cross referenced the proximal interactome with a recent large-scale analysis of iPOND-MS data that identified TLK1 and TLK2 as high confidence interactors with nascent DNA at active replication forks[26]. Several of the TLK2-WT hits, including RAD50, BRD4, CHD8, ASF1B, SCML2 and NACC1 were also found at active forks with high confidence in iPOND studies. We next compared our TLK2-WT proximal interactome to the SFARI (Simon’s Foundation Autism Research Initiative) and DECIPHER databases and identified 8 proteins: CHD7, CHD8, NACC1, CCNK, JMJD1C, RAD50, MSANTD2, and YEATS2. These data suggest potential functional links between TLK2 and a number of proteins involved in neurodevelopmental disorders with overlapping pathologies. Details about SFARI and OMIM classifications are provided in Supplementary table S9.
Figure 4. BioID based analysis of the proximal interactome of TLK2.
(A) Western blot of AD-293 lysates expressing BioID constructs: N-FLAG-BirA alone or fused to the indicated TLK2 allele. Detection with anti-TLK2, anti-FLAG or Streptavidin-HRP are shown. Ponceau stained nitrocellulose membrane is shown as a loading control. (B) Network clustering of all prey hits with a SAINT score of > or = to 0.7 in TLK2-WT samples. Physical interactions reported in Biogrid (solid lines) and functional interactions (dashed lines) reported in STRING are indicated[43, 44]. Functionally enriched clusters are indicated by color coding, Bait, TLK2 substrates, proteins found in the SFARI/DECIPHER gene database (yellow fill) or proteins enriched on nascent DNA/replication forks are indicated (red font). (C-D) Scatterplots of average spectral counts (Log2 transformed) of bait and prey proteins identified with the TLK2-D551G and TLK2-S617L alleles compared to TLK2-WT. Previously identified TLK2 interactors, as well as proteins enriched on replication forks or found in the SFARI/DECIPHER databases are indicated (see legend).
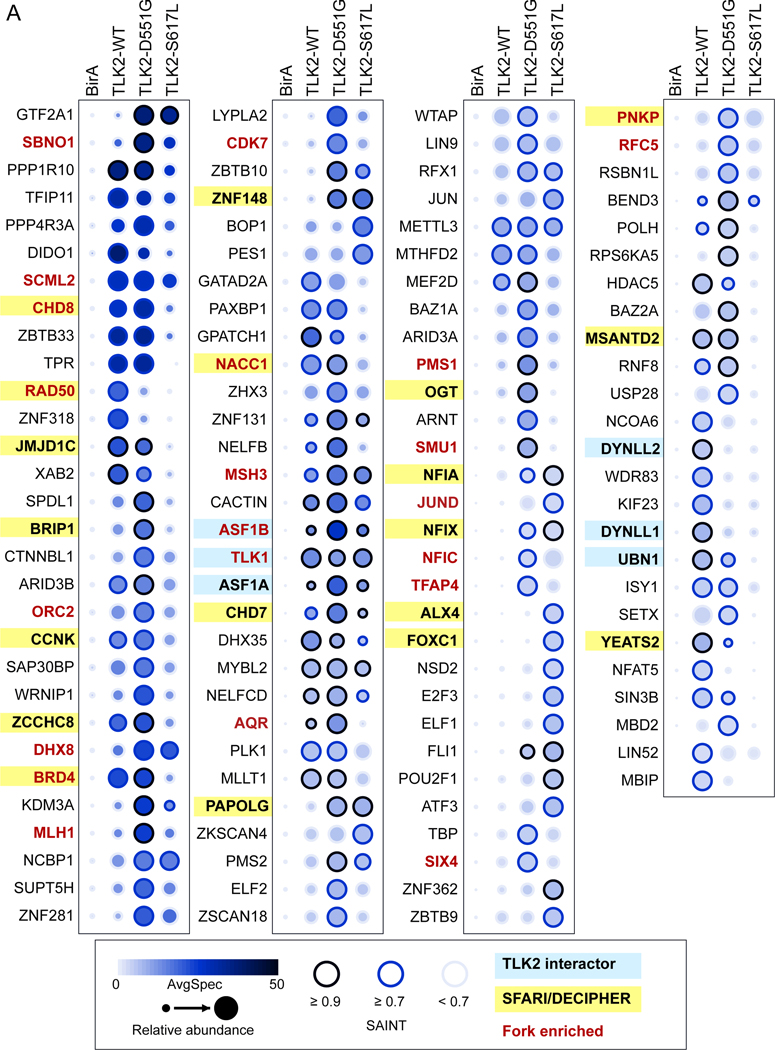
In parallel to TLK2-WT, we performed BioID analysis with TLK2-D551G and TLK2-S617L. Both mutants accumulated to higher levels than TLK2-WT, consistent with what we previously observed with other inactive TLK2 mutants[10]. Both mutants caused numerous alterations in the proximal interactions compared to TLK2-WT (figures 4C, D and 5). This included a reduction in several replication fork and ASD related proteins, including RAD50 and YEATS2 with both mutants, and JMJD1C, BRD4, CCNK, NACC1, MSANTD2 and CHD8 with TLK2-S617L (figures 4C, D and 5). In addition, some proteins, including ZNF148, NFIA, NFIX and PAPOLG, that are all in the SFARI database, were significantly enriched with both mutants, but not TLK2-WT (figures 4C, D and 5).
Figure 5. Missense variants alter the proximal interactome of TLK2.
