Abstract
Amyotrophic lateral sclerosis is a relentless neurodegenerative disease that is mostly fatal within 3–5 years and is diagnosed on evidence of progressive upper and lower motor neuron degeneration. Around 15% of those with amyotrophic lateral sclerosis also have frontotemporal degeneration, and gene mutations account for ∼10%. Amyotrophic lateral sclerosis is a variable heterogeneous disease, and it is becoming increasingly clear that numerous different disease processes culminate in the final degeneration of motor neurons. There is a profound need to clearly articulate and measure pathological process that occurs. Such information is needed to tailor treatments to individuals with amyotrophic lateral sclerosis according to an individual’s pathological fingerprint. For new candidate therapies, there is also a need for methods to select patients according to expected treatment outcomes and measure the success, or not, of treatments. Biomarkers are essential tools to fulfil these needs, and urine is a rich source for candidate biofluid biomarkers. This review will describe promising candidate urinary biomarkers of amyotrophic lateral sclerosis and other possible urinary candidates in future areas of investigation as well as the limitations of urinary biomarkers.
Keywords: ALS, urine, biomarker, proteins, metabolites
Finding objective fluid-based biomarkers for amyotrophic lateral sclerosis that reflect the pathological processes occurring in a phenotypically heterogeneous disease is essential for assessing if treatments are working or not. This requires evaluating alternate non-invasive biofluids such as urine, with caveats such as standardizing collection, processing and validating candidates.
Graphical Abstract
Graphical Abstract.
Introduction
Amyotrophic lateral sclerosis (ALS) is a relentlessly progressive disease resulting in the death of the upper and lower motor neurons, around 15% of those with ALS also have frontotemporal degeneration (ALS/FTD), and gene mutations account for ∼10% of ALS.1 Numerous different disease processes culminate in the final degeneration of upper and lower motor neurons that can be fatal within 2–5 years after diagnosis. Some people diagnosed with ALS survive longer than 4–5 years, and others experience a much more rapid disease progression.1,2 This phenotype variability reflects a highly complex disease, where tools to group people at diagnosis, those that respond to specific treatments and those that can predict outcomes in clinical trials are crucial. Such tools are biomarkers, and numerous types are needed to report on the intertwined pathological processes occurring in a particular patient with ALS and develop personalized treatments.2 The National Institute of Health originally defined biomarkers as ‘measurable characteristics, indicative of biological, pathological or pharmacodynamic responses to therapeutic interventions’.3 The Food and Drug Administration in conjunction with the National Institute of Health has since produced a guidance document that summarizes each type of biomarker tool (diagnostic, monitoring, response or pharmacodynamic, predictive and prognostic), and the ways to ensure the tool are adequate for its proposed purpose, i.e. validation.4 These tools can be physical, physiological, imaging, genetic or biofluid tests.
Although numerous candidate prognostic, predictive and pharmacodynamic biomarkers for use in ALS have been identified, only the revised ALS functional rating scale (ALSFRS-R) has progressed to use in clinical trials for ALS.5,6 The ALSFRS-R is a questionnaire-based scoring scale in which 12 activities of daily living are scored from 0 to 4 (4 is normal) and summed to produce a score between 48 (i.e. healthy) and 0.7,8 Candidate biomarkers include electrophysiological, neurophysiological, tissue sampling, imaging, genetic and biofluid biomarkers. Those present in biofluids show great potential as a source for objective candidates in ALS. They are generally advantageous over electrophysiological, neurophysiological, tissue analysis and imaging markers in that testing is often easier for the participant, may incur lower expense and allows for reproducible quantification.2,9-11 Biofluid candidates are being increasingly tested alongside candidate treatments in adaptive platform trials, such as the HEALEY ALS Platform trial for ALS,12 with the same protocol and a shared placebo group used across multiple treatments being tested.
Why urine? Components, comparison of urine to CSF and blood
The source of biofluid for ALS biomarker investigation is an important consideration. CSF is close to motor neuron injury, and neuroinflammation in ALS and for those reasons has been thought to have advantages as an ALS biomarker source, over systemic biomarkers such as blood and urine. However, if biomarkers found in urine and blood can be shown to be related to motor neuron degeneration and neurological status, they also should be investigated. This is shown in biomarkers such as neurofilament light (NfL)13-15 and interleukin (IL)-1816 being as useful in blood as they are as CSF biomarkers. Urine with a less complex proteome than blood and retaining the metabolome is an attractive and underinvestigated source of systemic biomarkers for ALS. Other advantages of urine are that large quantities can be collected, and it is less invasive for participants, when compared with CSF, especially for repeated collection.
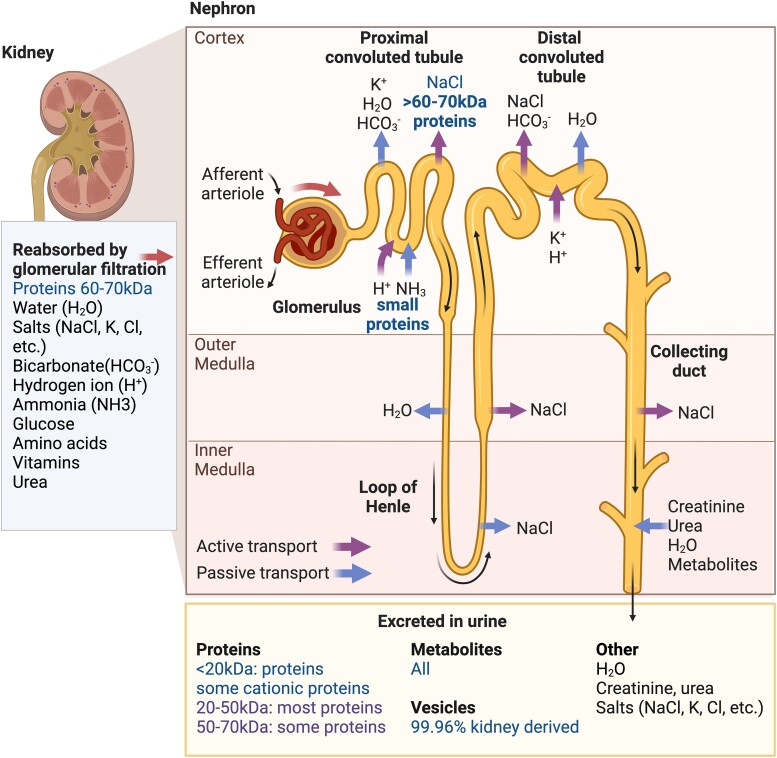
Urine consists of components from the circulation, which are filtered through the kidney into the bladder and then excreted via the urethra. The human kidney (Fig. 1) is composed of more than 1 million functional units called nephrons, which are subdivided into two parts: the glomerulus, filtering plasma from renal blood flow, and the renal tubule, playing a vital role in the reabsorption of nutrients, fluids and other compounds that the body needs back to the blood. Approximately 180 L of fluid a day is filtered from renal blood flow that allows for toxins, metabolic waste products and excess ions and electrolytes to be excreted while keeping essential substances in the blood.17,18 Clearance of solutes occurs through a combination of glomerular filtration, tubular secretion and tubular reabsorption while maintaining plasma homeostasis.17,19 Urine is collected in the bladder and voided via the urethra. Human urine comprises water (95%), urea (2%), creatinine (0.1%), uric acid (0.03%) and lower levels of chloride, sodium, potassium, sulphate, ammonium, phosphate, other ions and molecules (including metabolites and protein) and cells and extracellular/microvesicles.20-23 Physiologically, 20–30% of proteins present in the blood appear in urine, whereas metabolites are freely filtered into the urine in exact proportion to that present in blood24 (Fig. 1).
Figure 1.
Urinary biomarkers for ALS derived from normal kidney function. Urine is a rich source of candidate ALS biomarkers, whose solute content is determined by the size and charge of components passing through the nephrons. The functional unit of the kidney is the nephron that consists of the glomerulus, which filters blood, and the renal tubule, which reabsorbs necessary components back into the blood. These components include salts like sodium chloride (NaCl), potassium (K+) and chloride (Cl−) ions. Hydrogen (H+), ammonia (NH3+) and bicarbonate (HCO3−) are among other ions that are reabsorbed, alongside glucose, amino acids, vitamins and urea. All the blood metabolites and proteins less than 20 kDa in size pass through the nephron and into the urine. Most proteins of 20–50 kDa size are excreted, while only some 50–70 kDa proteins, especially cationic proteins, are excreted. Most proteins larger than 60–70 kDa are actively transported back into blood at the proximal convoluted tubule. Levels of other urinary components such as salts and urea are determined by their active and passive transport along the tubule, while most vesicles found in urine are derived from the urogenital system.
Urinary proteome
Although human urine is easy to obtain non-invasively, proteomic analysis and biomarker discovery have lagged that of serum/plasma, which has much more complex proteome and contains approximately 60–70 mg protein/ml.25 Over 99% of the blood proteome is composed of 22 high-abundance proteins (e.g. albumin, transferrin and immunoglobulins) unrelated to most disease states, making it difficult to identify low-abundance biomarkers by conventional mass spectrometry and enzyme-linked immunosorbent assay (ELISA).26 These high-abundance proteins are also found in the CSF.27 Conversely, human urine rarely exceeds 0.2 mg protein/ml17,18 because the renal system efficiently reabsorbs most proteins. In the final voided urine, protein does not exceed ∼150 mg/day, of which albumin accounts for 20 mg17 meaning it is often easier to identify low-abundance proteins.
An important consideration in surveying urine for biomarkers is our knowledge about how proteins enter the final urinary matrix (Fig. 1). Our understanding dating back to the 1960s indicates that the kidney’s filtration system progressively restricts the size of molecules excreted in the urine.28 All proteins with a molecular weight of 20 kDa or less easily cross the filtration barrier and those with a molecular weight of 20–50 kDa are mostly filtered into the urine. As the molecular mass of a protein increases, the fraction that is filtered progressively decreases such that compounds of 60–70 kDa are largely retained in the capillary lumen and reabsorbed. More recently, it has been shown that the charge of the proteins also impacts the size of the proteins excreted. Positively charged proteins (cationic) are more freely filtered than negatively charged (anionic) proteins as the glomerular barrier is anionically charged by glycosaminoglycan heparan sulphate in the glomerular basement membrane.29 Thus, anionic molecules of the same molecular weight are filtered to only 50% of those with cationic charge. Hence, the cut-off of 50 kDa may be larger if the protein is positively charged.30 Around 20–30% of proteins found in blood are also found in urine. There are also soluble proteins secreted by epithelial cells and extracellular vesicles (EVs) that all arise from the urogenital system and are excreted in urine.31 Despite the restriction on protein species that comes from the circulation, urine is a rich source of proteins/peptides for biomarker discovery. A caveat is the reported heterogeneity according to time of day, sex, age, diet, pH (4–8), proteolysis while the urine is stored in the bladder and degradation of collected urine samples upon storage.32-34 In a quantitative analysis of variability in the normal urinary proteome, inter-individual variability exceeded 47% and intra-individual variability exceeded 45%.32 Standardized protocols are essential to reduce variability in the urinary proteome, the most important being the time of collection and the time between collection and sample processing.32
As of 2023, there have been 4500 potential urinary protein groups identified and confirmed (UniProt search); many were found because of recent improvements in mass spectrometry detection and instrumentation. Practically, around 2000 protein groups in urine can be found using mass spectrometry.35,36 This can be contrasted with blood that contains an estimated possible 4500 protein groups,37 but only 300–600 are usually detectable, with a similar profile in CSF (proteinatlas.org/humanproteome). The number of groups of proteins identified in urine is clearly higher than in blood or CSF.32-34 This is mainly due to urine having a less complex proteome; it is not dominated by proteins with high abundance as is the case for blood26 or CSF27 but instead contains a wide range of low-abundance proteins.34 This source of biomarkers for ALS is relatively unexplored. Table 1 lists current ALS protein biomarkers, including those found in urine, such as size, relationship to pathology and neurological status and if upregulated or downregulated in ALS.
Table 1.
Protein biomarkers of ALS in biofluids
| Name, accession number | Pathological association | Size (kDa) | Biofluid | Change in ALS | Detection technique | Link to disease |
|---|---|---|---|---|---|---|
| p75ECD, P08138 | Neurodegeneration | 30–50 | Urine | Increase | WB, LC-MS, ELISA | Prognostic, correlated to ALSFRS-R, increases longitudinally over disease progression.38-40 |
| NfL, P07196 | Neurodegeneration | 61–68, 22 | CSF, serum, blood | Increase | Immunoassay, ELISA, SIMOA, WB | Prognostic, correlated to ALSFRS-R and progression rate.15,41-44 NA for whole blood WB.45 |
| pNfH, P12036 | Neurodegeneration | 190–210 | CSF, serum | Increase | ELISA, SIMOA | Prognostic, correlated to ALSFRS-R and progression rate.15,41,46,47 |
| Titin, Q8WZ42 | Muscle degeneration | ∼23 | Urine | Increase | ELISA | Prognostic, especially for lower limb ALS.48 |
| C9ORF72: poly-glycine-proline, NA | Disease specific | ND | CSF | Increase | ELISA | Related to repeats,49-51 not disease.50 |
| TDP-43, ratio pTDP: TDP, Q13148 | Disease specific | 45, 43–50 | Plasma, CSF | Increase or NA | SIMOA, WB, LC- MS, ELISA | Improves diagnosis in combination with NfL in SIMOA assay.52 Relationship to disease NA.53,54 Ratio pTDP: TDP, no correlation to disease.55 |
| FUS, P35637 | Disease specific | 53 | Plasma | Increase | WB | Relationship to disease not investigated.54 |
| SOD1, P00441 | Disease specific | 15.4 | Plasma | Increase | WB | Relationship to disease not investigated.54 |
| pTau and tTau, P10636 | Other | 40–60 | CSF | Increase | Immunoassay | tTau and pTau/tTau prognostic.56 |
| Collagen, P53420 | Other | 164 | Serum, urine | Decrease | Radioimmunoassay | Correlated to disease duration.57,58 |
| MCP-1, P13500 | Immune system | 11–13 | CSF, serum | Increase, NS | ELISA | CSF prognostic.16,59-61 |
| CXCL13, O43927 | Immune system | 13 | CSF | Decrease | Immunoassay | Relationship to prognosis and ALSFRS-R not investigated.62 |
| CHIT1, Q13231 | Immune system | 51 | CSF | Increase | LC-MS, ELISA | Prognostic, correlated to disease progression.59,63-66 |
| CHI3L1, P36222 | Immune system | 40 | CSF | Increase | LC-MS, ELISA | Prognostic, correlated to progression.59,65,66 |
| CHI3L2, Q15782 | Immune system | 39 | CSF | Increase | LC-MS | Correlates to disease progression.66 |
| C reactive protein, P02741 | Immune system | 22 | CSF, serum | Increase/NS | LC-M, ELISA/mesoscale | Significant in CSF.67 NS in CSF/serum.16 Serum levels increased in ALS;68 prognostic.69 |
| IL-6, P05231 | Immune system | 21–28 | CSF, serum | Increase/NS | ELISA/immunoassay | CSF correlated to duration70/NS.16 Increased in serum.71 |
| IL-18, Q14116 | Immune system | 23 | CSF | Increase | Immunoassay | Correlated to progression and changes over progression.16 |
| TNF-α, P01375 | Immune | 17 | CSF, serum | Increase | Immunoassay | Not consistently associated with ALS prognosis or progression.71,72 |
| Cystatin C, P01034 | Immune system | 13.3 | CSF | Decrease | ELISA | Correlated to survival, prognostic.73-75 |
| Native transthyretin, CysGly, P02766 | Immune system | 55 69 |
CSF CSF |
Decrease Increase |
LC-MS | Relationship to ALS not tested.76 CysGly—transthyretin does not change over ALS progression.67 |
| sCD14, P08571 | Immune system | 40 | CSF, serum, urine | Decrease Increase |
ELISA | Lower CSF CD14 associated with poor prognosis.77 Elevated in CSF, urine and serum. Serum sCD14 prognostic.68 |
LC, liquid chromatography; MS, mass spectrometry; NA, not applicable; NS, not significant; WB, western blot.
