Abstract
Objectives:
To test whether mitochondrial transplantation (MITO) mitigates damage in 2 models of acute kidney injury (AKI).
Background:
MITO is a process where exogenous isolated mitochondria are taken up by cells. As virtually any morbid clinical condition is characterized by mitochondrial distress, MITO may find a role as a treatment modality in numerous clinical scenarios including AKI.
Methods:
For the in vitro experiments, human proximal tubular cells were damaged and then treated with mitochondria or placebo. For the ex vivo experiments, we developed a non-survival ex vivo porcine model mimicking the donation after cardiac death renal transplantation scenario. One kidney was treated with mitochondria, although the mate organ received placebo, before being perfused at room temperature for 24 hours. Perfusate samples were collected at different time points and analyzed with Raman spectroscopy. Biopsies taken at baseline and 24 hours were analyzed with standard pathology, immunohistochemistry, and RNA sequencing analysis.
Results:
In vitro, cells treated with MITO showed higher proliferative capacity and adenosine 5′-triphosphate production, preservation of physiological polarization of the organelles and lower toxicity and reactive oxygen species production. Ex vivo, kidneys treated with MITO shed fewer molecular species, indicating stability. In these kidneys, pathology showed less damage whereas RNAseq analysis showed modulation of genes and pathways most consistent with mitochondrial biogenesis and energy metabolism and downregulation of genes involved in neutrophil recruitment, including IL1A, CXCL8, and PIK3R1.
Conclusions:
MITO mitigates AKI both in vitro and ex vivo.
Key Words: renal transplant, mitochondria transplant, organ reperfusion, regeneration, IRI, metabolism
In the United States, 5000 patients die annually while waiting for a renal transplant,1 whereas more than one fifth of the renal grafts that are procured from deceased donors are discarded. Strategies to increase the donor organ pool2 have resulted in the use of organs derived from unconventional donors that include, among others, expanded criteria, donation after cardiac death (DCD) and acute kidney injury (AKI) donors.3 The outcome of these transplants is characterized by a higher incidence of delayed graft function (DGF) when compared with standard criteria or living donors, occurring in the immediate aftermath of the surgery.4 As DGF is an important risk factor for acute rejection, increased resource utilization and overall costs, and diminished medium-term graft survival rates,5,6 there is an urgent need to develop therapies that minimize its incidence.7,8
DGF is a form of acute damage specific to the renal allograft, resulting from an insult that occurs during the ischemic phase and at reperfusion, in a bimodal manner.9 In the ischemic phase, removal of the organ from its vascular supply causes cellular dysfunction that may lead to irreversible damage or necrosis if the cell’s adaptive ability to utilize anaerobic metabolism is overwhelmed. Transient ischemia modifies mitochondrial volume, structure, and their altered function persists post reperfusion.10 In the kidney, tubular cells are very sensitive to ischemia and suffer almost immediately with the increase in mitochondrial permeability transition pores, the disruption of mitochondrial cristae, the translocation of cytochrome C and the subsequent activation of the Bcl-2 and caspase-3 pathways.11,12 At reperfusion, a burst in the reactive oxygen species (ROS) is the injury leading event, promoting further activation of various injurious intracellular pathways, cell death, as well as entailing activation of the innate and subsequently the adaptive immune system.12 Depending on the duration and extent of ischemia reperfusion injury (IRI), organ dysfunction or even irreversible failure may occur. Therefore, strategies that protect or preserve mitochondria during IRI are urgently needed.
To better understand the critical role played by the mitochondrion in this setting, it should be emphasized that the kidney is also one of the most energy-demanding organs of the human body, sharing the highest resting metabolic rate with the heart, and has the second highest mitochondrial content and oxygen consumption after the heart.11
In multicellular biological entities, mitochondrial transfer occurs when eukaryotic cells acquire new mitochondria from neighboring healthy donor cells, to restore respiration and euphysiology and ultimately ensure survival.13,14 During stress, mitochondria can be transferred between cells or isolated mitochondria can be transplanted into cells to restore mitochondrial function in recipient cells.13 As mitochondrial dysfunction is a common denominator in a myriad of pathologic conditions including renal IRI, isolated mitochondria transplantation (MITO) has recently emerged as a promising therapeutic approach.10,15 In fact, rather than targeting a specific step of the above mentioned chain of events, MITO has been proposed to address irreversible mitochondrial damage by providing viable mitochondria isolated from a healthy source10,13,16–20 and data already exists on the application of this approach to treat renal ischemic damage.21–24 In a clinically relevant model of IRI, damage was inflicted to the animals by occlusion of the renal arteries for 60 minutes.21 Once arterial patency was restored, autologous mitochondria were injected directly within the renal arteries of the study animals. Strikingly, during the 24-hour follow-up, the study animals showed a better renal function whereas postmortem pathology revealed a more pronounced damage in the control kidneys. The same team has been able to successfully translate MITO to the bedside, in a cohort of pediatric patients requiring postcardiotomy extracorporeal membrane oxygenation for severe refractory cardiogenic shock after IRI.25,26
Therefore, in this study, we tested the hypothesis that MITO may mitigate AKI in an in vitro model of tubular damage and in an ex vivo porcine model of DCD renal transplantation mimicking IRI.
METHODS
In vitro Experiments
Cell Culture
Human conditionally immortalized proximal tubular cells (ciPTECs) were purchased from Cell4Pharma (Oss, Nederland) and cultured in Dulbecco’s modified Eagle Medium F-12 (Thermo Fisher Scientific) supplemented with ITS, hydrocortisone, EGF, tri-iodothyronine (all from Sigma-Aldrich) and 10% Foeatal Bovine Serum (FBS) (Euroclone). ciPTECs were immortalized using a temperature sensitive SV40ts A58 and human telomerase genes and proliferate at 33°C but acquire a proximal tubular cell phenotype at 37°C. ciPTECs were expanded in 5% CO2 at 33°C and used for the experiments at least 1 week after culture at 37°C. Immortalized tubular HK-2 cells (from ATCC) were cultured in Dulbecco’s modified Eagle Medium low glucose plus 10% FBS (both from Euroclone) at 37°C in 5% CO2.
Mitochondrial Isolation
The mitochondria isolation protocol was adapted from Konari et al.27 Briefly, cells were detached and pelleted by centrifugation at 100g for 5 minutes. The cell pellet was resuspended in homogenization buffer containing 20 mM HEPES-KOH (pH 7.4), 220 mM mannitol, 70 mM sucrose and a protease inhibitor cocktail (Sigma-Aldrich) at a density of 1.0 × 107 cells/ml and incubated on ice for 5 min. Cells were ruptured by 60 strokes using a 26-gauge needle on ice and homogenate centrifuged 3 times at 400g for 5 minutes to remove unbroken cells. Isolated mitochondria were pelleted by centrifugation at 5800g for 5 minutes and resuspended in resuspension buffer (RB) (250 mmol/L sucrose, 20 mmol/L K+-HEPES buffer [pH7.2], 0.5 mmol/L K+-EGTA [pH 7.4]).
Analysis of Isolated Mitochondria
Purity of Isolated Mitochondria. Briefly, ciPTEC mitochondria were isolated as described above, incubated with MitoTracker green (Invitrogen)28 for 30 minutes, pelleted by centrifugation, and washed with twice with RB to remove excess dye. Purity of the mitochondria suspension was assessed using FACS Celesta flow cytometer (BD Biosciences). Ten thousand events were recorded for each acquisition, gated at 500 nm to 3 µm size after bead calibration (BD Biosciences).
Viability of Isolated Mitochondria. ciPTEC mitochondria were isolated and mitochondrial function evaluated using adenosine 5′-triphosphate (ATP) production (CellTiter-glo 2.0 kit and mitochondrial membrane potential28 (MitoTracker red CMXRos, Invitrogen) assays according to manufacturer’s instructions. Isolated mitochondria were incubated with MitoTracker red for 30 minutes, pelleted by centrifugation, and resuspended in fresh RB twice to remove excess dye.
Evaluation of MITO
ciPTECs were incubated with MitoTracker red CMXRos (Invitrogen) and CMXRos labeled mitochondria were isolated as described. For MITO, unlabeled ciPTECs were incubated without or with CMXRos labeled mitochondria for 24 hours. To assess CMXRos labeled mitochondrial uptake after MITO, ciPTECs were washed 3 times with PBS, detached using TrypLE (Thermo Fisher Scientific) and MITO evaluated by flow cytometry (FACScan, BD Biosciences). To further confirm MITO, HK-2 cells were infected with baculovirus encoding green fluorescent protein (GFP)-labeled E1alpha pyruvate dehydrogenase (BacMam2 delivery system, Life Technologies) and GFP labeled mitochondria were isolated as described above. Fluorescence microscopy (Nikon Eclipse 80i, Nikon Europe) was used to evaluate MITO in ciPTECs treated with 10 μg/mL of GFP-labeled HK-2 mitochondria for 24 hours.
