Acinetobacter baumannii is a major cause of hospital-acquired infections and among the top five pathogens associated with mortality.1 Due to its ability to rapidly acquire antimicrobial resistance traits, MDR isolates have been reported worldwide. Carbapenem-resistant A. baumannii (CRAB) are a particular concern, as only few treatment options, including colistin, tigecycline/eravacycline and cefiderocol, are currently available. For this reason, this pathogen has been listed by the WHO as ‘priority 1’ pathogen for research of new antimicrobials.2 In this context, rapid diagnostics is crucial to guide best antibiotic treatment3 and to prevent nosocomial transmission of CRAB. The most prevalent acquired carbapenemases in A. baumannii are class D oxacillinases, including the OXA-23, OXA-40 and OXA-58 groups. Other less frequently acquired carbapenemases include class A (e.g. carbapenemase variants of GES-type) and class B MBLs (e.g. NDM, VIM and IMP). Existing phenotypic methods are quite labour-intensive and exhibit variable performances in detecting carbapenemase production in A. baumannii.4,5 Molecular methods including PCR or loop-mediated isothermal amplification (LAMP) assays allow for accurate detection of most prevalent carbapenemase genes but require expensive equipment.6 Isothermal detection methods combined with lateral flow strips have been recently developed for rapid detection of the most prevalent carbapenemase genes in A. baumannii, but currently these assays are not commercially available.7 Immunochromatographic lateral flow assays (LFIAs) for the detection of carbapenemase-producing A. baumannii isolates are available on the market, but only allow detection of single carbapenemase types, such as OXA-23 (OXA-23 K-SeT, Coris BioConcept, Belgium) or metallo-carbapenemases (NG-Biotech, France).8 A recently developed type of LFIA for rapid detection of the most prevalent acquired carbapenemases in A. baumannii, including OXA-23, OXA-40/58 and NDM-types, is the ‘RESIST ACINETO’ assay (Coris BioConcept, Belgium).9 It is important here to note that although OXA-40 and OXA-58 belong to different families of OXA carbapenemases, their detection is combined in a single band and thus cannot be distinguished. This may represent a drawback for tracking certain types of outbreaks.
Here we evaluate retrospectively the diagnostic performance of this new assay using a collection of 131 A. baumannii clinical isolates (Figure 1). Fourteen strains were obtained from the Institut Pasteur’s strain collection (https://www.pasteur.fr/en/public-health/biobanks-and-collections/collection-institut-pasteur-cip), while the remaining 117 clinical isolates were derived from single patients between January 2014 and December 2022 in the routine diagnostic laboratory of the Institute of Medical Microbiology at the University of Zurich. Of these, 106 exhibited carbapenem-resistant profiles, while 25 were susceptible to carbapenems. β-Lactamase-genes were detected by WGS, which was performed using our in-house available Illumina MiSeq platform with paired-end 150-nt reads. Intrinsic oxacillinases, as well as acquired carbapenem resistance markers, including carbapenemases and ESBLs, were detected using Unicycler v0.4.8 assemblies10 in combination with ABRicate (https://github.com/tseemann/abricate) and the NCBI database.11 All strains were typed in Ridom SeqSphere+ by MLST according to the Pasteur (ST) scheme and in addition with core-genome MLST.12 RESIST ACINETO was performed on isolated colonies grown overnight on blood agar plates (tryptic soy agar with 5% sheep blood, bioMérieux, France) at 37°C according to the manufacturer’s instructions. All genomes were submitted to the ENA (https://www.ebi.ac.uk/ena/browser) under project number PRJEB62871.