Urinary protein candidates that have been identified as ALS biomarkers or candidates in other biofluids
Related to neurodegeneration
p75ECD
A neurodegeneration biomarker identified in urine is the extracellular domain (ECD) of the common neurotrophin receptor p75 (p75ECD). The full-length receptor, p75NTR, is the 16th member of the tumour necrosis factor (TNF) superfamily. It is a 75 kDa transmembrane protein that binds immature (pro) or mature neurotrophins (brain-derived growth factor, nerve growth factor, neurotrophin 3 and neurotrophin 4/5). This receptor is involved in promoting cell survival or apoptosis dependent on its ligands, how it complexes with other cell receptors (e.g. tyrosine kinase receptors A, B and C and sortilin), cell type and surrounding environment.78,79 The result is that p75NTR is involved in processes such as neuron development, pruning and maintenance of nerve cells. Importantly, for ALS, p75NTR is expressed by motor neurons during development, only to be downregulated in adulthood and only re-expressed by motor neurons and Schwann cells following neuronal damage such as that in ALS animal models80,81 and in people with ALS.80,82,83 Additionally, p75NTR undergoes regulated intramembrane proteolysis, in which the 50 kDa ECD of the 75 kDa receptor is cleaved by TNF-α-converting enzyme, α-secretase/ADAM10/17.79,84 In the 1980s, DiStefano et al.85 described the appearance of the ECD in rat urine via radioimmunoassay, post-bilateral sciatic nerve lesions. This observation was followed up in our laboratory using immunoprecipitation experiments on SOD1G93A mouse urine to show the appearance of ECD in an ALS animal model.38 Further immunoprecipitation experiments showed that humans with ALS also have ECD fragments in their urine at a molecular weight of 35–50 kDa, with the presence of ECD confirmed by mass spectrometry.38 An ELISA was then developed to measure mouse and human urinary p75ECD, and this assay showed increased levels of ECD in people with ALS compared with healthy controls.38 Using this ELISA, p75ECD was further found to be a candidate biomarker of disease progression in ALS and baseline p75ECD was prognostic for survival and correlated to disease severity described using the ALSFRS-R.39,40 Hence, urinary p75ECD can be classified as a monitoring and prognostic biomarker.
Recently, urinary p75ECD testing has been included as an exploratory biomarker in several clinical trials.86,87 Notably, in a Phase 2a safety trial of the antiretroviral Triumeq, although not designed to determine efficacy, urinary p75ECD levels over the treatment period showed potential as a biomarker.86 A follow-up randomized clinical trial is underway where urinary p75ECD has been included as a potential biomarker of efficacy (NCT05193994).
Neurofilament light and heavy chains, not yet identified in urine
Neurofilaments are important structural components of myelinated axons and help to increase axon diameter, allowing for faster nerve conductance. In the CNS, neurofilaments are protein polymers composed of NfL (61–68 kDa) and alpha-internexin (68–72 kDa), which form the neurofilament core and co-assemble with neurofilament medium chain (∼150 kDa) and phosphorylated neurofilament heavy chain (pNfH, 190–210 kDa).41,46,47 All four proteins contain conserved rod domains and unique amino-terminal (N-terminal) and carboxyl-terminal (C-terminal) domains.46 Of these proteins, NfL is a well-established marker in CSF and serum of both acute and chronic neuronal damage, is increased in ALS, frontotemporal degeneration (FTD) and a number of diseases involving neuronal damage13,42-44,88-90 and is recognized as a neurodegenerative biomarker for ALS.15,91,92 pNfH in CSF and serum has also been demonstrated as a neurodegenerative biomarker for ALS.15,41,46,47 The specificity and sensitivity of the single molecule array (SIMOA) in comparison with ELISA have allowed accurate measurement of NfL14,15,93,94 and recently pNfH.15 Furthermore, NfL has been validated as a biomarker for clinical trials of familial ALS.92,95
Although detectable by ELISA or SIMOA, interestingly, NfL has not been readily detected by conventional shotgun mass spectrometry methods in the CSF63 or blood96,97 of ALS patients. One explanation is that NfL is at very low concentrations in CSF (100–500 pg/ml range).44,98,99 The levels in peripheral blood (10–100 pg/ml range) are even lower13,15 and may be masked by the high-abundance proteins in serum that make up 99% of the proteome.26,100 This is evidenced by the detection of NfL by mass spectrometry as significantly upregulated in the blood of ALS patients after depleting high-abundance proteins and enriching tissue-derived proteins101 and upregulated in CSF after using isobaric tags.102
Another explanation as to why it is difficult to identify NfL in mass spectrometry of blood and CSF is that the stability and fragment state of NfL has not been considered and may influence detection. In a normal state, it has been widely recognized that full-length NfL is slow to turn over. This was demonstrated in rodents103 and in retinal ganglion cells.104 NfL is, however, degraded by the calcium protease calpain,105,106 and degradation is influenced by phosphorylation state.106 Protease activity and phosphorylation are important in axonal degradation, but there is no indication as to the proportion of full-length and other NfL species in CSF and blood in ALS. There may be multiple NfL species as axons degrade as suggested by a recent study using immunocapture coupled with mass spectrometry where multiple molecular weight species of NfL were identified in Alzheimer’s CSF, but no full-length NfL was detected.107 Intriguingly, Malaspina et al.45 also found the main NfL band in plasma samples from ALS patients on a western blot was 22 kDa (not 61 kDa). It could be speculated that accurate serum NfL measurement from ALS patients by SIMOA means that it is not necessary to consider half-life and degradation. However, if the level of phosphorylation and protease activity is driving NfL accumulation, it is important to determine the most important NfL fragments that relate to ALS. The immunocapture study in CSF from Alzheimer’s patients indicated that the antibody used in the commercial NfL SIMOA targets a small peptide of eight amino acids: NFL 324–331. It would be interesting to determine the truncated species present in ALS CSF and blood. It would be unlikely that the 61 kDa NfL could enter the urine, as its size means it would be restricted and most likely reabsorbed.28-30 However, fragments of NfL may be present in urine and require further investigation. Interestingly, the only known study where urinary NfL was measured by SIMOA found no differences between urinary NfL in 93 people with stroke/haemorrhage disease compared with 10 healthy controls when corrected for urinary dilution.108 The form of NfL fragments in urine should be investigated and compared with blood and CSF, to determine if fragments of NfL are important as a urinary ALS biomarker.
Related to muscle degeneration
N-terminal titin
Another promising urinary candidate protein is the N-terminal fragment of titin, which is a marker of muscle degeneration. Titin (originally known as connectin) is a major myofibrillar component of skeletal muscle.109 Titin is large (3200–4200 kDa) and responsible for passive muscle elasticity. N- and C-terminal fragments of titin are produced through cleavage by matrix metalloproteinases-9 or -2 and calpain and have been detected in urine by proteomic studies.110,111 Matrix metalloproteinases are associated with the degradation of cardiac titin and calpain with skeletal muscle titin. Interestingly, a wide range of titin peptides covering the whole molecule are found in the urine of Duchenne muscular dystrophy model mice.112 Human and mouse ELISAs have been developed for the N-terminal fragment (∼23 kDa)113,114 and have revealed that the N-terminal fragment is increased in cardiac patients,115 pathological conditions such as oxidative stress and muscle loss including extensive literature in Duchenne muscular dystrophy.116,117 An interesting observation in Duchenne muscular dystrophy mouse studies was that there was an increase as muscular dystrophy peaks in early life and then a sharp decrease with age and muscle regeneration, with either creatinine or specific gravity as urinary dilution correction factors.114,118
Urinary N-terminal titin was suggested as a novel biomarker for ALS with strong survival and prognostic potential.48 It was also suggested as a progression biomarker, but, unlike urinary p75ECD, where sampling was every 3 months across disease progression,39,40 there was only one follow-up sample at 6 months,48 and no rate of progression reported, so its role as a progression biomarker is not clear. An earlier study looking at fragments of titin via western blot showed that people with ALS, Charcot–Marie–Tooth disease, limb girdle muscular dystrophy and myotonic dystrophy patients had minimal reactive fragments when compared with Duchenne muscular dystrophy.119 A recent publication has demonstrated in an animal model that urinary N-terminal fragments of titin (ratio to creatinine) do not increase post-sciatic nerve injury and urinary measures may not reflect early muscle denervation events.120 This suggests that urinary N-terminal titin may not be a marker of early or minor muscle degeneration and further investigation is required into the muscle loss required before urinary titin is detected in ALS. The literature so far suggests that urinary N-terminal fragments of titin are possible prognostic biomarkers for ALS. However, further investigation into titin over ALS progression and a comparison of urinary correction factors (i.e. specific gravity, osmolality and creatinine) is required. This is because muscle loss may involve reduced urinary creatinine when measured over 24 h, but this is not so clear in spot urine samples.121-123
Extracellular vesicles
EVs are nanosized, are released from cells, consist of a lipid membrane and contain cargo including DNA, RNA, proteins, lipids, amino acids and metabolites (microvesicles.org). Upon their discovery in biofluids,124 EVs were thought to remove intracellular waste, but later studies found they contain mRNA and miRNA and recipient cells can translate EV-associated mRNAs into proteins.125-127 Larger EVs called microvesicles (also known as ectosomes, microparticles, oncosomes or shedding vesicles) have a diameter of ∼100–1000 nm and are shed by budding of the plasma membrane; smaller EVs or exosomes are ∼30–140 nm in size, originate in the endosome and are released through exocytosis.128,129 Apoptotic bodies (∼800–5000 nm) are released during cell death and contain nuclear material, cellular organelles and membrane and cytosolic contents128,129
EVs and their contents make interesting candidate biomarker sources. EV contents remain stable due to the phospholipid bilayer membrane, and EVs have been isolated from CSF,130 blood131 and urine.23 It is important to consider that urine EVs are mostly derived from the kidney, bladder and genital tissues. Collecting duct epithelial cells secrete exosomes, but there is no evidence that endogenous non-urinary-derived EVs reach the urine under physiological conditions.132 In a large study, it was found that only 0.04% of urinary EV (uEV) proteins were derived from outside the urinary tract in healthy individuals.132 Nevertheless, global changes in the contents of urinary exosomes because of genetic disease have been proposed. For example, an increase in urinary exosome-associated phosphorylated Ser-1292 leucine-rich repeat kinase 2 (LRRK2; 286 kDa) may be a biomarker for familial Parkinson’s disease, and an increase in urinary pS1292-LRRK2 may be associated with a higher risk of converting to Parkinson’s.133 Disease-specific proteins of ALS (as mentioned below) would not be expected to be in uEVs unless associated with familial ALS and expressed in epithelial cells. Other miRNA, DNA, lipids and metabolites are also found in uEVs but, unless there is a body system-wide change, would not be likely to be biomarkers of ALS.
Disease-specific proteins
TDP-43
Ubiquitinated hyperphosphorylated cytoplasmic inclusions of the 43 kDa transactive response DNA-binding protein (TDP-43) is the pathological hallmark of ALS in 96% of all ALS cases and 50% of cases of FTD. TDP-43 is present in cytoplasmic inclusions and is not normally excreted,134 and its measurement in CSF and blood has shown a limited value as a diagnostic and prognostic biomarker, showing high variability in the results of different studies using ELISA,135-137 but clearly elevated using SIMOA as a more sensitive assay.52 Clearance of pathological inclusions may include an accumulation of both phosphorylated and total TDP-43 in the CSF, and measurement by ELISA/SIMOA specific for phosphorylated TDP-43 in a ratio with total TDP-43 may be more reliable as a biomarker in plasma.55 A mass spectrometry method developed by Turner et al. indicated intracellular brain-derived TDP-43 as a possible ALS biomarker based on the ratio of C:N terminal peptide fragment detection.138 Since it is not normally excreted, an attractive explanation is that pathological TDP-43 is sequestered from cells via microvesicles or EVs.53,54,139 It may be the EVs that enable a prion-like propagation of TDP-43 inclusions as alluded to by Don Cleveland’s group.134 TDP-43 has not been documented in urine, but the full-length 43 kDa and the 35 and 25 kDa C-terminal fragments may be at very low concentrations, which requires enrichment strategies for mass spectrometry detection. Since there has been no definitive proof uEV (proteins derived from outside the urinary system with the vast majority from epithelial cells of the collecting duct),132 it would be unlikely that pathological TDP43 inclusions that derive from the CNS would be in uEVs. Nevertheless, uEvs could contain mutant TDP43, but since this protein is not highly expressed in adult tissue,140,141 it may be difficult to detect.
C9ORF72 dipeptide repeat proteins
Pathologically, C9orf72 is the most common gene implicated in ALS and FTD affecting 40% of familial ALS and 25% of familial behavioural-variant FTD. A pathological mechanism of C9orf72 gene expansion entails the translation of the expansion into dipeptide repeat proteins (DPRs): glycine-alanine, glycine-arginine, proline-alanine, proline-arginine and glycine-proline (GP). Production of poly-proline-arginine, poly-G and poly-glycine-alanine leads to neurotoxicity via impaired protein translation.49,142,143 Dipeptide repeat proteins may be associated with phase separation144 that influences toxicity. Poly-glycine-proline in CSF, measured by ELISA, has attracted attention as a potential biomarker in C9orf72 gene expansion carriers in both behavioural-variant FTD and ALS.49-51 Asymptomatic mutation carriers have also been found to have elevated CSF poly-glycine-proline, and levels are raised in those diagnosed with ALS, in most, but not all mutation carriers.50 The size of the dipeptide repeat proteins is, however, unknown,49 nor if present in urine.
FUS/SOD1
ALS-causing mutations such as fused in sarcoma (FUS) and superoxide dismutase 1 (SOD1) conspicuously lack TDP-43 proteinopathy in most cases.145 Mutations are found in the FUS C-terminal nuclear localization sequence, causing the mislocalization of the normally nuclear protein to the cytoplasm, leading to the accumulation of cytoplasmic FUS and FUS aggregation.146 FUS is a 53 kDa protein, present in cellular inclusions, and has not been reported in body fluids including urine.
It was reported that EVs most likely contain mutant SOD1.54 There have been over 200 SOD1 mutations identified in ALS, and familial and sporadic SOD mutations account for about 5% of ALS. This type of familial ALS results in the death of motor neurons, similar to sporadic ALS, and is not caused by a change in enzyme activity.2,147 SOD1 is a 15.4 kDa protein and is ubiquitously found in human urine and difficult to distinguish between kidney derived and that from mutant SOD1 ALS-associated protein in ALS urine. uEVs that derive from kidney epithelium132 may contain mutant SOD1 or FUS but have not been investigated.
Other proteins
Ratio of phosphorylated tau to tTau
Tau is a neuronal microtubule-associated protein that is present in several different isoforms (40–60 kDa) depending on post-translational modifications, including phosphorylation.56 Specific kinases mediate the phosphorylation of tau at threonine 181 (pTau), which lost its affinity for tubulin, leading to microtubule instability and disintegration. Total tau (tTau) and pTau have been proposed as biomarkers in neurodegenerative disorders. However, the clinical usefulness of tTau and pTau as diagnostic and prognostic biomarkers in ALS is still debated. There are various studies indicating that tau, ptau and tTau in CSF, is a candidate ALS biomarker148 or not.149,150 Studies have also shown CSF pTau/tTau ratio as an ALS biomarker,56,148,151 and this may be prognostic and diagnostic, differentiating patients with ALS from mimics.56 The type of tau in urine has not been investigated, but the size may restrict excretion.