In vitro Model of Tubular Damage and MITO of Healthy Mitochondria
To simulate AKI/IRI in renal tubules, ciPTECs were damaged using a chemically induced model of anoxia, as described by Dagher.29 In particular, the combination of antimycin A (mitochondrial electron transport chain complex III inhibitor) and 2-deoxyglucose (glycolysis inhibitor) prevents substrate oxidation and causes depletion of ATP stores, thus recapitulating typical IRI features. ciPTECs were treated with antimycin A (1 µM, Sigma-Aldrich) and 2-deoxyglucose (10 mM, Sigma-Aldrich) in serum-free complete medium. After 4 hours, damaged cells were washed twice in HBSS (Lonza) and incubated in complete medium without FBS in the presence of RB alone or in the presence of mitochondria (20 µg/mL RB) isolated from undamaged ciPTECs for mitochondrial transplantation. This dose of mitochondria was determined by preliminary dose-response experiments (data not shown). After 24 hours, untreated (NT), IRI damaged (IRI) and IRI damaged treated with MITO (IRI+MITO) ciPTECs were washed 3 times and cultured with complete medium plus 10% FBS for further 72 hours.
After 72 hours, IRI or IRI+MITO ciPTECs (1×106 cells) were lysed in 0.5 mL buffer (50 mM Tris-HCl, 100 mM KCl, 5 mM MgCl2, 1.8 mM ATP, 1 mM EDTA, pH7.2) supplemented with Protease Inhibitor Cocktail III (Sigma-Aldrich), 1 mM phenylmethylsulfonyl fluoride and 250 mM NaF. After centrifugation at 650g for 3 minutes at 4°C to remove unlysed cells, followed by centrifugation at 13,000g for 5 minutes at 4°C, a cytosolic fraction (supernatant) was used for cytosolic ROS measurements. The mitochondrial fraction (pellet) was washed once and resuspended in 0.25 mL of buffer containing 250 mM sucrose, 15 mM K2HPO4, 2 mM MgCl2, 0.5 mM EDTA. 50 µL aliquots were sonicated and used for the measurement of protein content by the BCA Protein Assay kit (Sigma-Aldrich); 10 µg samples were analyzed by SDS-PAGE and immunoblotting with an anti-porin antibody (Abcam; clone 20B12AF2) to confirm the presence of mitochondrial proteins in the extracts. The remaining 200 µL mitochondrial fraction was used to evaluate mitochondrial function in IRI damaged ciPTECs without or with MITO.
For gene expression analyses, total RNA was extracted from IRI ciPTECs without or with MITO using TRIzol Reagent (Ambion, Thermo Fisher Scientific) after the manufacturer’s instructions and subsequently quantified through NanoDrop2000 spectrophotometer (Thermo Fisher Scientific). cDNA was obtained from 200 ng of total RNA using the High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific) following the manufacturer’s instructions.
Experimental Endpoints in Undamaged ciPTECs and IRI Damaged ciPTECs without or with MITO
ciPTECs’ Viability. ciPTECs’ proliferation was evaluated using the BrdU proliferation assay (Sigma-Aldrich), after the manufacturer’s protocol. Cytotoxicity and cell viability were evaluated using the Apotox-Glo Triplex Assay kit (Promega) according to the manufacturer’s protocol. Apoptosis gene expression was measured by quantitative RT-PCR (StepOnePlus Real Time Machine, Thermo Fisher Scientific) in 20 µL containing 5 ng of cDNA template, 200 nM oligonucleotide primers, and Power SYBR Green PCR Master Mix (Thermo Fisher Scientific). The sequence-specific oligonucleotide primers used for qRT-PCR are GAPDH: Fw: 5′ -TGG AAG GAC TCA TGA CCA CAG T - 3′ and Rv: 5′ - CAT CAC GCC ACA GTT TCC C - 3′; SOD 1: Fw: 5′ - GGA TTC CAT GTT CAT GAG TTT GG - 3′ and Rv: 5′ - CCA ACA TGC CTC TCT TCA TCC T - 3′; Caspase 3: Fw: 5′ - CCA CAG CAC CTG GTT ATT ATT CTT G - 3′ and Rv: 5′ - AAC CCG .GGT AAG AAT GTG CAT A - 3′ (MWG-Biotech). Data were analyzed using the ΔΔCt method and GAPDH as housekeeping gene.
Mitochondrial ROS. The mitochondrial fraction (200 µL) from IRI damaged ciPTECs without or with MITO was treated for 30 minutes at 37°C with 5 µM of the ROS-sensitive fluorescent probe MitoSOX (Thermo Fisher Scientific). Mitochondrial ROS were measured using a HT Synergy 96-well micro-plate reader (Bio-Tek Instruments). The relative fluorescence units were converted into nanomoles ROS/mg mitochondrial protein, using a standard curve performed with serial dilutions of H2O2.
Mitochondrial Thiobarbituric Reactive Substances (TBARS). Oxidative damage was measured in the mitochondrial fraction (200 µL) from IRI damaged ciPTECs without or with MITO using the Lipid Peroxidation (4-HNE) Assay Kit (Abcam) that measures 4-hydroxy-nonenale as an index of lipid peroxidation. Results were expressed as nmol/mg mitochondrial protein.
Mitochondria Depolarization. 1×106 cells were washed with PBS, detached by gentle scraping and incubated for 30 minutes at 37°C with 2 μM of the fluorescent probe JC-1 (Biotium Inc.), then centrifuged at 13,000g for 5 minutes and re-suspended in 0.5 mL PBS. Red fluorescence (λ excitation: 550 nm, λ emission: 600 nm) and green fluorescence (λ excitation: 485 nm; λ emission: 535 nm) were measured (Synergy HT Multi-Mode Microplate Reader, Bio-Tek Instruments) and the relative fluorescence units used to calculate the percentage of green fluorescence (depolarized) versus red fluorescence (polarized) mitochondria, considered an index of damaged mitochondria.
Tricarboxylic Acid (TCA) Metabolism. The activities of citrate synthase (Citrate Synthase Assay Kit, Sigma-Aldrich), α-ketoglutarate dehydrogenase (Alpha Ketoglutarate (alpha KG) Assay Kit, Abcam), succinate dehydrogenase (Succinate Dehydrogenase Activity Colorimetric Assay Kit, BioVision) and malate dehydrogenase (Malate Dehydrogenase Assay Kit, Sigma-Aldrich) were assayed according to the manufacturer’s instructions. TCA activity in the mitochondrial fraction (200 µl) from basal (undamaged, NT), and IRI damaged ciPTECs without or with MITO are expressed as mU/mg protein (citrate synthase), nmoles NADH/min/mg protein (α-ketoglutarate dehydrogenase, malate dehydrogenase), and nmoles FADH2/min/mg protein (succinate dehydrogenase).
Electron Transport Chain Complex (ETC) Activity. To measure electron flux from complex I to complex III (an index of the mitochondrial respiration), isolated mitochondria were re-suspended in 0.2 mL buffer A (5 mM KH2PO4, 5 mM MgCl2, 5% w/v bovine serum albumin (BSA); pH 7.2) and followed by 0.1 mL buffer B (25% w/v saponin, 50 mM KH2PO4, 5 mM MgCl2, 5% w/v BSA, 0.12 mM oxidized cytochrome c, 0.2 mM NaN3, pH 7.5) for 5 minutes at room temperature to block complex IV and allow the accumulation of reduced cytochrome c. The cytochrome c reduction reaction was started by adding 0.15 mM NADH and was followed for 5 minutes at 37°C, reading the absorbance at 550 nm by a Packard microplate reader EL340 (Bio-Tek Instruments). Results were expressed as nanomoles of reduced cytochrome c/min/mg mitochondrial protein. ATP levels were measured with the ATP Bioluminescent Assay Kit (Merck, Darmstadt, Germany) according to the manufacturer’s instructions.
