Visual Abstract
Keywords: apolipoprotein L1 (APOL1), CKD, ESKD
Abstract
Significance Statement
African Americans are at increased risk of CKD in part due to high-risk (HR) variants in the apolipoprotein L1 (APOL1) gene, termed G1/G2. A different APOL1 variant, p.N264K, reduced the risk of CKD and ESKD among carriers of APOL1 HR variants to levels comparable with individuals with APOL1 low-risk variants in an analysis of 121,492 participants of African ancestry from the Million Veteran Program (MVP). Functional genetic studies in cell models showed that APOL1 p.N264K blocked APOL1 pore-forming function and ion channel conduction and reduced toxicity of APOL1 HR mutations. Pharmacologic inhibitors that mimic this mutation blocking APOL1-mediated pore formation may be able to prevent and/or treat APOL1-associated kidney disease.
Background
African Americans are at increased risk for nondiabetic CKD in part due to HR variants in the APOL1 gene.
Methods
We tested whether a different APOL1 variant, p.N264K, modified the association between APOL1 HR genotypes (two copies of G1/G2) and CKD in a cross-sectional analysis of 121,492 participants of African ancestry from the MVP. We replicated our findings in the Vanderbilt University Biobank (n=14,386) and National Institutes of Health All of Us (n=14,704). Primary outcome was CKD and secondary outcome was ESKD among nondiabetic patients. Primary analysis compared APOL1 HR genotypes with and without p.N264K. Secondary analyses included APOL1 low-risk genotypes and tested for interaction. In MVP, we performed sequential logistic regression models adjusting for demographics, comorbidities, medications, and ten principal components of ancestry. Functional genomic studies expressed APOL1 HR variants with and without APOL1 p.N264K in cell models.
Results
In the MVP cohort, 15,604 (12.8%) had two APOL1 HR variants, of which 582 (0.5%) also had APOL1 p.N264K. In MVP, 18,831 (15%) had CKD, 4177 (3%) had ESKD, and 34% had diabetes. MVP APOL1 HR, without p.N264K, was associated with increased odds of CKD (odds ratio [OR], 1.72; 95% confidence interval [CI], 1.60 to 1.85) and ESKD (OR, 3.94; 95% CI, 3.52 to 4.41). In MVP, APOL1 p.N264K mitigated the renal risk of APOL1 HR, in CKD (OR, 0.43; 95% CI, 0.28 to 0.65) and ESKD (OR, 0.19; CI 0.07 to 0.51). In the replication cohorts meta-analysis, APOL1 p.N264K mitigated the renal risk of APOL1 HR in CKD (OR, 0.40; 95% CI, 0.18 to 0.92) and ESKD (OR, 0.19; 95% CI, 0.05 to 0.79). In the mechanistic studies, APOL1 p.N264K blocked APOL1 pore-forming function and ion channel conduction and reduced toxicity of APOL1 HR variants.
Conclusions
APOL1 p.N264K is associated with reduced risk of CKD and ESKD among carriers of APOL1 HR to levels comparable with individuals with APOL1 low-risk genotypes.
Introduction
In the United States, African Americans have a four-fold increase in the risk of ESKD.1–3 Although some of this difference can be attributed to socioeconomic factors and other comorbidities, previous studies have also identified genetic variants in apolipoprotein L1 (APOL1) that significantly increase the risk of nondiabetic ESKD.4–6
The two APOL1 high-risk (HR) variants (termed G1 and G2) are common in individuals of West African descent as they confer resistance to lethal Trypanosoma brucei Rhodesiense infections, which is a pathogen that was endemic to West Africa and remains endemic to other areas of Africa.7,8 APOL1 is a circulating component of the innate immune system that protects against parasitic infection by forming nonselective cation channels in the plasma membrane leading to colloid osmotic swelling and eventual parasite lysis. Although just one copy of APOL1 G1 or G2 confers protection against human African trypanosomiasis, two copies increased the risk of kidney disease.5
Although the genetic link between APOL1 and kidney disease is well-established, the mechanism by which APOL1 induces kidney injury remains uncertain.4,6 The expression of APOL1 HR variants in kidney cells is toxic.9 In vitro studies have implicated multiple mechanisms in APOL1-mediated cell death, including cation influx through the APOL1 pore-forming function,10 mitochondrial dysfunction,11 activation of the inflammasome,12,13 and effects on the related protein APOL3.14 Uncertainty regarding the mechanism by which APOL1 induces toxicity to kidney cells has limited the development of therapies to treat patients with APOL1-mediated kidney disease (AMKD).
Several other rare coding variants in APOL115–18 have been proposed to modify the renal toxicity of APOL1 HR variants.15–17 One of these coding variants was described in a case report of a Ghanaian patient with trypanosomiasis who had two APOL1 HR variants but also two copies of APOL1 p.N264K. In vitro testing showed that p.N264K reduced the trypanolytic activity of APOL1 and increased susceptibility to trypanosomal infection.17 These studies raise the possibility that variants within APOL1 may act as genetic modifiers of APOL1 G1 and G2 variants.
In this study, we evaluate the role of APOL1 p.N264K as a genetic modifier of CKD risk conferred by the presence of APOL1 risk variants G1/G2 in humans. Prior attempts to characterize the interaction of APOL1 p.N264K with APOL1 HR variants in humans have been unrevealing, as the combination of APOL1 p.N264K with the APOL1 HR variants is present in only approximately 0.5% of individuals of African ancestry.17 We leveraged an extremely large national cohort of US veterans, the Million Veteran Program (MVP), with electronic health-linked genomic data and two independent replication cohorts, the Vanderbilt University Biobank (BioVU) and the National Institutes of Health (NIH) All of Us program, to evaluate the effect of APOL1 p.N264K on kidney disease risk. We complemented these genetic epidemiology studies with cellular functional studies to elucidate the mechanism of action of the APOL1 p.N264K variant.
Methods
Study Population, Design, and Oversight
The primary cohort was participants enrolled in the Department of Veterans Affairs (VA) MVP.19 Electronic health record data were obtained from the VA Corporate Data Warehouse and MVP Study Mart.20 The study population consisted of participants 18 years and older, with genetic information available, of African ancestry, and who regularly received care at the VA for the 2 years before enrollment. All individuals with enough data in the electronic health record to assess kidney function were included to preserve generalizability and include all potential known and unknown second hits. Patients signed informed consent when enrolled in the biobank.19 The MVP received ethical and study protocol approval by the VA Central Institutional Review Board and the Tennessee Valley Healthcare System R&D.
Replication was performed in the Vanderbilt BioVU biobank and in the NIH All of Us study (dataset version v5). The study population for BioVU and All of Us consisted of self-described non-Hispanic Black participants with available genetic information (Supplemental Tables 1 and 2). Studies in BioVU have demonstrated a high degree of correlation between self-described and genetic ancestry.21 BioVU received ethical and study protocol approval by the Vanderbilt University Medical Center Institutional Review Board. The All of Us study was approved by the IRB of the NIH All of Us Research Program. This study was conducted in accordance with the Declaration of Helsinki.
