Abstract
We determined and compared the method detection limits (MDLα) of a PCR and an immunofluorescence assay (IFA) for detection of Cryptosporidium parvum oocysts in soils. Based on the MDLα and the quantitative nature and stability of the IFA, PCR analysis is not a useful screening step for soil studies of oocyst transport.
Methods for detecting oocysts of Cryptosporidium parvum in soils have been applied, but not thoroughly evaluated, for plot scale studies of transport in runoff (3, 9). Methods must be stable and sensitive to produce credible information (especially when applying principles of mass balance to evaluate partitioning). Two methods, PCR and immunofluorescence assay (IFA), have been discussed in research focused on developing PCR for water sampling (8, 10). PCR sensitivity has been characterized by the lowest number of oocysts added to experimental samples that led to amplification. However, sensitivity has a rigorous definition when methods are applied for environmental investigations. The method detection limit (MDL) describes the reproducibility of results by using a complete sample processing and analytic protocol (4). It is determined from replicate analyses and is based upon statistical analyses which incorporate an acceptable level of risk (α) of false-positive or false-negative results (4).
Comparison of the MDLs of PCR and IFA is complicated by the different types of data obtained from the two types of analysis. Nonquantitative PCR yields dichotomous, categorical results (presence or absence of amplifiable DNA), and IFA yields interval data (numbers of oocysts present).
Inhibitory substances in sediments (such as humic acids [10]) affect PCR performance, leading to false-negative PCR results, as indicated by positive IFA results (8, 10). DNA may not be replicated in some or all aliquots from a single sample. By assuming that the expected proportion of successful amplifications (π) is a function of the concentrations of amplifiable DNA and interfering compounds, the MDL for PCR can be estimated in terms of the minimum number of oocysts that produce at least one positive result in a batch of aliquots. By using data obtained from trials with well-mixed, fixed masses of the soil that would be used for transport studies, the proportion can be modeled by logistic regression (2) to determine the MDL (confidence level α [MDLα]).
Soil characterization and seeding.
All experiments were performed with 1.00 g of oocyst-free Collamer silt loam (by weight, 13.4% sand, 64% silt, and 22.6% clay). The soil was dried and sieved to 2.36 mm and had the following characteristics: pH 5.61 (in water), 3.89% organic matter, 2.89% total carbon, 2.89% organic carbon, and a cation-exchange capacity (determined with NH4Cl) of 18.4 cmol/kg. Oocysts were obtained from the feces of naturally infected calves and stored as described by Jenkins et al. (7). Samples were processed immediately after seeding at levels reported in Tables 1 and 2.
TABLE 1.
Results of replicate PCR trials
| No. of oocysts/g of soil | No. of replicate sample aliquots amplified | Proportion successful |
|---|---|---|
| 0 | 9 | 0.00 |
| 40 | 10 | 0.40 |
| 45 | 10 | 0.40 |
| 50 | 5 | 0.40 |
| 68 | 7 | 0.57 |
| 90 | 7 | 0.57 |
| 113 | 10 | 0.70 |
| 150 | 10 | 0.80 |
| 200 | 5 | 1.00 |
| 500 | 5 | 1.00 |
TABLE 2.
Results of replicate IFA trials
| Estimated no. of oocysts/g of soil | No. of trials conducted | Estimated avg no. of oocysts recovered/g of soil |
|---|---|---|
| 240 | 5 | 120 |
| 460 | 5 | 150 |
| 1,030 | 5 | 500 |
| 2,360 | 5 | 800 |
| 4,640 | 5 | 1,720 |
| 10,300 | 5 | 3,520 |
Recovery of oocysts from soils.
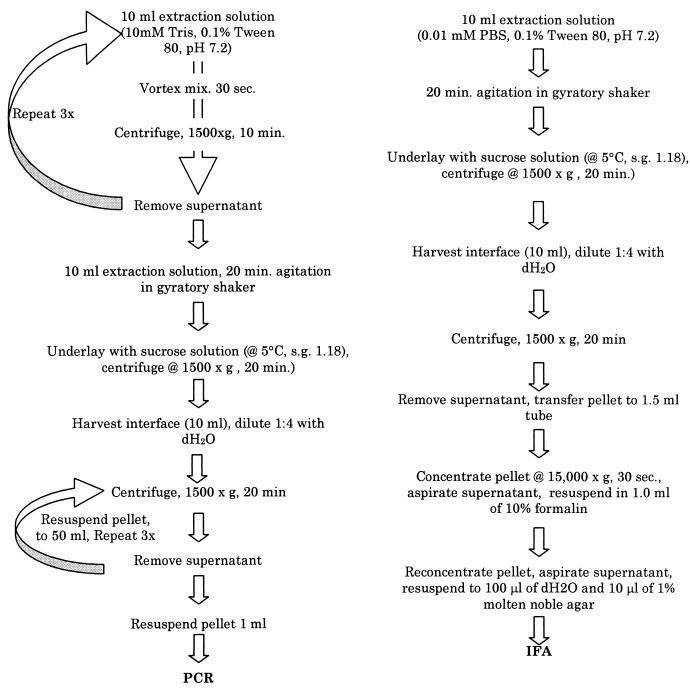
Oocysts were recovered from soil samples by using procedures adapted from Mawdsley et al. (9) (Fig. 1). The extraction procedure for PCR was intensive to provide highly purified extracts with little interfering debris.