Ex vivo Experiments
Study Design
Protocol was approved by our Institutional Animal Care and Use Committee (protocol #A21-053). Yorkshire pigs (n=4), weighing on average 25 kg, were purchased from a commercial source. The day of the experiment, the animals were euthanized and the kidneys were removed and placed on ice. A Luer-Lok connector was inserted into the renal artery, secured with 3/0 silk ties and the kidneys flushed with 250 mL of PBS by gravity. To mimic the DCD scenario and consequent warm ischemic damage, the isolated kidneys were submerged in a plastic bag in a 37°C bath for 30 minutes. After removal to room temperature, biopsy was taken for pathology, immunohistochemistry and RNA sequencing (RNASeq) analysis.
For mitochondrial transplantation, MITO kidneys were injected with 0.5 mL isolated mitochondria diluted to 10 mL in PBS, directly into the renal artery as a single bolus. In preliminary experiments, 10 mL volume resulted to be ideal for delivering mitochondria throughout the kidney without spilling of the preparation in the renal vein. The control (CTL) kidney received 10 mL of PBS. Four MITO and 3 CTL kidneys [one CTL kidney (pig#3) was unable to be cannulated/perfused and was removed from the study] were placed in separate, disposable cassettes in the CaVESWave perfusion system (Biomed Innovations, Regenerative Medicine Development Organization) that produces a pneumatically-actuated programmable flow with adjustable systolic/diastolic pressure and shapeable dicrotic notch with ascending/descending slopes. The MITO and CTL kidneys remained (unperfused) for 30 minutes at room temperature before starting perfusion with 1L of DEVOL perfusate at 25oC (Duke Ex-Vivo Organ Laboratory, Duke University), to allow mitochondrial uptake without incurring any risk to wash them out, as described.21 The DEVOL perfusate is fabricated to duplicate human plasma oncotic and osmotic pressure, acid-base balance, and critical electrolytes; kidneys were perfused through the renal artery and the venous outflow recirculated throughout the study period. During this time, resistances and flow rates were monitored, recorded and analyzed. Five 5 mL samples of the outflow from the CTL and MITO cassettes were collected at 0, 6, 12, 18, and 24 hours, stored at −20°C and analyzed by Raman spectroscopy and for relevant kidney injury markers NGAL and KIM-1 (for details, see Supplementary material, Supplemental Digital Content 1, http://links.lww.com/SLA/E728). At the end of the 24 hours period, a second biopsy sample was taken and sent for tissue analyses and RNAseq.
Mitochondria Isolation Protocol
Mitochondria were isolated from the psoas muscle of the same animal immediately after the removal of the kidneys. Muscle tissue has been previously successfully used as a rich mitochondrial source both in pigs and human studies,21,25 having a high capacity for ATP production.30 The isolation protocol was adapted from the work of Preble et al.31 Briefly, 1 g of muscle was isolated, placed in respiration buffer (300 mM sucrose, 10 mM K-HEPES, and 1 mM K-EGTA) and homogenized using a gentle MACS tissue dissociator (Miltenyi Biotec). Tissue debris was removed centrifugation at 750g and the cell suspension transferred to a 50 mL tube and 4mg/ml protease was added. After 10 minutes on ice, the cell digest was filtered through a 40 µm polyethylenterephthalat mesh filter into a fresh 50 mL tube and 20mg/mL BSA was added. After a second 40 µm filtration step and 2 passages through a 10 µm filter, mitochondria were isolated by centrifugation at 9000g for 10 minutes at 4°C. Mitochondrial pellets were suspended in 0.5 mL respiration buffer used immediately for mitochondrial transplantation as above. Mitochondria were not counted.
Experimental Endpoints in DCD Kidneys without or with MITO
Perfusate Analysis with Raman Spectroscopy and Bioinformatics Evaluation. Samples of perfusate from venous outflow were analyzed using Raman spectroscopy using an Agiltron (Woburn, MA) PeakSeeker PRO-785 dispersive Raman spectrometer as previously described.32–34 Briefly, samples were stored and scanned in flat bottom borosilicate glass vials (Thermo Fisher Scientific) using a 785 nm laser over a 200 to 2000 cm−1 range at 30 mW laser power, 30 s excitation time, 0.2 mm laser spot size, and manufacturer default spectral resolution (8 cm−1). A total of 10 scans per sample (intensity values per 1 cm−1) were obtained, spectra averaged and truncated to 400 to 2000 cm−1 and baseline correction was applied using the Goldindec algorithm.35 The resulting spectra were analyzed using the Rametrix LITE and PRO Toolboxes in MATLAB R1018a.36 Total spectral distance (TSD) calculations and statistical comparisons were performed using previously published methods.32,37 For TSD calculations, the absolute difference between a spectrum and a reference spectrum were calculated across all wavenumbers. These distance values were then summed to yield a TSD value. In this study, the spectrum from the initial time-point (T0) of pig kidney #1 was used as the reference. One-way analysis of variance (ANOVA) and pairwise comparisons using Tukey HSD method were applied to all TSD values to test experimental parameters for statistical significance (P < 0.05). In addition, principal component analysis and discriminant analysis of principal components were applied using the Rametrix Toolboxes,32,36 to determine discernible differences between Raman spectra of CTL and MITO samples. The model was validated by leave-one-out analysis over the entire dataset, and the accuracy, sensitivity, specificity, positive-predictive, and negative-predictive value of the left-out samples were calculated, as shown previously.32
Immunohistochemical Analysis. Biopsies collected at 0 and 24 hours were fixed in 4% paraformaldehyde, embedded in paraffin, and 5 mm sections fixed onto polylysine-coated slides. Apoptotic cells were identified using an in situ thymidine deoxyribose-mediated deoxy-UTP nick end labeling (TUNEL) DNA 3-end labeling kit (Apoptag plus peroxidase in situ apoptosis detection kit, Merck Millipore, #S7101) and expressed as number of apoptotic cells/field. Immunohistochemistry for the detection of 4-Hydroxy-2-nonenal (HNE) was performed using a rabbit, polyclonal antiserum anti-HNE (HNE-11S) (Cat # HNE11-S, Alpha Diagnostic International, St. Antonio, TX, USA). Quantification of HNE-positive area was performed using Fiji and ImageJ software and the results expressed as the HNE+ area/total area±SD.
RNA Extraction, Total RNA-Seq Library Construction, and Sequencing. Kidney biopsies stored in RNAlater (Thermo Fisher Scientific) were homogenized in QIAzol Lysis Reagent (Qiagen) using the TissueLyser LT (Qiagen) and RNA isolated using the Quick-zol RNA Mini Prep Kit (Zymo Research). Total RNA was analyzed on the 2100 Bioanalyzer using the Agilent RNA 6000 Nano Kit (Agilent Technologies) and RNA quantified using the Qubit RNA HS Assay Kit (Thermo Fisher Scientific).
Total RNA-Seq library construction was performed using the Swift Biosciences RNA Library Kit (Swift Biosciences), after rRNA depletion using Lexogen RiboCop Depletion Kit Human/Mouse/Rat v2 (Lexogen Inc., Greenland, NH). Final library concentration was measured through the Qubit 1X dsDNA HS Assay kit (Thermo Fisher Scientific) and library size was evaluated on the 4200 TapeStation using the Agilent D1000 ScreenTape System (Agilent Technologies). Multiplexed libraries were sequenced on a NovaSeq. 6000 (Illumina, San Diego, CA) using 75bp single-end sequencing. On average, about 50 million reads were generated from each sample. Details for bioinformatic analysis of RNA-Seq data are provided in Supplemental Materials, Supplemental Digital Content 1, http://links.lww.com/SLA/E728.
Statistical Analysis
Data are represented as the mean ± s.e.m. Unless otherwise noted, statistical analysis were performed using unpaired t test or ANOVA followed by Dunnett or Tukey range test for post hoc comparisons between groups. Statistical analysis was performed using GraphPad Prism. The differences were considered significant at P < 0.05.
RESULTS
In vitro Experiments
Isolated Mitochondria are Respiration-competent and are Transplanted into ciPTECs
The mitochondrial fraction from differentiated ciPTECs (proximal tubular cell phenotype) showed 93% positive for green fluorescence (Fig. 1A) and robust respiratory activity (Fig. 1B). Notably, increasing the amount of mitochondria (from 10 to 100 µg/mL) resulted in a dose dependent increase in ATP. Consistent with this, CMXRos labeling demonstrated mitochondrial membrane potential, a key indicator of biological activity in the isolated mitochondrial fraction (Fig. 1C) and, as detected by flow cytometry, MitoTracker red-labeled mitochondria were transplanted into ciPTECs (Fig. 1D). To further confirm mitochondrial internalization in ciPTECs, GFP-labeled mitochondria were isolated from HK-2 human renal proximal tubular cells expressing GFP-labeled E1 α-pyruvate dehydrogenase, a protein specifically expressed in the mitochondrial matrix. After MITO, GFP-labeled HK-2 mitochondria were detectable in ciPTECs for at least 3 hours after transplantation (Fig. 1E). Taken together, these results confirmed that mitochondria isolated from renal proximal tubular cells were biologically active and capable of ATP production. Furthermore, the isolated mitochondria were internalized by renal proximal tubular cells supporting viable MITO.