APOL1 Genotype
APOL1 HR variants G1 (rs73885319 p.S342G; rs60910145 p.I384M), G2 (rs71785313, a six base pair deletion that removes amino acids N388 and Y389), and APOL1 p.N264K (rs73885316) were directly genotyped on DNA that was extracted from whole blood.22,23 Participants were defined as HR if they had two HR alleles: homozygotes for G1/G1, homozygotes for G2/G2, or compound heterozygotes for G1/G2. All other participants were characterized as low risk. G1, G2, and rs73885316 genotypes were in Hardy–Weinberg equilibrium and present at frequencies comparable with previous reports and in population databases.24–27 Haplotypes using known protein-altering coding variants in the APOL1 gene were generated, as well as the distribution of N264K genotypes on HR and low-risk APOL1 groups. Haplotypes were estimated and reported at a minor allele frequency threshold >0.001 where there is a higher degree of confidence (Supplemental Tables 10 and 11).28
Outcomes
In this study, our primary outcome was CKD and our secondary outcome was ESKD in patients without diabetes. CKD was defined as GFR <60 ml/min per 1.73 m2 on at least two occasions separated by at least 3 months with no normal in between and with persistent GFR to <60 ml/min per 1.73 m (2) until their last observation, to prevent capturing slowly recovering AKI as CKD.29 ESKD was defined as having two diagnostic or procedural codes related to kidney replacement therapy, GFR <15 ml/min per 1.73 m2 on at least two occasions separated by at least 3 months, or having a diagnostic or procedural code for kidney transplantation (Supplemental Tables 3 and 4).30 The kidney outcomes definitions were harmonized across all cohorts.
Covariates
Characteristics reported include demographics, comorbidities, laboratory tests, and medications. Determination of African ancestry was completed by the MVP genetic core. In MVP, African ancestry assignment is derived from projecting the genetic principal components of MVP to those in the 1000 Genomes Project reference panel.22,23 Baseline covariates were assessed 2 years before CKD or ESKD. All covariate definitions are provided in Supplemental Table 4. GFR was creatinine-based and was calculated using the CKD Epidemiology Collaboration Equation.31
Statistical Analyses
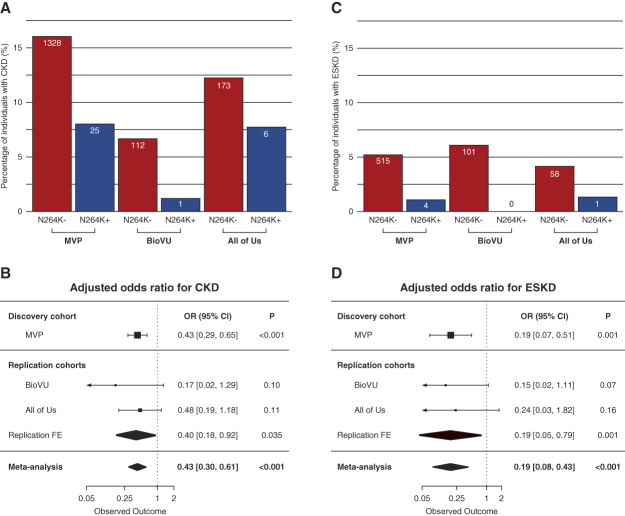
All prespecified primary analyses were restricted to individuals without diabetes. The primary analyses were conducted in APOL1 HR carriers. The reference group was participants with APOL1 HR variant without p.N264K, and we assessed the reduction in risk conferred by carrying p.N264K (Figure 1). We conducted logistic regression to evaluate the effect of APOL1 p.N264K on CKD (primary outcome) or ESKD (secondary outcome) in the APOL1 HR group. This analysis was adjusted for age, sex, and ten principal components of ancestry. The effects in our replication cohorts were combined in a meta-analysis (Figure 1).
Figure 1.
Association of APOL1 HR (homozygous for G1 or G2 or compound heterozygous for G1/G2) with and without p.N264K allele with CKD and ESKD among MVP participants, BioVU and the All of Us. APOL1, apolipoprotein L1; BioVU, Vanderbilt University Biobank; CI, confidence interval; FE, Fixed effect; HR, high risk; MVP, Million Veteran Program; OR, odds ratio.
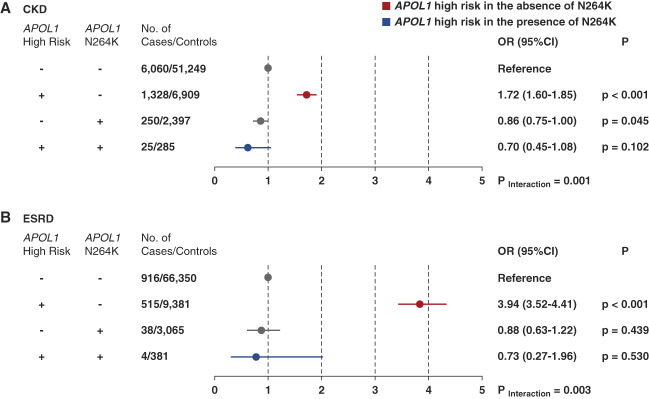
The secondary analysis was conducted only in the discovery cohort. In this analysis, we treated carriers of low-risk APOL1 genotypes as a reference group and formally tested for an additive interaction between APOL1 HR variants and p.N264K (Figure 2 and Supplemental Tables 5 and 6). To evaluate the modifying effect of the presence of APOL1 p.N264K, we performed an interaction analysis using an additive scale between APOL1 HR status and APOL1 p.N264K (reference group: APOL1 low-risk group without the APOL1 p.N264K variant).32 Sequential multivariable models were constructed. Model 1 was adjusted for age, sex, and ten principal ancestry components; model 2 was further adjusted for body mass index and renin/angiotensin/aldosterone inhibition (angiotensin-converting enzyme inhibitors or angiotensin II receptor blockers); while model 3 was further adjusted for hypertension; and model 4 adjusted for cancer (Supplemental Tables 5 and 6). Mendelian monogenic forms that were identified included autosomal dominant polycystic kidney disease (ADPKD) present in 0.35% and Alport syndrome present in 0.035% of the participants. There are 424 patients with ADPKD (0.35%); 341 are APOL1 low-risk genotype, and 83 are APOL1 HR genotype. There were 17 N264K heterozygous in the APOL1 low-risk group, and only two are N264K+ heterozygous in the APOL1 HR group. Model 5 adjust for ADPKD. Exploratory analysis was performed in patients with diabetes in the MVP. All regression models satisfied the Hosmer–Lemeshow test for goodness of fit. A table of clinical characteristics for homozygous carriers of N264K compared with other groups is presented in Supplemental Table 7.
Figure 2.
The presence of an allele p.N264K in patients with APOL1 HR group (homozygous for G1 or G2 or compound heterozygous for G1/G2) mitigates the risk of CKD and ESKD among MVP participants to values similar to those of participants in the APOL1 low-risk group. All OR are relative to the reference and reported as OR (95% CI). Reference group: APOL1 low-risk genotype, and N264K− APOL1 HR refers to two copies of the APOL1 HR variants G1 or G2 or G1 and G2. APOL1 LR, 0 or 1 total copy of the G1 or G2 HR variant. N264K+, carrying 1 or 2 copies of APOL1 N264K. N264K−, carrying 0 copies of APOL1 N264K. Logistic regression was used to evaluate the association of APOL1 HR and p.N264K allele genotype and CKD (A) and ESKD (B). ORs were adjusted for age, sex, ten principal components of ancestry, BMI, hypertension, and renin–angiotensin–aldosterone system blockade. Minimally adjusted models are presented in Supplemental Table 2. BMI, body mass index.