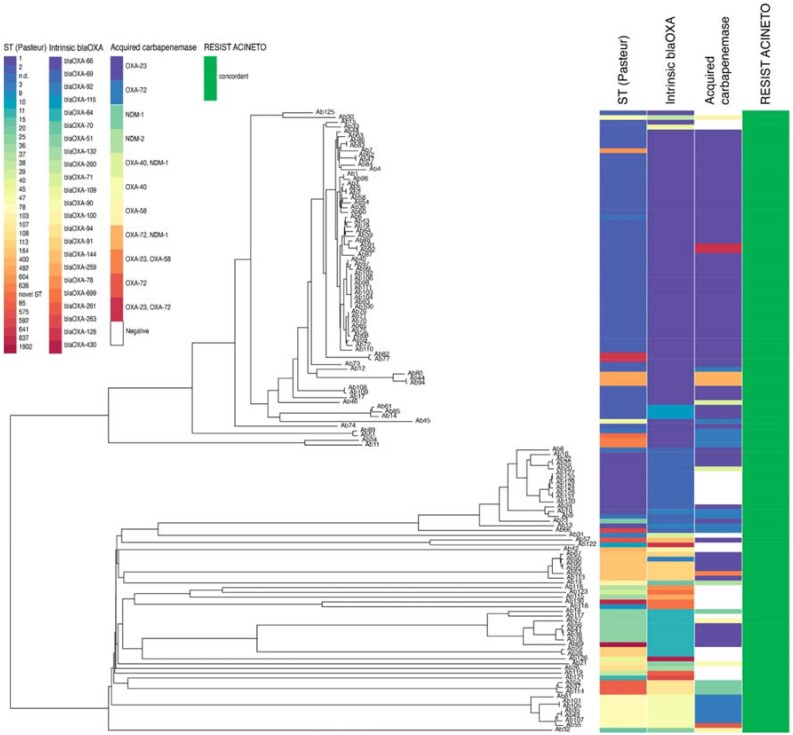
Figure 1.
Phylogenetic neighbour-joining tree of A. baumannii. The tree was generated in Ridom SeqSphere+ based on core genes with associated metadata in columns, from left to right: ST (Pasteur), intrinsic oxacillinases identified in the genome, acquired carbapenemases and RESIST ACINETO result (green for congruent result).
The strain collection comprised 25 carbapenemase-negative and 106 carbapenemase-producing isolates. Seventy-two of 106 isolates harboured blaOXA-23 (68%), 17 blaOXA-72 (16%), three blaOXA-58 and one blaOXA-40, while three isolates carried two oxacillinase genes (two blaOXA-23/blaOXA-58 and one blaOXA-23/blaOXA-72). Four isolates harboured blaNDM-1 and one blaNDM-2, while the remaining carbapenemase producers harboured a combination of genes coding for NDM-1 and an oxacillinase (two blaNDM-1/blaOXA-23, three blaNDM-1/blaOXA-72). The A. baumannii isolates belonged to 34 different STs, with ST2 being the most prevalent (58/131; 36.7%).
RESIST ACINETO correctly identified all six carbapenemase variants, including those from the isolates producing two carbapenemases, thus exhibiting excellent sensitivity (100%). Strong bands appeared within 5–10 min of incubation in all but one case, where a faint band corresponding to NDM emerged at 15 min incubation. Nonetheless this isolate was classified as a true positive. OXA-72 was identified as a member of the OXA-40 group of OXA β-lactamases. No false positive results, which might also arise due to cross-reactivity with one of the 23 detected intrinsic OXA-51-like oxacillinases, among which OXA-66 was the most prevalent (66/131, 50.4%), were observed (specificity 100%).
A limitation of our study is that the collection of A. baumannii isolates is biased and reflects the epidemiological situation of the Zurich region in Switzerland, with only six different carbapenemase variants identified so far. Also, while some types were abundantly present, such as OXA-23 (68%), other globally present types, such as OXA-40, were underrepresented (1%). Moreover, in this study the performance of the RESIST ACINETO was tested on A. baumannii colonies grown on blood agar plates. Considering that most laboratories identify A. baumannii on Columbia agar or MacConkey agar plates, further studies with a more diverse collection of carbapenemase variants and isolates grown on different media are warranted to fully evaluate the robustness of the method.
In conclusion, RESIST ACINETO provides a reliable test for the detection of the most prevalent carbapenemases in A. baumannii. The sensitivity and specificity from isolated colonies of overnight growth is excellent (each 100%).
Acknowledgements
We thank the technician team of the IMM for dedicated help and the University of Zurich for continuous support.
Contributor Information
Stefano Mancini, Institute of Medical Microbiology, University Zurich, Gloriastrasse 28/30, 8006 Zurich, Switzerland.
Helena M B Seth-Smith, Institute of Medical Microbiology, University Zurich, Gloriastrasse 28/30, 8006 Zurich, Switzerland.