FIGURE 1.
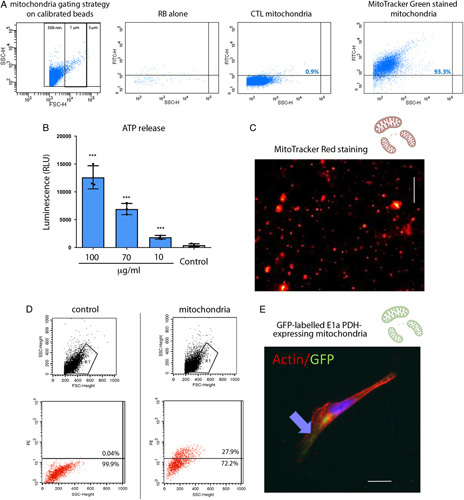
Isolation and internalization of viable mitochondria. A, Isolated mitochondria were suspended in resuspension buffer (RB) and stained with MitoTracker Green FM 200 nM. Calibrated beads were used to gate events of 0.5 to 3 μm. A representative cytofluorimetric analysis shows the absence of fluorescence in RB with MitoTracker Green or in unstained mitochondria. Fluorescence intensity of MitoTracker Green labeled mitochondria, reaching >90% events, was observed among the 0.5 to 1 μm gates. B, Adenosine 5′-triphosphate (ATP) production by isolated mitochondria resuspended in RB at different concentrations (100–10 µg/ml), showed a mitochondrial dose dependent increase in ATP concentration. Control: RB alone. n=3. One-way analysis of variance (ANOVA) with Dunnett multicomparison test was performed: ***P < 0.001 versus Control. C, A representative image of isolated viable mitochondria labeled with MitoTracker red. Scalebar: 5 μm. D, Representative cytofluorimetric analysis shows the acquisition of MitoTracker red labeled mitochondria in MITO human conditionally immortalized proximal tubular cell (ciPTECs). E, A representative image of mitochondria expressing the GFP-labeled E1α pyruvate dehydrogenase after MITO shows the presence of GFP-labeled mitochondria within ciPTECs. Actin is labeled in red. Scalebar: 5 μm. CTL indicates control.
MITO Significantly Enhances Proliferation and Decreases Cytotoxicity in a Cell Model of IRI
After validation of MITO in ciPTECs, we sought to determine the potential protective effect of MITO in an in vitro model of IRI. To mimic IRI, ciPTECs were exposed to a combination of antimycin A and 2-deoxyglucose in serum-free medium for 4 hours. IRI induced ciPTECs were treated without or with isolated mitochondria from non-IRI (undamaged, healthy) ciPTECs (Fig. 2A). After 24 hour MITO and 72hour recovery, we found MITO was able to increase in ATP production and enhance ciPTEC proliferation after IRI damage (Fig. 2B). We also found that MITO significantly reduced cell cytotoxicity as demonstrated by the significant increase in the cleavage of the cell-permeant peptide that depends on the protease activity, and therefore is restricted to intact viable cells (Fig. 2B). MITO mitigation of cytotoxicity was further confirmed by qRT-PCR that showed a significant decrease in the expression of 2 important markers of apoptosis, caspase 3 and superoxide dismutase 1 (Fig. 2C).
FIGURE 2.
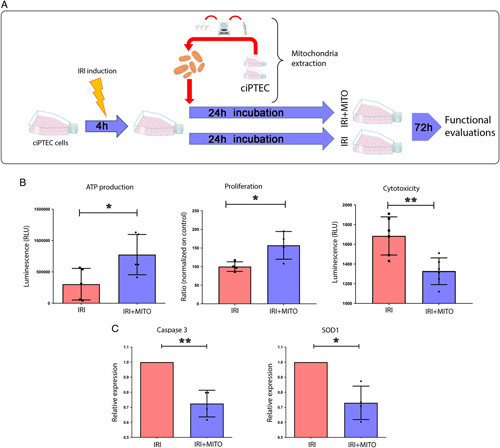
Mitochondrial transplantation (MITO) reduces reactive oxygen species and mitigates mitochondrial damage in IRI. A, Human conditionally immortalized proximal tubular cell (ciPTEC) were subjected to IRI and co-incubated without (IRI) or with mitochondria (20 µg/ml resuspension buffer, IRI+MITO) isolated from healthy ciPTECs. After 24 hours, cells were washed 3 times, cultured for further for 72 hours, and mitochondria isolated for functional evaluation. B, IRI damaged ciPTECs were compared with IRI damaged ciPTECs treated with MITO. MITO promoted ATP production, proliferation and reduced cytotoxicity compared with IRI damaged cells. C, Expression of apoptotic genes, Caspase-3 and superoxide dismutase (SOD1) was assessed in IRI ciPTECs and compared with IRI+MITO ciPTECs. qRT-PCR results showed a significant decrease in apoptosis in IRI+MITO ciPTECs (mean ± SD normalized to GAPDH and expressed relative to untreated IRI ciPTECs). Data are mean ±SD of 4 different experiments, Student t test was performed: *P< 0.05, **P < 0.01 vs IRI). ATP indicates adenosine 5′-triphosphate.
MITO Significantly Reduces ROS, Mitigates Mitochondrial Damage and Improves Mitochondrial Metabolism in IRI ciPTECs
Mitochondrial dysfunction is common in pathologic conditions including IRI. To test our hypothesis and characterize the mechanism of cell recovery after IRI, we evaluated mitochondrial metabolic activity in our cell model of IRI without and with MITO. We found oxidative stress was upregulated in IRI and was significantly reduced by MITO as demonstrated by the decrease of ROS generation together with the coherent decrease of TBARS production, a generic metric of ROS presence (Fig. 3A). Moreover, MITO ci-PTECs showed the restoration of the mitochondrial membrane potential after IRI damage, confirming previous data (Fig. 3A). Then, to further characterize the mechanism of MITO mediated cell recovery after IRI, we measured TCA cycle and electron transport chain enzymatic activity. We found that MITO reversed the effect of IRI on the activity of relevant enzymes of TCA cycles (citrate synthase, succinate, and malate dehydrogenase) and oxidative phosphorylation (Fig. 3B), supporting improved mitochondrial function. Consistent with these observations, MITO increased the activity of enzymes correlated with the electron transport chain and increased intracellular ATP level in ciPTEC after IRI (Fig. 3C). Recovery from IRI seem to be dose-dependent (Suppl. Fig.1A, Supplemental Digital Content 1, http://links.lww.com/SLA/E728), where MITO restored 2 other important pathways related with mitochondrial homeostasis, namely glutaminolysis, demonstrated by the significant restoration of the glutaminase activity, and the beta-oxidation pathway, as shown by the increase of palmitic acid oxidation (Suppl. Fig.1A, Supplemental Digital Content 1, http://links.lww.com/SLA/E728). Taken together, our results show that MITO is able to mitigate IRI damage in an in vitro model of renal tubular injury that is illustrated by a principal component analysis using all data from the in vitro IRI experimental model (Suppl. Fig.1B, Supplemental Digital Content 1, http://links.lww.com/SLA/E728).
FIGURE 3.
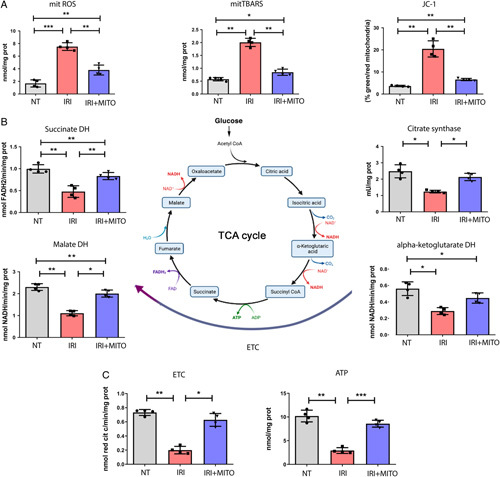
MITO improves mitochondrial metabolism after IRI. Normal healthy human conditionally immortalized proximal tubular cells (ciPTECs) (NT) or IRI damaged ciPTECs were treated without (IRI) or with mitochondrial transplantation (MITO) (20 µg/ml, IRI+MITO). A, MITO normalized reactive oxygen species (ROS) levels, mitochondrial thiobarbituric reactive substances and JC-1 membrane potential altered by IRI. B, Key tricarboxylic acid (TCA) cycle enzyme [succinate dehydrogenase (DH), citrate synthase, malate DH] activities in IRI ciPTECs were significantly increased by MITO (IRI+MITO). C, Electron transport chain (ETC) and adenosine 5′-triphosphate (ATP) concentrations in IRI ciPTECs were significantly increased with MITO treatment (IRI+MITO). Data are mean ±SD of 4 different experiments, one-way analysis of variance (ANOVA) with Tukey multicomparison test was performed: *P< 0.05, **P < 0.01, ***P < 0.001.