FIG. 1.
Extraction and concentration procedures used for seeded soil samples for PCR and IFA analysis. dH2O, distilled water; s.g., specific gravity; PBS, phosphate-buffered saline.
Detection of oocysts by IFA.
Four 10-μl aliquots of sample concentrate were withdrawn and spread within 1.0-cm-diameter wells, two per slide. After desiccation (24 h, 20°C), smears were incubated for 25 min with 30-μl volumes of monoclonal and secondary antibody solutions (diluted 1:10 in 0.01 mM phosphate-buffered saline) (EnSys, Inc., Research Triangle Park, N.C.). Between incubations, the smear was rinsed twice with 50 μl of 0.01 mM phosphate-buffered saline, which was removed by vacuum aspiration from the edge of each well. The stained smear was covered with 15 μl of fluorescence preservative-mounting fluid and a 22- by 22-mm coverslip, sealed to the slide with mounting cement. The smear was examined by using a Zeiss LSM-210 microscope with a 63×/1.4 plan-neofluar objective. Nodes of a grid formed by x and y separations of 500 μm (196 in total) were examined as described by Anguish and Ghiorse (1). Fluorescing objects of the appropriate size and shape were further examined by differential interference contrast microscopy. Results included confirmed and presumed oocysts (as defined for water sample analysis [12]). Negative controls, kit controls, and seeding solutions of oocysts were included for quality assurance-quality control.
DNA extraction and purification.
We added 300 μl of 100 mM NaPO4 (pH 8.0), 300 μl of lysis buffer (100 mM NaCl, 500 mM Tris [pH 8.0], 10% sodium dodecyl sulfate), and 300 μl of phenol (equilibrated, pH 7.8) to 2-ml microcentrifuge tubes containing 2.5 g of sterilized 0.1-mm-diameter zirconium beads (BioSpec Products, Bartlesville, Okla.) and sample concentrate. The mixture was homogenized by bead mill and centrifuged, and the phenol and aqueous phases were collected. After collection of a second rinse of 300 μl of distilled H2O, the extracts were concentrated by using butanol and SpinBind DNA extraction (FMC BioProducts, Rockland, Maine) in accordance with the manufacturer’s instructions.
PCR amplification.
PCRs were prepared by standard methods (6). The PCR mixture consisted of 5 μl of buffer (10 mM Tris [pH 8.8], 50 mM KCl; bovine serum albumin at 1 mg/ml, 0.5% Tween 20, 15 mM MgCl2), 1 μl each of the forward and reverse primers (20 μM), 0.5 μl (1 U) of Taq DNA polymerase (Promega, Madison, Wis.), and 29.5 μl of distilled H2O per 10-μl aliquot of DNA extract, overlaid with 50 μl of sterile mineral oil. The primer pair corresponded to nucleotides 601 to 621 and 1015 to 1035 of the C. parvum and C. muris 18S rRNA gene (GenBank accession no. L16996) (Integrated DNA Technologies, Inc., Coralville, Iowa). The tubes were heated to 80°C, and 2.5 μl of 1:1:1:1 solution of 100 μM dATP, dGTP, dCTP, and dTTP was added. Amplification consisted of initial denaturation (30 s, 98°C); 35 cycles of annealing (30 s, 55°C), extension (60 s, 74°C), and denaturation (30 s, 94°C); and final extension (10 min, 74°C). Negative and positive controls (1 ng of purified oocyst DNA) were included. The amplified product was separated by electrophoresis using 1.5% agarose gels in 1× TBE buffer (90 mM Tris-HCl, 90 mM boric acid, 2 mM Na-EDTA) at 5 V/cm.
Determination of MDLα for PCR and IFA.
We used logistic regression to model the PCR response and linear regression to model the IFA response (Minitab, release 11; Minitab Inc., State College, Pa.). We inverted prediction intervals about concentrations representing no response at various confidence levels (α) by using the t and χ2 distributions for the linear and logistic models, respectively (5, 11).
Results of trials.
Trial results are reported in Tables 1 and 2. The regression models of IFA (equation 1) and PCR (equation 2) performance were highly significant (P ≪ 0.001):
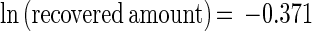 |
1 |
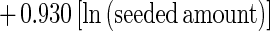 |
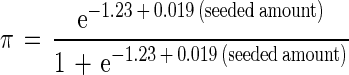 |
2 |
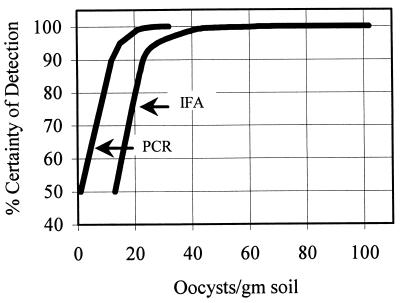
Table 3 and Fig. 2 present estimated MDLαs for PCR and IFA as the percent certainty of detection given the presence of oocysts at specific concentrations.
