Abstract
Inherited retinal degeneration is a term used to describe heritable disorders that result from the death of light sensing photoreceptor cells. Although we and others believe that it will be possible to use gene therapy to halt disease progression early in its course, photoreceptor cell replacement will likely be required for patients who have already lost their sight. While advances in autologous photoreceptor cell manufacturing have been encouraging, development of technologies capable of efficiently delivering genome editing reagents to stem cells using current good manufacturing practices (cGMP) are needed. Gene editing reagents were delivered to induced pluripotent stem cells (iPSCs) using a Zephyr microfluidic transfection platform (CellFE). CRISPR-mediated cutting was quantified using an endonuclease assay. CRISPR correction was confirmed via digital PCR and Sanger sequencing. The resulting corrected cells were also karyotyped and differentiated into retinal organoids. We describe use of a novel microfluidic transfection platform to correct, via CRISPR-mediated homology-dependent repair (HDR), a disease-causing NR2E3 mutation in patient-derived iPSCs using cGMP compatible reagents and approaches. We show that the resulting cell lines have a corrected genotype, exhibit no off-target cutting, retain pluripotency and a normal karyotype and can be differentiated into retinal tissue suitable for transplantation. The ability to codeliver CRISPR/Cas9 and HDR templates to patient-derived iPSCs without using proprietary transfection reagents will streamline manufacturing protocols, increase the safety of resulting cell therapies, and greatly reduce the regulatory burden of clinical trials.
Keywords: CRISPR genome editing, microfluidic transfection, cGMP, induced pluripotent stem cells
Graphical Abstract
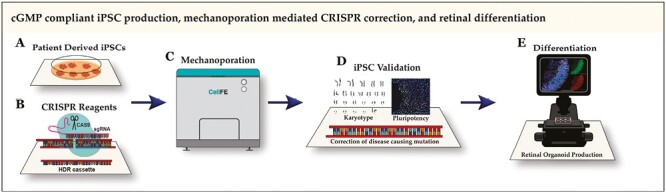
Significance Statement.
Photoreceptor cell replacement will likely be required for patients who have already lost their sight due to inherited retinal degeneration. We describe use of a novel microfluidic transfection platform to correct, via CRISPR-mediated homology dependent repair, a disease-causing NR2E3 mutation in patient-derived induced pluripotent stem cells using reagents and approaches compatible with current good manufacturing practices. The ability to deliver genome editing reagents to patient-derived induced pluripotent stem cells without using proprietary transfection reagents will streamline manufacturing protocols and increase the safety of resulting cell therapies.
Introduction
Inherited retinal degeneration is a blanket term used to describe a group of heritable disorders that result from death of retinal photoreceptor cells and irreversible blindness. While it is theorized that gene therapy could halt disease progression if performed early in disease course, this approach is unlikely to be effective for patients who have already lost most of their vision due to widespread photoreceptor cell death. In an attempt to develop treatments for these patients, we and others have devised pipelines for the manufacture of patient-derived photoreceptor cells intended for autologous cell replacement.1-10 While complicated in practice, the general approach can be described simply. First, somatic cells are collected from a patient and reprogrammed to induced pluripotent stem cells (iPSCs) via forced expression of several key transcription factors (eg, OCT4, SOX2, KLF4, and C-MYC, commonly referred to as the Yamanaka factors).11,12 While the resulting iPSCs could at this point be differentiated into patient-derived photoreceptor cells, they would still contain the mutation that initially caused the patient’s disease. Therefore, before differentiation, gene-modifying macromolecules would ideally be delivered to the iPSCs to correct the patient’s disease-causing genetic variants. Genome editing of cultured cells has been achieved using a variety of different approaches (eg, CRISPR, zinc finger nucleases, and TALENs), all of which require technology to facilitate their delivery to the cell nucleus.13-20 After the patient’s disease causing mutation(s) are corrected, the resulting iPSCs can be differentiated into photoreceptor cells for autologous transplantation.10,21-43
To be suitable for the correction of mutations in iPSCs whose progeny are destined for transplant, transfection technologies must be compatible with current good manufacturing practices (cGMP) and safe for use in human subjects. To be cGMP compatible, all reagents must be well defined and preferably xenofree. Furthermore, the transfection approach should be easily adapted to automated cell manufacturing systems, give high cell survival, and enable high-volume production. Current CRISPR technologies being considered for therapeutic genome editing work in 2 steps. First, small guide RNAs and a nuclease are used to induce a double-stranded DNA break at a prescribed location within the cell’s genome (typically within proximity to the patient’s disease-causing mutation). After inducing this break, the cell’s own DNA repair machinery will attempt to repair the damage via either non-homologous end joining (NHEJ), which is often imprecise, or the more accurate homology directed repair (HDR). By co-delivering wild-type sequence spanning the patient’s mutation as a template, HDR can result in precise correction of the disease causing mutation in the cell’s genome.44-46
Several technologies such as electroporation, lipofection, and viral transduction have been commercialized for delivering macromolecules to cells. Viral cargo size limitations preclude the use of clinically relevant viruses such as AAV for delivery of genome editing reagents and larger DNA repair templates. While lipofection and electroporation have been used by our group and others to successfully correct disease-causing mutations in patient iPSCs,13-15,45 the reagents used are both proprietary and not well defined, therefore poorly compatible with cGMP. Recently, our group demonstrated a novel transfection technology that uses mechanical perturbations to induce convective delivery of large macromolecules to cells.47,48 As this technique does not require the use of proprietary chemical reagents, it is well suited to clinical production (ie, iPSCs can be transfected directly in E8 maintenance media, for which clinically compatible CTS versions with accompanying FDA drug master files exist). In this work, we demonstrate the use of this technology to deliver CRISPR reagents and induce HDR-mediated mono- and bi-allelic correction of disease-causing NR2E3 mutations in patient-derived iPSCs. Resulting cell lines were found to be pluripotent, have a normal karyotype, and retain the ability to generate high-quality retinal organoids containing transplantable retinal progenitor cells from which cGMP-compliant cell therapies can be produced.