(A) Dotplot of prey proteins with a SAINT score of > or = to 0.7 with any of the 3 baits generated using ProHits-viz[45]. Average spectral counts (SC), relative abundance and SAINT score ranges are indicated, as well as proteins enriched on replication forks [26] or found in the SFARI/DECIPHER databases (see legend). Additional details are provided in table S8.
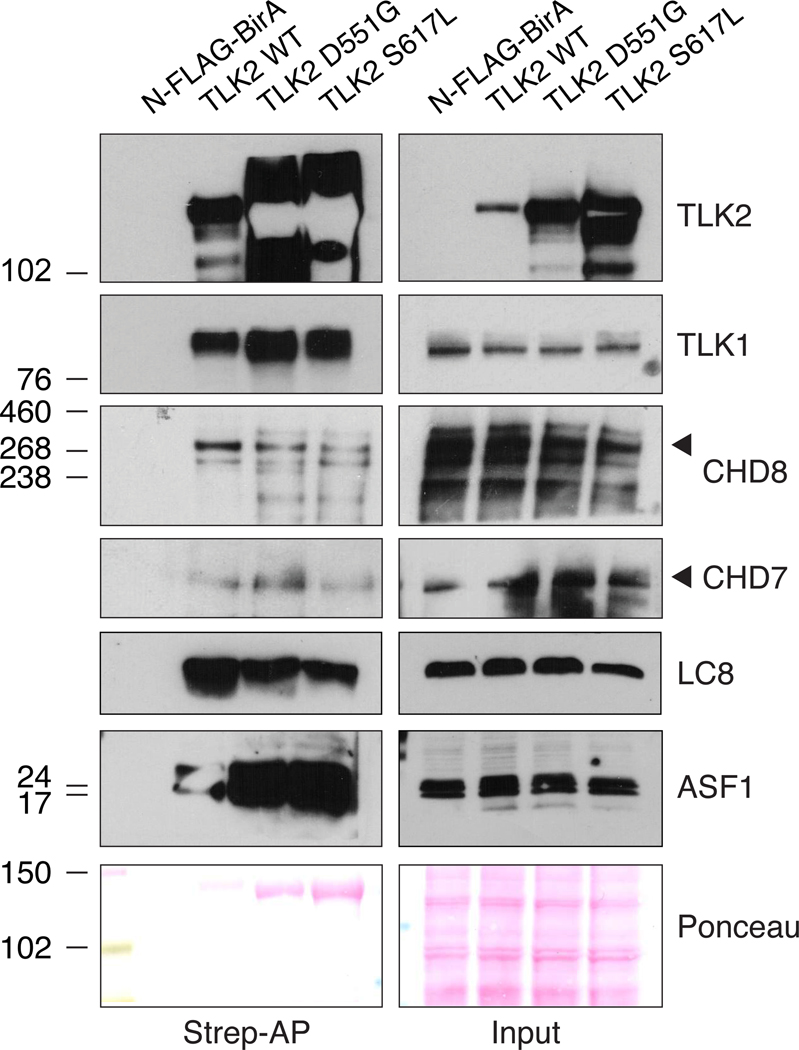
The CHD7 and CHD8 chromodomain helicases were of particular interest to us because they are mutated respectively in CHARGE syndrome[27] (OMIM 214800) and susceptibility to autism[28] (OMIM 615032) and implicated in modulating chromatin structure[29, 30]. In addition, CHD8 has been localized to active sites of DNA replication, like both TLK1 and TLK2[26]. CHD8 spectra were detected at similar levels between TLK2-WT and TLK2-D551G, that retains some activity, but were reduced with the kinase dead TLK2-S617L (figures 4C, D and 5). In contrast, CHD7 spectra were highest in cells expressing TLK2-D551G. We performed BioID-Westerns to validate these proximal interactions and their relative differences. Expression of the control N-FLAG-BirA and biotin supplementation led to no detectable CHD7 or CHD8 detected in Western blots of Strep-affinity purified protein lysates (figure 6A). In contrast, both CHD7 and CHD8 were clearly validated with all TLK2 alleles, although CHD8 levels were lowest with S617L and CHD7 highest in TLK2-D551G, consistent with the MS data. As expected from the BioID-MS, TLK1 co-purified with all TLK2 alleles to a similar degree. It was also notable that the substrates, ASF1A and ASF1B, were highest with D551G and similar between TLK2-WT and TLK2-S617L, despite the much higher level of TLK2-S617L expression. These results indicate the missense mutations have differential effects on both proximal interactions and TLK-ASF1 interactions.
Figure 6: Validation of proximal interactions with CHD7 and CHD8.
(A) Western blots of the indicated proteins from Strep-AP lysates from AD-293 cells transfected with the indicated BioID construct and supplemented with biotin. Input levels are shown and ponceau stained blots provided as a loading control. Representative data from 2 biological replicates is shown.
DISCUSSION
To our knowledge, at least 13 TLK2 missense variants have been reported in MRD57 cases [7, 9, 11], with mutations clustering in the PKD. Previously described patients showed ID in different range of severity, ASD, language delay, motor delay, gastro-intestinal issues and dysmorphic facies as prevalent features[7, 8]. In our cases, we confirmed a high incidence of ID (4/4), language delay (4/6) and dysmorphic facial features (6/6). On the contrary, comparing to the available clinical literature, we observed a under representation of gastro-intestinal problems (0/6) and ASD (1/6). We also observed additional features, including difficulties in reading and writing (3/6) or in memory and transcription (3/6) and skeletal anomalies of the hands (1/6) and the feet (1/6).