Collagen type IV
Collagen type IV makes up about 50% of all basement membrane components152,153 and provides support to epithelium, endothelium, muscles, fat cells, Schwann cells and axons.152 Type IV collagen has three polypeptide α-chains in triple helix form (540 kDa), and because of its size, the only passage of entry into the urine is through leaky tubules in kidney injury and diabetes.154 In ALS, abnormalities in the basement membrane and its surrounding structures were reported in SOD1G93A mice.155 In ALS patients, an immunohistochemical study found that there was decreased type IV collagen in the skin and in serum by radioimmunoassay.57 Interestingly, there was significantly less collagen IV in urine from a small study of 20 people with ALS compared with 20 healthy controls but no correction for creatinine.58 In ALS, a lower level of collagen IV compared with healthy controls contrasts with higher levels in kidney dysfunction and agrees with loss of collagen in the skin, and this is a systemic process. This is supported by not only a decrease in skin and urinary collagen but also less collagen metabolites such as the 486.5 Da glucosylgalactosyl hydroxylysine156 measured by reverse-phase high-performance liquid chromatography (HPLC) using a standard curve. Larger studies using ELISA and mass spectrometry HPLC should be undertaken to determine the diagnostic and prognostic ability of urinary collagen IV in ALS.
Additional proteins related to ALS pathology
Progranulin, vascular endothelial growth factor, TGF-beta and ferritin/transferrin
Progranulin is a conserved 593 amino acid, 88 kDa glycosylated, secreted protein. Pathogenic mutations of the progranulin gene have been found associated with FTD (but not ALS) and result in reduced progranulin in the CSF.157 It is not changed in CSF of ALS compared with healthy controls,158 and there is no correlation between CSF and serum progranulin.159 Progranulin and granulin peptides produced by proteolysis that promotes inflammatory activity are produced by many types of tissue.160 Taken together, including the size of the whole molecule, the usefulness of urinary progranulin as a biomarker for ALS is not clear. Other growth factors involved in neurodegeneration such as vascular endothelial growth factor72 in CSF (21–27 kDa) and transforming growth factor-beta (44 kDa) and receptors in CSF and serum100,161,162 have been suggested as ALS biomarkers but have not been measured in urine. Blood-based ferritin and hepcidin associated with iron metabolism have also been proposed as biomarkers of ferroptosis in ALS, a process associated with oxidative stress.163,164 However, the kidney is actively involved in iron homeostasis as it reabsorbs filtered iron to prevent loss in the kidney such that it would be expected to be difficult to distinguish the relationship between serum and urine ferroptosis in people with normal kidney function and blood iron status.165
Immune markers
The pathology of ALS results in damage to neurons, resulting in the innate and adaptive immune system responding to reduce the damage and then being overwhelmed. The innate system is triggered by aggregated proteins (e.g. TDP-43, FUS or SOD1) or danger signals (e.g. reactive oxygen species) produced by motor neurons. This results in reactive microglia/macrophages and astrocytes, the main components of the innate immune system in the CNS, responding in an anti-inflammatory manner, releasing, for example, neurotrophic factors. Intracellular nucleotide oligomerization domain-like receptor protein 3 inflammasome complexes in astrocytes also recognize and respond to misfolded proteins and mediate inflammatory responses. The adaptive immune system involving T cells is well described in ALS and Tregs, and M2 macrophages/microglia respond to danger signals by producing anti-inflammatory signals such as transforming growth factor-beta, IL-10, IGF-1 and IL-4, in the Th2 anti-inflammatory response and suppress T-helper type 1 cells in the case of Tregs.166 The adaptive response can thus be also observed in systemic change in ILs/chemokines. As the danger signals increase, the innate and adaptive anti-inflammatory response becomes overwhelmed, and there is a switch to a pro-inflammatory process, which results in the release of TNF-α, IL-6 and IL-1β and inflammasome-caspase 1, from glia and pro-inflammatory cytokines [e.g. ILs and interferon-gamma (IFN-γ)] from T-helper type 1 and 17 cells of the adaptive immune system that, in turn, worsens disease progression.167 Thus, there is an innate and adaptive response, and immune markers that reflect the stage of ALS if found in urine may be useful as prognostic, progression or predictive biomarkers for ALS.
MCP-1 and chemokines
A candidate biomarker for immune response in ALS is monocyte chemoattractant protein-1 (MCP-1) also known as chemokine C–C motif ligand 2. Chemokines are grouped into four classes based on the positioning of their N-terminal cysteine residues: CC, CXC, XC and CX3. MCP-1/chemokine C–C motif ligand 2 belongs to a sub-family of 27 CC chemokines with an N-terminal CC domain.168 It is a chemoattractant chemokine of 11–13 kDa that is involved in activating microglia and promoting the migration of peripheral immune cells such as monocytes/macrophages to inflammation sites.169,170 In the SOD1 mouse model of ALS, increased levels of MCP-1 are found in the spinal cord.171 Measurement of MCP-1 in CSF by ELISA has indicated that it is increased in ALS compared with disease mimics and healthy controls.59 However, levels are inconclusive in blood.16,60,61
Urinary MCP-1 is dysregulated in several kidney diseases and in diabetes.172 It has been suggested as a diagnostic biomarker for lupus nephritis173 and also for early kidney dysfunction and diabetic kidney disease.174 Urinary MCP-1 has also been suggested as a major indicator of pain175 and is increased in Alzheimer’s disease, but levels were also influenced by age and gender.176 Yet, there are no studies investigating MCP-1 in urine in ALS. A comparative study in CSF, blood and urine, in comparison with the ALSFRS-R, would determine the relevance of this marker for ALS prognosis and progression and as a predictive biomarker. Highlighting the need for other chemokines to be investigated is a recent report of chemokine CXCL-13 as a potential biomarker for ALS.62 Interestingly, the level of this marker declined in CSF and blood from ALS patients, sampled at various times from diagnosis. This interesting work could be repeated across biofluids at disease diagnosis, in comparison with the ALSFRS-R and over disease progression.
Chitinases
Chitin is a polysaccharide that is an essential structural component in numerous organisms. It is degraded by a chitinase, including chitotriosidase (CHIT1), an acidic mammalian chitinase, and several chitinase-like proteins: chitinase-3-like 1 (CHI3L1), chitinase-3-like 2 (CHI3L2), oviductin-specific glycoprotein and stabilin-1-interacting chitinase-like protein. CHIT1 (51 kDa), CHI3L1 (40 kDa) and CHI3L2 (39 kDa) have been investigated as biomarkers for ALS. CHIT1 and CHI3L2 are produced by macrophages, neutrophils and microglia, and CHI3L1 is also produced by reactive astrocytes.64,65,177-179 There has been quite extensive work examining these chemokines as prognostic biomarkers in CSF and blood but not urine.
CHIT1 was identified in CSF using mass spectrometry and found to be a candidate prognostic marker.63,64 A longitudinal mass spectrometry analysis indicated that CHIT1, CHI3L1 and CHI3L2 correlate with disease progression and with pNfH levels.66 Using commercially available ELISAs, Gille et al.59 showed that CHIT1 and CHI3L1 poorly discriminate between ALS and mimics and weakly correlate with disease progression. CSF CHI3L1 was independently associated with survival and suggested as a prognostic biomarker. However, it should also be noted that a CHIT1 polymorphism has been identified that reduces the CHIT1 levels in CSF of patients with ALS.180 A contrasting study using both mass spectrometry and ELISA showed that CSF CHIT1 was significantly higher in ALS compared with disease and healthy controls, while CHI3L1 was higher in ALS and disease controls than healthy controls, and the rate of increase in these biomarkers correlates to disease progression.65
Unlike CSF, there has been no significant increase in CHIT1 nor CHI3L1 in blood from ALS compared with controls nor association with disease.60,64,65 This suggests that CHIT1 and CHI3L1 may be site specific rather than systemic inflammatory markers. Neither CHIT1 nor CHI3L1 has been investigated in urine but may not be viable urinary candidates as neither is significantly elevated in the blood when compared with controls; this, along with the CHIT1 polymorphism, complicates the interpretation of results.
C-reactive protein
C-reactive protein (CRP) is an acute phase protein of 22 kDa produced in the liver and secreted into the bloodstream mostly during an inflammatory episode, largely in response to IL-6 (IL-6) signalling and, to a lesser extent, IL-1beta and other pro-inflammatory cytokines.181 CRP plays a variety of key roles during inflammation. It binds to damaged, necrotic and microbial cells, promotes phagocytosis by neutrophils and macrophages, and activates the complement system, which itself helps maintain inflammation. Rising CRP concentrations furthermore activate neutrophils and monocytes and promote the secretion of IL-6, IL-1β and TNF-α.181 Because of these effects, CRP has been classically regarded as a pro-inflammatory molecule. At the same time, CRP has anti-inflammatory effects: it stimulates the release of anti-inflammatory agents such as IL-10 and IL-1Rα, and, while activating the complement system, it also recruits several complement inhibitors, possibly in a time-dependent manner.182 As a result, the net effect of CRP in vivo appears to be weakly anti-inflammatory.183 In addition, IL-6 stimulating production of acute phase proteins such as CRP can occur in the absence of inflammation, rather than as in ALS as part of clearance of damaged cells.
There have been many apparent inconsistencies concerning the physiological roles of CRP including in ALS, which have been clarified by the discovery that this protein exists in two isoforms with different functions, a pentameric isoform synthesized by the liver (pCRP) that is largely anti-inflammatory and a monomeric isoform (mCRP) that is activated by local cues of inflammation and tissue injury and is pro-inflammatory.183 mCRP stimulates secretion of pro-inflammatory cytokines, induces the M1 phenotype in macrophages and promotes the release of reactive oxygen species, which function not only to debilitate pathogens but also to exact collateral damage on host tissue.183,184 While mCRP activates the complement, it also blocks the final stages of the cascade in the presence of certain inhibitory factors. This mechanism may permit a tightly controlled activation of the complement during the non-inflammatory removal of damaged cells.182
There have been a number of studies investigating CRP as a biomarker for ALS showing significant increases in CSF67 and serum68,185,186 or no significance in CSF and serum.16 A systematic review has since shown serum CRP is prognostic for ALS.69 CRP has been used to group patients in a Phase 2B clinical trial of an immune modulator NP001, where it was postulated that high CRP would mean a faster progression rate, but this criteria grouped those whom had a slower progression but were responsive to anti-inflammatory therapy.187 None of the literature looks at specifically looks at mCRP and pCRP, so it is difficult to distinguish pro-inflammatory or anti-inflammatory states. Since serum CRP is a routine general non-specific inflammatory marker, urinary CRP may not be useful.
ILs and other cytokines
ILs are one of the most well-reported indicators of systemic anti- and pro-inflammation. They are mainly synthesized by T cells, macrophages and endothelial cells, promoting the development and differentiation of T and B cells, and hematopoietic cells and range in size from 12–30 kDa. A number of these have been reported as changed in ALS, including IL-1β, IL-1Rα, IL-1, IL-2, IL-4, IL-5, IL-6, IL-7, IL-8, IL-9, IL-10, IL-12p70, IL-13, IL-15, IL-17, IL-17A, IL-18 and IL-21.61,71 It has not been clearly demonstrated that the serum/plasma levels of these markers increase (or decrease) at baseline in ALS.64,65 However, some ILs found in CSF may have some utility in the separation of ALS from disease mimics. IL-6 and IL-18 have been found upregulated in the CSF of ALS patients,70 and in a large progression study, IL-18 (but not IL-6) was consistently elevated across progression in the CSF of ALS patients.16 TNF-α is a 17 kDa major pro-inflammatory cytokine secreted by activated macrophages and is involved in the induction of cytokine production, phagocyte cell activation, activation or expression of adhesion molecules, and growth stimulation.188 TNF-α is reported upregulated in the CSF,72 and in serum,71 but not consistently associated with disease state. Increased urinary ILs IL6ST and IL-19 have been reported in a small study in Alzheimer’s,189 although the levels were not adjusted for dilution. However, there are no reports of urinary ILs as a biomarker in ALS. One reason may be that urinary IL levels are not a clear surrogate for blood levels. This is supported by a study showing no correlation between the levels of 13 cytokines in urine and plasma among a group of healthy, reproductive-aged women.190
Cst C and transthyretin
Cystatin C (Cst C) is a potent extracellular inhibitor of cysteine protease of ∼13.3 kDa and is generally considered a ubiquitously expressed protein191 used as a biomarker of kidney function. A decline in urine accompanies a rise in blood and failed kidney function. Independent of its inhibition of protease, it has also been implicated in apoptosis and inflammation.192 Cst C and transferrin are found in Bunina bodies, which are inclusion bodies found in lower motor neurons in ALS193,194 and may be present with but distinct from those with TDP-43 inclusions.195 Cst C levels are decreased in the CSF of ALS patients73,74 and correlated with the survival time implying that it is prognostic and may be a potent neuroprotective in ALS.75 The neuroprotective effect of Cst C has been shown in cell culture experiments where mutant SOD1 was expressed.196 However, Cst C level is a common signature of neuron vulnerabilities and neurodegeneration and is not specific for ALS.197 Urinary Cst C in ALS has not been investigated but would be difficult to distinguish from being a marker of kidney function.192 Transthyretin is a 55 kDa protein that is primarily synthesized by the liver and the choroid plexus.198 Reduced levels of native transthyretin76 and increased levels of oxidized CysGly–transthyretin are found in the CSF in ALS compared with controls67 and may be involved in dealing with TDP-43 inclusions.199 There are also several genetic mutations of transthyretin resulting in amyloid neuropathy that can mimic motor symptoms of ALS.198 Like Cst C, it is not specific for ALS. Urinary transthyretin has not been investigated, but the 55 kDa size means a large proportion may be largely retained in the capillary lumen of the kidney tubules and reabsorbed.28
Soluble CD14
Monocytes/microglia express CD14 early in ALS, preceding onset.200 There is also increased CD14 in spinal cord tissue in ALS patients.171 Soluble CD14 (sCD14) is a 40 kDa part of the CD14 receptor that is cleaved from the cell surface and released after monocyte activation. In a small study, CSF, blood and urinary sCD14 from ALS patients was found elevated when compared with healthy controls and blood sCD14 could be prognostic in ALS and elevated compared with FTD, Alzheimer’s disease or immune-mediated neuropathy.68 Urinary dilution was not considered when quantifying sCD14 in urine. In another study, sCD14 in the CSF of patients at baseline was reduced, and lower levels were prognostic.77 Larger studies are needed to determine the usefulness of urinary sCD14 as an ALS biomarker.
Urinary metabolome
Considerations regarding metabolite candidates as ALS biomarkers
Metabolites are small molecules of less than 1.5 kDa201 that are end products of cellular or organ processes. Since the kidneys do an extraordinary job of concentrating certain metabolites from the blood, some compounds that are far below the limit of detection in the blood are well above the detection limit in urine.201 Although there are variations in concentration of these components, due to diet, sex and time of day,202 relative to other biofluids such as CSF203 or saliva,204 urine contains significantly more compounds (5–10×) and exhibits significantly more chemical diversity (2–3×). Table 2 shows current metabolomic biomarkers for ALS including urinary markers.
Table 2.