Analyses using APOL1 low-risk N264K, as the reference, were also conducted in BioVU and All of US; however, interaction was not tested, given small sample size (Supplemental Tables 8 and 9). All statistical tests were two-sided, and P = 0.05 was considered statistically significant. All analyses were conducted using R version 3.6.1.
Podocyte Viability Assay
The human telomerase reverse transcriptase-immortalized kidney podocyte cell line was procured from the laboratory of Dr. Moin Saleem at the University of Bristol, the United Kingdom. Coding sequences of APOL1 G0, G1, and G2 with and without the p.N264K variant with a C-terminal HiBiT tag under the control of a doxycycline-inducible promoter were introduced by lentivirus, and stable pools were generated.
For the viability assay, cells of each genotype were incubated with a range of doxycycline concentrations for 96 hours and viability was assessed using the CellTiter-Glo reagent (Promega, G7570). Data are shown from a single concentration of doxycycline (500 ng/ml) that produced comparable APOL1 expression across the clonal cell lines as determined by the measurement of the HiBiT luminescence signal.
Calcium Uptake Assay in Human Embryonic Kidney Cells
Stable human embryonic kidney cell lines were generated with the genetically encoded calcium sensor GCaMP6 and doxycycline-inducible expression of APOL1 G2 with and without N264K. To measure APOL1-dependent calcium influx, the expression of APOL1 was induced with 50 ng/ml doxycycline for 6 hours. Cells were then exposed to varying concentrations of extracellular calcium, and GCaMP6 fluorescence was measured over 3 minutes.
All genetic engineering of APOL1 was performed on the background of the most common African haplotype (E150K, I384M, K255R) (Supplemental Summary 1), which has previously been associated with higher levels of cytotoxicity (Supplemental Table 10).9 More detailed methods are provided in the Supplemental Appendix.
Results
Cohort Characteristics
Among 121,492 MVP participants of African ancestry, 104,756 (86.2%) were men. The median age was 59 years (interquartile range, 51–66 years), and 40,842 (33.6%) had diabetes at enrollment. In total, 18,831 (15%) had CKD, and 4177 (3.4%) had ESKD. Consistent with previous reports in MVP and other cohorts, 12.8% (n=15,604) of the participants had two HR APOL1 gene variants24,25 and 5254 (4.3%) had at least one copy of the APOL1 p.N264K allele. The APOL1 p.N264K variant occurred most commonly on a haplotype in combination with the low-risk APOL1 G0 variant (n=3,627, 1.5%); however, a recombinant haplotype, containing p.N264K and the G2 risk allele, was observed (n=1,595, 0.6%) (Supplemental Table 10). Among MVP participants, 4672 individuals carried at least one p.N264K allele in combination with a low-risk APOL1 genotype, and 582 individuals carried at least one p.N264K allele in combination with a HR APOL1 genotype (n=381 G1/G2 and n=201 G2/G2) (Table 1 and Supplemental Table 11). A small group of homozygous p.N264K carriers was also identified (n=51 with low-risk APOL1 genotypes and n=3 with HR APOL1 genotype; Supplemental Table 7). There were no clinically notable differences in the baseline clinical characteristics across these genetic strata of participants (Table 1).
Table 1.
Demographics and clinical characteristics of Million Veteran Program cohort
| Characteristics | No APOL1 p.N264K Alleles | ≥1 APOL1 p.N264K Alleles | ||
|---|---|---|---|---|
|
APOL1 Low Risk |
APOL1 HR |
APOL1 Low Risk |
APOL1 HR |
|
| Characteristics | N=101,216 | N=15,022 | N=4672 | N=582 |
| Age, median (IQR), yr | 59.0 (51.0–66.0) | 59.0 (51.0–66.0) | 59.0 (51.0–66.0) | 60.0 (53.0–66.0) |
| Male, n (%) | 87,266 (86.2) | 12,922 (86.0) | 4059 (86.9) | 509 (87.5) |
| eGFR, ml/min per 1.73 m2, median (IQR) | 87.0 (71.4–103) | 84.5 (69.0–101) | 86.7 (71.6–102) | 88.5 (73.1–103) |
| BMI, median (IQR) | 29.7 (26.0–33.8) | 29.8 (26.2–34.1) | 29.5 (26.1–33.9) | 29.4 (25.8–33.4) |
| Clinical variables | ||||
| Diabetes, n (%) | 33,950 (33.5) | 5126 (34.1) | 1569 (33.6) | 197 (33.8) |
| Hypertension, n (%) | 67,652 (66.8) | 10,447 (69.5) | 3124 (66.9) | 403 (69.2) |
| Systolic BP, mm Hg, median (IQR) | 130 (120–139) | 130 (120–140) | 130 (120–139) | 131 (120–140) |
| Diastolic BP, mm Hg, median (IQR) | 79.0 (72.0–86.0) | 79.0 (72.0–86.0) | 79.0 (72.0–86.0) | 79.0 (72.0–86.0) |
| Medications | ||||
| Renin–angiotensin–aldosterone system inhibition | 40,480 (40.0) | 6201 (41.3) | 1877 (40.2) | 234 (40.2) |
| No. of antihypertensives | ||||
| 1 or 2, n (%) | 37,516 (37.1) | 5685 (37.8) | 1726 (36.9) | 216 (37.1) |
| 3 or more, n (%) | 28,256 (27.9) | 4421 (29.4) | 1307 (28.0) | 174 (29.9) |
| Kidney disease outcomes | ||||
| CKD, n (%) | 15,179 (15) | 2896 (19.3) | 685 (3.6) | 71 (12.2) |
| ESKD, n (%) | 2973 (2.94) | 1054 (7.02) | 136 (2.91) | 14 (2.41) |
| Outpatient dipstick proteinuria ≥2+, n (%) | 5340 (7.69) | 1117 (10.8) | 218 (6.81) | 26 (6.27) |
| Nephrotic syndrome, n (%) | 684 (0.68) | 191 (1.27) | 31 (0.66) | 4 (0.69) |
| FSGS, n (%) | 83 (0.08) | 53 (0.35) | 3 (0.06) | 1 (0.17) |
| ADPKD | 324 (0.32) | 81 (0.54) | 17 (0.36) | 2 (0.34) |
Apolipoprotein L1 low risk, carrying 0 or 1 total copy of the apolipoprotein L1 risk variants G1 or G2. apolipoprotein L1 high risk, carrying two apolipoprotein L1 risk variants, either two copies of the apolipoprotein L1 G1 risk mutation, two copies of the apolipoprotein L1 G2 risk mutation, or one copy of apolipoprotein L1 G1 and one copy of apolipoprotein L1 G2. APOL1, apolipoprotein L1; HR, high risk; IQR, interquartile range; BMI, body mass index; ADPKD, autosomal dominant polycystic kidney disease; CKD, stage 3/stage 4 CKD.
