Abstract
Cyclospora cayetanensis causes diarrheal disease worldwide without a confirmed mode of transmission. Wastewater was examined for the presence of this organism. Oocysts were detected microscopically, and their identity was confirmed by molecular techniques. These findings verify that current techniques can isolate Cyclospora oocysts and suggest that fecally contaminated water may act as a vehicle of transmission.
Cyclospora cayetanensis has emerged as an important cause of acute and chronic gastroenteritis worldwide (2). The environmentally resistant form of the organism, an oocyst, is shed in feces and subsequently sporulates to the infectious form after 7 to 12 days of incubation at ambient temperature (2, 5, 6). While the mode of transmission has not been entirely elucidated, C. cayetanensis oocysts have been detected on vegetables sold in the San Juan of Miraflores shantytown (Lima, Peru), where cyclosporiasis is endemic (5a), and consumption of Guatemalan raspberries has been epidemiologically linked with recent C. cayetanensis outbreaks in the United States (2). Although highly suspected, cyclosporiasis via contaminated water has not been confirmed (2).
This study was conducted from March to June 1997 in the San Juan of Miraflores shantytown. Eleven water samples were collected from the primary oxidation lagoon and analyzed for C. cayetanensis by fluorescent microscopy and PCR specific for C. cayetanensis.
Eight water samples, ranging in volume from 1 to 5 liters, were collected with the Envirochek capsule (Gelman Sciences, Ann Arbor, Mich.). Particulate matter was eluted by hand with 100 ml of eluting solution and concentrated in accordance with the manufacturer’s recommendations (Gelman Sciences). The eluting solution consisted of 1% sodium dodecyl sulfate, 1% Tween 80, NaCl, KH2PO4, Na2HPO4 · 12H2O, KCl, and Antifoam A (Sigma Chemical Co.). Elution by hand consisted of horizontal agitation for 10 min, releasing trapped particles. Suspended particulate matter was decanted into a 250-ml centrifuge bottle and centrifuged at 1,050 × g for 10 min. Two additional washings were performed, with rotation of the capsule 90° each time. The centrifuged particulate matter was added together as a final centrifugate.
Three samples, 378 liters each, were collected with the Hannifin polypropylene cartridge filter (Commercial Filters Parker Hannifin Corp., Lebanon, Ind.). These samples were eluted by hand according to standard procedures (1). The woven filter was sliced to the core, and the fibers were divided into six equal portions. Fibers were successively washed in three 1-liter volumes of eluting solution, with wringing of the fibers between containers. The eluate suspensions were combined, and a 100-ml aliquot was centrifuged at 1,050 × g for 10 min.