Functional characterization has been reported only for four variants in the PKD, (p.(His493Arg), p.(His518Arg), p.(Asp629Asn), p.(Arg720Ala)) and has shown at least a 50% reduction in enzymatic activity compared to wild type protein[10]. Our data further expands the characterization of MRD57 missense mutations and reinforces the prevailing hypothesis that the majority of these impair TLK2 kinase activity. Both TLK2-D551G and TLK2-S617L showed profoundly impaired kinase activity, as well as altered subcellular localization, the significance of which remains unclear (figure 3, S3 and S4). As also suggested by in silico tools (table S5), TLK2-S617L caused a more severe impairment of protein activity and localization compared to TLK2-D551G mutant. It is interesting to note, that case carrying p.(Asp617Leu) was characterized by a mild-severe intellectual disability, with a IQ (verbal IQ 63, non verbal IQ 74) that was lower than the one observed in case carrying p.(Asp551Gly) variant and comparable to the one observed in patients carrying p.(Glu475Ter) variant (table S6). Basing on the limited available clinical information, we could therefore suggest that the higher impairment of the protein activity mediated by p.(Ser617Leu) variant could be reflected on the clinical side by a more severe phenotype.
Both mutants overexpressed in AD-293 cells showed higher expression compared to WT. Accordingly, LCLs carrying p.(Asp551Gly) variant showed significantly higher TLK2 protein levels compared to control. Surprisingly, the higher protein expression was counterbalanced by drastically reduced mRNA levels, suggesting a potential negative feedback control, further causing reduced TLK2 total mRNA levels.
Despite the severe effects on kinase activity caused by the heterozygous missense mutations identified in MRD57 patients to date, monoallelic Tlk2 loss did not cause overt phenotypes in mice, but neurodevelopment and behaviour were not assessed in these animals[5]. Given that TLK2 dimerizes with both TLK1 and TLK2 and this is important for its activity, it is also likely that kinase impaired mutants exert some dominant negative effects that contribute to an overall phenotype that is more severe than haploinsufficiency[31]. The recent identification of multiple MRD57 cases with TLK2 haploinsufficiency suggests that an in-depth evaluation of neurodevelopment is needed in mice with reduced Tlk2 levels to determine if they represent a model of MRD57. The placental issues observed in mice with homozygous deletion of Tlk2 were not identified in Tlk2 heterozygous mice, but human gestation is considerably longer, and more subtle placental issues could be present. Recent work showed that total TLK depletion leads to an innate immune secretory response in cancer cells and mice[2, 6, 32]. Maternal immune activation (MIA) has been implicated in ASD and associated with placental defects in mice, suggesting that impaired chromatin maintenance and epigenetic dysregulation could potentially underlie the pathological effects of TLK2 haploinsufficiency[2, 33, 34]. This is consistent with the increased chromatin accessibility reported here in TLK2-D551G patient cells (figure 2B-E), as well as the strong enrichment of chromatin proteins in the TLK2 interactome in the SFARI and DECIPHER databases.
Many genes encoding proteins involved in chromatin remodelling are associated with neurodevelopmental disorders. TLK2, as well as the missense mutants we tested, showed proximal interactions with many of them, including CHD8 and CHD7, that are mutated in ASD and CHARGE syndrome. In addition to these proteins, both TLK2 missense mutants showed altered interactions with additional proteins implicated in neurodevelopment. This included RAD50, a part of the MRE11-RAD50-NBS1 DNA repair complex that localizes to replication forks and plays a key role in DNA-double-strand break repair[35]. RAD50 mutations, present in the DECIPHER database, underlie Nijmegen breakage syndrome-like disorder (OMIM 613078). This condition is characterized by microcephaly, which is also commonly observed in many patients with TLK2 variants. Further, RAD50 proximal interactions were reduced with both missense mutants, potentially suggesting reduced localization to replication forks (figure 5)[36]. Similarly, YEATS2, a chromatin reader component, is suggested as an ASD-associated gene by de novo genetic risk analysis and GWAS (SFARI database criteria 3.1, suggestive evidence)[37, 38], was linked to epilepsy and was enriched with TLK2-WT compared to either missense mutant[39–41]. In contrast, ZNF148 and PAPOLG, that are also associated with neurodevelopmental disorders, were strongly enriched with both missense mutants and detected at very low levels with TLK2-WT, while other SFARI genes, including BRD4, JMJD1C, MSANTD2, CCNK and NACC1 were reduced specifically with the less active TLK2-S617L variant. In future work, it will be of interest to examine the potential functional relevance of these interactions to determine if their alterations underlie the altered chromatin state we observed in LCLs from case 1 or other phenotypes associated with TLK2 loss of function. This approach may detect new candidate genes involved in neurodevelopmental disorders or help us understand the involvement of this network of SFARI genes in isolated or syndromic ASD and ID.
The knowledge of altered protein interactomes is important to understand the molecular impact of disease mutations and could be helpful in identifying pharmacological treatments to mitigate more severe phenotypes, such as epileptic seizures. It is attractive to imagine the possibility of repurposing drugs able to modulate the functions of some genes to influence their impact on disease pathology[42].