Materials and Methods
Patient-derived iPSCs
This study was approved by the Institutional Review Board of the University of Iowa (project approval #200202022) and adhered to the tenets set forth in the Declaration of Helsinki. Patient iPSCs were generated from an individual with molecularly confirmed enhanced S cone syndrome (ESCS) as described previously.14,21
Delivery of NR2E3 Cas9 RNP
Ribonucleoprotein (RNP) complex was formed by combining 15 μg/mL single guide RNA (sgRNA) (IDT, Coralville, IA; TGCTGCTGTCTCCGCACACG) and 25 μg/mL HiFi cas9 nuclease V3 (IDT; Cat#1081060) for 15 minutes at room temperature. Patient iPSCs were passaged as single cells using TrypLE and a 40-µm cell strainer. RNPs were added to iPSCs (1 × 106 cells/mL) in normal cell growth media (Essential 8 (E8); Thermo Fisher Scientific, Waltham, MA) and 0.5 mL was added to each inlet of the transfection device. Different device gap sizes (ie, 7.2, 9, and 10.4 μm, each of which are commercially available) and inlet pressures (50 and 90 PSI) were tested. Cleavage was assessed via a T7 endonuclease 1 (T7E1) assay using NR2E3-specific primers (F2: GCGTGGGTTCGTTCAAATG; R2: TCCAGCTTAGCACAGGTTTC) as described previously.14,45 Densitometry was performed using ImageJ.
Homology-directed Repair (HDR) in NR2E3 Patient-Specific IPSCs
An HDR construct containing a puromycin resistance cassette was synthesized by GenScript as described previously.14 Use of recombinant DNA was approved by the University of Iowa Institutional Biosafety Committee, rDNA #220122. All work with rDNA were performed in accordance with the National Institutes of Health guidelines. Transfection was done as above using the device with a 10.4-µm gap size (CellFE) at 50 or 90 PSI with the addition of 40–80 µg/mL of plasmid HDR construct. Puromycin selection was performed, and surviving colonies (which are not necessarily monoclonal) were picked and subjected to digital PCR to identify mono- vs biallelic-HDR. Digital PCR results were sequence confirmed using Sanger sequencing and off-target editing was analyzed as described previously.15
Digital PCR
Custom probe-based assays were designed against wild type or the c.119-2A>C mutation by Integrated DNA Technologies. Several locked nucleic acids (indicated with “+”) were incorporated into the probes: WT (CC+CT+C+A+GGCG) and mutant (CCCT+C+C+GGCG). Digital PCR was performed using NR2E2-specific primers (F2: GCGTGGGTTCGTTCAAATG; R2: TCCAGCTTAGCACAGGTTTC) and the Applied Biosystems 3D PCR platform (ThermoFisher) as described previously.49
Karyotype
iPSCs were karyotyped at the Pediatric Cytogenetics Laboratory at the University of Iowa Hospitals and Clinics. Briefly, iPSCs were arrested in the metaphase of cell division using colcemid and chromosomes were stained by the G-banding method. Chromosome number was determined by microscopic analysis and these cells were examined for the presence or absence of detectable structural rearrangements. Karyotypes were prepared from computer assisted digital images of these metaphases.
Retinal Differentiation
Retinal differentiation was performed as described previously with some modifications.14,50 Briefly, iPSCs were cultured on Laminin-521-coated plates in E8 medium. Embryoid bodies (EBs) were lifted with ReLeSR and transitioned from E8 to neural induction medium (NIM-DMEM/F12 (1:1), 1% N2 supplement, 1% non-essential amino acids, 1% Glutamax, 2 μg/mL heparin (Sigma), and Primocin (Invivogen)) over a 4-day time period. On day 6, NIM was supplemented with 1.5 nM BMP4 (R&D Systems). On day 7, EBs were adhered to CellStart coated plates (Thermo Fisher Scientific). BMP4 was gradually transitioned out of the NIM over 7 days. On day 16, the media was changed to retinal differentiation medium (RDM—DMEM/F12 (3:1), 2% B27 supplement, 1% non-essential amino acids, 1% Glutamax, and 0.2% Primocin). On days 25–30, the entire EB outgrowth was mechanically lifted using a cell scraper and transferred to ultra-low attachment flasks in 3D-RDM (RDM plus 10% fetal bovine serum (FBS; Atlas Biologicals), 100 µM taurine (Sigma), 1:1000 chemically defined lipid supplement (Thermo Fisher Scientific), and 1 µM all-trans retinoic acid (until day 100; Sigma)). The cells were fed 3 times per week with 3D-RDM until harvest.
Immunocytochemistry
iPSCs and day 40 organoids were fixed with 4% paraformaldehyde for 30-60 minutes at room temperature. Organoids were equilibrated to 15% sucrose in PBS, followed by 30% sucrose. Organoids were cryopreserved in 50:50 solution of 30% sucrose/PBS: tissue freezing medium (Electron Microscopy Sciences, Hatfield, PA) and cryosectioned (15 μm). Cells were blocked with 5% normal donkey serum, 3% bovine serum albumin, and 0.1% Triton-X, and stained overnight with the following primary antibodies: NANOG (R&D; Cat# AF1997), OCT4 (Stemgent; Cat#09-0023), LHX2 (Abcam; Cat#AB184337), CHX10 (Exalpha; Cat# X1179P), OTX2 (R&D Systems, Minneapolis, MN; Cat# AF1979), and Recoverin (EMD Millipore, Burlington, MA; Cat# AB5585). The following secondary antibodies (Thermo Fisher Scientific) were incubated for 1 hour: donkey anti-goat 488 (Cat# R37118), donkey anti-rabbit 488 (Cat#R37118), donkey anti-sheep 647 (Cat# A21448), and donkey anti-rabbit 647 (Cat# A31573). Cell nuclei were counterstained using DAPI (Thermo Fisher Scientific; Cat# 62248). Images were acquired using a Leica TCS SPE upright confocal microscope system (Leica Microsystems, Wetzlar, Germany).