Metabolite (less than 1.5 kDa) biomarkers of ALS in biofluids
| Name of sub-pathway or metabolite | Pathological association | Biofluid | Change in ALS | Detection technique | Link to disease |
|---|---|---|---|---|---|
| Glutamate metabolism | Neurodegeneration | Plasma | Increase | LC-MS | Plasma amino acids Linked to early ALS.205 |
| Nucleotide metabolites | Neurodegeneration | Serum | Increase Decrease |
LC-MS | Higher in ALS than healthy controls.206 |
| Energy metabolites: glycolysis, gluconeogenesis | Energy metabolism | Serum/CSF | Increase | Colorimetric/LC-MS | Higher in ALS than healthy controls in CSF207 and serum.206 |
| Creatine metabolites | Muscle degeneration | Serum | Increase | LC-MS | Higher in ALS than healthy controls.206 |
| Creatinine | Muscle degeneration | Plasma | Increase | Colorimetric assay | Prognostic/not prognostic.208-212 |
| Diacylglycerols | Lipid metabolism | Serum | Increase | LC-MS | Higher in ALS than healthy controls.206,213 |
| Phosphatidylcholines: phosphatidylethanolamine | Lipid metabolism | Serum | Decrease | LC-MS | Related to progression213 and maybe diagnostic.214 |
| Ceramide | Lipid metabolism | Serum | Decrease | LC-MS | Higher in ALS than healthy controls.206,213 |
| 8-Hydroxydeoxyguanosine | Oxidative stress | Serum, CSF, urine | Increase | ELISA HPLC/colorimetric electrodes |
Serum: no clear association.163 Urine: correlated to ALSFRS-R.215 not correlated to ALSFRS-R.216 |
| Uric acid | Oxidative stress | Serum | Increase | Enzymatic-colorimetric method | High Uric acid linked to slow ALS progression.217 |
| Total antioxidant status | Oxidative stress | Serum | Increase | Colorimetric assay | Higher in ALS than controls.218 |
| Glutathione metabolites | Oxidative stress | Whole blood | Increase | Colorimetric assay | Higher in ALS than controls.218 |
| 4-Hydroxynonenal | Oxidative stress | CSF, serum | Increase | ELISA, colorimetric assay | Correlated to disease219 and ALSFRS-R.163 |
| Neopterin | Immune system | Urine | Increase | HPLC-UV, ELISA, | Prognostic.220 Not prognostic.40 Correlated to ALSFRS-R and increases longitudinally over disease progression.40 |
HPLC, high-performance liquid chromatography; LC, liquid chromatography; MS, mass spectrometry; UV, ultraviolet.
Metabolites related to ALS pathology
Metabolites dysregulated in CSF and or plasma/serum and linked to ALS pathology have been described, but these are rarely investigated in urine (Table 2). Those in CSF and serum/plasma include altered amino acid metabolism, e.g. excitatory amino acid glutamate,205,218 and those associated with muscle loss (e.g. creatinine kinase) altered carbohydrate metabolism (e.g. glycan metabolites and energy metabolites such as glucose), short-chain fatty acids (e.g. acyl carnitine),206,207 co-factors and vitamin and nucleotide metabolites.221 Other groups of metabolites altered in ALS include those related to antioxidant defence such as glutathione metabolites206 and those altered in lipid metabolism.213
A major issue with most of the studies is a discordance in collection protocols and how each is related to the clinical stage of disease, including time from diagnosis, and relationship to ALSFRS-R.222 Meta-analysis including CSF and serum showed 16 pathways altered in ALS,221 including caffeine metabolism, aminoacyl-tRNA biosynthesis and valine, leucine and isoleucine biosynthesis. However, it is not clear in the metabolomics studies if the samples were all taken from ALS patients with similar ALSFRS-R and time from diagnosis. Notable exceptions are single metabolite studies focused on oxidative stress such as urate and 8-hydroxydeoxyguanosine (8-OHdG), where correlation to ALSFRS-R and relationship to prognosis and progression are reported.215,217
Creatinine (113.12 Da) is a breakdown product of creatine phosphate and reflects the creatine pool and the impaired uptake of creatine into muscle cells.223 Creatinine is released from muscle at a constant rate, resulting in a stable plasma concentration and is freely filtered at the glomerulus and secreted by the proximal tubules.223,224 Creatinine clearance is commonly determined from a 24-h collection of urine.223 Alternative excretory pathways through the gut may occur224 especially when the kidney is not working. Total creatinine excretion in the steady state is dependent on muscle mass, and day-to-day creatinine excretion remains constant for an individual and is related to lean body weight, age, sex and ethnicity. In general, men excrete 20–25 mg creatinine/kg body weight/day, whereas women excrete 15–20 mg/kg/day and may be affected by diet, for example, eating a large meal of cooked meat, which may also result in a substantial increase in serum creatinine.224
Studies have also indicated that plasma creatinine levels may be a simple biomarker for ALS muscle wasting and decreased serum creatinine at baseline is prognostic for poor survival and associated with an aggravation of the disease progression.208-211 A meta-analysis of 14 studies of plasma creatinine concluded that mortality was higher if creatinine was lower than the median, but, noted caution, because confounding factors such as age, sex and body mass index were not uniformly included in some analyses and are known to be associated with ALS and plasma creatinine levels.225 Another study that included all the covariates did not find any relationship between plasma creatinine and survival nor progression rate but did find a correlation with the ALSFRS-R at baseline.212 Interestingly, Mitsumoto et al.210 expected there to be higher levels of urinary creatinine as muscle waste, but to date, this has not been found.39 It is possible the reduction in serum creatinine is reflective of muscle mass rather than muscle breakdown, but the variability and the known covariates make it difficult to determine reliability as a progression and prognostic biomarker.
A recent study focusing on lipids showed that triglycerides were increased and ceramides were decreased in ALS compared with controls, and phosphatidylethanolamine decreased over disease progression but there was again no reported relationship to ALSFRS-R.213 Covariates, such as BMI, sex, time of sample collection and presence of diabetes, are not uniformly used in every analysis, which could account for differences in top significant metabolites across reported literature.206,218,221 There is one study that looked at blood taken 5 years before diagnosis214 from over 200 000 people including 260 individuals who developed ALS, which found no significant metabolite associated with ALS diagnosis, once all the covariates were included in the analysis, and taking into account multiple comparisons. However, there was a trend in decreasing metabolites such as nucleosides, triacylglycerols, urate and phosphatidylcholines214 associated with developing ALS. Interestingly, there was no correlation to metabolites previously detected in ALS patients such as in the meta-analysis of Blasco et al.221 Metabolites dysregulated in the urine of ALS patients include 8-OHdG215 and neopterin.40,220
Metabolites related to oxidative stress in ALS
Several metabolites have been suggested as biomarkers of oxidative stress for ALS. An oxidized nucleoside of DNA, 8-OHdG (283.3 Da), is a widely studied oxidized metabolite of DNA damage present in urine, as well as CSF and blood. In an ALS study, urinary 8-OHdG (corrected by creatinine) was increased in compared with healthy controls, was correlated to the ALSFRS-R and was increased at a measurable rate over a 1-year study.215 Although higher in ALS than in healthy individuals, another study found no association of urinary 8-OHdG with the ALSFRS-R nor change over progression.216 Other research has found higher levels in the plasma of ALS patients compared with healthy controls and no difference between slow and fast progressors over time.163 Interestingly, a study of healthy individuals found that there was a correlation between urinary 8-OHdG and older age and gender.226 Urinary 8-OHdG has been used in a pre-clinical study as a pharmacodynamic biomarker227 but not in clinical trials. Further work needs to be done on a larger sample population to determine if urinary 8-OHdG is prognostic for survival in ALS and could be used as a predictive biomarker for treatments targeting oxidative stress.
High uric acid (168.11 Da) in serum, which is a final product of purine metabolism and an oxidative stress marker, has been linked to slower disease progression in ALS,217 and low levels of uric acid at diagnosis are prognostic in ALS, especially for males. A Phase 2 randomized, double-blind placebo-controlled trial using inosine to increase urate successfully increased serum urate, but no improvement was seen in the ALSFRS-R; however, this was a small safety and tolerability trial.228 4-Hydroxynonenal (156.22 Da), an oxidative stress marker produced by lipid peroxidation, also showed promise,163,219 with higher baseline levels correlated to worse symptoms 18 months later, faster progressing patients having higher levels of 4-hydroxynonenal. Urinary uric acid and 4-hydroxynonenal have not yet been reported in ALS patients.
Metabolites related to immune dysfunction in ALS
A downstream product of cytokine signalling, urinary neopterin, is a small metabolite of 253.21 Da and a promising pro-inflammation marker in ALS.40 Neopterin derives from guanosine triphosphate and is part of the pteridine family,229,230 which are pyrazino-pyrimidine compounds whose biological activity is dependent on chain substituents as well as the oxidation state of the ring. Tetrahydrobiopterin, a reduced pteridine that exhibits biological activity, functions as a co-factor of inducible nitric oxide synthase production230 and regulates apoptotic death by nitric oxide synthesis.231 However, tetrahydrobiopterin is unstable and easily oxidized to dihydrobiopterin and then biopterin.230,232 Neopterin is the oxidized product of 7,8-dihydroneopterin, and both biopterin and neopterin are stable in urine.230,232 Other pterdines include xanthopterin, isoxanthopterin, 6,7-dimethylpterin, 6-biopterin, 6-xydroxymethylpterin, pterin and pterin-6-carboxylic acid, which have not been extensively investigated as biomarkers in urine and not implicated in ALS.
In 1967, Sakurai and Goto233 isolated 25 mg of neopterin from 500 L of human urine. Neopterin is fluorescent in urine and detectable by HPLC at 353 nm excitation and 438 nm emission wavelengths.234 Neopterin [2-amino-4-oxo-6-(d-erythro-1,2,3,trihydroxypropyl)-pteridine] and its reduced form, 7,8-dihydroneopterin, are produced in large amounts by activated monocytes, macrophages and dendritic cells,235,236 after stimulation with IFN-γ and, to a lesser extent, TNF-α.237 Since microglia are the resident macrophages of the CNS, it has been assumed that they produce neopterin, as they respond to IFN-γ,238 and neopterin has been detected in the CSF.239,240 There is some in vitro evidence that microglia and neurons release neopterin. The concentration of neopterin reflects the presence of IFN-γ in body fluids, which makes it a sensitive marker of cell-mediated immunity.241
Evidence from animal models242,243 suggests that in response to ALS pathology, including protein aggregation, there is an anti-inflammatory response, which then shifts to pro-inflammatory as the anti-inflammatory process is overwhelmed with coping with accumulating protein aggregation. Microglia become pro-inflammatory as part of the activated microglial response244 and induce the release of neurotoxic factors from astrocytes that can kill motor neurons.245,246 In non-neuronal cells outside of the CNS, the pro-inflammatory state is evidenced by a switch to T-helper types 1 and 17 cells, as well as induction of cytotoxic CD8 cells, inflammatory monocytes and natural killer cells.247 This cascade also results in T-helper type 1 cell release of pro-inflammatory cytokines such as ILs and IFN-γ.248
Neopterin was first detected in some serum and CSF samples from ALS patients in 1993 and in 2020 in urine via ultraviolet HPLC,220 where it was suggested to be a prognostic marker. In a more recent analysis, undertaken by the authors, using an ELISA, and a smaller number of samples, neopterin was not prognostic, although correlated to the ALSFRS-R.40 Interestingly, neopterin increased at a measurable rate over disease progression,40 and it was suggested to be a candidate predictive marker of pro-inflammation in ALS.40 Further large studies to determine if neopterin is a valid predictive biomarker useful in clinical trials that influence the inflammatory state in ALS are required.
Reflections and limitations
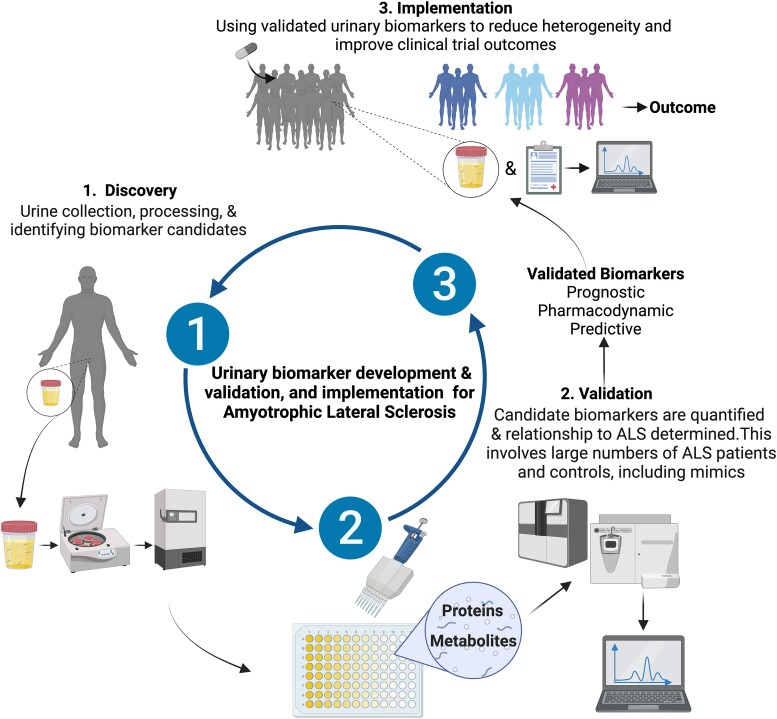
The relationship between pathological features in ALS and what is happening to motor neurons and support cells should be reflected in a candidate biomarker. Since the CSF is close to the site of injury, it has been expected to be the most useful biomarker, although other biomarkers are deemed valid if correlated to that found in CSF.91 Urinary biomarkers that reflect pathological processes in ALS should be examined, as obtaining urine is less invasive for ALS patients than CSF. Multiple components of urine are ripe for investigation as biomarkers for MND (Fig. 2) that can improve the chance of finding treatments. However, as a biomarker source, urine is not without limitations.
Figure 2.
Urinary biomarker development and implementation for ALS. The urinary biomarker development includes discovery, validation and utilization. Urine samples are collected from people with ALS and controls for discovery-type experiments. Biomarkers are isolated, measured and compared with clinical characteristics and pathology to determine if a possible candidate biomarker. The biomarkers are then validated across larger cohorts of ALS and controls, including mimic diseases. Validated prognostic, pharmacodynamic and predictive biomarkers can then be utilized to reduce heterogeneity and determine efficacy in clinical trials and determine outcomes (drug was a success or not).
Heterogeneity
Urinary protein and metabolite biomarkers can be affected by the time of collection, hydration status, urinary pH, kidney function, sex, age, diet and other disease states.32-34 Degradation and proteolysis can occur while the urine is stored in the bladder and in urine samples upon storage.32-34 For example, in a quantitative report on the urinary proteome, inter-individual variability exceeded 47% and intra-individual variability exceeded 45%.32 Metabolites in normal urine can vary even more than the proteome202; for example, using the large data from the National Health and Nutrition Examination Survey (2698 individuals), urinary phytoestrogen concentration was highly variable across the population, even where age, sex, lifestyle, race and poverty were used to reduce variability.249
The type of urine collection could be another source of variability. Although 24-h urine samples are the gold standard for biomarker measurement, it is often not practical, spot urine samples, when corrected for urinary dilution is seen as an adequate surrogate.250 Hydration is a source of urine variability in spot urine samples, and the use of creatinine as a hydration correction factor251 has been in clinical medicine for over 40 years.252 This is because creatinine has a steady state of excretion, but creatinine is effected by muscle mass, diabetes and meat (protein) intake.224 Nevertheless, the World Health Organization has defined cut-off values for very dilute (less than 0.3 mg/ml) and very concentrated (higher than 3.0 mg/ml) in urine252,253 In our 2017 and 2022 studies,39,40 applying these limits for spot urine creatinine for urinary dilution protection to biomarkers did not preclude detection of prognostic value or association with ALSFRS-R and progression. In addition, there was no diurnal variation of the candidate biomarkers. However, osmolality, which is not related to muscle mass nor diabetes status, should be considered as a correction factor, and in a large population study, osmolality was shown to be acceptable.254
Recommendations for standardizing spot urine collection, processing and use in biomarker analysis
Standardized protocols are essential255 to reduce the variability of the urinary proteome and metabolome in spot samples. Urine from those with renal or bladder abnormalities or uncontrolled diabetes should be screened out of the study. Our experience38-40 and the literature36,255,256 suggest those interested in urinary biomarkers for ALS use the following procedures:
Urine collection should be mid-stream.