TABLE 3.
Estimated percent certainty of detection at specific concentrations of oocysts per gram of soil for IFA and PCR
| Certainty of detection (%) | No. of oocysts/g detected
|
|
|---|---|---|
| IFA | PCR | |
| 50 | 13 | 1a |
| 90 | 23 | 12 |
| 95 | 28 | 15 |
| 99 | 41 | 21 |
| 99.5 | 46 | 23 |
| 99.9 | 65 | 27 |
| 99.95 | 76 | 29 |
| 99.99 | 102 | 32 |
The estimate was a fraction of an oocyst—the value presented is a rounding up of the original estimate.
FIG. 2.
Graphical comparison of MDLs at various certainties of detection given the presence of oocysts in soil samples.
Average recovery from soils with IFA.
Recovery from 30 trials with seeded soils by IFA averaged 43% ± 5.7% (95% confidence interval).
Comparison of MDLαs for IFA and PCR applied for soil analysis.
IFA yields quantitative estimates of the number oocysts per gram of soil. The recovery efficiency is stable across orders of magnitude of seeding levels. A coefficient could be used to estimate the actual number of oocysts present from observed amounts (e.g., observed number/0.43 ≈ actual number present), which would be useful for applying the principle of mass balance in plot studies of transport. A substantial difference between the MDLαs of PCR and IFA would suggest that PCR should be used to screen soil samples prior to applying IFA for quantification. Although PCR has an MDLα consistently lower than that of IFA, the difference is not sufficient to recommend qualitative screening for plot studies because of the small return of information from the expense and effort.
Acknowledgments
This work was supported in part by funds provided by the USDA Competitive Research Grants Program (award 93-37102-8958) and the New York City Department of Environmental Protection through the Watershed Agricultural Program for Watershed Protection.
We are grateful to D. Bowman for providing oocysts for experimental work and to L. Anguish for laboratory support.
REFERENCES
- 1.Anguish L, Ghiorse W. Computer-assisted laser scanning and video microscopy for analysis of Cryptosporidium parvum oocysts in soil, sediment and feces. Appl Environ Microbiol. 1997;63:724–733. doi: 10.1128/aem.63.2.724-733.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown P J. Measurement, regression and calibration. 1st ed. Oxford statistical series. Vol. 12. New York, N.Y: Clarendon Press; 1993. [Google Scholar]
- 3.Brush C F. Ph.D. thesis. Ithaca, N.Y: Cornell University; 1997. [Google Scholar]
- 4.Clesceri L, Greenberg A, Trussel R, editors. Standard methods for the examination of water and wastewater. 17th ed. Section 1030E. Method detection limit. Washington, D.C: American Public Health Association; 1989. [Google Scholar]
- 5.Hauck W W. A note on confidence bands for the logistic response curve. Am Statistician. 1983;37:158–160. [Google Scholar]
- 6.Innis M A, Gelfand D H. Optimization of PCRs. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols: a guide to methods and applications. San Diego, Calif: Academic Press, Inc.; 1990. pp. 3–12. [Google Scholar]
- 7.Jenkins M B, Anguish L J, Bowman D D, Walker M J, Ghiorse W C. Assessment of a dye permeability assay for determination of inactivation rates of Cryptosporidium parvum oocysts. Appl Environ Microbiol. 1997;63:3844–3850. doi: 10.1128/aem.63.10.3844-3850.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson D W, Pieniazek N J, Griffin D W, Misener L, Rose J B. Development of a PCR protocol for sensitive detection of Cryptosporidium oocysts in water samples. Appl Environ Microbiol. 1995;61:3849–3855. doi: 10.1128/aem.61.11.3849-3855.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mawdsley J L, Brooks A E, Merry R J. Movement of the protozoan pathogen Cryptosporidium parvum through three contrasting soil types. Biol Fertil Soils. 1996;21:30–36. [Google Scholar]
- 10.Mayer C, Palmer C. Evaluation of PCR, nested PCR, and fluorescent antibodies for detection of Giardia and Cryptosporidium species in wastewater. Appl Environ Microbiol. 1996;62:2081–2085. doi: 10.1128/aem.62.6.2081-2085.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neter J, Wasserman W, Kutner M H. Applied linear statistical models. 3rd ed. Homewood, Ill: Richard D. Irwin, Inc.; 1990. [Google Scholar]
- 12.U.S. Environmental Protection Agency. National primary drinking water regulations: monitoring requirements for public drinking water supplied: Cryptosporidium, Giardia, viruses, disinfection byproducts, water treatment plant data and other information requirements. Fed Regist. 1994;59:6332–6444. [Google Scholar]