Results
The manufacture of iPSC-derived cell therapies for inherited retinal degenerative blindness will require cGMP-compatible transfection technologies suitable for use with automated cell manufacturing systems. To meet this need, we and others have developed microfluidic transfection platforms, which facilitate the delivery of macromolecules to cells via the application of mechanical forces.47,48,51-53 While our initial results were promising, adoption of these technologies by the wider community required the development of a mature, commercial transfection platform. The CellFE Zephyr transfection device consists of a pressure control system, into which single use, disposable consumables (Fig. 1A-C) can be inserted. To deliver payload to the interior of cells, the cargo is simply suspended with the target cells in appropriate cell media and loaded into the consumable. Once placed into the desktop system, pressure is applied at the consumable’s inlets, forcing the cell/payload mixture through microfluidic channels. Within the channels, the uptake of the cargo is accomplished through rapid deformation of the cells, as depicted in Fig. 1B and described previously.47,48 The use of pre-sterilized, single use consumables eliminates the risk of cross-contamination of samples isolated from different patients, and the transfection does not depend on any reagents other than an appropriate cell growth media, such as E8 or mTseR, both of which are available as CTS versions that have FDA drug master files.
Figure 1.
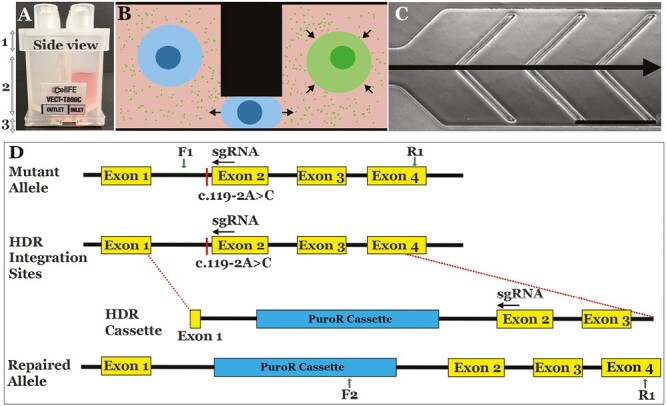
Overview. (A) Single use, disposable transfection consumable capable of processing eight independent samples simultaneously. (B, C) Within the consumable, pressure-driven flow is used to force cells under periodic constrictions, causing rapid deformations which lead to the uptake of macromolecules from the surrounding buffer. Scale bar = 500 μm. (D) Schematics depicting genomic disease-causing homozygous c.199-2A>C mutation and genotype following CRISPR-based homology-directed repair.
To verify that the Zephyr system is capable of delivering payloads relevant to the manufacture of patient-specific cell therapies for inherited retinal degenerative blindness, we used the system to deliver pre-complexed spCas9/sgRNA RNPs along with plasmid-based HDR cassettes to correct a disease-causing mutation present in the NR2E3 gene of patient-derived iPSCs. This approach was described previously and illustrated in Fig. 1D.14
Before we could use the CellFE Zephyr platform to repair disease-causing mutations in patient-derived iPSCs, appropriate operating conditions for the device had to be determined. While there are many aspects of the microchannel design which can be tuned, we focused on applied pressure and ridge gap size, which are the 2 parameters with the largest effect on delivery efficiency and cell viability. Based on the measured size of iPSCs and our previous experience, we attempted to deliver pre-complexed CRISPR RNPs to our patient-derived iPSCs using channels with 7.2, 9, and 10.4 µm gaps at both 50 PSI and 90 PSI inlet pressure. After transfection, the cells were plated on freshly coated LN521-coated culture plates and allowed to recover for 2 days followed which time they were harvested and their gDNA was collected. We then examined the efficiency of the cas9-induced cutting using a T7E1 assay while simultaneously using the amount of gDNA recovered from each culture as a measure of relative viability. As shown in Fig. 2A, the amount of DNA isolated decreased sharply with decreasing gap size at 90 PSI, while viability at all gaps improved when the inlet pressure was lowered to 50 PSI. Given that there was not a substantial difference in cutting efficiency between devices with different gaps (Fig. 2B and C), we decided to attempt to induce HDR using the 10.4 µm gap device as the larger gaps apply less force on the cells, leading to higher cell survival.
Figure 2.
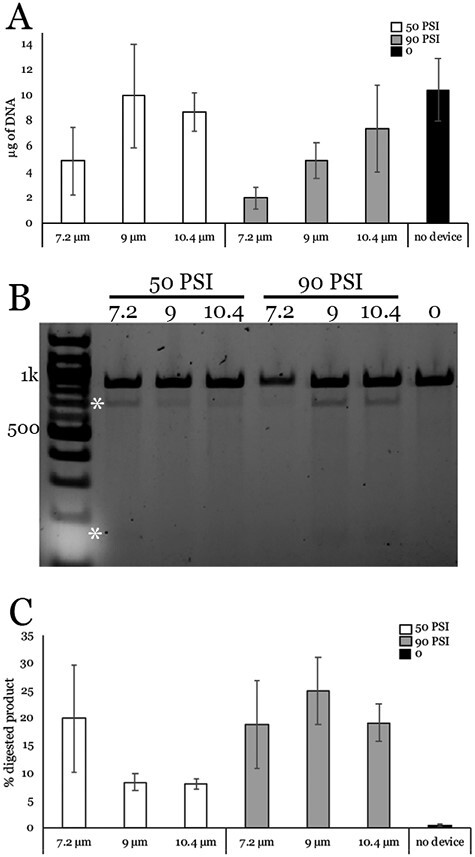
Analysis of CRISPR-Cas9 sgRNA-mediated cleavage efficiency using the CellFE Zephyr platform. (A) Amount of DNA isolated from iPSCs harvested from one well of a 6-well plate that were transfected 2 days prior using the given gap size and pressure. (B) Representative gel image showing T7E1 assay to analyze cleavage efficiency. Asterisks indicate cleavage products. (C) Densitometric analysis of digested products. Error bars represent standard error of the mean (SEM) from 3 independent experiments.