Ex vivo Experiments
MITO Mitigates IRI in a Model of DCD Renal Transplant
An ex vivo model mimicking the DCD renal transplant setting (Fig. 4A) was used to test the ability of mitochondrial transplantation to repair an IRI damaged kidney. The histologic evaluation of the kidney tissue revealed the presence of macroscopic damage in both MITO and control kidneys (Fig. 4B); however, we observed a significant decrease in the presence of apoptotic cells in MITO kidneys (Fig. 4C). Likewise, HNE, a marker of lipid peroxidation and reactive lipid generation, was significantly reduced in MITO kidneys. These results are consistent with the hypothesis that MITO may be beneficial in reducing the pathophysiology associated with DCD kidney transplantation.
FIGURE 4.
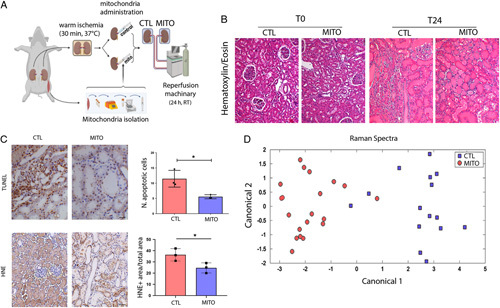
MITO improves IRI in a model of donation after cardiac death (DCD) renal transplant. A, An ex vivo model of DCD kidneys was used to test the hypothesis that mitochondrial transplantation (MITO) improves the pathophysiology associated with IRI. Kidneys were subjected to warm ischemia for 30 minutes, followed by injection with vehicle (CTL) or mitochondria isolated from 1g of autologous psoas muscle (MITO). After 30 min, kidneys were perfused for 24 hours. (n=4 animals, 5 kidneys (one CTL discarded), 2 experimental groups, CTL and MITO). B, The presence of macroscopic damage in control (CTL) and mitochondrial treated (MITO) kidneys was shown by hematoxylin and eosin staining. C, thymidine deoxyribose-mediated deoxy-UTP nick end labeling (TUNEL) assay and 4-hydroxy-2-nonenal (HNE) showed a decrease of apoptotic cells and lipid peroxidation, respectively, in MITO kidneys (representative images) that was confirmed by quantification. Student t test was performed: *P < 0.05. D, Raman spectra, collected at sequential timepoints from control and MITO kidneys were baseline normalized and then analyzed using principal component analysis and discriminate analysis of principal components. The canonical plot matrix shows differences in the molecular composition of perfusate from control and MITO kidneys appearing as definitive separated clusters. MITO treated kidneys shed fewer molecules/molecular species over 24 hours than CTL kidneys, indicating greater stability/viability [shown statistically by calculation of Total Spectral Distance vs Analytical Standard (Surine) using the RametrixTM Toolboxes]. CTL indicates control.
Raman spectrometry was used to metabolically profile the perfusate from MITO and control kidneys. Principle component analysis of the perfusate chemical composition showed definitive separated clusters of MITO and control metabolic profiles (Fig. 4D). Analysis of perfusate samples collected over time showed a significant difference (P < 0.05) between the initial time-point (T0) and 18 hours for both control and MITO groups, with less statistically significant total spectral distance values in MITO samples (Table 1 and Suppl. Table 1 and 2, Supplemental Digital Content 1, http://links.lww.com/SLA/E728). Analyses of perfusate chemical composition from the different animals in the 2 experimental groups showed significantly different Raman spectra within the control group, whereas none were statistically significant for the MITO group (Table 1 and Suppl. Tables 3 and 4, Supplemental Digital Content 1, http://links.lww.com/SLA/E728). Taken together, results from analyses of perfusate chemical composition support that MITO may have a stabilizing effect on kidney function in an IRI model of DCD kidneys. No differences between the 2 groups were observed in terms of resistance and flow rates by the software of the CaVESWave perfusion system. Markers of renal damage NGAL and KIM1, assessed in the perfusate of MITO-treated kidneys in respect to control, also reduced a trend of reduction although it did not reach statistical significance (Suppl. Fig. 2, Supplemental Digital Content 1, http://links.lww.com/SLA/E728).
TABLE 1.
Summary of Total Spectral Distance Calculations, ANOVA, and Pairwise Comparisons
| Group | % different samples with time (all kidneys) | % different samples with kidney (all time points) | Stat. analysis |
|---|---|---|---|
| CTL | 50 | 67 | P<0.05 |
| MITO | 30 | 0 | P<0.05 |
Statistical differences in total spectral distance (indicative of metabolite concentrations) in pairwise comparisons of perfusate over time and in pairwise comparison of kidneys within groups (over all time points), revealed that perfusate samples from MITO kidneys showed fewer significant differences than the control group. The single values are reported in Suppl. Tables 1-4.
ANOVA indicates one-way analysis of variance; CTL, control; MITO, mitochondrial transplantation.
MITO Modulates Gene Expression for Key Cellular Signaling Pathways in the DCD Kidney Model
To better understand the mechanisms involved in the MITO, we performed RNAseq analysis on tissue samples collected from control and MITO kidneys at baseline (T0) and at 24 hours (T24). The differentially expressed genes (DEGs) are shown in the Venn Diagram (Suppl. Fig. 3, Supplemental Digital Content 1, http://links.lww.com/SLA/E728). Bioinformatic analyses of the transcripts revealed significant modulation of genes and pathways associated with mitochondrial biogenesis (PPAR pathway) and mitochondrial metabolism (IL-17, Ca2+, cAMP, and CREB signaling) in MITO samples versus control (Fig. 5 and Suppl. Table 5, Supplemental Digital Content 1, http://links.lww.com/SLA/E728). DEGs involved in the activation of intracellular pathways, as detected by ingenuity pathway analysis, are shown in Figure 6. In particular, induction of numerous signaling pathway mediators was detected, including G proteins/adenylyl cyclases/cAMP signaling, MAPK/ERK signaling, and PI3K/AKT signaling, coupled to increased levels of ELK1 and CREB transcription factors.
FIGURE 5.
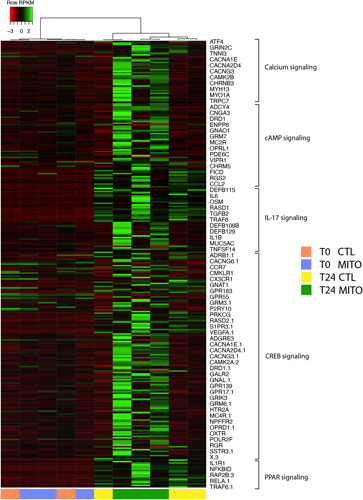
Mitochondrial transplantation (MITO) modulates mitochondrial biogenesis and mitochondrial metabolism pathways in a donation after cardiac death kidney model. Pathway analysis was performed using Ingenuity Pathway Analysis software (IPA, Qiagen). Selection of the pathways was based on the significant modulation of transcripts between MITO T24 versus control (CTL) T24 (fold change >1.5 or < −1.5; P < 0.05). The hierarchical clustering heatmap shows calcium, cAMP, IL-17, CREB, and PPAR signaling pathway specific transcript modulation in control (CTL) and treated (MITO) at T0 and T24. Values for differentially expressed genes in these 5 pathways are reported in Suppl. Table 5, Supplemental Digital Content 1, http://links.lww.com/SLA/E728.
FIGURE 6.