In conclusion, we provided the clinical description of six new cases carrying likely pathogenic and pathogenic TLK2 variants and we presented new insights into the impact of TLK2 missense variants, observing impairment in kinase activity, localization, and interaction (Figure S4). Our work offers a deep characterization of two missense variants localized in a key domain of the TLK2 protein, where most mutations related to MRD57disorder occur, providing new insights into the potential role TLK2 in neurodevelopmental disorders.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to the patients and their families for their precious collaboration. This research received funding specifically appointed to Department of Medical Sciences from the Italian Ministry for Education, University and Research (Ministero dell’Istruzione, dell’Università e della Ricerca - MIUR) under the programme “Dipartimenti di Eccellenza 2018 – 2022” Project code D15D18000410001. M.V.P. was funded by an FPI fellowship from the Ministry of Science, Innovation and Universities (MCIU) and M.S.C. by a Masters fellowship from the BIST and support from the IRB Barcelona. T.H.S. was funded by the MCIU (PGC2018095616-B-I00/GINDATA and FEDER). M.T. was funded by Fondazione Bambino Gesù (Vite Coraggiose). The whole exome sequencing was performed as part of the Autism Sequencing Consortium and was supported by the NIMH (MH111661). Thanks to the MSPCF of IRB Barcelona, a principal unit in Proteored, PRB3, supported by PT17/0019 of the PE I+D+i 2013–2016, funded by SCIII and ERDF. This study makes use of data generated by the DECIPHER community. A full list of centres that contributed to the generation of the data is available from http://decipher.sanger.ac.uk and via email from decipher@sanger.ac.uk. Funding for the project was provided by the Wellcome Trust.
Footnotes
WEB RESOURCES
Autism Sequencing Consortium exome analysis browser, https://asc.broadinstitute.org/
BioGRID Database, https://thebiogrid.org/
ClinVar, https://www.ncbi.nlm.nih.gov/clinvar/
Constrained Coding Regions Browser, https://s3.us-east-2.amazonaws.com/ccrs/ccr.html
DECIPHER, https://decipher.sanger.ac.uk/
gnomAD Browser, v2.1.1, https://gnomad.broadinstitute.org/
Human Splicing Finder, v3.1, http://www.umd.be/HSF/
Modeller, v9.25, https://salilab.org/modeller/
OMIM, https://omim.org/
Modeller, https://salilab.org/modeller/
SFARI gene, https://gene.sfari.org/
STRING Database, https://string-db.org
Varsome, https://varsome.com/
Worldwide Protein Data Bank, https://www.wwpdb.org/
Additional information: None of the authors has any competing financial or non-financial interest to disclose.
REFERENCES
- 1.Lee SB, Segura-Bayona S, Villamor-Payà M, Saredi G, Todd MAM, Attolini CSO, Chang TY, Stracker TH, Groth A. Tousled-like kinases stabilize replication forks and show synthetic lethality with checkpoint and PARP inhibitors. Sci Adv Published Online First: 2018. doi: 10.1126/sciadv.aat4985 [DOI] [PMC free article] [PubMed]
- 2.Segura-Bayona S, Stracker TH. The Tousled-like kinases regulate genome and epigenome stability: implications in development and disease. Cell. Mol. Life Sci. 2019. doi: 10.1007/s00018-019-03208-z [DOI] [PMC free article] [PubMed]
- 3.Bruinsma W, Berg J, Aprelia M, Medema RH. Tousled‐like kinase 2 regulates recovery from a DNA damage‐induced G2 arrest. EMBO Rep Published Online First: 2016. doi: 10.15252/embr.201540767 [DOI] [PMC free article] [PubMed]
- 4.Klimovskaia IM, Young C, Strømme CB, Menard P, Jasencakova Z, Mejlvang J, Ask K, Ploug M, Nielsen ML, Jensen ON, Groth A. Tousled-like kinases phosphorylate Asf1 to promote histone supply during DNA replication. Nat Commun Published Online First: 2014. doi: 10.1038/ncomms4394 [DOI] [PMC free article] [PubMed]
- 5.Segura-Bayona S, Knobel PA, Gonzalez-Buron H, Youssef SA, Peña-Blanco A, Coyaud E, LopezRovira T, Rein K, Palenzuela L, Colombelli J, Forrow S, Raught B, Groth A, De Bruin A, Stracker TH. Differential requirements for Tousled-like kinases 1 and 2 in mammalian development. Cell Death Differ Published Online First: 2017. doi: 10.1038/cdd.2017.108 [DOI] [PMC free article] [PubMed]
- 6.Sandra Segura-Bayona Marina Villamor-Payà, Camille Stephan-Otto Attolini Travis H. Stracker. Tousled-like kinase activity is required for transcriptional silencing and suppression of innate immune signaling. 2019.