After determining appropriate operating conditions for the transfection system, we next determined if it could be used to correct disease causing mutations via HDR. We used the 10.4 µm gap device to co-deliver a plasmid HDR cassette along with pre-complexed spCas9/sgRNA RNPs at 2 pressures. As shown in Fig. 3A-C, our treated cells showed good survival and cell morphology 2 days post-transfection. As we described previously, we treated cells with puromycin to select for cells that incorporated the HDR cassette.14 We first analyzed the bulk population of cells with junction PCR (using a forward primer in the puromycin resistance cassette and a reverse primer outside the HDR cassette (Fig. 1)) and detected integration of the HDR cassette at both pressures (data not shown). We proceeded to isolate colonies and found that 16 of the 31 colonies amplified the expected PCR product when subjected to junction PCR. We then performed digital PCR on the positive colonies to determine what percentage of sequences showed correction of c.119-2A>C. Several colonies had ~50% of sequences correct, indicating monoallelic correction, while 1 colony (C34) had ~100% correct, indicating biallelic correction (Fig. 3D). We used Sanger sequencing to confirm monoallelic genomic correction of colony 11 (40 of 90 PCR products correct, 44.4%) and biallelic genomic correction of colony 34 (84 of 87 PCR products correct, 97%) (Fig. 3E, F). Colonies with >50% and <100% correction were determined to be polyclonal (ie, contained iPSCs with one and 2 alleles corrected). No off-target sequence modifications were observed in either colony (Supplementary material).
Figure 3.
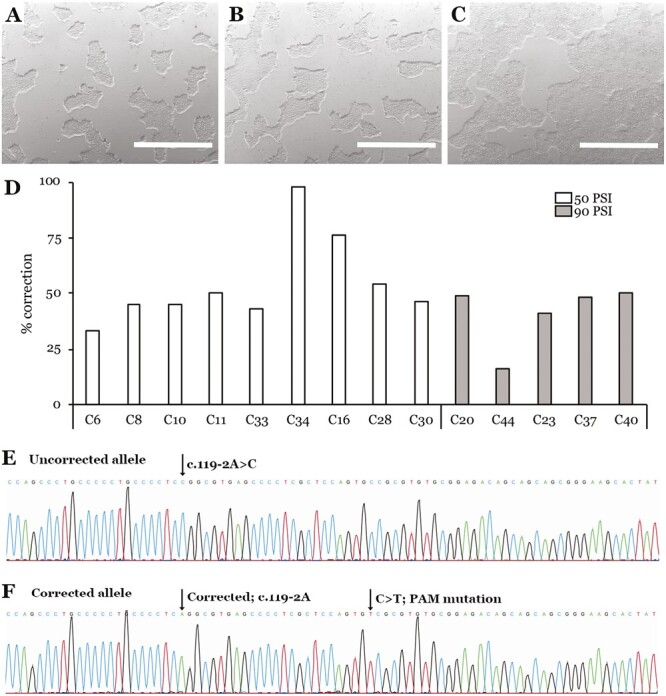
CRISPR-based homology-directed repair of the c.119-2A>C mutation in patient-derived iPSCs. (A, C) Bright field images of cells 2 days post-transfection using 50PSI (A), 90PSI (B), or no device (C). Scale bar = 1 mm. (D) Digital PCR analysis of colonies after puromycin selection using wild type or c.119-2A>C probes to determine percent correction. Note—colonies that deviate from 50% (monoallelic) or 100% (biallelic) correction are polyclonal. Representative Sanger sequencing chromatograms of uncorrected sequence (E) and corrected and PAM mutation (F).
We selected 2 colonies (one with monoallelic correction and one with biallelic correction) to subject to further analysis to assess their suitability for use in the manufacture of photoreceptor replacement therapies. As shown in Fig. 4, both colonies were successfully expanded into cell lines with normal karyotype (A and D), morphology (B and E), and expression of the pluripotency markers NANOG and OCT4 (C and F). To further demonstrate cell potency, patient iPSCs were differentiated into retinal organoids. CRISPR corrected cell lines had a similar propensity to generate retinal organoids as their founding uncorrected parental iPSC line (Fig. 5A-C). In addition, CRISPR corrected organoids showed characteristic morphology (Fig. 5D and G) and staining of early retinal progenitor markers LHX2, CHX10, OTX2, and Recoverin (Fig. 5E, F, H and I). Collectively, these data indicate that the mechanical transfection protocol developed was sufficient to induce HDR in patient-derived iPSCs without deleterious off-target edits, disrupting chromosomal structure, or altering cellular potency. As this approach does not use specialized transfection reagents and can be done using readily available cGMP qualified reagents, it is ideal for incorporation into a clinical cell manufacturing pipeline.
Figure 4.