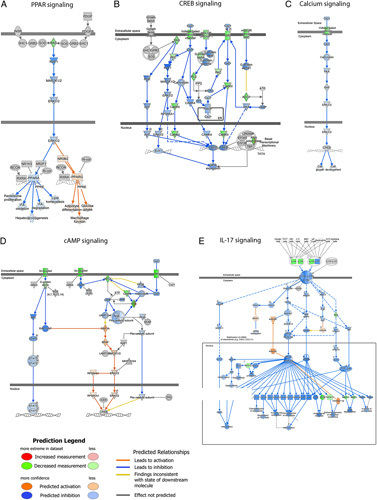
Mitochondrial transplantation (MITO) increases intracellular signaling pathway genes in a donation after cardiac death kidney model. Pathway analysis for transcript modulations after 24 hour perfusion between treatment (MITO T24) versus control (CTRL T24) (fold change >1.5 or < −1.5; P <0.05) was performed using Ingenuity Pathway Analysis software (IPA, Qiagen). Increased expression of signaling pathway genes was observed, including G proteins/adenylyl cyclases/cAMP signaling mediators, MAPK/ERK and PI3K/AKT genes, and increased levels of ELK1 and CREB transcription factors. Results show the interaction of common activated signaling pathway genes after MITO. Upregulated genes in dataset are represented in blue.
DEGs were further analyzed with Advaita Bioinformatics’ iPathwayGuide, to gain a deeper insight into the associated Gene Ontologies (GO). Clustering of differentially expressed genes based on the Biological Process subcategory derived from GO annotations highlighted the themes of ion transport and metabolism. The specific GOs along with their rank (based on ascending p value) and number of DEGs are presented in Suppl. Fig. 4A, Supplemental Digital Content 1, http://links.lww.com/SLA/E728. A similar theme was observed upon clustering DEGs by cellular components, with most DEGs belonging to transporter complex, ribosome, respirasome, and extracellular organelles (Suppl. Fig. 4B, Supplemental Digital Content 1, http://links.lww.com/SLA/E728). When DEGs were clustered by Molecular Function, the themes of transporter and enzyme activity emerged (Suppl. Fig. 4C, Supplemental Digital Content 1, http://links.lww.com/SLA/E728). Furthermore, the pathways that were predicted to be most impacted by these DEGs were found to be congruent with the themes highlighted by GO and were mainly related to ROS, metabolic pathways, cAMP and calcium signaling pathways (ranked 2, 21, 10, and 24 based on p-value) (Suppl. Fig. 4D, Supplemental Digital Content 1, http://links.lww.com/SLA/E728 and Suppl. Table 5, Supplemental Digital Content 1, http://links.lww.com/SLA/E728).
DISCUSSION
Our dual in vitro and ex vivo pilot study support that MITO is effective at mitigating stress in damaged renal cells. These data are of prospective interest to the renal transplant community due to the intuitive implications that they may have in devising strategies to repair and regenerate kidneys that have suffered from severe acute damage. These are, among others, organs procured from DCD donors with prolonged warm ischemia time or from AKI donors with a severe impairment of the renal function, or even kidneys procured from uncontrolled DCD (uDCD) donors. This latter donor category is of paramount interest because of its potential to dramatically change the organ donor availability. In fact, it has been estimated that the United States alone could produce about ten thousands uDCD donors each year, so potentially adding 20 more thousand transplantable renal allografts38 to the almost 25,000 renal transplants that were done in 2022 (https://optn.transplant.hrsa.gov/data/view-data-reports/national-data/#, accessed on January 26, 2023). These uDCD allografts, however, are expected to sustain a more pronounced ischemic damage than their controlled counterpart, highlighting the need to develop strategies to mitigate AKI/IRI.
As MITO bears immense potential to meet this urgent need, we designed a dual pilot study to test the hypothesis that MITO mitigates acute renal cell damage both in vitro and ex vivo. We found that, in vitro, cells treated with mitochondrial transplantation showed a significant decrease in the IRI induced mitochondrial membrane potential alteration and a reduction of ROS and TBARS, which are byproducts of ROS augmentation. At the same time, the assessment of the activity of mitochondrial-related enzymes showed the significant increase in the activity of TCA cycle related enzymes (citrate synthase, succinate dehydrogenase, and malate dehydrogenase) and of enzymes involved in oxidative phosphorylation. Consistent with these observations, an increased intracellular ATP concentration in mitochondria-treated cells was observed in the treated cells when compared with the controls. Taken together, these data demonstrate that MITO is able to mitigate the acute damage inflicted to tubular cells in vitro, while rescuing the proliferative capacity of the cells and minimizing cytotoxicity. MITO restores the partially disrupted mitochondrial homeostasis, enhances the activity of mitochondrial-related enzymes, reinstates the mitochondrial membrane potential, and resets ATP production.
On the basis of in vitro results, we used a more clinically relevant model to test if MITO could mitigate IRI in isolated kidneys. We developed the ex vivo model mimicking the DCD renal transplant scenario, for 3 reasons. First, because the ex vivo model of renal transplantation may provide critical information for translation without necessarily having to rely on in vivo data.39 Second, the DCD allograft features a more pronounced damage than the non-DCD allograft, thus offering a valuable platform for testing the efficacy of MITO. Finally, as 40% of the renal transplants performed in the Unite States are DCD renal transplants, alternative, more effective approaches that may improve the function and the overall outcome of “less than optimal graft” are urgently needed. Our transplant program has a broad, longstanding experience in the use of such allografts including DCD kidneys40–55 and our patients would see potential benefit from treatments that mitigate DCD kidney damage. Overall, our ex vivo data confirmed the beneficial effects of MITO observed in vitro. In fact, kidneys treated with mitochondria featured a less pronounced tubular damage and overall cytotoxicity, both at standard pathology and histochemistry. Considering that these changes were apparent already after 24 hours, it can be inferred that the effects exerted by MITO are almost immediate and that, in terms of in vivo survival and in patients, may affect delayed graft function based on the evidence that the degree of the damage correlates to the degree of impairment of renal allograft function and its ability to recover.56
For the purposes of the present study, we elected to use Raman spectroscopy to monitor the organ. Indeed, over the past few years, our group has developed and validated a novel approach to molecular urinalysis, using a combination of Raman spectroscopic, computational, and physicochemical analytical methods. This new method of urinalysis, termed Raman chemometric urinalysis or Rametrix, was successfully applied to determining the molecular characteristics and physical properties of urine specimens from both healthy human volunteers32 and patients affected by various urological conditions including chronic kidney disease.31,33,57,58 This method can be applied to virtually all solids and liquids specimens, including perfusates, to determine its molecular profiling (or ‘fingerprinting’) to provide metabolomic information in near real-time. When applied to analyzing perfusate, Raman spectroscopy can be used as an online indicator of organ health. Given our results and PCA of the data, MITO kidneys separated quite strikingly from the control organs which shed more molecular species, as an indirect evidence of more pronounced damage.
Transcriptome analysis of kidneys treated with MITO showed modulation of genes and pathways most consistent with mitochondrial biogenesis and energy metabolism. The predicted upregulation of signaling involving PPAR, cAMP, and CREB machinery suggests increased mitochondrial fatty acid oxidation59,60 and energy production. This mechanism is highly plausible in light of the reduced HNE in mitochondria-treated organs, as it is an important metabolic process for removing the toxic lipids that could contribute to acute and/or chronic kidney injury.61 Overall, the transcriptional fingerprint of kidneys treated with MITO was consistent with our histologic findings featuring less tubular damage. Ultimately, deeper understanding of the underlying mechanism driving the MITO induced renoprotection in AKI could improve outcomes in transplant patients when utilizing DCD kidneys.
In conclusion, our study has important in clinical translational implications. We found that MITO may have the potential to become a valuable therapy in the clinical setting of acute renal damage and that could transform the transplant field. The next step should be the validation of our findings in a clinically relevant survival model of DCD renal transplantation.
Supplementary Material
ACKNOWLEDGMENTS
The authors acknowledge the contributions of Andrew Barbas, MD PhD, and Nasser Abraham, PhD, (Duke Ex-Vivo Organ Laboratory, DEVOL perfusate), Carrie DiMarzio and Charles Ardeena (Biomed Innovations, LLC, CaVESWave perfusion system), and Barbara K. Yoza, PhD (Dept. of Surgery, Wake Forest School of Medicine, manuscript preparation).