Among 14,386 BioVU participants of African ancestry, the median age was 45 years, 38% were male, 3979 (28%) had diabetes, 978 (28%) had CKD, and 416 (2.8%) had ESKD. A total of 2205 (15.3%) participants had two HR APOL1 gene variants, 703 (4.9%) had at least one copy of the p.N264K allele, and 100 (0.7%) had two copies of HR APOL1 gene variants and at least one copy of the APOL1 p.N264K allele (Supplemental Table 1). For the All of Us cohort, we included 14,704 participants of African ancestry with eGFR data, the median age was 54.8 years, 5399 (36.7%) were male, 4187 (28.5%) had diabetes, 2077 (13.6%) had CKD, and 360 (2.4%) had ESKD. A total of 2049 (13.9%) had two HR APOL1 gene variants, 705 (4.8%) had at least one copy of the p.N264K allele, and 97 (0.66%) had two copies of HR APOL1 gene variants and at least one copy of the APOL1 p.N264K allele (Supplemental Table 2).
Association between APOL1 Genetic Variants with CKD and ESKD in MVP and Replication Cohorts
In MVP, carriers of two APOL1 HR variants without p.N264K had a 1.7-fold increase in the odds of CKD (95% confidence interval [CI], 1.60 to 1.85, P < 0.001 versus low-risk without p.N264K) and a 3.9-fold increase in the odds of ESKD (95% CI, 3.52 to 4.41, P < 0.001 versus low-risk without N264K) among MVP participants without diabetes in the adjusted models consistent with previous studies.4
In our primary analysis, we quantify the protective effect of APOL1 p.N264K in participants with APOL1 HR genotypes in our discovery cohort and in the meta-analysis of our two replication cohorts. In the MVP, APOL1 p.N264K substantially mitigated the renal risk of APOL1 HR variants in both CKD, our primary outcome (odds ratio [OR], 0.43; 95% CI, 0.28 to 0.65; P < 0.001) and ESKD, our secondary outcome (OR, 0.19; 95% CI, 0.07 to 0.51, P = 0.001) (Figure 1). We replicated these findings in the meta-analysis of nondiabetic patients from BioVU (n=11,971) and All of Us (n=12,105). In a meta-analysis across the replication cohorts, APOL1 p.N264K mitigated the renal risk of APOL1 HR variants for CKD (OR, 0.40; 95% CI, 0.18 to 0.92; P = 0.035) and ESKD (OR, 0.19; 95% CI, 0.05 to 0.79; P = 0.01). In a meta-analysis across all cohorts, APOL1 p.N264K mitigated the renal risk of APOL1 HR variants for CKD (OR, 0.43; 95% CI, 0.30 to 0.61; P < 0.001) and ESKD (OR, 0.19; 95% CI, 0.08 to 0.43; P < 0.001). These data demonstrate that APOL1 p.N264K substantially reduces the risk for multiple kidney outcomes in APOL1 HR carriers across multiple cohorts.
In our secondary analyses, we treated the carriers of low-risk APOL1 genotypes as a reference group (APOL1 low-risk and N264K−) and formally tested for an additive interaction between APOL1 HR variants and p.N264K. When APOL1 HR carriers also inherited the APOL1 p.N264K allele (Figure 2), it eliminated the increased risk of CKD (OR, 0.70; 95% CI, 0.45 to 1.08; P = 0.102 compared with low-risk N264K−). In participants with low-risk APOL1 genotypes, carrying the APOL1 p.N264K variant led to a trend toward reduced odds of CKD (OR, 0.86; 95% CI, 0.75 to 1.00; P = 0.05 versus low-risk, N264K−). There was a significant interaction, with the degree of protection conferred by APOL1 p.N264K greater in the APOL1 HR genotype background (Pinteraction = 0.001). The results were highly concordant across sequentially adjusted models from minimal to fully adjusted (Figure 2A and Supplemental Table 5). These data suggest that the APOL1 p.N264K variant reduces the risk of developing CKD in humans and specifically abrogates the increased susceptibility to CKD in APOL1 HR carriers.
We also evaluated the interaction between APOL1 HR and p.N264K in the progression to ESKD. Among MVP participants without diabetes, two APOL1 HR variants were associated with a 3.9-fold increase in the odds of ESKD (95% CI, 3.52 to 4.41; P < 0.001 versus low risk). Conversely, individuals with both two APOL1 HR alleles and the APOL1 p.N264K allele had no increased odds of ESKD (OR, 0.73; 95% CI, 0.27 to 1.96; P = 0.53 versus low risk). The protective effect of the APOL1 p.N264K variant for ESKD was greater in those participants with APOL1 HR variants (Pinteraction = 0.003) (Figure 2B). The results were concordant across sequentially adjusted models (Supplemental Table 6). Taken together, these analyses indicate that in individuals with APOL1 HR variants, APOL1 p.N264K attenuates risk for CKD and ESKD to levels comparable with carriers of APOL1 low-risk variants in a large population cohort. Comparison between heterozygous and homozygous carriers of p.N264K was not performed because there were only 54 participants homozygous for p.N264K (27 with G0/G0, 24 with G0/G2, and 3 with G2/G2).
Sensitivity Analysis of APOL1 Genetic Variants with Kidney Outcomes in MVP Diabetics
The role of APOL1 risk variants in diabetic kidney diseases remains uncertain. We, therefore, considered the role of APOL1 p.N264K in diabetic patients in a separate subgroup analysis. In MVP participants with diabetes, we found that APOL1 HR alleles increased the risk of both CKD (OR, 1.3; 95% CI, 1.21 to 1.41; P < 0.001) and ESKD (OR, 1.8; 95% CI, 1.66 to 2.04; P < 0.001) in the adjusted models, albeit with a smaller risk estimate. However, individuals with both the APOL1 HR genotype and the p.N264K allele had a risk for CKD comparable with carriers of low-risk APOL1 variants (OR, 0.73; 95% CI, 0.51 to 1.06; P = 0.101 versus low-risk). Similarly, APOL1 HR carriers with p.N264K had no increased risk for ESKD (OR, 0.8; 95% CI, 0.43 to 1.56; P = 0.553 versus low risk). As with nondiabetic MVP participants, the protective effect of p.N264K was greater in carriers of the HR APOL1 genotype for both CKD (Pinteraction = 0.003) and ESKD (Pinteraction = 0.014). Thus, the protection from AMKD observed for APOL1 p.N64K in nondiabetic patients extends to patients with diabetes.
Experimental Characterization of APOL1 p.N264K Function
To better understand the robust reduction in AMKD risk found in carriers of the APOL1 p.N264K variant, we sought to characterize the mechanism of action for this variant. We hypothesized that the protective effect of p.N264K on kidney phenotypes might be related to differences in APOL1 function in kidney cells. We first evaluated how the p.N264K mutation influenced APOL1-induced toxicity in immortalized human podocytes, a critical cell type for AMKD.13,33 Inducible expression of either APOL1 G1 or G2, but not G0, resulted in podocyte cell death (Figure 3A). However, expression at similar levels of the risk variants with the p.N264K mutation had substantially reduced toxicity (Figure 3B and Supplemental Figure 1).
Figure 3.