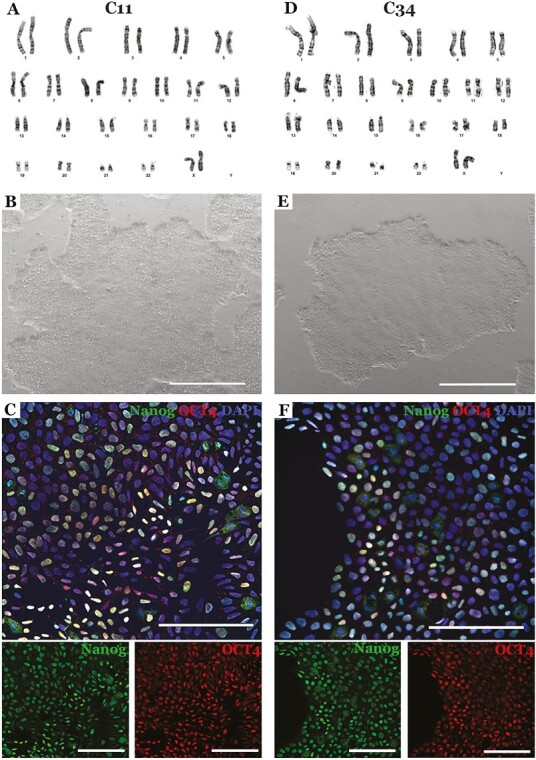
Characterization of iPSC colonies following Zephyr-mediated mono- (C11) and biallelic (C34) CRISPR correction. (A, D) Sample karyotypes demonstrating that CRISPR corrected cells obtained via mechanoporation retained a normal karyotype (A = C11, D = C34). (B, E) Brightfield images demonstrating that CRISPR corrected cells generated via mechanoporation maintained a normal iPSC morphology (B = C11, E = C34). (C, F) Immunohistochemical analysis demonstrating that following Zephyr-mediated CRISPR correction patients iPSCs retained expression of the pluripotency markers NANOG and OCT4 (C = C11, F = C34). DAPI = nuclear stain. Scale bar in B and E = 1 mm. Scale bar in C and F = 100 μm.
Figure 5.
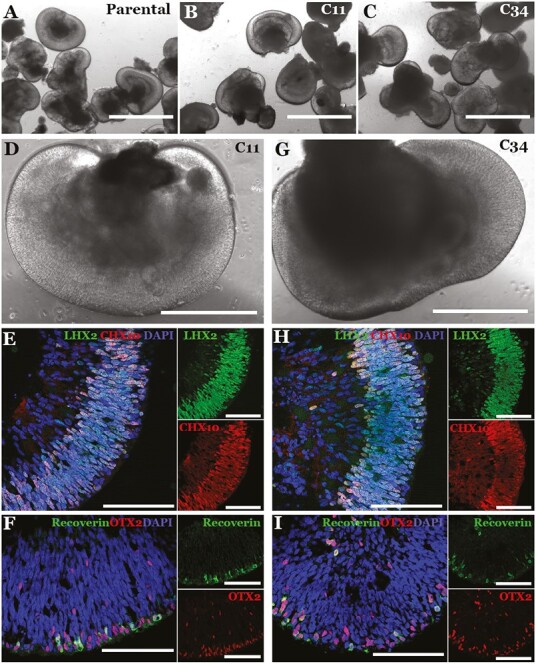
Retinal differentiation of patient iPSCs following Zephyr-mediated mono- (C11) and biallelic (C34) CRISPR correction. (A-I) Brightfield (A-D, G) and confocal microscopy (E-F, H-I) of iPSC-derived 40-day 3D retinal organoids following Zephyr-mediated monoallelic (C11) and biallelic (C34) CRISPR correction. For confocal microscopy, antibodies targeted against the retinal progenitor cell markers LHX2 and CHX10 (E, H) and the photoreceptor precursor cell markers Recoverin (green) and OTX2 (red) (F, I) were used. DAPI = nuclear stain. Scale bar in A-C = 1000 µm. Scale bar in D and G = 400 µm. Scale bar in E, F, H, and I = 100 µm.
Discussion
While we have shown that microfluidic transfection systems can deliver gene-editing reagents to iPSCs, it is important to consider the advantages and disadvantages of these new approaches as compared to conventional transfection strategies such as electroporation and lipofection. One limitation shared by all these approaches is the need to optimize the transfection procedure for each cell population to be processed. Performance of microfluidic transfection devices such as the ones used in this work are sensitive to the diameter of cells being treated and could fail to deliver cargo to cells that are too small or simply clog if used with cells that are too large. Therefore, working with different cell populations may require the use of multiple devices with varying channel geometry. However, this limitation is not unique to mechanoporation given that lipofection and electroporation also must be optimized on a population-to-population basis.
Once the transfection procedure is optimized, we can see qualitative differences in the experience of using the different approaches. Microfluidic transfection using the Zephyr is a simple procedure to perform. One prepares a cell suspension containing the desired cargo, loads it into a device and performs the transfection at the optimized pressure. While the experience of preparing the delivery suspension, loading it into a consumable and performing the transfection on a piece of dedicated equipment is very similar to that of electroporation, the difference is that microfluidic transfection can be performed in complete media, rather than a proprietary buffer. This has the advantage of easier adaptation to cGMP and automated cell manufacturing systems.
One current limitation of the microfluidic transfection strategy presented in this work is the high concentrations of cas9 RNPs and plasmid HDR construct required to achieve gene editing. For example, the results presented in this paper required the use of 12.5 µg of cas9, 7.5 µg of sgRNA, and 20 μg of plasmid HDR construct to perform a 500-µL transfection of ~500,000 cells. Meanwhile protocols currently in use in our group require 5.9 µg of cas9, 1.4 µg of sgRNA, and 1 μg of plasmid HDR construct per 100 µL electroporation reaction of approximately 1 million cells. While reagent consumption can be reduced by simply decreasing the volume of delivery suspension used, this obviously decreases the total yield of transfected cells. This is not of great concern for iPSC-derived therapies as the transfected cells can simply be expanded after processing, but this could be an issue for other workflows.
One issue that should be considered regardless of transfection method used, is genetic purity of the resulting cell line following CRISPR correction, colony isolation, and cell line expansion. As demonstrated in Fig. 3, several of the iPSC colonies that were screened via digital PCR deviated from the 50% and 100% correction that would be expected had the colony contained a true monoclonal cell population. For colonies such as 16 (ie, Figure 3D, C16), a mixture of cells that had monoallelic correction (ie, 50% of alleles edited) and those that had biallelic correction (ie, 100% of alleles edited) were present. While we could readily obtain monoclonal cell lines by increasing the number of colonies screened, these data indicate that subcloning and single-cell expansion could be used to generate additional cell lines with monoallelic and biallelic correction as needed. One caveat, however, is that true single iPSC clonal expansion results in significantly increased selection pressure, which can lead to selection of cell lines with karyotypic abnormalities (eg, karyotypic anomalies that bestow a significant growth advantage are common54-57). As such, CRISPR corrected clonally expanded cell lines should be considered independent of the parental cell line and karyotyped to ensure genomic integrity (Figure 4).