Footnotes
B.B. conceived, designed, and wrote the study project, supervised all investigations relative to the in vitro experiments, collected results, interpreted data relative to the in vitro and ex vivo experiments, and wrote the manuscript. She was responsible for the primary undertaking, completion, and supervision of all experiments with G.O. G.O. conceived, designed, and wrote the study project, supervised all investigations relative to the ex vivo experiments, collected results, interpreted data relative to the in vitro and ex vivo experiments, and wrote the manuscript. With B.B., he was responsible for the primary undertaking, completion, and supervision of all experiments. A.R. performed all in vitro experiments and some of the ex vivo experiments. The manuscript generated from his PhD thesis that he successfully defended at the end of 2021. He collected results, and interpreted data relative to the in vitro and ex vivo experiments. A.A. contributed to the design of the research, supervised most of the ex vivo experiments, collected results, interpreted data relative to the in vitro and ex vivo experiments, and wrote the manuscript. L.N.B. conducted most of the ex vivo experiments, collected and analyzed data, and wrote the manuscript. C.R., C.G., A.D., and P.E.P. contributed to the design of the in vitro experiments, conducted and supervised the in vitro experiments, and interpreted data relative to the in vitro experiments. S.S., S.D.S., M.E.T., and L.P. contributed to the study design relative to the ex vivo experiments, performed RNA sequencing analysis, collected and interpreted the relative data, and wrote the manuscript. J.R. and R.S.S. contributed to the study design, performed Raman spectroscopy analysis, collected and interpreted the relative data, and wrote the manuscript. B.A. provided input on mitochondrial physiology and data interpretation, edited the manuscript, and revised it critically. J.M. contributed to the study design, provided input on mitochondrial transfer, provided guidance on the implementation of the ex vivo experiments, edited the manuscript, and revised it critically. F.M. performed all surgeries, contributed to all ex vivo experiments, collected and analyzed data. C.P. assisted to all surgeries, contributed to all ex vivo experiments, collected and analyzed data. All the authors gave final approval of the final version of the manuscript.
This study was supported, in part, by the National Center for Advancing Translational Sciences (NCATS), National Institutes of Health, through Grant Award Number UL1TR001420 (G.O.), and the Ministry of Health and Research (MIUR), local research (B.B.).
J.R. and R.S.S. are co-founders of Rametrix Technologies, on the board of which J.R. serves as President and CEO, whereas R.S.S. as Chief Technology Officer. The remaining authors report no conflicts of interest.
Supplemental Digital Content is available for this article. Direct URL citations are provided in the HTML and PDF versions of this article on the journal's website, www.annalsofsurgery.com.
Contributor Information
Andrea Rossi, Email: andrea.issor.1992@gmail.com.
Amish Asthana, Email: aasthana@wakehealth.edu.
Chiara Riganti, Email: chiara.riganti@unito.it.
Sargis Sedrakyan, Email: ssedrakyan@chla.usc.edu.
Lori Nicole Byers, Email: lbyers@wakehealth.edu.
John Robertson, Email: drbob@vt.edu.
Ryan S. Senger, Email: senger@vt.edu.
Filippo Montali, Email: fmontali@ausl.pr.it.
Cristina Grange, Email: cristina.grange@unito.it.
Alessia Dalmasso, Email: alessia.dalmasso@unito.it.
Paolo E. Porporato, Email: paolo.porporato@unito.it.
Chris Palles, Email: cpalles@ufl.edu.
Matthew E. Thornton, Email: Matthew.thornton@med.usc.edu.
Stefano Da Sacco, Email: SDaSacco@chla.usc.edu.
Laura Perin, Email: LPerin@chla.usc.edu.
Bumsoo Ahn, Email: bahn@wakehealth.edu.
James McCully, Email: James.McCully@childrens.harvard.edu.
Giuseppe Orlando, Email: gorlando@wfubmc.edu;gorlando@wakehealth.edu.
Benedetta Bussolati, Email: benedetta.bussolati@unito.it.
REFERENCES
- 1. Brennan C, Husain SA, King KL, et al. A donor utilization index to assess the utilization and discard of deceased donor kidneys perceived as high risk. Clin J Am Soc Nephrol. 2019;14:1634–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Orlando G, Soker S, Stratta RJ, et al. Will regenerative medicine replace transplantation? Cold Spring Harb Perspect Med. 2013;3:a015693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rao PS, Ojo A. The alphabet soup of kidney transplantation: SCD, DCD, ECD--fundamentals for the practicing nephrologist. Clin J Am Soc Nephrol. 2009;4:1827–1831. [DOI] [PubMed] [Google Scholar]
- 4. Gill J, Rose C, Lesage J, et al. Use and outcomes of kidneys from donation after circulatory death donors in the United States. J Am Soc Nephrol. 2017;28:3647–3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tapiawala SN, Tinckam KJ, Cardella CJ, et al. Delayed graft function and the risk for death with a functioning graft. J Am Soc Nephrol. 2010;21:153–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gill J, Dong J, Rose C, et al. The risk of allograft failure and the survival benefit of kidney transplantation are complicated by delayed graft function. Kidney Int. 2016;89:1331–1336. [DOI] [PubMed] [Google Scholar]
- 7. Orlando G, Murphy SV, Bussolati B, et al. Rethinking regenerative medicine from a transplant perspective (and vice versa). Transplantation. 2019;103:237–249. [DOI] [PubMed] [Google Scholar]
- 8. Edgar L, Pu T, Porter B, et al. Regenerative medicine, organ bioengineering and transplantation. Br J Surg. 2020;107:793–800. [DOI] [PubMed] [Google Scholar]
- 9. Kalogeris T, Baines CP, Krenz M, et al. Cell biology of ischemia/reperfusion injury. Int Rev Cell Mol Biol. 2012;298:229–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. McCully JD, Del Nido PJ, Emani SM. Mitochondrial transplantation for organ rescue. Mitochondrion. 2022;64:27–33. [DOI] [PubMed] [Google Scholar]
- 11. Bhargava P, Schnellmann RG. Mitochondrial energetics in the kidney. Nat Rev Nephrol. 2017;13:629–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang X, Agborbesong E, Li X. The role of mitochondria in acute kidney injury and chronic kidney disease and its therapeutic potential. Int J Mol Sci. 2021;22:11253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang ZH, Chen L, Li W, et al. Mitochondria transfer and transplantation in human health and diseases. Mitochondrion. 2022;65:80–87. [DOI] [PubMed] [Google Scholar]
- 14. Liu Z, Sun Y, Qi Z, et al. Mitochondrial transfer/transplantation: an emerging therapeutic approach for multiple diseases. Cell Biosci. 2022;12:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Doulamis IP, McCully JD. Mitochondrial transplantation for ischemia reperfusion injury. Methods Mol Biol. 2021;2277:15–37. [DOI] [PubMed] [Google Scholar]
- 16. McCully JD, Levitsky S, Del Nido PJ, et al. Mitochondrial transplantation for therapeutic use. Clin Transl Med. 2016;5:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang L, Liu Q, Hu H, et al. Progress in mesenchymal stem cell mitochondria transfer for the repair of tissue injury and treatment of disease. Biomed Pharmacother. 2022;153:113482. [DOI] [PubMed] [Google Scholar]
- 18. Tan YL, Eng SP, Hafez P, et al. Mesenchymal stromal cell mitochondrial transfer as a cell rescue strategy in regenerative medicine: a review of evidence in preclinical models. Stem Cells Transl Med. 2022;11:814–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fairley LH, Grimm A, Eckert A. Mitochondria transfer in brain injury and disease. Cells. 2022;11:3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen Y, Yang F, Chu Y, et al. Mitochondrial transplantation: opportunities and challenges in the treatment of obesity, diabetes, and nonalcoholic fatty liver disease. J Transl Med. 2022;20:483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Doulamis IP, Guariento A, Duignan T, et al. Mitochondrial transplantation by intra-arterial injection for acute kidney injury. Am J Physiol Renal Physiol. 2020;319:F403–F413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jabbari H, Roushandeh AM, Rostami MK, et al. Mitochondrial transplantation ameliorates ischemia/reperfusion-induced kidney injury in rat. Biochim Biophys Acta Mol Basis Dis. 2020;1866:165809. [DOI] [PubMed] [Google Scholar]