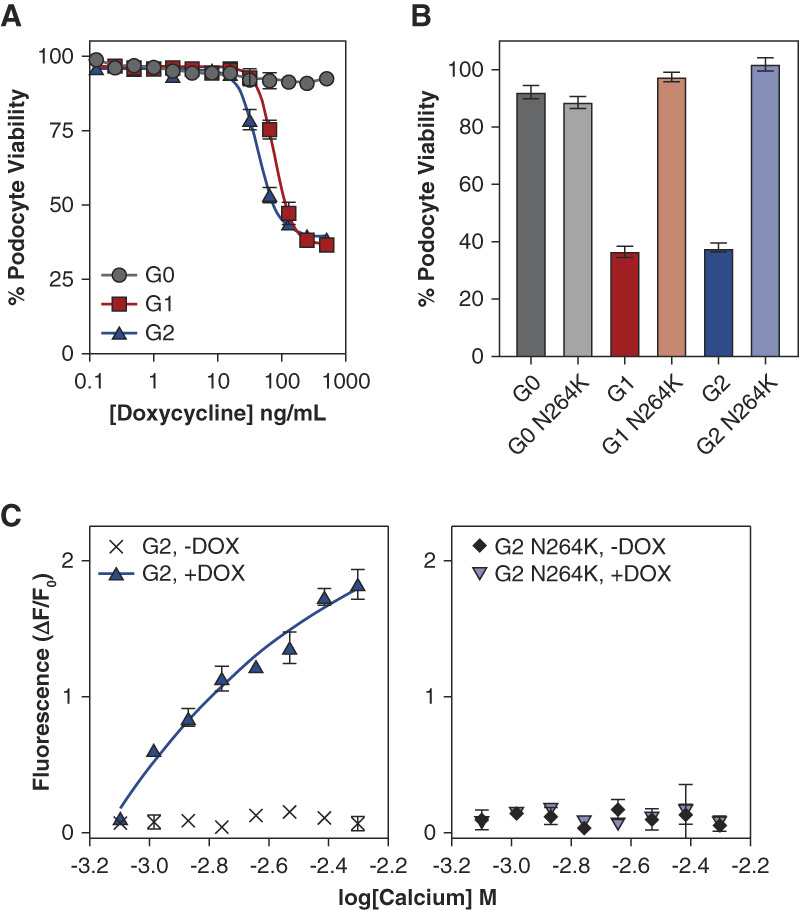
Effects of APOL1 p.N264K on protein function. (A) APOL1 disease variants, G1 and G2, are toxic when overexpressed in human immortalized podocytes. APOL1 G0 overexpression does not affect podocyte cell viability. Error bars represent SEM. (B) The cytotoxicity of APOL1 G1 and G2 risk variants is attenuated by the N264K mutation. Error bars represent SEM. (C) G2 APOL1-mediated calcium transit is blocked by the N264K mutation in HEK cells. The expression of APOL1 G2 or APOL1 G2 with p.N264K was induced with doxycycline, and fluorescence was measured in the presence of increasing concentrations of calcium. The fluorescence level in (C) is a measure of intracellular calcium. Error bars represent SD. HEK, human embryonic kidney.
To investigate the molecular mechanism underlying the observed reduced toxicity, we evaluated the ion conductance of APOL1 G2 with and without p.N264K in kidney cells. Induction of APOL1 G2 resulted in a robust influx of calcium (Figure 3C and Supplemental Figure 2, A–C). Strikingly, we found that cells expressing the comparable levels of APOL1 G2 p.N264K completely lacked detectable pore function (Figure 3C and Supplemental Figure 2, D and E). These data suggest that the APOL1 p.N264K variant suppresses the cellular toxicity of APOL1 HR variants in the kidney by reducing ion conductance through the APOL1 pore.
Discussion
We report a robust protective interaction between the APOL1 p.N264K variant and the APOL1 HR alleles among African ancestry participants in three large independent cohorts. The presence of a single copy of the APOL1 p.N264K mutation mitigated the increased risk conferred by two APOL1 HR variants across both CKD and ESKD end points, in both diabetics and nondiabetics, to levels comparable with the carriers of APOL1 low-risk variants. In kidney cells, the p.N264K variant blocked the ion conductance of APOL1 and reduced its toxicity. Our results suggest that APOL1 ion conductance is essential to its cellular toxicity and to the pathogenesis of AMKD. These insights have important clinical implications for therapeutics and diagnostics.
Identification and functionalization of a genetic variant that suppresses AMKD provides a hypothesis to guide therapeutic discovery. Multiple candidate mechanisms of AMKD have been proposed and investigated in vitro.34 Our data support ion conductance of APOL1 as a critical contributor to AMKD and suggest that pharmacologic interventions that mimic the effects of p.N264K by blocking APOL1 ion conductance could be therapeutic for AMKD.35 Indeed, several small molecule inhibitors of APOL1 ion conductance have been recently reported to reduce albuminuria in mouse models of AMKD. In a small clinical trial in patients with FSGS and APOL1 HR variants, a small molecule inhibitor of APOL1 ion conductance was reported to reduce proteinuria over 12 weeks of treatment.36 The concordant effects of the p.N264K variant and pharmacologic interventions that act by the same mechanism36 illustrate the value of identifying and characterizing genetic modifiers of disease and their potential effect in human populations as we accomplish in this study.
Genetic testing to identify the carriers of APOL1 HR alleles is under consideration as part of precision health strategies.37 APOL1 risk variant testing may have a role in identifying people at greater risk of developing kidney disease for more aggressive management of risk factors, counseling prospective kidney donors, and selecting candidates for therapy directed at APOL1.38 Our results suggest that people with a HR genotype who also carry p.N264K have a lower risk for kidney disease. Therefore, determination of p.N264K status may be an important component of APOL1 diagnostic testing and subsequent clinical decision making.
Strengths of our study include a uniquely large national dataset of APOL1 genetic data and carefully curated electronic health record with a wealth of phenotypic information. The definitions were harmonized in the discovery and the two replication cohorts. This dataset is an order of magnitude larger than any prior published cohorts. This scale of data is required to formally assess the genetic interaction between APOL1 HR loci and the APOL1 p.N264K allele, the combination of which are present in only 1:200 individuals. The wealth of phenotype data enabled rigorous adjudication of end points and clinical covariates from electronic health record data.
Limitations include reduced generalizability as our MVP discovery cohort was approximately 86% male and the limited availability of histologic data. We did not study whether APOL1 p.N264K allele was protective for other phenotypes associated with APOL1 such as sepsis,13,39 preeclampsia,40 FSGS, or AKI.22 In patients with diabetes, we were not able to confirm the diagnosis of diabetic nephropathy, which would require a histologic diagnosis. In addition, the sample size of patients with diabetes in BioVU and All of Us was too small to provide an accurate estimate. Despite these limitations, our estimates were robust and were consistent across nested multivariable models that included known comorbidities, and our findings were replicated in two independent cohorts, BioVU and the NIH All of Us.
In conclusion, APOL1 p.N264K mutation impaired ion conductance and protected individuals with APOL1 HR variants from CKD and ESKD. This human genetic observation supports that pharmacologic inhibitors that mimics this genetic mutation by blocking the APOL1 pore formation and ion channel conduction may be able to prevent and/or treat APOL1-associated kidney disease.
Supplementary Material
Acknowledgments
The human genetic association study was designed and supervised by the first and last authors in consultation with other authors. Mechanistic studies were designed and supervised by the second to last author. All the authors contributed to the collection and analysis of the data and vouch for the accuracy and completeness of the data. The first and last authors wrote the manuscript, which was reviewed and approved by all the authors. This work is part of KidPhenGen, a collaborative team of researchers with focus in understanding the genetic basis of CKD progression and translation to therapeutic development.
Footnotes
H.-C.C., V.A.A., and Z.Y. contributed equally to this work.
A complete list of investigators and staff in the VA Million Veteran Program is provided in Supplemental Summary 1.