- 7.Reijnders MRF, Miller KA, Alvi M, Goos JAC, Lees MM, de Burca A, Henderson A, Kraus A, Mikat B, de Vries BBA, Isidor B, Kerr B, Marcelis C, Schluth-Bolard C, Deshpande C, Ruivenkamp CAL, Wieczorek D, Baralle D, Blair EM, Engels H, Lüdecke HJ, Eason J, Santen GWE, Clayton-Smith J, Chandler K, Tatton-Brown K, Payne K, Helbig K, Radtke K, Nugent KM, Cremer K, Strom TM, Bird LM, Sinnema M, Bitner-Glindzicz M, van Dooren MF, Alders M, Koopmans M, Brick L, Kozenko M, Harline ML, Klaassens M, Steinraths M, Cooper NS, Edery P, Yap P, Terhal PA, van der Spek PJ, Lakeman P, Taylor RL, Littlejohn RO, Pfundt R, Mercimek-Andrews S, Stegmann APA, Kant SG, McLean S, Joss S, Swagemakers SMA, Douzgou S, Wall SA, Küry S, Calpena E, Koelling N, McGowan SJ, Twigg SRF, Mathijssen IMJ, Nellaker C, Brunner HG, Wilkie AOM. De Novo and Inherited Loss-of-Function Variants in TLK2: Clinical and Genotype-Phenotype Evaluation of a Distinct Neurodevelopmental Disorder. Am J Hum Genet 2018;102:1195–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Töpf A, Oktay Y, Balaraju S, Yilmaz E, Sonmezler E, Yis U, Laurie S, Thompson R, Roos A, MacArthur DG, Yaramis A, Güngör S, Lochmüller H, Hiz S, Horvath R. Severe neurodevelopmental disease caused by a homozygous TLK2 variant. Eur J Hum Genet Published Online First: 2019. doi: 10.1038/s41431-019-0519-x [DOI] [PMC free article] [PubMed]
- 9.Lelieveld SH, Reijnders MRF, Pfundt R, Yntema HG, Kamsteeg EJ, De Vries P, De Vries BBA, Willemsen MH, Kleefstra T, Löhner K, Vreeburg M, Stevens SJC, Van Der Burgt I, Bongers EMHF, Stegmann APA, Rump P, Rinne T, Nelen MR, Veltman JA, Vissers LELM, Brunner HG, Gilissen C. Meta-analysis of 2,104 trios provides support for 10 new genes for intellectual disability. Nat Neurosci 2016;19:1194–6. [DOI] [PubMed] [Google Scholar]
- 10.Mortuza GB, Hermida D, Pedersen AK, Segura-Bayona S, López-Méndez B, Redondo P, Rüther P, Pozdnyakova I, Garrote AM, Muñoz IG, Villamor-Payà M, Jauset C, Olsen JV., Stracker TH, Montoya G. Molecular basis of Tousled-Like Kinase 2 activation. Nat Commun Published Online First: 2018. doi: 10.1038/s41467-018-04941-y [DOI] [PMC free article] [PubMed]
- 11.Satterstrom FK, Kosmicki JA, Wang J, Breen MS, De Rubeis S, An JY, Peng M, Collins R, Grove J, Klei L, Stevens C, Reichert J, Mulhern MS, Artomov M, Gerges S, Sheppard B, Xu X, Bhaduri A, Norman U, Brand H, Schwartz G, Nguyen R, Guerrero EE, Dias C, Aleksic B, Anney R, Barbosa M, Bishop S, Brusco A, Bybjerg-Grauholm J, Carracedo A, Chan MCY, Chiocchetti AG, Chung BHY, Coon H, Cuccaro ML, Curró A, Dalla Bernardina B, Doan R, Domenici E, Dong S, Fallerini C, Fernández-Prieto M, Ferrero GB, Freitag CM, Fromer M, Gargus JJ, Geschwind D, Giorgio E, González-Peñas J, Guter S, Halpern D, Hansen-Kiss E, He X, Herman GE, Hertz-Picciotto I, Hougaard DM, Hultman CM, Ionita-Laza I, Jacob S, Jamison J, Jugessur A, Kaartinen M, Knudsen GP, Kolevzon A, Kushima I, Lee SL, Lehtimäki T, Lim ET, Lintas C, Lipkin WI, Lopergolo D, Lopes F, Ludena Y, Maciel P, Magnus P, Mahjani B, Maltman N, Manoach DS, Meiri G, Menashe I, Miller J, Minshew N, Montenegro EMS, Moreira D, Morrow EM, Mors O, Mortensen PB, Mosconi M, Muglia P, Neale BM, Nordentoft M, Ozaki N, Palotie A, Parellada M, Passos-Bueno MR, Pericak-Vance M, Persico AM, Pessah I, Puura K, Reichenberg A, Renieri A, Riberi E, Robinson EB, Samocha KE, Sandin S, Santangelo SL, Schellenberg G, Scherer SW, Schlitt S, Schmidt R, Schmitt L, Silva IMW, Singh T, Siper PM, Smith M, Soares G, Stoltenberg C, Suren P, Susser E, Sweeney J, Szatmari P, Tang L, Tassone F, Teufel K, Trabetti E, Trelles M del P, Walsh CA, Weiss LA, Werge T, Werling DM, Wigdor EM, Wilkinson E, Willsey AJ, Yu TW, Yu MHC, Yuen R, Zachi E, Agerbo E, Als TD, Appadurai V, Bækvad-Hansen M, Belliveau R, Buil A, Carey CE, Cerrato F, Chambert K, Churchhouse C, Dalsgaard S, Demontis D, Dumont A, Goldstein J, Hansen CS, Hauberg ME, Hollegaard MV., Howrigan DP, Huang H, Maller J, Martin AR, Martin J, Mattheisen M, Moran J, Pallesen J, Palmer DS, Pedersen CB, Pedersen MG, Poterba T, Poulsen JB, Ripke S, Schork AJ, Thompson WK, Turley P, Walters RK, Betancur C, Cook EH, Gallagher L, Gill M, Sutcliffe JS, Thurm A, Zwick ME, Børglum AD, State MW, Cicek AE, Talkowski ME, Cutler DJ, Devlin B, Sanders SJ, Roeder K, Daly MJ, Buxbaum JD. Large-Scale Exome Sequencing Study Implicates Both Developmental and Functional Changes in the Neurobiology of Autism. Cell Published Online First: 2020. doi: 10.1016/j.cell.2019.12.036 [DOI] [PMC free article] [PubMed]