Conclusion
Patient-specific iPSC-derived cell therapies hold great promise for the treatment of many conditions, including inherited retinal degenerative blindness. While mature protocols have been developed by many groups to produce cell therapies from patient-derived iPSCs, these protocols often require the use of transfection technologies such as lipofection to repair disease causing mutations, which are not compatible with cGMP. In this work, we used a microfluidic transfection system to co-deliver cas9 RNPs with an HDR cassette to repair a disease-causing mutation in the NR2E3 gene of a patient-derived iPSC line. This approach, which used well defined reagents in a cGMP compatible workflow, produced several corrected lines, each of which exhibited normal cell morphology and karyotype that could produce high quality retinal tissue suitable for use as a cell therapy. These results demonstrate that microfluidic transfection is now a mature, readily available technology capable of enabling the development of cGMP compatible, clinical cell manufacturing protocols.
Supplementary Material
Contributor Information
Laura R Bohrer, Institute for Vision Research, Carver College of Medicine, University of Iowa, Iowa City, IA, USA; Department of Ophthalmology and Visual Sciences, Carver College of Medicine, University of Iowa, Iowa City, IA, USA.
Nicholas E Stone, Institute for Vision Research, Carver College of Medicine, University of Iowa, Iowa City, IA, USA; Department of Ophthalmology and Visual Sciences, Carver College of Medicine, University of Iowa, Iowa City, IA, USA.
Allison T Wright, Institute for Vision Research, Carver College of Medicine, University of Iowa, Iowa City, IA, USA; Department of Ophthalmology and Visual Sciences, Carver College of Medicine, University of Iowa, Iowa City, IA, USA.
Sewoon Han, CellFE, Inc., Alameda, CA, USA.
Ian Sicher, CellFE, Inc., Alameda, CA, USA.
Todd A Sulchek, George W. Woodruff School of Mechanical Engineering, Georgia Institute of Technology, Atlanta, GA, USA.
Robert F Mullins, Institute for Vision Research, Carver College of Medicine, University of Iowa, Iowa City, IA, USA; Department of Ophthalmology and Visual Sciences, Carver College of Medicine, University of Iowa, Iowa City, IA, USA.
Budd A Tucker, Institute for Vision Research, Carver College of Medicine, University of Iowa, Iowa City, IA, USA; Department of Ophthalmology and Visual Sciences, Carver College of Medicine, University of Iowa, Iowa City, IA, USA.
Conflict of Interest
S.H. and I.S. are employees, hold intellectual property, and are equity holders of CellFE, Inc., a company developing microfluidic-based cell engineering products. T.S. receives income, holds intellectual property, and is an equity holder of CellFE, Inc.
Data Availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Funding
This research was funded by the National Institutes of Health (R01 EY026008) and the National Science Foundation (2134701).
Ethics Approval
This study was approved by the Institutional Review Board of the University of Iowa (project approval #200202022) and adhered to the tenets set forth in the Declaration of Helsinki.
Author Contributions
L.R.B., N.E.S., B.A.T. planned and performed the experiments, analyzed the resulting data, and wrote the manuscript. S.H. and I.S. assisted with initial device optimization. A.T.W. performed cell culture and ICC. T.A.S. and R.F.M. assisted in planning experiments and editing manuscript. All authors reviewed and approved the manuscript. First author order was determined alphabetically.
References
- 1. Tucker BA, Park IH, Qi SD, et al. Transplantation of adult mouse iPS cell-derived photoreceptor precursors restores retinal structure and function in degenerative mice. PLoS One. 2011;6(4):e18992. 10.1371/journal.pone.0018992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lamba DA, McUsic A, Hirata RK, et al. Generation, purification and transplantation of photoreceptors derived from human induced pluripotent stem cells. PLoS One. 2010;5(1):e8763. 10.1371/journal.pone.0008763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zerti D, Hilgen G, Dorgau B, et al. Transplanted pluripotent stem cell-derived photoreceptor precursors elicit conventional and unusual light responses in mice with advanced retinal degeneration. Stem Cells. 2021;39(7):882-896. 10.1002/stem.3365 [DOI] [PubMed] [Google Scholar]
- 4. Lin B, McLelland BT, Aramant RB, et al. Retina organoid transplants develop photoreceptors and improve visual function in RCS rats with RPE dysfunction. Invest Ophthalmol Vis Sci. 2020;61(11):34. 10.1167/iovs.61.11.34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Santos-Ferreira TF, Borsch O, Ader M.. Rebuilding the missing part-A review on photoreceptor transplantation. Front Syst Neurosci. 2016;10:105. 10.3389/fnsys.2016.00105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Goureau O, Orieux G.. Photoreceptor cell transplantation for future treatment of retinitis pigmentosa. Med Sci MS. 2020;36(6-7):600-606. [DOI] [PubMed] [Google Scholar]
- 7. Gagliardi G, Ben M’Barek K, Goureau O.. Photoreceptor cell replacement in macular degeneration and retinitis pigmentosa: A pluripotent stem cell-based approach. Prog Retin Eye Res. 2019;71:1-25. 10.1016/j.preteyeres.2019.03.001 [DOI] [PubMed] [Google Scholar]
- 8. Mandai M, Watanabe A, Kurimoto Y, et al. Autologous induced stem-cell-derived retinal cells for macular degeneration. N Engl J Med. 2017;376(11):1038-1046. 10.1056/NEJMoa1608368 [DOI] [PubMed] [Google Scholar]