The time between collection and sample processing is no more than 4 h, with samples stored on ice.
Urinalysis or urine dipsticks should be used to test for blood, high glucose, high bilirubin, high pH and the presence of high leukocytes.255 Abnormalities should be reported to the physician immediately and preclude the urine being used.
Although stabilizers such as boric acid and sodium azide can be added, literature36,256 and our experience38-40 suggest that centrifugation at 2000g at 4°C is preferable to remove cellular debris that may interfere with assays. If undertaking mass spectrometry analysis, a list of contaminant marker proteins can be used to screen out samples.256
After centrifugation, the urinary supernatant should be aliquoted into storage vials and stored at −70°C to reduce the necessity of going through freeze–thaw cycles.
Each biomarker once identified should be tested for stability at room temperature and at 4°C over at least 72 h. Diurnal stability and effect of ultraviolet light should also be undertaken with the goal of having as less variability as possible.
Once standardized protocols are in place, each biomarker should be checked for association with age and sex and, in the case of metabolome, diet.202
Consider using osmolality to correct for urinary dilution.
Conclusions
Standardizing urine collection and protocols for testing biomarkers will increase the usefulness of urine as potential prognostic, progression (monitoring) and predictive biomarkers for ALS. As listed above, there are many possible biomarkers related to ALS pathology, found in other biofluids, that can be investigated in urine. At present, the most promising urinary biomarker candidates include p75ECD, neopterin, titin and 8-OHdG. Each candidate should be validated in large cohorts that include healthy controls and disease mimics and candidates validated across laboratories. The recent inclusion of urine from ALS patients in large biobanks is encouraging. For example, the National Institute of Health funded Clinical Research in ALS and Related Disorders for Therapeutic Development biobank (https://create.rarediseasesnetwork.org/resources/researchers-clinicians/create-biorepository). After validation, the biomarkers can then be classified as prognostic, pharmacodynamic or predictive (or a mixture). For clinical trials, urinary biomarkers and panels of biomarkers for ALS can then be utilized to reduce heterogeneity and to determine if a potential ALS treatment is useful or not (Fig. 2). Ideal candidates should also be able to describe the pathological processes and to be used to tailor treatments (when available) for individuals with ALS. An example is a pro-inflammatory biomarker that could be used to detect those that may respond to an anti-inflammatory treatment.
Acknowledgements
We would like to acknowledge the consortium premodiALS (a premotor disease signature for ALS), funded in the scope of the Joint Programming Neurodegenerative Diseases (JPND) in the 2021 initiative as supporter of this work. All figures and graphical abstract created with Biorender.com.
Contributor Information
Mary-Louise Rogers, Flinders Health and Medical Research Institute, College of Medicine and Public Health, Flinders University, Adelaide 5042, South Australia, Australia.
David W Schultz, Neurology Department and MND Clinic, Flinders Medical Centre, Adelaide 5042, South Australia, Australia.
Vassilios Karnaros, Flinders Health and Medical Research Institute, College of Medicine and Public Health, Flinders University, Adelaide 5042, South Australia, Australia.
Stephanie R Shepheard, Flinders Health and Medical Research Institute, College of Medicine and Public Health, Flinders University, Adelaide 5042, South Australia, Australia.
Funding
The study was supported by the National institute of Health (# U01 NS107027, Clinical Research in ALS and Related Disorders for Therapeutic Development Consortium), Motor Neurone Disease Research Australia (# IG 2048, BG 2106, IG 2237 and IG:2335) and FightMND (# 05_2022_CIG_Rogers; 11_IMPACT_2020_Rogers; and Flinders University MND Drug Efficacy Testing Facility).
Competing interests
The authors report no competing interests.
Data availability
Data sharing is not applicable to this article as no new data were created or analysed in this study.
References
- 1. Genge A, Chio A. The future of ALS diagnosis and staging: Where do we go from here? Amyotroph Lateral Scler Frontotemporal Degener. 2023;24(3–4):165–174. [DOI] [PubMed] [Google Scholar]
- 2. Kiernan MC, Vucic S, Talbot K, et al. Improving clinical trial outcomes in amyotrophic lateral sclerosis. Nat Rev Neurol. 2021;17(2):104–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Atkinson A, Colburn W, DeGruttola V, et al. Biomarkers and surrogate endpoints: Preferred definitions and conceptual framework*. Clin Pharmacol Ther. 2001;69(3):89–95. [DOI] [PubMed] [Google Scholar]
- 4. BEST F-NBWG. FDA-NIH Biomarker Working Group . BEST (Biomarkers, EndpointS, and other tools) resource [internet]. In: 2016 FaDAU, ed. last Updated November 2021 ed. Silver Spring, Bethesda (MD). Co-published by National Institutes of Health (US). 2021:45. [Google Scholar]
- 5. Rooney J, Burke T, Vajda A, Heverin M, Hardiman O. What does the ALSFRS-R really measure? A longitudinal and survival analysis of functional dimension subscores in amyotrophic lateral sclerosis. J Neurol Neurosurg Psychiatry. 2017;88(5):381–385. [DOI] [PubMed] [Google Scholar]
- 6. Hannaford A, Byth K, Pavey N, et al. Clinical and neurophysiological biomarkers of disease progression in amyotrophic lateral sclerosis. Muscle Nerve. 2023;67(1):17–24. [DOI] [PubMed] [Google Scholar]
- 7. Kaufmann P, Levy G, Thompson JLP, et al. The ALSFRSr predicts survival time in an ALS clinic population. Neurology. 2005;64(1):38–43. [DOI] [PubMed] [Google Scholar]
- 8. Bakers JNE, de Jongh AD, Bunte TM, et al. Using the ALSFRS-R in multicentre clinical trials for amyotrophic lateral sclerosis: Potential limitations in current standard operating procedures. Amyotroph Lateral Scler Frontotemporal Degener. 2022;23(7–8):500–507. [DOI] [PubMed] [Google Scholar]
- 9. Holdom CJ, Steyn FJ, Henderson RD, McCombe PA, Rogers M-L, Ngo ST. Biofluid biomarkers of amyotrophic lateral sclerosis (ALS). In: Peplow PV Martinez B and Gennarelli TA, eds. Neurodegenerative diseases biomarkers: Towards translating research to clinical practice. Springer US; 2022:263–306. [Google Scholar]
- 10. Morello G, Salomone S, D'Agata V, Conforti FL, Cavallaro S. From multi-omics approaches to precision medicine in amyotrophic lateral sclerosis. Front Neurosci. 2020;14:577755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Katzeff JS, Bright F, Phan K, et al. Biomarker discovery and development for frontotemporal dementia and amyotrophic lateral sclerosis. Brain. 2022;145(5):1598–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Paganoni S, Berry JD, Quintana M, et al. Adaptive platform trials to transform amyotrophic lateral sclerosis therapy development. Ann Neurol. 2021;91(2):165–175. [DOI] [PubMed] [Google Scholar]
- 13. Lu CH, Macdonald-Wallis C, Gray E, et al. Neurofilament light chain: A prognostic biomarker in amyotrophic lateral sclerosis. Neurology. 2015;84(22):2247–2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kuhle J, Barro C, Andreasson U, et al. Comparison of three analytical platforms for quantification of the neurofilament light chain in blood samples: ELISA, electrochemiluminescence immunoassay and Simoa. Clin Chem Lab Med. 2016;54(10):1655–1661. [DOI] [PubMed] [Google Scholar]
- 15. Benatar M, Zhang L, Wang L, et al. Validation of serum neurofilaments as prognostic and potential pharmacodynamic biomarkers for ALS. Neurology. 2020;95(1):e59–e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huang F, Zhu Y, Hsiao-Nakamoto J, et al. Longitudinal biomarkers in amyotrophic lateral sclerosis. Ann Clin Transl Neurol. 2020;7(7):1103–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Eaton DC, Pooler JP. Clearance and measures of renal function. Vander’s renal physiology, 9e. McGraw-Hill Education; 2018. [Google Scholar]
- 18. Ogobuiro I, Tuma F. Physiology, renal. Statpearls. StatPearls PublishingCopyright © 2022, StatPearls Publishing LLC.; 2022. [PubMed] [Google Scholar]
- 19. Cortes GA, Flores JL. Physiology, urination. Statpearls. StatPearls PublishingCopyright © 2022, StatPearls Publishing LLC.; 2022. [PubMed] [Google Scholar]
- 20. Folin O. Laws governing the chemical composition of urine. Am J Physiology Legacy Content. 1905;13(1):66–115. [Google Scholar]
- 21. Putnam DF. Composition and concentrative properties of human urine. In: Administration NAaS, ed. Mcdonnell douglas astronautics company advanced biotechnology and power department huntington beach, California. National Aeronautics and Space Administration Washington, D.C. 20546; 1971:109. [Google Scholar]
- 22. Thongboonkerd V, McLeish KR, Arthur JM, Klein JB. Proteomic analysis of normal human urinary proteins isolated by acetone precipitation or ultracentrifugation. Kidney Int. 2002;62(4):1461–1469. [DOI] [PubMed] [Google Scholar]
- 23. Pisitkun T, Shen RF, Knepper MA. Identification and proteomic profiling of exosomes in human urine. Proc Natl Acad Sci U S A. 2004;101(36):13368–13373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Eaton DC, Pooler JP. Renal functions, basic processes, and anatomy. Vander’s renal physiology. McGraw-Hill Education; 2018. [Google Scholar]
- 25. Hellstern P. Rossi's principles of transfusion medicine. Vol. 6. John Wiley & Sons Ltd; 2022:200–208. [Google Scholar]
- 26. Tirumalai RS, Chan KC, Prieto DA, Issaq HJ, Conrads TP, Veenstra TD. Characterization of the low molecular weight human serum proteome. Mol Cell Proteomics. 2003;2(10):1096–1103. [DOI] [PubMed] [Google Scholar]
- 27. Jankovska E, Svitek M, Holada K, Petrak J. Affinity depletion versus relative protein enrichment: A side-by-side comparison of two major strategies for increasing human cerebrospinal fluid proteome coverage. Clin Proteomics. 2019;16:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Graham RC, Jr., Karnovsky MJ. Glomerular permeability. Ultrastructural cytochemical studies using peroxidases as protein tracers. J Exp Med. 1966;124(6):1123–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kanwar YS, Linker A, Farquhar MG. Increased permeability of the glomerular basement membrane to ferritin after removal of glycosaminoglycans (heparan sulfate) by enzyme digestion. J Cell Biol. 1980;86(2):688–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Julian BA, Suzuki H, Suzuki Y, Tomino Y, Spasovski G, Novak J. Sources of urinary proteins and their analysis by urinary proteomics for the detection of biomarkers of disease. Proteomics Clin Appl. 2009;3(9):1029–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jia L, Zhang L, Shao C, et al. An attempt to understand kidney's protein handling function by comparing plasma and urine proteomes. PLoS One. 2009;4(4):e5146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nagaraj N, Mann M. Quantitative analysis of the intra- and inter-individual variability of the normal urinary proteome. J Proteome Res. 2011;10(2):637–645. [DOI] [PubMed] [Google Scholar]
- 33. Shao C, Zhao M, Chen X, et al. Comprehensive analysis of individual variation in the urinary proteome revealed significant gender differences. Mol Cell Proteomics. 2019;18(6):1110–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dayon L, Cominetti O, Affolter M. Proteomics of human biological fluids for biomarker discoveries: Technical advances and recent applications. Expert Rev Proteomics. 2022;19(2):131–151. [DOI] [PubMed] [Google Scholar]
- 35. Muntel J, Xuan Y, Berger ST, et al. Advancing urinary protein biomarker discovery by data-independent acquisition on a quadrupole-orbitrap mass spectrometer. J Proteome Res. 2015;14(11):4752–4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Swensen AC, He J, Fang AC, et al. A comprehensive urine proteome database generated from patients with various renal conditions and prostate cancer. Front Med (Lausanne). 2021;8:548212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shao D, Huang L, Wang Y, et al. HBFP: A new repository for human body fluid proteome. Database (Oxford). 2021;2021:baab065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shepheard SR, Chataway T, Schultz DW, Rush RA, Rogers M-L. The extracellular domain of neurotrophin receptor p75 as a candidate biomarker for amyotrophic lateral sclerosis. PLoS One. 2014;9(1):e87398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shepheard SR, Wuu J, Cardoso M, et al. Urinary p75(ECD): A prognostic, disease progression, and pharmacodynamic biomarker in ALS. Neurology. 2017;88(12):1137–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shepheard SR, Karnaros V, Benyamin B, et al. Urinary neopterin: A novel biomarker of disease progression in amyotrophic lateral sclerosis. Eur J Neurol. 2022;29(4):990–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lu CH, Petzold A, Topping J, et al. Plasma neurofilament heavy chain levels and disease progression in amyotrophic lateral sclerosis: Insights from a longitudinal study. J Neurol Neurosurg Psychiatry. 2015;86(5):565–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tortelli R, Ruggieri M, Cortese R, et al. Elevated cerebrospinal fluid neurofilament light levels in patients with amyotrophic lateral sclerosis: A possible marker of disease severity and progression. Eur J Neurol. 2012;19(12):1561–1567. [DOI] [PubMed] [Google Scholar]
- 43. Tortelli R, Copetti M, Ruggieri M, et al. Cerebrospinal fluid neurofilament light chain levels: Marker of progression to generalized amyotrophic lateral sclerosis. Eur J Neurol. 2015;22(1):215–218. [DOI] [PubMed] [Google Scholar]
- 44. Weydt P, Oeckl P, Huss A, et al. Neurofilament levels as biomarkers in asymptomatic and symptomatic familial amyotrophic lateral sclerosis. Ann Neurol. 2016;79(1):152–158. [DOI] [PubMed] [Google Scholar]
- 45. Lombardi V, Carassiti D, Giovannoni G, Lu CH, Adiutori R, Malaspina A. The potential of neurofilaments analysis using dry-blood and plasma spots. Sci Rep. 2020;10(1):97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Petzold A. Neurofilament phosphoforms: Surrogate markers for axonal injury, degeneration and loss. J Neurol Sci. 2005;233(1–2):183–198. [DOI] [PubMed] [Google Scholar]
- 47. Boylan KB, Glass JD, Crook JE, et al. Phosphorylated neurofilament heavy subunit (pNF-H) in peripheral blood and CSF as a potential prognostic biomarker in amyotrophic lateral sclerosis. J Neurol Neurosurg Psychiatry. 2013;84(4):467–472. [DOI] [PubMed] [Google Scholar]
- 48. Yamada S, Hashizume A, Hijikata Y, et al. Ratio of urinary N-terminal titin fragment to urinary creatinine is a novel biomarker for amyotrophic lateral sclerosis. J Neurol Neurosurg Psychiatry. 2021;92(10):1072–1079. [DOI] [PubMed] [Google Scholar]