Natalia Kolesnik-Goldmann, Institute of Medical Microbiology, University Zurich, Gloriastrasse 28/30, 8006 Zurich, Switzerland.
Vladimira Hinic, Institute of Medical Microbiology, University Zurich, Gloriastrasse 28/30, 8006 Zurich, Switzerland.
Tim Roloff, Institute of Medical Microbiology, University Zurich, Gloriastrasse 28/30, 8006 Zurich, Switzerland.
Frank Imkamp, Institute of Medical Microbiology, University Zurich, Gloriastrasse 28/30, 8006 Zurich, Switzerland.
Adrian Egli, Institute of Medical Microbiology, University Zurich, Gloriastrasse 28/30, 8006 Zurich, Switzerland.
Funding
This work was supported by the University of Zurich, Zurich, Switzerland.
Transparency declarations
All authors: no conflicts of interest to declare.
References
- 1. Resistance C A. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 2022; 399: 629–55. 10.1016/S0140-6736(21)02724-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tacconelli E, Carrara E, Savoldi A et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis 2018; 18: 318–27. 10.1016/S1473-3099(17)30753-3 [DOI]
- 3. Palombo M, Bovo F, Amadesi S et al. Synergistic activity of cefiderocol in combination with piperacillin-tazobactam, fosfomycin, ampicillin-sulbactam, imipenem-relebactam and ceftazidime-avibactam against carbapenem-resistant Gram-negative bacteria. Antibiotics (Basel) 2023; 12: 858. 10.3390/antibiotics12050858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mitteregger D, Wessely J, Barisic I et al. A variant carbapenem inactivation method (CIM) for Acinetobacter baumannii group with shortened time-to-result: rCIM-A. Pathogens 2022; 11: 482. 10.3390/pathogens11040482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Simner PJ, Opene BNA, Chambers KK et al. Carbapenemase detection among carbapenem-resistant glucose-nonfermenting Gram-negative bacilli. J Clin Microbiol 2017; 55: 2858–64. 10.1128/JCM.00775-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Haldorsen BC, Janice J, Samuelsen O. Evaluation of the Amplex eazyplex(R) SuperBug Acineto test for detection of acquired OXA and NDM carbapenemases in Acinetobacter spp. J Glob Antimicrob Resist 2021; 24: 340–1. 10.1016/j.jgar.2021.01.019 [DOI] [PubMed] [Google Scholar]
- 7. Hu S, Niu L, Zhao F et al. Identification of Acinetobacter baumannii and its carbapenem-resistant gene blaOXA-23-like by multiple cross displacement amplification combined with lateral flow biosensor. Sci Rep 2019; 9: 17888. 10.1038/s41598-019-54465-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mertins S, Higgins PG, Rodriguez MG et al. Generation and selection of antibodies for a novel immunochromatographic lateral flow test to rapidly identify OXA-23-like-mediated carbapenem resistance in Acinetobacter baumannii. J Med Microbiol 2019; 68: 1021–32. 10.1099/jmm.0.001015 [DOI] [PubMed] [Google Scholar]
- 9. Mertins S, Higgins PG, Thunissen C et al. Development of an immunochromatographic lateral flow assay to rapidly detect OXA-23-, OXA-40-, OXA-58- and NDM-mediated carbapenem resistance determinants in Acinetobacter baumannii. J Med Microbiol 2023; 72. 10.1099/jmm.0.001681 [DOI] [PubMed] [Google Scholar]
- 10. Wick RR, Judd LM, Gorrie CL et al. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 2017; 13: e1005595. 10.1371/journal.pcbi.1005595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Feldgarden M, Brover V, Haft DH et al. Validating the AMRFinder tool and resistance gene database by using antimicrobial resistance genotype-phenotype correlations in a collection of isolates. Antimicrob Agents Chemother 2019; 63: e00483-19. 10.1128/AAC.00483-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Higgins PG, Prior K, Harmsen D et al. Development and evaluation of a core genome multilocus typing scheme for whole-genome sequence-based typing of Acinetobacter baumannii. PLoS One 2017; 12: e0179228. 10.1371/journal.pone.0179228 [DOI] [PMC free article] [PubMed] [Google Scholar]