All centrifugates were resuspended in a 2.5% potassium dichromate solution. A centrifugate between 0.1 and 0.5 ml was mixed with zinc sulfate (specific gravity, 1.2) for selective centrifugation for 1 min at 600 × g. Particles with a specific gravity of less than 1.2 were harvested for microscopic examination. Wet mount preparations were scanned at a magnification of ×200 with UV epifluorescence (excitation filter, 355 to 425 nm; dichroic beam-splitting mirror, 455 nm; and suppression filter, 470 nm). Objects that autofluoresed and had the same size and shape as C. cayetanensis oocysts were examined at a magnification of ×1,000 with phase-contrast microscopy for confirmation. All samples were subjected to a nested PCR protocol amplifying a 294-bp portion of the 18S rDNA segment (7). The PCR protocol included a 50-μl subsample from a 1-ml concentrated water sample suspension in potassium dichromate. The subsample was washed four times with 1× PCR buffer by centrifugation at 14,000 × g for 3 min. The packed pellet was resuspended in 25 μl of 1× PCR buffer (The 1× PCR buffer is made originally as a 10× PCR buffer, and from this a 1× buffer was made by a 1:10 dilution with double-distilled sterile water. The 10× buffer contains the following components: 500 mM KCl, 100 mM Tris-HCl [pH 9.0, 25°C], 1% Triton X-100 [Fisher, Petersberg, Pa.], and 15 mM MgCl2.) and 25 μl of 6% resin matrix (Instagene; Biorad, Hercules, Calif.). The resin matrix removes inhibitors from the DNA preparation suspension. The suspension was subjected to seven freeze-thaw cycles of 2 min in a dry ice-ethanol bath followed by 2 min in a 98°C water bath (4). The suspension was vortexed and centrifuged at 14,000 × g for 3 min. Twenty-five microliters of supernatant was transferred to a microcentrifuge tube for PCR amplification. The PCR assay and primers were as described by Yoder et al. (7) except that the annealing temperature of 52°C was changed to 50°C and a cytosine replaced an adenosine at position 14 in CYC3FE. Amplification products were confirmed by restriction fragment length polymorphisms (RFLP) with one unit of the restriction endonuclease MnlI (Amersham Life Sciences Inc., Arlington Heights, Ill.) (3). The predicted restriction fragment sizes are 140 bp, 106 bp, and 48 bp for C. cayetanensis and 127 bp, 106 bp, and 62 bp for Eimeria spp. Microscopically confirmed C. cayetanensis positive control oocysts were obtained from Universidad Peruana Cayetano Heredia. C. cayetanensis DNA was liberated by the simple freeze-thaw technique and purified by phenol-chloroform extraction. The Eimeria sp. positive control of avian origin was obtained from Ynes Ortega. Eimeria DNA was obtained by the same protocol as for Cyclospora spp. and amplified by the nested PCR assay. Ten microliters was analyzed for amplified product by electrophoresis in a 1.2% agarose gel stained with ethidium bromide. Restriction endonuclease digests were analyzed in a 4% MetaPhor agarose gel (FMC BioProducts, Rockland, Maine) stained with ethidium bromide.
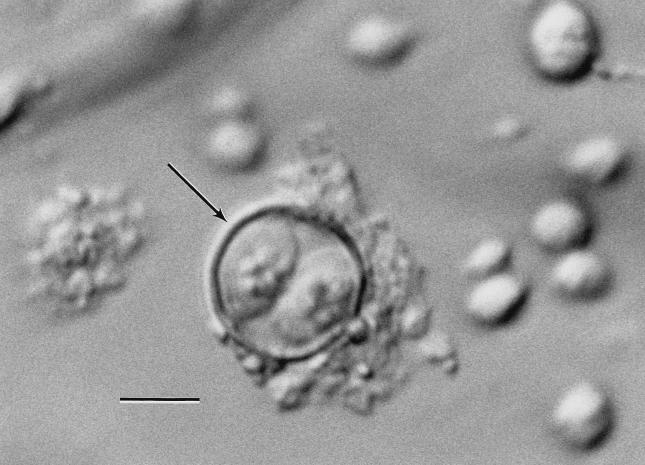
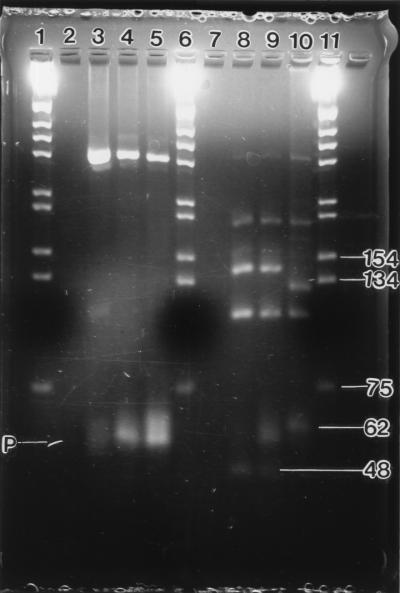
Unsporulated oocysts were observed (1 oocyst per liter) in four of the eight samples collected with the Envirochek capsule and in all three samples collected with the Hannifin filter. On direct examination, 2 of the 11 samples contained sporulated oocysts (Fig. 1). Following a 2-week incubation period at ambient temperature, sporulated oocysts were detected in one additional sample. Amplified product in one microscopy-positive sample was confirmed as C. cayetanensis by RFLP fragment digestion (Fig. 2). The PCR detection limit was determined to be 10 oocysts per inoculated fecal sample (unpublished results).