- 49. Su Z, Zhang Y, Gendron TF, et al. Discovery of a biomarker and lead small molecules to target r(GGGGCC)-associated defects in c9FTD/ALS. Neuron. 2014;83(5):1043–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lehmer C, Oeckl P, Weishaupt JH, et al. Poly-GP in cerebrospinal fluid links C9orf72-associated dipeptide repeat expression to the asymptomatic phase of ALS/FTD. EMBO Mol Med. 2017;9(7):859–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Querin G, Biferi MG, Pradat PF. Biomarkers for C9orf7-ALS in symptomatic and pre-symptomatic patients: State-of-the-art in the new era of clinical trials. J Neuromuscul Dis. 2022;9(1):25–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kasai T, Kojima Y, Ohmichi T, et al. Combined use of CSF NfL and CSF TDP-43 improves diagnostic performance in ALS. Ann Clin Transl Neurol. 2019;6(12):2489–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Feneberg E, Steinacker P, Lehnert S, et al. Limited role of free TDP-43 as a diagnostic tool in neurodegenerative diseases. Amyotroph Lateral Scler Frontotemporal Degener. 2014;15(5–6):351–356. [DOI] [PubMed] [Google Scholar]
- 54. Sproviero D, La Salvia S, Giannini M, et al. Pathological proteins are transported by extracellular vesicles of sporadic amyotrophic lateral sclerosis patients. Front Neurosci. 2018;12:487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ren Y, Li S, Chen S, et al. TDP-43 and phosphorylated TDP-43 levels in paired plasma and CSF samples in amyotrophic lateral sclerosis. Front Neurol. 2021;12:663637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Agnello L, Colletti T, Lo Sasso B, et al. Tau protein as a diagnostic and prognostic biomarker in amyotrophic lateral sclerosis. Eur J Neurol. 2021;28(6):1868–1875. [DOI] [PubMed] [Google Scholar]
- 57. Ono S, Imai T, Takahashi K, et al. Decreased type IV collagen of skin and serum in patients with amyotrophic lateral sclerosis. Neurology. 1998;51(1):114–120. [DOI] [PubMed] [Google Scholar]
- 58. Ono S, Imai T, Matsubara S, et al. Decreased urinary concentrations of type IV collagen in amyotrophic lateral sclerosis. Acta Neurol Scand. 1999;100(2):111–116. [DOI] [PubMed] [Google Scholar]
- 59. Gille B, De Schaepdryver M, Dedeene L, et al. Inflammatory markers in cerebrospinal fluid: Independent prognostic biomarkers in amyotrophic lateral sclerosis? J Neurol Neurosurg Psychiatry. 2019;90(12):1338–1346. [DOI] [PubMed] [Google Scholar]
- 60. Hu Y, Cao C, Qin XY, et al. Increased peripheral blood inflammatory cytokine levels in amyotrophic lateral sclerosis: A meta-analysis study. Sci Rep. 2017;7(1):9094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Moreno-Martinez L, Calvo AC, Munoz MJ, Osta R. Are circulating cytokines reliable biomarkers for amyotrophic lateral sclerosis? Int J Mol Sci. 2019;20(11):2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Trolese MC, Mariani A, Terao M, et al. CXCL13/CXCR5 signalling is pivotal to preserve motor neurons in amyotrophic lateral sclerosis. EBioMedicine. 2020;62:103097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Thompson AG, Gray E, Thezenas ML, et al. Cerebrospinal fluid macrophage biomarkers in amyotrophic lateral sclerosis. Ann Neurol. 2018;83(2):258–268. [DOI] [PubMed] [Google Scholar]
- 64. Steinacker P, Verde F, Fang L, et al. Chitotriosidase (CHIT1) is increased in microglia and macrophages in spinal cord of amyotrophic lateral sclerosis and cerebrospinal fluid levels correlate with disease severity and progression. J Neurol Neurosurg Psychiatry. 2018;89(3):239–247. [DOI] [PubMed] [Google Scholar]
- 65. Vu L, An J, Kovalik T, Gendron T, Petrucelli L, Bowser R. Cross-sectional and longitudinal measures of chitinase proteins in amyotrophic lateral sclerosis and expression of CHI3L1 in activated astrocytes. J Neurol Neurosurg Psychiatry. 2020;91(4):350–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Thompson AG, Turner MR. Untangling neuroinflammation in amyotrophic lateral sclerosis. J Neurol Neurosurg Psychiatry. 2019;90(12):1303–1304. [DOI] [PubMed] [Google Scholar]
- 67. Ryberg H, An J, Darko S, et al. Discovery and verification of amyotrophic lateral sclerosis biomarkers by proteomics. Muscle Nerve. 2010;42(1):104–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Beers DR, Zhao W, Neal DW, et al. Elevated acute phase proteins reflect peripheral inflammation and disease severity in patients with amyotrophic lateral sclerosis. Sci Rep. 2020;10(1):15295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kharel S, Ojha R, Preethish-Kumar V, Bhagat R. C-reactive protein levels in patients with amyotrophic lateral sclerosis: A systematic review. Brain Behav. 2022;12(3):e2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ono S, Hu J, Shimizu N, Imai T, Nakagawa H. Increased interleukin-6 of skin and serum in amyotrophic lateral sclerosis. J Neurol Sci. 2001;187(1–2):27–34. [DOI] [PubMed] [Google Scholar]
- 71. Lu CH, Allen K, Oei F, et al. Systemic inflammatory response and neuromuscular involvement in amyotrophic lateral sclerosis. Neurol Neuroimmunol Neuroinflamm. 2016;3(4):e244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Chen X, Hu Y, Cao Z, Liu Q, Cheng Y. Cerebrospinal fluid inflammatory cytokine aberrations in Alzheimer's disease, Parkinson's disease and amyotrophic lateral sclerosis: A systematic review and meta-analysis. Front Immunol. 2018;9:2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Pasinetti GM, Ungar LH, Lange DJ, et al. Identification of potential CSF biomarkers in ALS. Neurology. 2006;66(8):1218–1222. [DOI] [PubMed] [Google Scholar]
- 74. Tsuji-Akimoto S, Yabe I, Niino M, Kikuchi S, Sasaki H. Cystatin C in cerebrospinal fluid as a biomarker of ALS. Neurosci Lett. 2009;452(1):52–55. [DOI] [PubMed] [Google Scholar]
- 75. Wilson ME, Boumaza I, Lacomis D, Bowser R. Cystatin C: A candidate biomarker for amyotrophic lateral sclerosis. PLoS One. 2010;5(12):e15133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ranganathan S, Williams E, Ganchev P, et al. Proteomic profiling of cerebrospinal fluid identifies biomarkers for amyotrophic lateral sclerosis. J Neurochem. 2005;95(5):1461–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Süssmuth SD, Sperfeld AD, Hinz A, et al. CSF glial markers correlate with survival in amyotrophic lateral sclerosis. Neurology. 2010;74(12):982–987. [DOI] [PubMed] [Google Scholar]
- 78. Ibanez CF, Simi A. P75 neurotrophin receptor signaling in nervous system injury and degeneration: Paradox and opportunity. Trends Neurosci. 2012;35(7):431–440. [DOI] [PubMed] [Google Scholar]
- 79. Conroy JN, Coulson EJ. High-affinity TrkA and p75 neurotrophin receptor complexes: A twisted affair. J Biol Chem. 2022;298(3):101568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Lowry K, Murray S, McLean C, et al. A potential role for the p75 low-affinity neurotrophin receptor in spinal motor neuron degeneration in murine and human amyotrophic lateral sclerosis. Amyotroph Lateral Scler. 2001;2(3):127–134. [DOI] [PubMed] [Google Scholar]
- 81. Copray J, Jaarsma D, Kust B, et al. Expression of the low affinity neurotrophin receptor p75 in spinal motoneurons in a transgenic mouse model for amyotrophic lateral sclerosis. Neuroscience. 2003;116(3):685–694. [DOI] [PubMed] [Google Scholar]
- 82. Seeburger J, Tarras S, Natter H, Springer J. Spinal cord motoneurons express p75NGFR and p145trkB mRNA in amyotrophic lateral sclerosis. Brain Res. 1993;621(1):111–115. [DOI] [PubMed] [Google Scholar]
- 83. Kerkhoff H, Jennekens FGI, Troost D, Veldman H. Nerve growth factor receptor immunostaining in the spinal cord and peripheral nerves in amyotrophic lateral sclerosis. Acta Neuropathol. 1991;81(6):649–656. [DOI] [PubMed] [Google Scholar]
- 84. Weskamp G, Schlöndorff J, Lum L, et al. Evidence for a critical role of the tumor necrosis factor alpha convertase (TACE) in ectodomain shedding of the p75 neurotrophin receptor (p75NTR). J Biol Chem. 2004;279(6):4241–4249. [DOI] [PubMed] [Google Scholar]
- 85. DiStefano PS, Johnson EM. Identification of a truncated form of the nerve growth factor receptor. Proc Natl Acad Sci USA. 1988;85(1):270–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Gold J, Rowe DB, Kiernan MC, et al. Safety and tolerability of Triumeq in amyotrophic lateral sclerosis: The lighthouse trial. Amyotroph Lateral Scler Frontotemporal Degener. 2019;20(7–8):595–604. [DOI] [PubMed] [Google Scholar]
- 87. Vucic S, Henderson RD, Mathers S, et al. Safety and efficacy of dimethyl fumarate in ALS: Randomised controlled study. Ann Clin Transl Neurol. 2021;8(10):1991–1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Gaiani A, Martinelli I, Bello L, et al. Diagnostic and prognostic biomarkers in amyotrophic lateral sclerosis: Neurofilament light chain levels in definite subtypes of disease. JAMA Neurol. 2017;74(5):525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Benatar M, Wuu J, Andersen PM, Lombardi V, Malaspina A. Neurofilament light: A candidate biomarker of presymptomatic amyotrophic lateral sclerosis and phenoconversion. Ann Neurol. 2018;84(1):130–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Verde F, Otto M, Silani V. Neurofilament light chain as biomarker for amyotrophic lateral sclerosis and frontotemporal dementia. Front Neurosci. 2021;15:679199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Sturmey E, Malaspina A. Blood biomarkers in ALS: Challenges, applications and novel frontiers. Acta Neurol Scand. 2022;146(4):375–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Benatar M, Wuu J, Turner MR. Neurofilament light chain in drug development for amyotrophic lateral sclerosis: A critical appraisal. Brain. 2023;146(7):2711–2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Rissin DM, Fournier DR, Piech T, et al. Simultaneous detection of single molecules and singulated ensembles of molecules enables immunoassays with broad dynamic range. Anal Chem. 2011;83(6):2279–2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Hendricks R, Baker D, Brumm J, et al. Establishment of neurofilament light chain Simoa assay in cerebrospinal fluid and blood. Bioanalysis. 2019;11(15):1405–1418. [DOI] [PubMed] [Google Scholar]
- 95. Meyer T, Schumann P, Weydt P, et al. Neurofilament light-chain response during therapy with antisense oligonucleotide tofersen in SOD1-related ALS: Treatment experience in clinical practice. Muscle Nerve. 2023;67(6):515–521. [DOI] [PubMed] [Google Scholar]
- 96. Brettschneider J, Lehmensiek V, Mogel H, et al. Proteome analysis reveals candidate markers of disease progression in amyotrophic lateral sclerosis (ALS). Neurosci Lett. 2010;468(1):23–27. [DOI] [PubMed] [Google Scholar]
- 97. Bereman MS, Beri J, Enders JR, Nash T. Machine learning reveals protein signatures in CSF and plasma fluids of clinical value for ALS. Sci Rep. 2018;8(1):16334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Petzold A. CSF biomarkers for improved prognostic accuracy in acute CNS disease. Neurol Res. 2007;29(7):691–708. [DOI] [PubMed] [Google Scholar]
- 99. Zetterberg H, Jacobsson J, Rosengren L, Blennow K, Andersen PM. Cerebrospinal fluid neurofilament light levels in amyotrophic lateral sclerosis: Impact of SOD1 genotype. Eur J Neurol. 2007;14(12):1329–1333. [DOI] [PubMed] [Google Scholar]
- 100. Zubiri I, Lombardi V, Bremang M, et al. Tissue-enhanced plasma proteomic analysis for disease stratification in amyotrophic lateral sclerosis. Mol Neurodegener. 2018;13(1):60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Leoni E, Bremang M, Mitra V, et al. Combined tissue-fluid proteomics to unravel phenotypic variability in amyotrophic lateral sclerosis. Sci Rep. 2019;9(1):4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Oeckl P, Weydt P, Thal DR, Weishaupt JH, Ludolph AC, Otto M. Proteomics in cerebrospinal fluid and spinal cord suggests UCHL1, MAP2 and GPNMB as biomarkers and underpins importance of transcriptional pathways in amyotrophic lateral sclerosis. Acta Neuropathol. 2020;139(1):119–134. [DOI] [PubMed] [Google Scholar]
- 103. Millecamps S, Gowing G, Corti O, Mallet J, Julien JP. Conditional NF-L transgene expression in mice for in vivo analysis of turnover and transport rate of neurofilaments. J Neurosci. 2007;27(18):4947–4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Nixon RA, Logvinenko KB. Multiple fates of newly synthesized neurofilament proteins: Evidence for a stationary neurofilament network distributed nonuniformly along axons of retinal ganglion cell neurons. J Cell Biol. 1986;102(2):647–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Zimmerman UJP, Schlaepfer WW. Characterization of a brain calcium-activated protease that degrades neurofilament proteins. Biochemistry. 1982;21(17):3977–3983. [DOI] [PubMed] [Google Scholar]
- 106. Pant HC. Dephosphorylation of neurofilament proteins enhances their susceptibility to degradation by calpain. Biochem J. 1988;256(2):665–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Budelier MM, He Y, Barthelemy NR, et al. A map of neurofilament light chain species in brain and cerebrospinal fluid and alterations in Alzheimer's disease. Brain Commun. 2022;4(2):fcac045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Kohlhase K, Frank F, Wilmes C, et al. Brain-specific biomarkers in urine as a non-invasive approach to monitor neuronal and glial damage. Eur J Neurol. 2023;30(3):729–740. [DOI] [PubMed] [Google Scholar]
- 109. Maruyama K, Natori R, Nonomura Y. New elastic protein from muscle. Nature. 1976;262(5563):58–60. [DOI] [PubMed] [Google Scholar]
- 110. Rouillon J, Zocevic A, Leger T, et al. Proteomics profiling of urine reveals specific titin fragments as biomarkers of Duchenne muscular dystrophy. Neuromuscul Disord. 2014;24(7):563–573. [DOI] [PubMed] [Google Scholar]
- 111. Sun S, Henriksen K, Karsdal MA, et al. Measurement of a MMP-2 degraded titin fragment in serum reflects changes in muscle turnover induced by atrophy. Exp Gerontol. 2014;58:83–89. [DOI] [PubMed] [Google Scholar]
- 112. Gargan S, Dowling P, Zweyer M, Swandulla D, Ohlendieck K. Identification of marker proteins of muscular dystrophy in the urine proteome from the mdx-4cv model of dystrophinopathy. Mol Omics. 2020;16(3):268–278. [DOI] [PubMed] [Google Scholar]
- 113. Maruyama N, Asai T, Abe C, et al. Establishment of a highly sensitive sandwich ELISA for the N-terminal fragment of titin in urine. Sci Rep. 2016;6:39375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Shirakawa T, Ikushima A, Maruyama N, et al. A sandwich ELISA kit reveals marked elevation of titin N-terminal fragment levels in the urine of mdx mice. Animal Model Exp Med. 2022;5(1):48–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Yoshihisa A, Kimishima Y, Kiko T, et al. Usefulness of urinary N-terminal fragment of titin to predict mortality in dilated cardiomyopathy. Am J Cardiol. 2018;121(10):1260–1265. [DOI] [PubMed] [Google Scholar]
- 116. Misaka T, Yoshihisa A, Takeishi Y. Titin in muscular dystrophy and cardiomyopathy: Urinary titin as a novel marker. Clin Chim Acta. 2019;495:123–128. [DOI] [PubMed] [Google Scholar]