- 9. Sharma R, Khristov V, Rising A, et al. Clinical-grade stem cell-derived retinal pigment epithelium patch rescues retinal degeneration in rodents and pigs. Sci Transl Med. 2019;11(475). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wiley LA, Burnight ER, DeLuca AP, et al. cGMP production of patient-specific iPSCs and photoreceptor precursor cells to treat retinal degenerative blindness. Sci Rep. 2016;6:30742. 10.1038/srep30742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Takahashi K, Yamanaka S.. Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell. 2006;126(4):663-676. 10.1016/j.cell.2006.07.024 [DOI] [PubMed] [Google Scholar]
- 12. Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861-872. 10.1016/j.cell.2007.11.019 [DOI] [PubMed] [Google Scholar]
- 13. Burnight ER, Gupta M, Wiley LA, et al. Using CRISPR-Cas9 to generate gene-corrected autologous iPSCs for the treatment of inherited retinal degeneration. Mol Ther. 2017;25(9):1999-2013. 10.1016/j.ymthe.2017.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bohrer LR, Wiley LA, Burnight ER, et al. Correction of NR2E3 associated enhanced S-cone syndrome patient-specific iPSCs using CRISPR-Cas9. Genes. 2019;10(4):278. 10.3390/genes10040278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Giacalone JC, Sharma TP, Burnight ER, et al. CRISPR-Cas9-based genome editing of human induced pluripotent stem cells. Curr Protoc Stem Cell Biol. 2018;44:5B.7.1-5B.7.22. 10.1002/cpsc.46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pruett-Miller SM, Connelly JP, Maeder ML, Joung JK, Porteus MH.. Comparison of zinc finger nucleases for use in gene targeting in mammalian cells. Mol Ther. 2008;16(4):707-717. 10.1038/mt.2008.20 [DOI] [PubMed] [Google Scholar]
- 17. Maeder ML, Thibodeau-Beganny S, Osiak A, et al. Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol Cell. 2008;31(2):294-301. 10.1016/j.molcel.2008.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Christian M, Cermak T, Doyle EL, et al. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186(2):757-761. 10.1534/genetics.110.120717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Oceguera-Yanez F, Kim SI, Matsumoto T, et al. Engineering the AAVS1 locus for consistent and scalable transgene expression in human iPSCs and their differentiated derivatives. Methods. 2016;101:43-55. 10.1016/j.ymeth.2015.12.012 [DOI] [PubMed] [Google Scholar]
- 20. Karakikes I, Termglinchan V, Cepeda DA, et al. A comprehensive TALEN-based knockout library for generating human-induced pluripotent stem cell–based models for cardiovascular diseases. Circ Res. 2017;120(10):1561-1571. 10.1161/CIRCRESAHA.116.309948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wiley LA, Anfinson KR, Cranston CM, et al. Generation of Xeno-Free, cGMP-compliant patient-specific iPSCs from skin biopsy. Curr Protoc Stem Cell Biol. 2017;42(1):4A.12.1-4A.12.14. 10.1002/cpsc.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tucker BA, Mullins RF, Streb LM, et al. Patient-specific iPSC-derived photoreceptor precursor cells as a means to investigate retinitis pigmentosa. eLife. 2013;2:e00824. 10.7554/eLife.00824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhong X, Gutierrez C, Xue T, et al. Generation of three dimensional retinal tissue with functional photoreceptors from human iPSCs. Nat Commun. 2014;5:4047. 10.1038/ncomms5047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Meyer JS, Howden SE, Wallace KA, et al. Optic vesicle-like structures derived from human pluripotent stem cells facilitate a customized approach to retinal disease treatment. Stem Cells. 2011;29(8):1206-1218. 10.1002/stem.674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Meyer JS, Shearer RL, Capowski EE, et al. Modeling early retinal development with human embryonic and induced pluripotent stem cells. Proc Natl Acad Sci USA. 2009;106(39):16698-16703. 10.1073/pnas.0905245106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kallman A, Capowski EE, Wang J, et al. Investigating cone photoreceptor development using patient-derived NRL null retinal organoids. Commun Biol. 2020;3(1):82. 10.1038/s42003-020-0808-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wahlin KJ, Maruotti J, Zack DJ.. Modeling retinal dystrophies using patient-derived induced pluripotent stem cells. Adv Exp Med Biol. 2014;801:157-164. 10.1007/978-1-4614-3209-8_20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ueda K, Onishi A, Ito SI, Nakamura M, Takahashi M.. Generation of three-dimensional retinal organoids expressing rhodopsin and S- and M-cone opsins from mouse stem cells. Biochem Biophys Res Commun. 2018;495(4):2595-2601. 10.1016/j.bbrc.2017.12.092 [DOI] [PubMed] [Google Scholar]
- 29. Collin J, Zerti D, Queen R, et al. CRX expression in pluripotent stem cell-derived photoreceptors marks a transplantable subpopulation of early cones. Stem Cells. 2019;37(5):609-622. 10.1002/stem.2974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Reichman S, Terray A, Slembrouck A, et al. From confluent human iPS cells to self-forming neural retina and retinal pigmented epithelium. Proc Natl Acad Sci USA. 2014;111(23):8518-8523. 10.1073/pnas.1324212111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mellough CB, Sernagor E, Moreno-Gimeno I, Steel DHW, Lako M.. Efficient stage-specific differentiation of human pluripotent stem cells toward retinal photoreceptor cells. Stem Cells Dayt Ohio. 2012;30(4):673-686. 10.1002/stem.1037 [DOI] [PubMed] [Google Scholar]
- 32. Rowland TJ, Buchholz DE, Clegg DO.. Pluripotent human stem cells for the treatment of retinal disease. J Cell Physiol. 2012;227(2):457-466. 10.1002/jcp.22814 [DOI] [PubMed] [Google Scholar]