FIG. 1.
Photomicrograph of C. cayetanensis in wastewater by Nomarski interference-contrast microscopy; bar = 5 μm. The arrow indicates an oocyst with two sporocysts.
FIG. 2.
C. cayetanensis and Eimeria amplified products from nested PCR (lanes 2 to 5) and RFLP fragments from MnlI digestion of those PCR amplified products (lanes 7 to 10). Lanes 1, 6, and 11, molecular size standards; lanes 2 and 7, negative control; lanes 3 and 8, Universidad Peruana Cayetano Heredia C. cayetanensis positive control; lanes 4 and 9, C. cayetanensis from wastewater; lanes 5 and 10, Eimeria spp. (U of A) positive control. Primers migrated to point P.
This is the first report to confirm the detection of oocysts of C. cayetanensis in wastewater. Contact with C. cayetanensis-contaminated water at some point may be the source of the oocysts detected on vegetables in Peru and the epidemiological link with raspberries from Guatemala. Our findings provide evidence that C. cayetanensis has the potential to be transmitted via contamination of drinking or irrigation water with wastewater.
Acknowledgments
We are indebted to Field Willingham and Carrie Hancock for their assistance during this study.
REFERENCES
- 1.American Public Health Association. Standard methods for the examination of water and wastewater. 19th ed. Washington, D.C: American Public Health Association; 1995. Immunofluorescence method for Giardia and Cryptosporidium spp. (PROPOSED) pp. 9–111. [Google Scholar]
- 2.Herwaldt B L, Ackers M-L the Cyclospora Working Group. An outbreak in 1996 of cyclosporiasis associated with imported raspberries. N Engl J Med. 1997;336:1548–1556. doi: 10.1056/NEJM199705293362202. [DOI] [PubMed] [Google Scholar]
- 3.Jinneman K C, Wetherington J H, Adams A M, Johnson J M, Tenge B J, Dang N-L, Hill W E. Differentiation of Cyclospora sp. and Eimeria spp. by using the polymerase chain reaction amplification products and restriction fragment length polymorphisms. Laboratory Information Bulletin No. 4044. U.S. Washington, D.C: Food and Drug Administration; 1996. [Google Scholar]
- 4.Johnson D W, Pieniazek N J, Griffin D W, Misener L, Rose J B. Development of a PCR protocol for sensitive detection of Cryptosporidium oocysts in water samples. Appl Environ Microbiol. 1995;61:3849–3855. doi: 10.1128/aem.61.11.3849-3855.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ortega Y R, Gilman R H, Sterling C R. A new coccidian parasite (Apicomplexa: Eimeriidae) from humans. J Parasitol. 1993;80:625–629. [PubMed] [Google Scholar]
- 5a.Ortega Y R, Roxas C R, Gilman R H, Miller N, Cabrera L, Taquiri C, Sterling C. Isolation of Cryptosporidium parvum and Cyclospora cayetanensis from vegetables collected in markets of an endemic region in Peru. Am J Trop Med Hyg. 1997;57:683–686. doi: 10.4269/ajtmh.1997.57.683. [DOI] [PubMed] [Google Scholar]
- 6.Ortega Y R, Sterling C R, Gilman R H, Cama V A, Diaz F. Cyclospora species—a new protozoan pathogen of humans. N Engl J Med. 1993;328:1308–1312. doi: 10.1056/NEJM199305063281804. [DOI] [PubMed] [Google Scholar]
- 7.Yoder K E, Sethabutr O, Relman D A. PCR-based detection of the intestinal pathogen Cyclospora. In: Persing D H, editor. PCR protocols for emerging infectious diseases, a supplement to Diagnostic molecular microbiology: principles and applications. Washington, D.C: ASM Press; 1996. pp. 169–176. [Google Scholar]