E.M.G. supervised the functional genomics work.
A.M.H. and A.G.B. jointly supervised this work.
Contributor Information
Collaborators: Sumitra Muralidhar, Jennifer Moser, Jennifer E. Deen, Philip S. Tsao, J. Michael Gaziano, Elizabeth Hauser, Amy Kilbourne, Shiuh-Wen Luoh, Michael Matheny, Dave Oslin, Lori Churby, Stacey B. Whitbourne, Jessica V. Brewer, Shahpoor (Alex) Shayan, Luis E. Selva, Saiju Pyarajan, Kelly Cho, Scott L. DuVall, Mary T. Brophy, Brady Stephens, Todd Connor, Dean P. Argyres, Tim Assimes, Adriana Hung, Henry Kranzler, Samuel Aguayo, Sunil Ahuja, Kathrina Alexander, Xiao M. Androulakis, Prakash Balasubramanian, Zuhair Ballas, Jean Beckham, Sujata Bhushan, Edward Boyko, David Cohen, Louis Dellitalia, L. Christine Faulk, Joseph Fayad, Daryl Fujii, Saib Gappy, Frank Gesek, Jennifer Greco, Michael Godschalk, Todd W. Gress, Samir Gupta, Salvador Gutierrez, John Harley, Kimberly Hammer, Mark Hamner, Robin Hurley, Pran Iruvanti, Frank Jacono, Darshana Jhala, Scott Kinlay, Jon Klein, Michael Landry, Peter Liang, Suthat Liangpunsakul, Jack Lichy, C. Scott Mahan, Ronnie Marrache, Stephen Mastorides, Elisabeth Mates, Kristin Mattocks, Paul Meyer, Jonathan Moorman, Timothy Morgan, Maureen Murdoch, James Norton, Olaoluwa Okusaga, Kris Ann Oursler, Ana Palacio, Samuel Poon, Emily Potter, Michael Rauchman, Richard Servatius, Satish Sharma, River Smith, Peruvemba Sriram, Patrick Strollo, Jr., Neeraj Tandon, Philip Tsao, Gerardo Villareal, Agnes Wallbom, Jessica Walsh, John Wells, Jeffrey Whittle, Mary Whooley, Allison E. Williams, Peter Wilson, Junzhe Xu, and Shing Shing Yeh
Disclosures
V.A. Assimon reports Employer: Cartography Biosciences, Maze Therapeutics, and Soteria Biotherapeutics; Consultancy: Capstan Therapeutics and Soteria Biotherapeutics; Ownership Interest: Cartography Biosciences, Cleave Therapeutics, Denali Therapeutics, Maze Therapeutics, and Soteria Biotherapeutics; Research Funding: Cartography Biosciences, Maze Therapeutics, and Soteria Biotherapeutics; and Patents or Royalties: Maze Therapeutics, Soteria Biotherapeutics, and University of California San Francisco. A.G. Bick reports Ownership Interest: TenSixteen Bio. H. Chan reports Employer: Maze Therapeutics; and Ownership Interest: Cytokinetics, Gilead, Illumina, Nektar, and Vertex.S. Chandrasekar reports Employer: Apple Inc., Arcus Biosciences, and Maze Therapeutics; and Ownership Interest: Apple Inc., Arcus Biosciences, and Maze Therapeutics. C.P. Chung reports Advisory or Leadership Role: Journals: Arthritis Care & Research, Clinical Pharmacology & Therapeutics, and Clinical Rheumatology. K. Estrada reports Employer: Maze TX; and Ownership Interest: BioMarin, Maze TX, and Regeneron. R.R. Graham reports Employer: Maze Therapeutics; Ownership Interest: Maze Therapeutics; and Patents or Royalties: Maze Therapeutics. E.M. Green reports Employer: Maze Therapeutics; Ownership Interest: Maze Therapeutics; Patents or Royalties: Maze Therapeutics; and Advisory or Leadership Role: Maze Therapeutics. M. Hoek reports Employer: Maze Therapeutics; Ownership Interest: Maze Therapeutics and Merck & Co.; Research Funding: Maze Therapeutics and Merck & Co.; and Patents or Royalties: Maze Therapeutics and Merck & Co. A.M. Hung reports Consultancy: NHLBI consultant for Gene and life interaction grant; Research Funding: Vertex Grant to VUMC and VHA CSR&D Merit “Genetics of Kidney Disease and Hypertension, Risk Prediction and Drug Response”; and Advisory or Leadership Role: Ad-hoc Scientific Review Committee CSR&D, Ad-Scientific Review Committee KNOD, Ad-hoc SRC NHLBI, Co-Chair Million Veteran Program Publications and Presentation committee, Co-Chair Pharmacogenomics for COVID-19 Million Veteran Program, Section Editor, Clinical Nephrology; Standing member SRC HSR&D bioinformatics, and Veterans Affairs. M.E. Matheny reports Consultancy: Food and Drug Administration (FDA) and NIH-VA-DoD Pain Management Grant Consortium (PMC3); and Advisory or Leadership Role: SMRB Study Section, Steering Committee—Indianapolis VA HSR&D COIN Center, Steering Committee—VA HSR&D VIREC, Steering Committee—Salt Lake City VA HSR&D COIN Center, VA HSR&D, Informatics & Methods Section, and VA ORD Million Veterans Program Executive Steering Committee. S.V. Mozaffari reports Employer: Maze Therapeutics; and Ownership Interest: Maze Therapeutics; 23andMe. C. Robinson-Cohen reports Advisory or Leadership Role: BMC Nephrology Editorial Board Member, CJASN Editorial Board Member, and Clinical Nephrology, Genetics Section Editor. S.C. Shah reports Consultancy: Medscape, Phathom Pharmaceuticals, and RedHill Biopharma; and Research Funding: American Gastroenterological Association, National Institute of Health, US Department of Veterans Affairs. E.D. Siew reports Ownership Interest: Amazon stock and Apple stock; Patents or Royalties: Author for UptoDate (royalties); Advisory or Leadership Role: Editorial board of CJASN. Personal fees; and Other Interests or Relationships: Consultancy Agreement with Novartis (no fees received during past 24 months). K. Susztak reports Consultancy: Astra Zeneca, GSK, Novo Nordisk, and Pfizer; Ownership Interest: Jnana; Research Funding: Astra Zeneca, Bayer, Boehringer Ingelheim, Calico, Gilead, GSK, Jnana, Kyowa Kirin Genentech, Maze, Novartis, Novo Nordisk, ONO Pharma, Regeneron, and Variant Bio; Honoraria: AstraZeneca, Bayer, Jnana, Maze, and Pfizer; and Advisory or Leadership Role: Editorial board; Cell Metabolism, EBioMedicine, JASN, Jnana, Journal of Clinical Investigation, Kidney International, Med, and Pfizer. T. Thompson reports the following: Ownership Interest: Tenacious Products, LLC. L. Wheless reports Research Funding: Dermatology Foundation and VA ClinicalSciences R&D; and Advisory or Leadership Role: Board of Directors, International Immunosuppression and Transplant Skin Cancer Collaborative. R. Tao reports Employer: Vanderbilt University Medical Center, and Genentech. All remaining authors have nothing to disclose.
Because Katalin Susztak is an editor of the Journal of the American Society of Nephrology, she was not involved in the peer review process for this manuscript. A guest editor oversaw the peer review and decision-making process for this manuscript.