- 12.Iossifov I, O’Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, Stessman HA, Witherspoon KT, Vives L, Patterson KE, Smith JD, Paeper B, Nickerson DA, Dea J, Dong S, Gonzalez LE, Mandell JD, Mane SM, Murtha MT, Sullivan CA, Walker MF, Waqar Z, Wei L, Willsey AJ, Yamrom B, Lee YH, Grabowska E, Dalkic E, Wang Z, Marks S, Andrews P, Leotta A, Kendall J, Hakker I, Rosenbaum J, Ma B, Rodgers L, Troge J, Narzisi G, Yoon S, Schatz MC, Ye K, McCombie WR, Shendure J, Eichler EE, State MW, Wigler M. The contribution of de novo coding mutations to autism spectrum disorder. Nature 2014;515:216–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O’Roak BJ, Deriziotis P, Lee C, Vives L, Schwartz JJ, Girirajan S, Karakoc E, MacKenzie AP, Ng SB, Baker C, Rieder MJ, Nickerson DA, Bernier R, Fisher SE, Shendure J, Eichler EE. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat Genet Published Online First: 2011. doi: 10.1038/ng.835 [DOI] [PMC free article] [PubMed]
- 14.De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE, Kou Y, Liu L, Fromer M, Walker S, Singh T, Klei L, Kosmicki J, Fu SC, Aleksic B, Biscaldi M, Bolton PF, Brownfeld JM, Cai J, Campbell NG, Carracedo A, Chahrour MH, Chiocchetti AG, Coon H, Crawford EL, Crooks L, Curran SR, Dawson G, Duketis E, Fernandez BA, Gallagher L, Geller E, Guter SJ, Hill RS, Ionita-Laza I, Gonzalez PJ, Kilpinen H, Klauck SM, Kolevzon A, Lee I, Lei J, Lehtimäki T, Lin CF, Ma’ayan A, Marshall CR, McInnes AL, Neale B, Owen MJ, Ozaki N, Parellada M, Parr JR, Purcell S, Puura K, Rajagopalan D, Rehnström K, Reichenberg A, Sabo A, Sachse M, Sanders SJ, Schafer C, Schulte-Rüther M, Skuse D, Stevens C, Szatmari P, Tammimies K, Valladares O, Voran A, Wang LS, Weiss LA, Willsey AJ, Yu TW, Yuen RKC, Cook EH, Freitag CM, Gill M, Hultman CM, Lehner T, Palotie A, Schellenberg GD, Sklar P, State MW, Sutcliffe JS, Walsh CA, Scherer SW, Zwick ME, Barrett JC, Cutler DJ, Roeder K, Devlin B, Daly MJ, Buxbaum JD. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature Published Online First: 2014. doi: 10.1038/nature13772 [DOI] [PMC free article] [PubMed]
- 15.Kopanos C, Tsiolkas V, Kouris A, Chapple CE, Albarca Aguilera M, Meyer R, Massouras A. VarSome: the human genomic variant search engine. Bioinformatics Published Online First: 2019. doi: 10.1093/bioinformatics/bty897 [DOI] [PMC free article] [PubMed]
- 16.Desmet FO, Hamroun D, Lalande M, Collod-Bëroud G, Claustres M, Béroud C. Human Splicing Finder: An online bioinformatics tool to predict splicing signals. Nucleic Acids Res Published Online First: 2009. doi: 10.1093/nar/gkp215 [DOI] [PMC free article] [PubMed]
- 17.Flex E, Martinelli S, Van Dijck A, Ciolfi A, Cecchetti S, Coluzzi E, Pannone L, Andreoli C, Radio FC, Pizzi S, Carpentieri G, Bruselles A, Catanzaro G, Pedace L, Miele E, Carcarino E, Ge X, Chijiwa C, Lewis MES, Meuwissen M, Kenis S, Van der Aa N, Larson A, Brown K, Wasserstein MP, Skotko BG, Begtrup A, Person R, Karayiorgou M, Roos JL, Van Gassen KL, Koopmans M, Bijlsma EK, Santen GWE, Barge-Schaapveld DQCM Ruivenkamp CAL, Hoffer MJV Lalani SR, Streff H Craigen WJ, Graham BH, van den Elzen APM, Kamphuis DJ, Õunap K, Reinson K, Pajusalu S, Wojcik MH, Viberti C, Di Gaetano C, Bertini E, Petrucci S, De Luca A, Rota R, Ferretti E, Matullo G, Dallapiccola B, Sgura A, Walkiewicz M, Kooy RF, Tartaglia M. Aberrant Function of the CTerminal Tail of HIST1H1E Accelerates Cellular Senescence and Causes Premature Aging. Am J Hum Genet Published Online First: 2019. doi: 10.1016/j.ajhg.2019.07.007 [DOI] [PMC free article] [PubMed]
- 18.Silva J, Aivio S, Knobel PA, Bailey LJ, Casali A, Vinaixa M, Garcia-Cao I, Coyaud É, Jourdain AA, Pérez-Ferreros P, Rojas AM, Antolin-Fontes A, Samino-Gené S, Raught B, González-Reyes A, Ribas De Pouplana L, Doherty AJ, Yanes O, Stracker TH. EXD2 governs germ stem cell homeostasis and lifespan by promoting mitoribosome integrity and translation. Nat Cell Biol Published Online First: 2018. doi: 10.1038/s41556-017-0016-9 [DOI] [PubMed]
- 19.Firth HV, Richards SM, Bevan AP, Clayton S, Corpas M, Rajan D, Vooren S Van, Moreau Y, Pettett RM, Carter NP. DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. Am J Hum Genet Published Online First: 2009. doi: 10.1016/j.ajhg.2009.03.010 [DOI] [PMC free article] [PubMed]