- 117. Matsuo M, Awano H, Maruyama N, Nishio H. Chapter one—Titin fragment in urine: A noninvasive biomarker of muscle degradation. In: Makowski GS, ed. Advances in clinical chemistry. Vol 90: Elsevier; 2019:1–23. [DOI] [PubMed] [Google Scholar]
- 118. Robertson AS, Majchrzak MJ, Smith CM, et al. Dramatic elevation in urinary amino terminal titin fragment excretion quantified by immunoassay in Duchenne muscular dystrophy patients and in dystrophin deficient rodents. Neuromuscul Disord. 2017;27(7):635–645. [DOI] [PubMed] [Google Scholar]
- 119. Matsumura K, Shimizu T, Sunada Y, et al. Degradation of connectin (titin) in Fukuyama type congenital muscular dystrophy: Immunochemical study with monoclonal antibodies. J Neurol Sci. 1990;98(2–3):155–162. [DOI] [PubMed] [Google Scholar]
- 120. Tanihata J, Minamisawa S. Urinary titin is not an early biomarker of skeletal muscle atrophy induced by muscle denervation in mice. PLoS One. 2023;18(8):e0289185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Proctor DN, O’Brien PC, Atkinson EJ, Nair KS. Comparison of techniques to estimate total body skeletal muscle mass in people of different age groups. Am J Physiol Endocrinol Metab. 1999;277(3):E489–E495. [DOI] [PubMed] [Google Scholar]
- 122. Volbeda M, Hessels L, Posma RA, Bakker SJ, Nijsten MW. Time courses of urinary creatinine excretion, measured creatinine clearance and estimated glomerular filtration rate over 30 days of ICU admission. J Crit Care. 2021;63:161–166. [DOI] [PubMed] [Google Scholar]
- 123. Pandhi P, Streng KW, Anker SD, et al. The value of spot urinary creatinine as a marker of muscle wasting in patients with new-onset or worsening heart failure. J Cachexia Sarcopenia Muscle. 2021;12(3):555–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Chargaff E, West R. The biological significance of the thromboplastic protein of blood. J Biol Chem. 1946;166(1):189–197. [PubMed] [Google Scholar]
- 125. Kleijmeer MJ, Raposo G, Geuze HJ. Characterization of MHC class II compartments by immunoelectron microscopy. Methods. 1996;10(2):191–207. [DOI] [PubMed] [Google Scholar]
- 126. Ratajczak J, Miekus K, Kucia M, et al. Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: Evidence for horizontal transfer of mRNA and protein delivery. Leukemia. 2006;20(5):847–856. [DOI] [PubMed] [Google Scholar]
- 127. Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–659. [DOI] [PubMed] [Google Scholar]
- 128. Kim G, Chen X, Yang Y. Pathogenic extracellular vesicle (EV) signaling in amyotrophic lateral sclerosis (ALS). Neurotherapeutics. 2022;19(4):1119–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Merchant ML, Rood IM, Deegens JKJ, Klein JB. Isolation and characterization of urinary extracellular vesicles: Implications for biomarker discovery. Nat Rev Nephrol. 2017;13(12):731–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Saman S, Kim W, Raya M, et al. Exosome-associated tau is secreted in tauopathy models and is selectively phosphorylated in cerebrospinal fluid in early Alzheimer disease. J Biol Chem. 2012;287(6):3842–3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Caby MP, Lankar D, Vincendeau-Scherrer C, Raposo G, Bonnerot C. Exosomal-like vesicles are present in human blood plasma. Int Immunol. 2005;17(7):879–887. [DOI] [PubMed] [Google Scholar]
- 132. Svenningsen P, Sabaratnam R, Jensen BL. Urinary extracellular vesicles: Origin, role as intercellular messengers and biomarkers; efficient sorting and potential treatment options. Acta Physiol (Oxf). 2020;228(1):e13346. [DOI] [PubMed] [Google Scholar]
- 133. Fraser KB, Moehle MS, Alcalay RN, West AB. Urinary LRRK2 phosphorylation predicts parkinsonian phenotypes in G2019S LRRK2 carriers. Neurology. 2016;86(11):994–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Ling SC, Polymenidou M, Cleveland DW. Converging mechanisms in ALS and FTD: Disrupted RNA and protein homeostasis. Neuron. 2013;79(3):416–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Kasai T, Tokuda T, Ishigami N, et al. Increased TDP-43 protein in cerebrospinal fluid of patients with amyotrophic lateral sclerosis. Acta Neuropathol. 2009;117(1):55–62. [DOI] [PubMed] [Google Scholar]
- 136. Suarez-Calvet M, Dols-Icardo O, Llado A, et al. Plasma phosphorylated TDP-43 levels are elevated in patients with frontotemporal dementia carrying a C9orf72 repeat expansion or a GRN mutation. J Neurol Neurosurg Psychiatry. 2013;85(6):684–691. [DOI] [PubMed] [Google Scholar]
- 137. Junttila A, Kuvaja M, Hartikainen P, et al. Cerebrospinal fluid TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis patients with and without the C9ORF72 hexanucleotide expansion. Dement Geriatr Cogn Dis Extra. 2016;6(1):142–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Feneberg E, Charles PD, Finelli MJ, et al. Detection and quantification of novel C-terminal TDP-43 fragments in ALS-TDP. Brain Pathol. 2021;31(4):e12923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Iguchi Y, Eid L, Parent M, et al. Exosome secretion is a key pathway for clearance of pathological TDP-43. Brain. 2016;139(Pt 12):3187–3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Huang C, Xia PY, Zhou H. Sustained expression of TDP-43 and FUS in motor neurons in rodent's lifetime. Int J Biol Sci. 2010;6(4):396–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Pacetti M, De Conti L, Marasco LE, et al. Physiological tissue-specific and age-related reduction of mouse TDP-43 levels is regulated by epigenetic modifications. Dis Model Mech. 2022;15(4):dmm049032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Gendron TF, van Blitterswijk M, Bieniek KF, et al. Cerebellar c9RAN proteins associate with clinical and neuropathological characteristics of C9ORF72 repeat expansion carriers. Acta Neuropathol. 2015;130(4):559–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Lee YB, Baskaran P, Gomez-Deza J, et al. C9orf72 poly GA RAN-translated protein plays a key role in amyotrophic lateral sclerosis via aggregation and toxicity. Hum Mol Genet. 2017;26(24):4765–4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. White MR, Mitrea DM, Zhang P, et al. C9orf72 poly(PR) dipeptide repeats disturb biomolecular phase separation and disrupt nucleolar function. Mol Cell. 2019;74(4):713–728.e716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Mackenzie IRA, Bigio EH, Ince PG, et al. Pathological TDP-43 distinguishes sporadic amyotrophic lateral sclerosis from amyotrophic lateral sclerosis withSOD1 mutations. Ann Neurol. 2007;61(5):427–434. [DOI] [PubMed] [Google Scholar]
- 146. Vance C, Rogelj B, Hortobagyi T, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323(5918):1208–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Brenner D, Freischmidt A. Update on genetics of amyotrophic lateral sclerosis. Curr Opin Neurol. 2022;35(5):672–677. [DOI] [PubMed] [Google Scholar]
- 148. Grossman M, Elman L, McCluskey L, et al. Phosphorylated tau as a candidate biomarker for amyotrophic lateral sclerosis. JAMA Neurol. 2014;71(4):442–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Brettschneider J, Petzold A, Süssmuth SD, Ludolph AC, Tumani H. Axonal damage markers in cerebrospinal fluid are increased in ALS. Neurology. 2006;66(6):852–856. [DOI] [PubMed] [Google Scholar]
- 150. Paladino P, Valentino F, Piccoli T, Piccoli F, La Bella V. Cerebrospinal fluid tau protein is not a biological marker in amyotrophic lateral sclerosis. Eur J Neurol. 2009;16(2):257–261. [DOI] [PubMed] [Google Scholar]
- 151. Schreiber S, Spotorno N, Schreiber F, et al. Significance of CSF NfL and tau in ALS. J Neurol. 2018;265(11):2633–2645. [DOI] [PubMed] [Google Scholar]
- 152. Grant DS, Leblond CP. Immunogold quantitation of laminin, type IV collagen, and heparan sulfate proteoglycan in a variety of basement membranes. J Histochem Cytochem. 1988;36(3):271–283. [DOI] [PubMed] [Google Scholar]
- 153. LeBleu VS, Macdonald B, Kalluri R. Structure and function of basement membranes. Exp Biol Med (Maywood). 2007;232(9):1121–1129. [DOI] [PubMed] [Google Scholar]
- 154. Tashiro K, Koyanagi I, Ohara I, et al. Levels of urinary matrix metalloproteinase-9 (MMP-9) and renal injuries in patients with type 2 diabetic nephropathy. J Clin Lab Anal. 2004;18(3):206–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Garbuzova-Davis S, Haller E, Saporta S, Kolomey I, Nicosia SV, Sanberg PR. Ultrastructure of blood-brain barrier and blood-spinal cord barrier in SOD1 mice modeling ALS. Brain Res. 2007;1157:126–137. [DOI] [PubMed] [Google Scholar]
- 156. Ono S, Shimizu N, Imai T, Rodriguez GP. Urinary collagen metabolite excretion in amyotrophic lateral sclerosis. Muscle Nerve. 2001;24(6):821–825. [DOI] [PubMed] [Google Scholar]
- 157. Van Damme P, Van Hoecke A, Lambrechts D, et al. Progranulin functions as a neurotrophic factor to regulate neurite outgrowth and enhance neuronal survival. J Cell Biol. 2008;181(1):37–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Feneberg E, Steinacker P, Volk AE, et al. Progranulin as a candidate biomarker for therapeutic trial in patients with ALS and FTLD. J Neural Transm (Vienna). 2016;123(3):289–296. [DOI] [PubMed] [Google Scholar]
- 159. Schreiber S, Debska-Vielhaber G, Abdulla S, et al. Peripheral nerve atrophy together with higher cerebrospinal fluid progranulin indicate axonal damage in amyotrophic lateral sclerosis. Muscle Nerve. 2018;57(2):273–278. [DOI] [PubMed] [Google Scholar]
- 160. Terryn J, Verfaillie CM, Van Damme P. Tweaking progranulin expression: Therapeutic avenues and opportunities. Front Mol Neurosci. 2021;14:713031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Peters S, Zitzelsperger E, Kuespert S, et al. The TGF-beta system as a potential pathogenic player in disease modulation of amyotrophic lateral sclerosis. Front Neurol. 2017;8:669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Galbiati M, Crippa V, Rusmini P, et al. Multiple roles of transforming growth factor beta in amyotrophic lateral sclerosis. Int J Mol Sci. 2020;21(12):4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Devos D, Moreau C, Kyheng M, et al. A ferroptosis–based panel of prognostic biomarkers for amyotrophic lateral sclerosis. Sci Rep. 2019;9(1):423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Jesus P, Blasco H, Patin F, et al. Ferritin and LDL-cholesterol as biomarkers of fat-free mass loss in ALS. Amyotroph Lateral Scler Frontotemporal Degener. 2019;20(5–6):441–444. [DOI] [PubMed] [Google Scholar]
- 165. van Swelm RPL, Wetzels JFM, Swinkels DW. The multifaceted role of iron in renal health and disease. Nat Rev Nephrol. 2020;16(2):77–98. [DOI] [PubMed] [Google Scholar]
- 166. Puentes F, Malaspina A, van Noort JM, Amor S. Non-neuronal cells in ALS: Role of glial, immune cells and blood-CNS barriers. Brain Pathol. 2016;26(2):248–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Béland L-C, Markovinovic A, Jakovac H, et al. Immunity in amyotrophic lateral sclerosis: Blurred lines between excessive inflammation and inefficient immune responses. Brain Commun. 2020;2(2):fcaa124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Hughes CE, Nibbs RJB. A guide to chemokines and their receptors. FEBS J. 2018;285(16):2944–2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Barna BP, Pettay J, Barnett GH, Zhou P, Iwasaki K, Estes ML. Regulation of monocyte chemoattractant protein-1 expression in adult human non-neoplastic astrocytes is sensitive to tumor necrosis factor (TNF) or antibody to the 55-kDa TNF receptor. J Neuroimmunol. 1994;50(1):101–107. [DOI] [PubMed] [Google Scholar]
- 170. Yoshimura T, Yuhki N, Moore SK, Appella E, Lerman MI, Leonard EJ. Human monocyte chemoattractant protein-1 (MCP-1) full-length cDNA cloning, expression in mitogen-stimulated blood mononuclear leukocytes, and sequence similarity to mouse competence gene JE. FEBS Lett. 1989;244(2):487–493. [DOI] [PubMed] [Google Scholar]
- 171. Henkel JS, Engelhardt JI, Siklós L, et al. Presence of dendritic cells, MCP-1, and activated microglia/macrophages in amyotrophic lateral sclerosis spinal cord tissue. Ann Neurol. 2004;55(2):221–235. [DOI] [PubMed] [Google Scholar]
- 172. Sauriasari R, Safitri DD, Azmi NU. Current updates on protein as biomarkers for diabetic kidney disease: A systematic review. Ther Adv Endocrinol Metab. 2021;12:20420188211049612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Lee YH, Song GG. Urinary MCP-1 as a biomarker for lupus nephritis: A meta-analysis. Z Rheumatol. 2017;76(4):357–363. [DOI] [PubMed] [Google Scholar]
- 174. Thakur V, Chattopadhyay M. Early urinary markers for diabetic and other kidney diseases. Curr Drug Targets. 2018;19(7):825–831. [DOI] [PubMed] [Google Scholar]
- 175. Desireddi NV, Campbell PL, Stern JA, et al. Monocyte chemoattractant protein-1 and macrophage inflammatory protein-1α as possible biomarkers for the chronic pelvic pain syndrome. J Urol. 2008;179(5):1857–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Xu Y, Shen YY, Zhang XP, et al. Diagnostic potential of urinary monocyte chemoattractant protein-1 for Alzheimer's disease and amnestic mild cognitive impairment. Eur J Neurol. 2020;27(8):1429–1435. [DOI] [PubMed] [Google Scholar]
- 177. Varghese AM, Sharma A, Mishra P, et al. Chitotriosidase—A putative biomarker for sporadic amyotrophic lateral sclerosis. Clin Proteomics. 2013;10(1):19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Gaur N, Huss E, Prell T, et al. Monocyte-derived macrophages contribute to chitinase dysregulation in amyotrophic lateral sclerosis: A pilot study. Front Neurol. 2021;12:629332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Pinteac R, Montalban X, Comabella M. Chitinases and chitinase-like proteins as biomarkers in neurologic disorders. Neurol Neuroimmunol Neuroinflamm. 2021;8(1):e921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Oeckl P, Weydt P, Steinacker P, et al. Different neuroinflammatory profile in amyotrophic lateral sclerosis and frontotemporal dementia is linked to the clinical phase. J Neurol Neurosurg Psychiatry. 2019;90(1):4–10. [DOI] [PubMed] [Google Scholar]
- 181. Black S, Kushner I, Samols D. C-reactive protein. J Biol Chem. 2004;279(47):48487–48490. [DOI] [PubMed] [Google Scholar]
- 182. Mihlan M, Blom AM, Kupreishvili K, et al. Monomeric C-reactive protein modulates classic complement activation on necrotic cells. FASEB J. 2011;25(12):4198–4210. [DOI] [PubMed] [Google Scholar]
- 183. Eisenhardt SU, Thiele JR, Bannasch H, Stark GB, Peter K. C-reactive protein: How conformational changes influence inflammatory properties. Cell Cycle. 2009;8(23):3885–3892. [DOI] [PubMed] [Google Scholar]
- 184. Thiele JR, Habersberger J, Braig D, et al. Dissociation of pentameric to monomeric C-reactive protein localizes and aggravates inflammation: In vivo proof of a powerful proinflammatory mechanism and a new anti-inflammatory strategy. Circulation. 2014;130(1):35–50. [DOI] [PubMed] [Google Scholar]
- 185. Keizman D, Rogowski O, Berliner S, et al. Low-grade systemic inflammation in patients with amyotrophic lateral sclerosis. Acta Neurol Scand. 2009;119(6):383–389. [DOI] [PubMed] [Google Scholar]