Funding
This research is based on data from the Million Veteran Program which is funded by the Office of Research and Development, Veterans Health Administration CX001897 to A.M. Hung. Vanderbilt BioVU is supported by institutional funding and NIH CTSA grant UL1TR002243. Genetic association studies were funded by the VA Clinical Science Research and Development investigator-initiated grant CX001897 (A.M. Hung) titled Genetic of Kidney Disease and Hypertension-Risk Prediction and Drug Response. C. Robinson-Cohen was supported by NIH grant R01DK122075. J.L. Triozzi is supported by 5T32DK007569-34. C. Mechanistic studies were funded and executed by MAZE Therapeutics. The funding agencies had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication. This publication does not represent the views of the Department of Veteran Affairs or the United States Government.
Author Contributions
Conceptualization: Alexander G. Bick, Adriana M. Hung, Edward D. Siew, Jefferson L. Triozzi.
Data curation: Caitlyn Vlasschaert, Otis Wilson.
Formal analysis: Victoria A. Assimon, Saranya Chandrasekar, Hua-Chang Chen, Adriana M. Hung, Sahar V. Mozaffari, Ran Tao, Caitlyn Vlasschaert, Zhihong Yu.
Funding acquisition: Adriana M. Hung.
Investigation: Victoria A. Assimon, Alexander G. Bick, Helen Chan, Saranya Chandrasekar, Hua-Chang Chen, Cecilia P. Chung, Karol Estrada, J. Michael Gaziano, Robert R. Graham, Eric M. Green, Maarten Hoek, Adriana M. Hung, Taralynn Mack, Michael E. Matheny, Sahar V. Mozaffari, Cassianne Robinson-Cohen, Shailja C. Shah, Edward D. Siew, Katalin Susztak, Trevor Thompson, Jefferson L. Triozzi, Philip Tsao, Lee Wheless, Otis Wilson.
Methodology: Victoria A. Assimon, Helen Chan, Saranya Chandrasekar, Cecilia P. Chung, Karol Estrada, J. Michael Gaziano, Robert R. Graham, Eric M. Green, Maarten Hoek, Adriana M. Hung, Michael E. Matheny, Sahar V. Mozaffari, Shailja C. Shah, Katalin Susztak, Ran Tao, Philip Tsao, Trevor Thompson, Lee Wheless, Zhihong Yu.
Project administration: Philip Tsao.
Resources: Helen Chan, Karol Estrada, Taralynn Mack.
Supervision: Alexander G. Bick, Eric M. Green, Adriana M. Hung.
Visualization: Cassianne Robinson-Cohen, Edward D. Siew, Jefferson L. Triozzi.
Writing – original draft: Alexander G. Bick, Adriana M. Hung.
Writing – review & editing: Alexander G. Bick, Hua-Chang Chen, Cecilia P. Chung, J. Michael Gaziano, Eric M. Green, Maarten Hoek, Adriana M. Hung, Taralynn Mack, Michael E. Matheny, Cassianne Robinson-Cohen, Shailja C. Shah, Katalin Susztak, Ran Tao, Trevor Thompson, Caitlyn Vlasschaert, Lee Wheless, Otis Wilson, Zhihong Yu.
Data Sharing Statement
The protocol and statistical code are available from Dr. A.M. Hung. However, data will require approval from the VA Office of Research Development and the Million Veteran Program.
Supplemental Material
This article contains the following supplemental material online at http://links.lww.com/JSN/E526.
Supplemental Appendix. Methods used in the mechanistic studies and functional genomics.
Supplemental Summary 1. MVP core acknowledgment.
Supplemental Table 1. BioVU cohort characteristics stratified by APOL1 genotype.
Supplemental Table 2. NIH All of Us cohort characteristics stratified by APOL1 genotype.
Supplemental Table 3. ESKD definition.
Supplemental Table 4. Definitions and procedure codes for dialysis and comorbidities.
Supplemental Table 5. MVP APOL1 high risk and p.N264K association with CKD.
Supplemental Table 6. MVP APOL1 high risk and p.N264K association with end stage.
Supplemental Table 7. MVP cohort characteristics stratified by APOL1 N264K genotype.
Supplemental Table 8. BioVU APOL1 high risk and p.N264K association with kidney disease outcomes among nondiabetic individuals.
Supplemental Table 9. All of Us APOL1 high risk and p.N264K association with kidney disease outcomes among nondiabetic individuals.
Supplemental Table 10. Haplotype frequency of APOL1 protein-altering variants in African populations with more than 20 individuals for the haplotype in the Million Veteran Program.
Supplemental Table 11 Distribution of N264K genotypes in APOL1 low-risk and high-risk genotypes.
Supplemental Figure 1. APOL1 protein expression in G0, G1, and G2 podocyte cell lines.
Supplemental Figure 2. Additional characterization of APOL1 cell lines used in the calcium uptake assay.
References
- 1.McClellan WM Warnock DG Judd S, et al. Albuminuria and racial disparities in the risk for ESRD. J Am Soc Nephrol. 2011;22(9):1721–1728. doi: 10.1681/ASN.2010101085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saran R Robinson B Abbott KC, et al. US renal data system 2016 annual data report: epidemiology of kidney disease in the United States. Am J Kidney Dis. 2017;69(3):A7–A8. doi: 10.1053/j.ajkd.2016.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johansen KL Chertow GM Gilbertson DT, et al. US renal data system 2021 annual data report: epidemiology of kidney disease in the United States. Am J Kidney Dis. 2022;79(4):A8–A12. doi: 10.1053/j.ajkd.2022.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parsa A Kao WL Xie D, et al. APOL1 risk variants, race, and progression of chronic kidney disease. N Engl J Med. 2013;369(23):2183–2196. doi: 10.1056/nejmoa1310345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Genovese G Friedman DJ Ross MD, et al. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science. 2010;329(5993):841–845. doi: 10.1126/science.1193032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kopp JB Nelson GW Sampath K, et al. APOL1 genetic variants in focal segmental glomerulosclerosis and HIV-associated nephropathy. J Am Soc Nephrol. 2011;22(11):2129–2137. doi: 10.1681/ASN.2011040388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brun R, Blum J, Chappuis F, Burri C. Human african trypanosomiasis. Lancet. 2010;375(9709):148–159. doi: 10.1016/s0140-6736(09)60829-1 [DOI] [PubMed] [Google Scholar]
- 8.CDC. African Trypanosomiasis, Also Known as “Sleeping Sickness”. Global Health, Division of Parasitic Diseases and Malaria; 2022. https://www.cdc.gov/parasites/sleepingsickness/epi.html [Google Scholar]