- 33. Tucker BA, Scheetz TE, Mullins RF, et al. Exome sequencing and analysis of induced pluripotent stem cells identify the cilia-related gene male germ cell-associated kinase (MAK) as a cause of retinitis pigmentosa. Proc Natl Acad Sci USA. 2011;108(34):E569-E576. 10.1073/pnas.1108918108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jin ZB, Okamoto S, Osakada F, et al. Modeling retinal degeneration using patient-specific induced pluripotent stem cells. PLoS One. 2011;6(2):e17084. 10.1371/journal.pone.0017084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Osakada F, Jin ZB, Hirami Y, et al. In vitro differentiation of retinal cells from human pluripotent stem cells by small-molecule induction. J Cell Sci. 2009;122(Pt 17):3169-3179. 10.1242/jcs.050393 [DOI] [PubMed] [Google Scholar]
- 36. Boucherie C, Mukherjee S, Henckaerts E, et al. Brief report: self-organizing neuroepithelium from human pluripotent stem cells facilitates derivation of photoreceptors. Stem Cells. 2013;31(2):408-414. 10.1002/stem.1268 [DOI] [PubMed] [Google Scholar]
- 37. Fligor CM, Huang KC, Lavekar SS, VanderWall KB, Meyer JS.. Differentiation of retinal organoids from human pluripotent stem cells. Methods Cell Biol. 2020;159:279-302. 10.1016/bs.mcb.2020.02.005 [DOI] [PubMed] [Google Scholar]
- 38. Zerti D, Dorgau B, Felemban M, et al. Developing a simple method to enhance the generation of cone and rod photoreceptors in pluripotent stem cell-derived retinal organoids. Stem Cells. 2020;38(1):45-51. 10.1002/stem.3082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hallam D, Hilgen G, Dorgau B, et al. Human-induced pluripotent stem cells generate light responsive retinal organoids with variable and nutrient-dependent efficiency. Stem Cells. 2018;36(10):1535-1551. 10.1002/stem.2883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Eiraku M, Takata N, Ishibashi H, et al. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature. 2011;472(7341):51-56. 10.1038/nature09941 [DOI] [PubMed] [Google Scholar]
- 41. Deng WL, Gao ML, Lei XL, et al. Gene correction reverses ciliopathy and photoreceptor loss in iPSC-derived retinal organoids from retinitis pigmentosa patients. Stem Cell Rep. 2018;10(4):1267-1281. 10.1016/j.stemcr.2018.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kim S, Lowe A, Dharmat R, et al. Generation, transcriptome profiling, and functional validation of cone-rich human retinal organoids. Proc Natl Acad Sci. 2019;116(22):10824-10833. 10.1073/pnas.1901572116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Homma K, Usui S, Kaneda M.. Knock-in strategy at 3ʹ-end of Crx gene by CRISPR/Cas9 system shows the gene expression profiles during human photoreceptor differentiation. Genes Cells. 2017;22(3):250-264. 10.1111/gtc.12472 [DOI] [PubMed] [Google Scholar]
- 44. Jang YY, Ye Z.. Gene correction in patient-specific iPSCs for therapy development and disease modeling. Hum Genet. 2016;135(9):1041-1058. 10.1007/s00439-016-1691-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Burnight ER, Giacalone JC, Cooke JA, et al. CRISPR-Cas9 genome engineering: Treating inherited retinal degeneration. Prog Retin Eye Res. 2018;65:28-49. 10.1016/j.preteyeres.2018.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Brookhouser N, Raman S, Potts C, Brafman DA.. May I cut in? Gene editing approaches in human induced pluripotent stem cells. Cells. 2017;6(1):5. 10.3390/cells6010005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Liu A, Islam M, Stone N, et al. Microfluidic generation of transient cell volume exchange for convectively driven intracellular delivery of large macromolecules. Mater Today. 2018;21(7):703-712. 10.1016/j.mattod.2018.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Liu A, Yu T, Young K, et al. Cell mechanical and physiological behavior in the regime of rapid mechanical compressions that lead to cell volume change. Small. 2019 Nov;16(2):1903857-1903857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mullin NK, Anfinson KR, Riker MJ, et al. Sensitive quantification of m.3243A>G mutational proportion in non-retinal tissues and its relationship with visual symptoms. Hum Mol Genet. 2022;31(5):775-782. 10.1093/hmg/ddab289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Capowski EE, Samimi K, Mayerl SJ, et al. Reproducibility and staging of 3D human retinal organoids across multiple pluripotent stem cell lines. Development. 2019;146(1):dev171686. 10.1242/dev.171686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Han X, Liu Z, Jo M, et al. CRISPR-Cas9 delivery to hard-to-transfect cells via membrane deformation. Sci Adv. 2015;1(7):e1500454. 10.1126/sciadv.1500454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hur J, Chung AJ.. Microfluidic and nanofluidic intracellular delivery. Adv Sci. 2021;8(15):e2004595. 10.1002/advs.202004595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sharei A, Zoldan J, Adamo A, et al. A vector-free microfluidic platform for intracellular delivery. Proc Natl Acad Sci USA. 2013;110(6):2082-2087. 10.1073/pnas.1218705110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sobol M, Raykova D, Cavelier L, et al. Methods of reprogramming to induced pluripotent stem cell associated with chromosomal integrity and delineation of a chromosome 5q candidate region for growth advantage. Stem Cells Dev. 2015;24(17):2032-2040. 10.1089/scd.2015.0061 [DOI] [PubMed] [Google Scholar]
- 55. Taapken SM, Nisler BS, Newton MA, et al. Karyotypic abnormalities in human induced pluripotent stem cells and embryonic stem cells. Nat Biotechnol. 2011;29(4):313-314. 10.1038/nbt.1835 [DOI] [PubMed] [Google Scholar]
- 56. Assou S, Girault N, Plinet M, et al. Recurrent genetic abnormalities in human pluripotent stem cells: definition and routine detection in culture supernatant by targeted droplet digital PCR. Stem Cell Rep. 2020;14(1):1-8. 10.1016/j.stemcr.2019.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Vitillo L, Anjum F, Hewitt Z, et al. The isochromosome 20q abnormality of pluripotent cells interrupts germ layer differentiation. Stem Cell Rep. 2023;18(3):782-797. 10.1016/j.stemcr.2023.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.