- 20.Havrilla JM, Pedersen BS, Layer RM, Quinlan AR. A map of constrained coding regions in the human genome. Nat Genet 2019;51. doi: 10.1038/s41588-018-0294-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manning KS, Cooper TA. The roles of RNA processing in translating genotype to phenotype. Nat. Rev. Mol. Cell Biol. 2017. doi: 10.1038/nrm.2016.139 [DOI] [PMC free article] [PubMed]
- 22.Siwaszek A, Ukleja M, Dziembowski A. Proteins involved in the degradation of cytoplasmic mRNA in the major eukaryotic model systems. RNA Biol. 2014. doi: 10.4161/rna.34406 [DOI] [PMC free article] [PubMed]
- 23.Treiber DK, Shah NP. Ins and outs of kinase DFG motifs. Chem. Biol. 2013. doi: 10.1016/j.chembiol.2013.06.001 [DOI] [PubMed]
- 24.Vijayan RSK, He P, Modi V, Duong-Ly KC, Ma H, Peterson JR, Dunbrack RL, Levy RM. Conformational analysis of the DFG-out kinase motif and biochemical profiling of structurally validated type II inhibitors. J Med Chem Published Online First: 2015. doi: 10.1021/jm501603h [DOI] [PMC free article] [PubMed]
- 25.Roux KJ, Kim DI, Burke B, May DG. BioID: A Screen for Protein-Protein Interactions. Curr Protoc Protein Sci 2018;91:19.23.1–19.23.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wessel SR, Mohni KN, Luzwick JW, Dungrawala H, Cortez D. Functional Analysis of the Replication Fork Proteome Identifies BET Proteins as PCNA Regulators. Cell Rep Published Online First: 2019. doi: 10.1016/j.celrep.2019.08.051 [DOI] [PMC free article] [PubMed]
- 27.Lalani SR, Safiullah AM, Fernbach SD, Harutyunyan KC, Thaller C, Peterson LE, McPherson JD, Gibbs RA, White LD, Hefner M, Davenport SLH, Graham JM, Bacino CA, Glass NL, Towbin JA, Craigen WJ, Neish SR, Lin AE, Belmont JW. Spectrum of CHD7 mutations in 110 individuals with CHARGE syndrome and genotype-phenotype correlation. Am J Hum Genet Published Online First: 2006. doi: 10.1086/500273 [DOI] [PMC free article] [PubMed]
- 28.Bernier R, Golzio C, Xiong B, Stessman HA, Coe BP, Penn O, Witherspoon K, Gerdts J, Baker C, Vulto-Van Silfhout AT, Schuurs-Hoeijmakers JH, Fichera M, Bosco P, Buono S, Alberti A, Failla P, Peeters H, Steyaert J, Vissers LELM, Francescatto L, Mefford HC, Rosenfeld JA, Bakken T, O’Roak BJ, Pawlus M, Moon R, Shendure J, Amaral DG, Lein E, Rankin J, Romano C, De Vries BBA, Katsanis N, Eichler EE. Disruptive CHD8 mutations define a subtype of autism early in development. Cell Published Online First: 2014. doi: 10.1016/j.cell.2014.06.017 [DOI] [PMC free article] [PubMed]
- 29.Manning BJ, Yusufzai T. The ATP-dependent chromatin remodeling enzymes CHD6, CHD7, and CHD8 exhibit distinct nucleosome binding and remodeling activities. J Biol Chem Published Online First: 2017. doi: 10.1074/jbc.M117.779470 [DOI] [PMC free article] [PubMed]
- 30.Murawska M, Brehm A. CHD chromatin remodelers and the transcription cycle. Transcription. 2011. doi: 10.4161/trns.2.6.17840 [DOI] [PMC free article] [PubMed]
- 31.Sunavala-Dossabhoy G, Li Y, Williams B, De Benedetti A. A dominant negative mutant of TLK1 causes chromosome missegregation and aneuploidy in normal breast epithelial cells. BMC Cell Biol Published Online First: 2003. doi: 10.1186/1471-2121-4-16 [DOI] [PMC free article] [PubMed]
- 32.Liu H, Dowdle JA, Khurshid S, Sullivan NJ, Bertos N, Rambani K, Mair M, Daniel P, Wheeler E, Tang X, Toth K, Lause M, Harrigan ME, Eiring K, Sullivan C, Sullivan MJ, Chang SW, Srivastava S, Conway JS, Kladney R, McElroy J, Bae S, Lu Y, Tofigh A, Saleh SMI, Fernandez SA, Parvin JD, Coppola V, Macrae ER, Majumder S, Shapiro CL, Yee LD, Ramaswamy B, Hallett M, Ostrowski MC, Park M, Chamberlin HM, Leone G. Discovery of Stromal Regulatory Networks that Suppress Ras-Sensitized Epithelial Cell Proliferation. Dev Cell Published Online First: 2017. doi: 10.1016/j.devcel.2017.04.024 [DOI] [PMC free article] [PubMed]
- 33.Lombardo M V, Moon HM, Su J, Palmer TD, Courchesne E, Pramparo T. Maternal immune activation dysregulation of the fetal brain transcriptome and relevance to the pathophysiology of autism spectrum disorder. Mol Psychiatry Published Online First: 2018. doi: 10.1038/mp.2017.15 [DOI] [PMC free article] [PubMed]
- 34.Carpentier PA, Dingman AL, Palmer TD. Placental TNF-α signaling in illness-induced complications of pregnancy. Am J Pathol Published Online First: 2011. doi: 10.1016/j.ajpath.2011.02.042 [DOI] [PMC free article] [PubMed]
- 35.Taylor AMR. Chromosome instability syndromes. Best Pract Res Clin Haematol Published Online First: 2001. doi: 10.1053/beha.2001.0158 [DOI] [PubMed]