- 9.Lan X Jhaveri A Cheng K, et al. APOL1 risk variants enhance podocyte necrosis through compromising lysosomal membrane permeability. Am J Physiol Renal Physiol. 2014;307(3):F326–F336. doi: 10.1152/ajprenal.00647.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olabisi OA Zhang JY VerPlank L, et al. APOL1 kidney disease risk variants cause cytotoxicity by depleting cellular potassium and inducing stress-activated protein kinases. Proc Natl Acad Sci U S A. 2016;113(4):830–837. doi: 10.1073/pnas.1522913113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma L Chou JW Snipes JA, et al. APOL1 renal-risk variants induce mitochondrial dysfunction. J Am Soc Nephrol. 2017;28(4):1093–1105. doi: 10.1681/ASN.2016050567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wakashin H Heymann J Roshanravan H, et al. APOL1 renal risk variants exacerbate podocyte injury by increasing inflammatory stress. BMC Nephrol. 2020;21(1):371. doi: 10.1186/s12882-020-01995-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu J Ma Z Raman A, et al. APOL1 risk variants in individuals of African genetic ancestry drive endothelial cell defects that exacerbate sepsis. Immunity. 2021;54(11):2632–2649.e6. doi: 10.1016/j.immuni.2021.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Uzureau S Lecordier L Uzureau P, et al. APOL1 C-terminal variants may trigger kidney disease through interference with APOL3 control of actomyosin. Cell Rep. 2020;30(11):3821–3836.e13. doi: 10.1016/j.celrep.2020.02.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Limou S, Dummer PD, Nelson GW, Kopp JB, Winkler CA. APOL1 toxin, innate immunity, and kidney injury. Kidney Int. 2015;88(1):28–34. doi: 10.1038/ki.2015.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lannon H Shah SS Dias L, et al. Apolipoprotein L1 (APOL1) risk variant toxicity depends on the haplotype background. Kidney Int. 2019;96(6):1303–1307. doi: 10.1016/j.kint.2019.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cuypers B Lecordier L Meehan CJ, et al. Apolipoprotein L1 variant associated with increased susceptibility to trypanosome infection. mBio. 2016;7(2):e02198. doi: 10.1128/mbio.02198-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schaub C Verdi J Lee P, et al. Cation channel conductance and pH gating of the innate immunity factor APOL1 are governed by pore-lining residues within the C-terminal domain. J Biol Chem. 2020;295(38):13138–13149. doi: 10.1074/jbc.ra120.014201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaziano JM Concato J Brophy M, et al. Million Veteran Program: a mega-biobank to study genetic influences on health and disease. J Clin Epidemiol. 2016;70:214–223. doi: 10.1016/j.jclinepi.2015.09.016 [DOI] [PubMed] [Google Scholar]
- 20.Fihn SD Francis J Clancy C, et al. Insights from advanced analytics at the veterans health administration. Health Aff. 2014;33(7):1203–1211. doi: 10.1377/hlthaff.2014.0054 [DOI] [PubMed] [Google Scholar]
- 21.Dumitrescu L Ritchie MD Brown-Gentry K, et al. Assessing the accuracy of observer-reported ancestry in a biorepository linked to electronic medical records. Genet Med. 2010;12(10):648–650. doi: 10.1097/gim.0b013e3181efe2df [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hung AM Shah SC Bick AG, et al. APOL1 risk variants, acute kidney injury and death in African ancestry participants hospitalized with COVID-19 from the Million Veterans Program. JAMA Intern Med. 2022;182(4):386–395. doi: 10.1001/jamainternmed.2021.8538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hunter-Zinck H Shi Y Li M, et al. Genotyping array design and data quality control in the million veteran program. Am J Hum Genet. 2020;106(4):535–548. doi: 10.1016/j.ajhg.2020.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bick AG Akwo E Robinson-Cohen C, et al. Association of APOL1 risk alleles with cardiovascular disease in Blacks in the million veteran program. Circulation. 2019;140(12):1031–1040. doi: 10.1161/circulationaha.118.036589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grams ME Rebholz CM Chen Y, et al. Race, APOL1 risk, and eGFR decline in the general population. J Am Soc Nephrol. 2016;27(9):2842–2850. doi: 10.1681/ASN.2015070763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen TK Katz R Estrella MM, et al. Association between APOL1 genotypes and risk of cardiovascular disease in MESA (Multi-Ethnic study of atherosclerosis). J Am Heart Assoc. 2017;6(12):e007199. doi: 10.1161/jaha.117.007199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Friedman DJ, Kozlitina J, Genovese G, Jog P, Pollak MR. Population-based risk assessment of APOL1 on renal disease. J Am Soc Nephrol. 2011;22(11):2098–2105. doi: 10.1681/ASN.2011050519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomson R Genovese G Canon C, et al. Evolution of the primate trypanolytic factor APOL1. Proc Natl Acad Sci U S A. 2014;111(20):E2130–E2139. doi: 10.1073/pnas.1400699111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levey AS Eckardt KU Tsukamoto Y, et al. Definition and classification of chronic kidney disease: a position statement from Kidney Disease: improving Global Outcomes (KDIGO). Kidney Int. 2005;67(6):2089–2100. doi: 10.1111/j.1523-1755.2005.00365.x [DOI] [PubMed] [Google Scholar]
- 30.Waikar SS Wald R Chertow GM, et al. Validity of international classification of diseases, ninth revision, clinical modification codes for acute renal failure. J Am Soc Nephrol. 2006;17(6):1688–1694. doi: 10.1681/ASN.2006010073 [DOI] [PubMed] [Google Scholar]
- 31.Levey AS Stevens LA Schmid CH, et al. A new equation to estimate glomerular filtration rate. Ann Intern Med. 2009;150(9):604–612. doi: 10.7326/0003-4819-150-9-200905050-00006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knol MJ, VanderWeele TJ, Groenwold RH, Klungel OH, Rovers MM, Grobbee DE. Estimating measures of interaction on an additive scale for preventive exposures. Eur J Epidemiol. 2011;26(6):433–438. doi: 10.1007/s10654-011-9554-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beckerman P Bi-Karchin J Park AS, et al. Transgenic expression of human APOL1 risk variants in podocytes induces kidney disease in mice. Nat Med. 2017;23(4):429–438. doi: 10.1038/nm.4287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Freedman BI, Kopp JB, Sampson MG, Susztak K. APOL1 at 10 years: progress and next steps. Kidney Int. 2021;99(6):1296–1302. doi: 10.1016/j.kint.2021.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hung AM Assimon VA Chen HC, et al. Genetic Inhibition of APOL1 Pore Forming Function Prevents APOL1 Kidney Disease. FR-PO307. Kidney Week 2022; 2022, Orlando, FL, USA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Egbuna O, Chertow GM. Inaxaplin for Proteinuric Kidney Disease in Persons with Two APOL1 Variants. Reply. N Engl J Med; 2023;388:2491. [DOI] [PubMed] [Google Scholar]
- 37.Kopp JB, Winkler CA. Genetic testing for APOL1 genetic variants in clinical practice: finally starting to arrive. Clin J Am Soc Nephrol. 2020;15(1):126–128. doi: 10.2215/CJN.01810219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Freedman BI, Poggio ED. APOL1 genotyping in kidney transplantation: to do or not to do, that is the question? (pro). Kidney Int. 2021;100(1):27–30. doi: 10.1016/j.kint.2020.11.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chaudhary NS Moore JX Zakai NA, et al. APOL1 nephropathy risk alleles and risk of sepsis in Blacks. Clin J Am Soc Nephrol. 2019;14(12):1733–1740. doi: 10.2215/CJN.04490419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sedor JR, Bruggeman LA, O'Toole JF. APOL1 and preeclampsia: intriguing links, uncertain causality, troubling implications. Am J Kidney Dis. 2021;77(6):863–865. doi: 10.1053/j.ajkd.2021.01.013 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The protocol and statistical code are available from Dr. A.M. Hung. However, data will require approval from the VA Office of Research Development and the Million Veteran Program.