- 186. Lunetta C, Lizio A, Maestri E, et al. Serum C-reactive protein as a prognostic biomarker in amyotrophic lateral sclerosis. JAMA Neurol. 2017;74(6) 660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Miller RG, Zhang R, Bracci PM, et al. Phase 2B randomized controlled trial of NP001 in amyotrophic lateral sclerosis: Pre-specified and post hoc analyses. Muscle Nerve. 2022;66(1):39–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Vahsen BF, Gray E, Thompson AG, et al. Non-neuronal cells in amyotrophic lateral sclerosis - from pathogenesis to biomarkers. Nat Rev Neurol. 2021;17(6):333–348. [DOI] [PubMed] [Google Scholar]
- 189. Saiyed N, Yilmaz A, Vishweswariah S, et al. Urinary cytokines as potential biomarkers of mild cognitive impairment and Alzheimer's disease: A pilot study. J Alzheimers Dis Rep. 2023;7(1):649–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Nobles C, Bertone-Johnson ER, Ronnenberg AG, et al. Correlation of urine and plasma cytokine levels among reproductive-aged women. Eur J Clin Invest. 2015;45(5):460–465. [DOI] [PubMed] [Google Scholar]
- 191. Abrahamson M, Barrett AJ, Salvesen G, Grubb A. Isolation of six cysteine proteinase inhibitors from human urine. Their physicochemical and enzyme kinetic properties and concentrations in biological fluids. J Biol Chem. 1986;261(24):11282–11289. [PubMed] [Google Scholar]
- 192. Zi M, Xu Y. Involvement of cystatin C in immunity and apoptosis. Immunol Lett. 2018;196:80–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Tomonaga M. Selective appearance of Bunina bodies in amyotrophic lateral sclerosis. A study of the distribution in midbrain and sacral cord. J Neurol. 1980;223(4):259–267. [DOI] [PubMed] [Google Scholar]
- 194. Okamoto K, Mizuno Y, Fujita Y. Bunina bodies in amyotrophic lateral sclerosis. Neuropathology. 2008;28(2):109–115. [DOI] [PubMed] [Google Scholar]
- 195. Kimura T, Jiang H, Konno T, et al. Bunina bodies in motor and non-motor neurons revisited: A pathological study of an ALS patient after long-term survival on a respirator. Neuropathology. 2014;34(4):392–397. [DOI] [PubMed] [Google Scholar]
- 196. Watanabe S, Hayakawa T, Wakasugi K, Yamanaka K. Cystatin C protects neuronal cells against mutant copper-zinc superoxide dismutase-mediated toxicity. Cell Death Dis. 2014;5(10):e1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Vijayakumar UG, Milla V, Cynthia Stafford MY, Bjourson AJ, Duddy W, Duguez SM. A systematic review of suggested molecular strata, biomarkers and their tissue sources in ALS. Front Neurol. 2019;10:400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Vieira M, Saraiva MJ. Transthyretin: A multifaceted protein. Biomol Concepts. 2014;5(1):45–54. [DOI] [PubMed] [Google Scholar]
- 199. Chu YP, Jin LW, Wang LC, Ho PC, Wei WY, Tsai KJ. Transthyretin attenuates TDP-43 proteinopathy by autophagy activation via ATF4 in FTLD-TDP. Brain. 2023;146(5):2089–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Zondler L, Muller K, Khalaji S, et al. Peripheral monocytes are functionally altered and invade the CNS in ALS patients. Acta Neuropathol. 2016;132(3):391–411. [DOI] [PubMed] [Google Scholar]
- 201. Wishart DS, Feunang YD, Marcu A, et al. HMDB 4.0: The human metabolome database for 2018. Nucleic Acids Res. 2017;46(D1):D608–D617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Bouatra S, Aziat F, Mandal R, et al. The human urine metabolome. PLoS One. 2013;8(9):e73076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Mandal R, Guo AC, Chaudhary KK, et al. Multi-platform characterization of the human cerebrospinal fluid metabolome: A comprehensive and quantitative update. Genome Med. 2012;4(4):38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Takeda I, Stretch C, Barnaby P, et al. Understanding the human salivary metabolome. NMR Biomed. 2009;22(6):577–584. [DOI] [PubMed] [Google Scholar]
- 205. Cecchi M, Messina P, Airoldi L, et al. Plasma amino acids patterns and age of onset of amyotrophic lateral sclerosis. Amyotroph Lateral Scler Frontotemporal Degener. 2014;15(5–6):371–375. [DOI] [PubMed] [Google Scholar]
- 206. Goutman SA, Boss J, Guo K, et al. Untargeted metabolomics yields insight into ALS disease mechanisms. J Neurol Neurosurg Psychiatry. 2020;91(12):1329–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Gray E, Larkin JR, Claridge TD, Talbot K, Sibson NR, Turner MR. The longitudinal cerebrospinal fluid metabolomic profile of amyotrophic lateral sclerosis. Amyotroph Lateral Scler Frontotemporal Degener. 2015;16(7–8):456–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Ikeda K, Hirayama T, Takazawa T, Kawabe K, Iwasaki Y. Relationships between disease progression and serum levels of lipid, urate, creatinine and ferritin in Japanese patients with amyotrophic lateral sclerosis: A cross-sectional study. Intern Med. 2012;51(12):1501–1508. [DOI] [PubMed] [Google Scholar]
- 209. Chiò A, Calvo A, Bovio G, et al. Amyotrophic lateral sclerosis outcome measures and the role of albumin and creatinine: A population-based study. JAMA Neurol. 2014;71(9):1134. [DOI] [PubMed] [Google Scholar]
- 210. Mitsumoto H, Garofalo DC, Santella RM, et al. Plasma creatinine and oxidative stress biomarkers in amyotrophic lateral sclerosis. Amyotroph Lateral Scler Frontotemporal Degener. 2020;21(3–4):263–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Hertel N, Kuzma-Kozakiewicz M, Gromicho M, et al. Analysis of routine blood parameters in patients with amyotrophic lateral sclerosis and evaluation of a possible correlation with disease progression-a multicenter study. Front Neurol. 2022;13:940375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Morgadinho J, Pronto-Laborinho AC, Conceição VA, et al. Plasma creatinine level does not predict respiratory function in amyotrophic lateral sclerosis. J Neuromuscul Dis. 2021;8(5):795–799. [DOI] [PubMed] [Google Scholar]
- 213. Phan K, He Y, Bhatia S, et al. Multiple pathways of lipid dysregulation in amyotrophic lateral sclerosis. Brain Commun. 2023;5(1):fcac340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Bjornevik K, Zhang Z, O'Reilly EJ, et al. Prediagnostic plasma metabolomics and the risk of amyotrophic lateral sclerosis. Neurology. 2019;92(18):e2089–e2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Bogdanov M, Brown RH, Matson W, et al. Increased oxidative damage to DNA in ALS patients. Free Radic Biol Med. 2000;29(7):652–658. [DOI] [PubMed] [Google Scholar]
- 216. Mitsumoto H, Santella RM, Liu X, et al. Oxidative stress biomarkers in sporadic ALS. Amyotroph Lateral Scler. 2008;9(3):177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Paganoni S, Zhang M, Quiroz Zárate A, et al. Uric acid levels predict survival in men with amyotrophic lateral sclerosis. J Neurol. 2012;259(9):1923–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Blasco H, Garcon G, Patin F, et al. Panel of oxidative stress and inflammatory biomarkers in ALS: A pilot study. Can J Neurol Sci. 2017;44(1):90–95. [DOI] [PubMed] [Google Scholar]
- 219. Simpson EP, Henry YK, Henkel JS, Smith RG, Appel SH. Increased lipid peroxidation in sera of ALS patients: A potential biomarker of disease burden. Neurology. 2004;62(10):1758–1765. [DOI] [PubMed] [Google Scholar]
- 220. Lunetta C, Lizio A, Gerardi F, et al. Urinary neopterin, a new marker of the neuroinflammatory status in amyotrophic lateral sclerosis. J Neurol. 2020;7(5–6):639. [DOI] [PubMed] [Google Scholar]
- 221. Blasco H, Patin F, Madji Hounoum B, et al. Metabolomics in amyotrophic lateral sclerosis: How far can it take us? Eur J Neurol. 2016;23(3):447–454. [DOI] [PubMed] [Google Scholar]
- 222. Nakazato T, Kanai K, Kataura T, Nojiri S, Hattori N, Saiki S. Plasma taurine is an axonal excitability-translatable biomarker for amyotrophic lateral sclerosis. Sci Rep. 2022;12(1):9155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. Prabhakar SS. Medical secrets. Vol. 5. Elsevier Mosby; 2012:190–219. [Google Scholar]
- 224. Kashani K, Rosner MH, Ostermann M. Creatinine: From physiology to clinical application. Eur J Intern Med. 2020;72:9–14. [DOI] [PubMed] [Google Scholar]
- 225. Lanznaster D, Bejan-Angoulvant T, Patin F, et al. Plasma creatinine and amyotrophic lateral sclerosis prognosis: A systematic review and meta-analysis. Amyotroph Lateral Scler Frontotemporal Degener. 2019;20(3–4):199–206. [DOI] [PubMed] [Google Scholar]
- 226. Mukli P, Wu DH, Csipo T, et al. Urinary biomarkers of oxidative stress in aging: Implications for prediction of accelerated biological age in prospective cohort studies. Oxid Med Cell Longev. 2022;2022:6110226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227. Amante DJ, Kim J, Carreiro ST, et al. Uridine ameliorates the pathological phenotype in transgenic G93A-ALS mice. Amyotroph Lateral Scler. 2010;11(6):520–530. [DOI] [PubMed] [Google Scholar]
- 228. Walk D, Nicholson K, Locatelli E, et al. Randomized trial of inosine for urate elevation in amyotrophic lateral sclerosis. Muscle Nerve. 2023;67(5):378–386. [DOI] [PubMed] [Google Scholar]
- 229. Nichol CA, Smith GK, Duch DS. Biosynthesis and metabolism of tetrahydrobiopterin and molybdopterin. Annu Rev Biochem. 1985;54(1):729–764. [DOI] [PubMed] [Google Scholar]
- 230. Kośliński P, Bujak R, Daghir E, Markuszewski MJ. Metabolic profiling of pteridines for determination of potential biomarkers in cancer diseases. Electrophoresis. 2011;32(15):2044–2054. [DOI] [PubMed] [Google Scholar]
- 231. Hoffmann G, Kenn S, Wirleitner B, et al. Neopterin induces nitric oxide-dependent apoptosis in rat vascular smooth muscle cells. Immunobiology. 1998;199(1):63–73. [DOI] [PubMed] [Google Scholar]
- 232. Basu P, Burgmayer SJN. Pterin chemistry and its relationship to the molybdenum cofactor. Coord Chem Rev. 2011;255(9):1016–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. Sakurai A, Goto M. Neopterin: Isolation from human urine. J Biochem. 1967;61(1):142–145. [DOI] [PubMed] [Google Scholar]
- 234. Schroecksnadel K, Winkler C, Fuchs D. Method for urinary neopterin measurements by HPLC. J Biochem Biophys Methods. 2006;66(1–3):99–100. [DOI] [PubMed] [Google Scholar]
- 235. Werner-Felmayer G, Werner ER, Fuchs D, Hausen A, Reibnegger G, Wachter H. Characteristics of interferon induced tryptophan metabolism in human cells in vitro. Biochim Biophys Acta. 1989;1012(2):140–147. [DOI] [PubMed] [Google Scholar]
- 236. Fuchs D, Weiss G, Reibnegger G, Wachter H. The role of neopterin as a monitor of cellular immune activation in transplantation, inflammatory, infectious, and malignant diseases. Crit Rev Clin Lab Sci. 1992;29(3–4):307–344. [DOI] [PubMed] [Google Scholar]
- 237. Berdowska A, Zwirska-Korczala K. Neopterin measurement in clinical diagnosis. J Clin Pharm Ther. 2001;26(5):319–329. [DOI] [PubMed] [Google Scholar]
- 238. Kann O, Almouhanna F, Chausse B. Interferon gamma: A master cytokine in microglia-mediated neural network dysfunction and neurodegeneration. Trends Neurosci. 2022;45(12):913–927. [DOI] [PubMed] [Google Scholar]
- 239. Millner MM, Franthal W, Thalhammer GH, et al. Neopterin concentrations in cerebrospinal fluid and serum as an aid in differentiating central nervous system and peripheral infections in children. Clin Chem. 1998;44(1):161–167. [PubMed] [Google Scholar]
- 240. Yan J, Kothur K, Mohammad S, et al. CSF Neopterin, quinolinic acid and kynurenine/tryptophan ratio are biomarkers of active neuroinflammation. EBioMedicine. 2023;91:104589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241. Murr C, Widner B, Wirleitner B, Fuchs D. Neopterin as a marker for immune system activation. Curr Drug Metab. 2002;3(2):175–187. [DOI] [PubMed] [Google Scholar]
- 242. Zhao W, Beers DR, Henkel JS, et al. Extracellular mutant SOD1 induces microglial-mediated motoneuron injury. Glia. 2010;58(2):231–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243. Liao B, Zhao W, Beers DR, Henkel JS, Appel SH. Transformation from a neuroprotective to a neurotoxic microglial phenotype in a mouse model of ALS. Exp Neurol. 2012;237(1):147–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244. McCauley ME, Baloh RH. Inflammation in ALS/FTD pathogenesis. Acta Neuropathol. 2019;137(5):715–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245. Nagai M, Re DB, Nagata T, et al. Astrocytes expressing ALS-linked mutated SOD1 release factors selectively toxic to motor neurons. Nat Neurosci. 2007;10(5):615–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246. Brettschneider J, Toledo JB, Van Deerlin VM, et al. Microglial activation correlates with disease progression and upper motor neuron clinical symptoms in amyotrophic lateral sclerosis. PLoS One. 2012;7(6):e39216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247. Zhao W, Beers DR, Hooten KG, et al. Characterization of gene expression phenotype in amyotrophic lateral sclerosis monocytes. JAMA Neurol. 2017;74(6):677–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248. Malaspina A, Puentes F, Amor S. Disease origin and progression in amyotrophic lateral sclerosis: An immunology perspective. Int Immunol. 2015;27(3):117–129. [DOI] [PubMed] [Google Scholar]
- 249. Rybak ME, Sternberg MR, Pfeiffer CM. Sociodemographic and lifestyle variables are compound- and class-specific correlates of urine phytoestrogen concentrations in the U.S. population. J Nutr. 2013;143(6):986S–994S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250. Wilson T, Garcia-Perez I, Posma JM, et al. Spot and cumulative urine samples are suitable replacements for 24-hour urine collections for objective measures of dietary exposure in adults using metabolite biomarkers. J Nutr. 2019;149(10):1692–1700. [DOI] [PubMed] [Google Scholar]
- 251. Barratt TM, McLaine PN, Soothill JF. Albumin excretion as a measure of glomerular dysfunction in children. Arch Dis Child. 1970;45(242):496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252. Barr DB, Wilder LC, Caudill SP, Gonzalez AJ, Needham LL, Pirkle JL. Urinary creatinine concentrations in the U.S. population: Implications for urinary biologic monitoring measurements. Environ Health Perspect. 2005;113(2):192–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253. Mikheev MI, Lowry LK. WHO global project on biological monitoring of chemical exposure at the workplace. Int Arch Occup Environ Health. 1996;68(6):387–388. [DOI] [PubMed] [Google Scholar]
- 254. Yeh H-C, Lin Y-S, Kuo C-C, et al. Urine osmolality in the US population: Implications for environmental biomonitoring. Environ Res. 2015;136:482–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255. Kowalewski NN, Forster CS. Collection, processing, and storage consideration for urinary biomarker research. J Vis Exp. 2021;176. [DOI] [PubMed] [Google Scholar]
- 256. Virreira Winter S, Karayel O, Strauss MT, et al. Urinary proteome profiling for stratifying patients with familial Parkinson's disease. EMBO Mol Med. 2021;13(3):e13257. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analysed in this study.