- 36.Waltes R, Kalb R, Gatei M, Kijas AW, Stumm M, Sobeck A, Wieland B, Varon R, Lerenthal Y, Lavin MF, Schindler D, Dörk T. Human RAD50 Deficiency in a Nijmegen Breakage Syndrome-like Disorder. Am J Hum Genet Published Online First: 2009. doi: 10.1016/j.ajhg.2009.04.010 [DOI] [PMC free article] [PubMed]
- 37.Ruzzo EK, Pérez-Cano L, Jung JY, Wang L kai, Kashef-Haghighi D, Hartl C, Singh C, Xu J, Hoekstra JN, Leventhal O, Leppä VM, Gandal MJ, Paskov K, Stockham N, Polioudakis D, Lowe JK, Prober DA, Geschwind DH, Wall DP. Inherited and De Novo Genetic Risk for Autism Impacts Shared Networks. Cell 2019;178:850–866.e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anney R, Klei L, Pinto D, Almeida J, Bacchelli E, Baird G, Bolshakova N, Bölte S, Bolton PF, Bourgeron T, Brennan S, Brian J, Casey J, Conroy J, Correia C, Corsello C, Crawford EL, De jonge M, Delorme R, Duketis E, Duque F, Estes A, Farrar P, Fernandez BA, Folstein SE, Fombonne E, Gilbert J, Gillberg C, Glessner JT, Green A, Green J, Guter SJ, Heron EA, Holt R, Howe JL, Hughes G, Hus V, Igliozzi R, Jacob S, Kenny GP, Kim C, Kolevzon A, Kustanovich V, Lajonchere CM, Lamb JA, Law-Smith M, Leboyer M, Le couteur A, Leventhal BL, Liu XQ, Lombard F, Lord C, Lotspeich L, Lund SC, Magalhaes TR, Mantoulan C, McDougle CJ, Melhem NM, Merikangas A, Minshew NJ, Mirza GK, Munson J, Noakes C, Nygren G, Papanikolaou K, Pagnamenta AT, Parrini B, Paton T, Pickles A, Posey DJ, Poustka F, Ragoussis J, Regan R, Roberts W, Roeder K, Roge B, Rutter ML, Schlitt S, Shah N, Sheffield VC, Soorya L, Sousa I, Stoppioni V, Sykes N, Tancredi R, Thompson AP, Thomson S, Tryfon A, Tsiantis J, Van Engeland H, Vincent JB, Volkmar F, Vorstman JAS, Wallace S, Wing K, Wittemeyer K, Wood S, Zurawiecki D, Zwaigenbaum L, Bailey AJ, Battaglia A, Cantor RM, Coon H, Cuccaro ML, Dawson G, Ennis S, Freitag CM, Geschwind DH, Haines JL, Klauck SM, Mcmahon WM, Maestrini E, Miller J, Monaco AP, Nelson SF, Nurnberger JI, Oliveira G, Parr JR, Pericak-Vance MA, Piven J, Schellenberg GD, Scherer SW, Vicente AM, Wassink TH, Wijsman EM, Betancur C, Buxbaum JD, Cook EH, Gallagher L, Gill M, Hallmayer J, Paterson AD, Sutcliffe JS, Szatmari P, Vieland VJ, Hakonarson H, Devlin B. Individual common variants exert weak effects on the risk for autism spectrum disorders. Hum Mol Genet Published Online First: 2012. doi: 10.1093/hmg/dds301 [DOI] [PMC free article] [PubMed]
- 39.Mi W, Guan H, Lyu J, Zhao D, Xi Y, Jiang S, Andrews FH, Wang X, Gagea M, Wen H, Tora L, Dent SYR, Kutateladze TG, Li W, Li H, Shi X. YEATS2 links histone acetylation to tumorigenesis of non-small cell lung cancer. Nat Commun Published Online First: 2017. doi: 10.1038/s41467-017-01173-4 [DOI] [PMC free article] [PubMed]
- 40.Zhao D, Guan H, Zhao S, Mi W, Wen H, Li Y, Zhao Y, Allis CD, Shi X, Li H. YEATS2 is a selective histone crotonylation reader. Cell Res. 2016. doi: 10.1038/cr.2016.49 [DOI] [PMC free article] [PubMed]
- 41.Yeetong P, Pongpanich M, Srichomthong C, Assawapitaksakul A, Shotelersuk V, Tantirukdham N, Chunharas C, Suphapeetiporn K, Shotelersuk V. TTTCA repeat insertions in an intron of YEATS2 in benign adult familial myoclonic epilepsy type 4. Brain Published Online First: 2019. doi: 10.1093/brain/awz267 [DOI] [PubMed]
- 42.Tranfaglia MR, Thibodeaux C, Mason DJ, Brown D, Roberts I, Smith R, Guilliams T, Cogram P. Repurposing available drugs for neurodevelopmental disorders: The fragile X experience. Neuropharmacology. 2019. doi: 10.1016/j.neuropharm.2018.05.004 [DOI] [PubMed]
- 43.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ, Von Mering C. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res Published Online First: 2019. doi: 10.1093/nar/gky1131 [DOI] [PMC free article] [PubMed]
- 44.Oughtred R, Stark C, Breitkreutz BJ, Rust J, Boucher L, Chang C, Kolas N, O’Donnell L, Leung G, McAdam R, Zhang F, Dolma S, Willems A, Coulombe-Huntington J, Chatr-Aryamontri A, Dolinski K, Tyers M. The BioGRID interaction database: 2019 update. Nucleic Acids Res Published Online First: 2019. doi: 10.1093/nar/gky1079 [DOI] [PMC free article] [PubMed]
- 45.Knight JDR, Choi H, Gupta GD, Pelletier L, Raught B, Nesvizhskii AI, Gingras AC. ProHits-viz: A suite of web tools for visualizing interaction proteomics data. Nat. Methods. 2017. doi: 10.1038/nmeth.4330 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.