- 23. Yuan Y, Yuan L, Li L, et al. Mitochondrial transfer from mesenchymal stem cells to macrophages restricts inflammation and alleviates kidney injury in diabetic nephropathy mice via PGC-1α activation. Stem Cells. 2021;39:913–928. [DOI] [PubMed] [Google Scholar]
- 24. Kubat GB, Ozler M, Ulger O, et al. The effects of mesenchymal stem cell mitochondrial transplantation on doxorubicin-mediated nephrotoxicity in rats. J Biochem Mol Toxicol. 2021;35:e22612. [DOI] [PubMed] [Google Scholar]
- 25. Guariento A, Piekarski BL, Doulamis IP, et al. Autologous mitochondrial transplantation for cardiogenic shock in pediatric patients following ischemia-reperfusion injury. J Thorac Cardiovasc Surg. 2021;162:992–1001. [DOI] [PubMed] [Google Scholar]
- 26. Emani SM, Piekarski BL, Harrild D, et al. Autologous mitochondrial transplantation for dysfunction after ischemia-reperfusion injury. J Thorac Cardiovasc Surg. 2017;154:286–289. [DOI] [PubMed] [Google Scholar]
- 27. Konari N, Nagaishi K, Kikuchi S, et al. Mitochondria transfer from mesenchymal stem cells structurally and functionally repairs renal proximal tubular epithelial cells in diabetic nephropathy in vivo. Sci Rep. 2019;9:5184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Poot M, Zhang YZ, Krämer JA, et al. Analysis of mitochondrial morphology and function with novel fixable fluorescent stains. J Histochem Cytochem. 1996;44:1363–1372. [DOI] [PubMed] [Google Scholar]
- 29. Dagher PC. Modeling ischemia in vitro: selective depletion of adenine and guanine nucleotide pools. Am J Physiol Cell Physiol. 2000;279:C1270–C1277. [DOI] [PubMed] [Google Scholar]
- 30. Kappler L, Hoene M, Hu C, et al. Linking bioenergetic function of mitochondria to tissue-specific molecular fingerprints. Am J Physiol Endocrinol Metab. 2019;317:E374–E387. [DOI] [PubMed] [Google Scholar]
- 31. Preble JM, Pacak CA, Kondo H, et al. Rapid isolation and purification of mitochondria for transplantation by tissue dissociation and differential filtration. J Vis Exp. 2014;91:e51682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Huttanus HM, Vu T, Guruli G, et al. Raman chemometric urinalysis (Rametrix) as a screen for bladder cancer. PLoS One. 2020;15:e0237070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Senger RS, Kavuru V, Sullivan M, et al. Spectral characteristics of urine specimens from healthy human volunteers analyzed using Raman chemometric urinalysis (Rametrix). PLoS One. 2019;14:e0222115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Senger RS, Sullivan M, Gouldin A, et al. Spectral characteristics of urine from patients with end-stage kidney disease analyzed using Raman Chemometric Urinalysis (Rametrix). PLoS One. 2020;15:e0227281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu J, Sun J, Huang X, et al. Goldindec: A novel algorithm for raman spectrum baseline correction. Appl Spectrosc. 2015;69:834–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Senger RS, Robertson JL. The Rametrix™ PRO Toolbox v1.0 for MATLAB®. PeerJ. 2020;8:e8179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tanniche I, Collakova E, Denbow C, et al. Characterizing glucose, illumination, and nitrogen-deprivation phenotypes of Synechocystis PCC6803 with Raman spectroscopy. PeerJ. 2020;8:e8585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Boyarsky BJ, Jackson KR, Kernodle AB, et al. Estimating the potential pool of uncontrolled DCD donors in the United States. Am J Transplant. 2020;20:2842–2846. [DOI] [PubMed] [Google Scholar]
- 39. Brasile L, Henry N, Orlando G, et al. Potentiating renal regeneration using mesenchymal stem cells. Transplantation. 2019;103:307–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Farney AC, Hines MH, al-Geizawi S, et al. Lessons learned from a single center’s experience with 134 donation after cardiac death donor kidney transplants. J Am Coll Surg. 2011;212:440–453. [DOI] [PubMed] [Google Scholar]
- 41. Garner M, Jay CL, Sharda B, et al. Long-term outcomes of kidney transplantation from deceased donors with terminal acute kidney injury: Single center experience and literature review. Clin Transplant. 2023;37:e14886. [DOI] [PubMed] [Google Scholar]
- 42. Harriman DI, Kazakov H, Rogers J, et al. Does prolonged cold ischemia affect outcomes in donation after cardiac death donor kidney transplants? Clin Transplant. 2022;36:e14628. [DOI] [PubMed] [Google Scholar]
- 43. Moore PS, Farney AC, Hartmann EL, et al. Experience with deceased donor kidney transplantation in 114 patients over age 60. Surgery. 2007;142:514–523 e5232. [DOI] [PubMed] [Google Scholar]
- 44. Moore PS, Farney AC, Sundberg AK, et al. Dual kidney transplantation: a case-control comparison with single kidney transplantation from standard and expanded criteria donors. Transplantation. 2007;83:1551–1556. [DOI] [PubMed] [Google Scholar]
- 45. Khan MA, El-Hennawy H, Farney AC, et al. Analysis of local versus imported expanded criteria donor kidneys: a single-center experience with 497 ECD kidney transplants. Clin Transplant. 2017;31. 10.1111/ctr.13029 [DOI] [PubMed] [Google Scholar]
- 46. Rogers J, Farney AC, Orlando G, et al. Dual kidney transplantation from donors at the extremes of age. J Am Coll Surg. 2019;228:690–705. [DOI] [PubMed] [Google Scholar]
- 47. Stratta RJ, Farney AC, Orlando G, et al. Dual kidney transplants from adult marginal donors successfully expand the limited deceased donor organ pool. Clin Transplant. 2016;30:380–392. [DOI] [PubMed] [Google Scholar]
- 48. Stratta RJ, Harriman D, Gurram V, et al. The use of marginal kidneys in dual kidney transplantation to expand kidney graft utilization. Curr Opin Organ Transplant. 2022;27:75–85. [DOI] [PubMed] [Google Scholar]
- 49. Stratta RJ, Harriman D, Gurram V, et al. Dual kidney transplants from adult marginal donors: Review and perspective. Clin Transplant. 2022;36:e14566. [DOI] [PubMed] [Google Scholar]
- 50. Stratta RJ, Moore PS, Farney AC, et al. Influence of pulsatile perfusion preservation on outcomes in kidney transplantation from expanded criteria donors. J Am Coll Surg. 2007;204:873–884. [DOI] [PubMed] [Google Scholar]
- 51. Stratta RJ, Rohr MS, Sundberg AK, et al. Intermediate-term outcomes with expanded criteria deceased donors in kidney transplantation: a spectrum or specter of quality? Ann Surg. 2006;243:594–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Stratta RJ, Sundberg AK, Rohr MS, et al. Optimal use of older donors and recipients in kidney transplantation. Surgery. 2006;139:324–333. [DOI] [PubMed] [Google Scholar]
- 53. Farney AC, Singh RP, Hines MH, et al. Experience in renal and extrarenal transplantation with donation after cardiac death donors with selective use of extracorporeal support. J Am Coll Surg. 2008;206:1028–1037. [DOI] [PubMed] [Google Scholar]
- 54. Orlando G, Khan MA, El-Hennawy H, et al. Is prolonged cold ischemia a contraindication to using kidneys from acute kidney injury donors? Clin Transplant. 2018;32:e13185. [DOI] [PubMed] [Google Scholar]
- 55. Zuckerman JM, Singh RP, Farney AC, et al. Single center experience transplanting kidneys from deceased donors with terminal acute renal failure. Surgery. 2009;146:686–695. [DOI] [PubMed] [Google Scholar]
- 56. Zagni M, Croci GA, Cannavò A, et al. Histological evaluation of ischemic alterations in donors after cardiac death: a useful tool to predict post-transplant renal function. Clin Transplant. 2022;36:e14622. [DOI] [PubMed] [Google Scholar]
- 57. Senger RS, Sayed Issa A, Agnor B, et al. Disease-associated multimolecular signature in the urine of patients with lyme disease detected using raman spectroscopy and chemometrics. Appl Spectrosc. 2022;76:284–299. [DOI] [PubMed] [Google Scholar]
- 58. Carswell W, Robertson JL, Senger RS. Raman spectroscopic detection and quantification of macro- and microhematuria in human urine. Appl Spectrosc. 2022;76:273–283. [DOI] [PubMed] [Google Scholar]
- 59. Manerba M, Govoni M, Manet I, et al. Metabolic activation triggered by cAMP in MCF-7 cells generates lethal vulnerability to combined oxamate/etomoxir. Biochim Biophys Acta Gen Subj. 2019;1863:1177–1186. [DOI] [PubMed] [Google Scholar]
- 60. Than TA, Lou H, Ji C, et al. Role of cAMP-responsive element-binding protein (CREB)-regulated transcription coactivator 3 (CRTC3) in the initiation of mitochondrial biogenesis and stress response in liver cells. J Biol Chem. 2011;286:22047–22054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Weinberg JM. Mitochondrial biogenesis in kidney disease. J Am Soc Nephrol. 2011;22:431–436. [DOI] [PubMed] [Google Scholar]


