Abstract
The mitochondrial electron transport chain (ETC) is a multi-component pathway that mediates the transfer of electrons from metabolic reactions that occur in the mitochondrion to molecular oxygen (O2). The ETC contributes to numerous cellular processes, including the generation of cellular ATP through oxidative phosphorylation, serving as an electron sink for metabolic pathways such as de novo pyrimidine biosynthesis and for maintaining mitochondrial membrane potential. Proper functioning of the mitochondrial ETC is necessary for the growth and survival of apicomplexan parasites including Plasmodium falciparum, a causative agent of malaria. The mitochondrial ETC of P. falciparum is an attractive target for antimalarial drugs, due to its essentiality and its differences from the mammalian ETC. To identify novel P. falciparum ETC inhibitors, we have established a real-time assay to assess ETC function, which we describe here. This approach measures the O2 consumption rate (OCR) of permeabilized P. falciparum parasites using a Seahorse XFe96 flux analyzer and can be used to screen compound libraries for the identification of ETC inhibitors and, in part, to determine the targets of those inhibitors.
Key features
• With this protocol, the effects of candidate inhibitors on mitochondrial O2 consumption in permeabilized asexual P. falciparum parasites can be tested in real time.
• Through the sequential injection of inhibitors and substrates into the assay, the molecular targets of candidate inhibitors in the ETC can, in part, be determined.
• The assay is applicable for both drug discovery approaches and enquiries into a fundamental aspect of parasite mitochondrial biology.
Keywords: Plasmodium falciparum, Malaria, Seahorse XFe96, Mitochondria, Electron transport chain, Antimalarial drugs, Drug discovery
Graphical overview
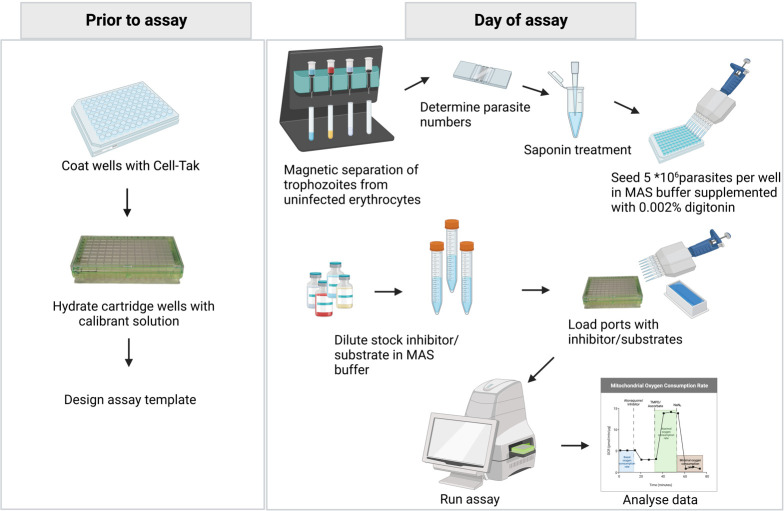
Seahorse assay experimental workflow. Prior to the assay, coat the cell culture microplate with Cell-Tak to help adhere the parasites to the wells; hydrate the cartridge wells to ensure proper sensor functionality and design the assay template using the Agilent Seahorse Wave Desktop software (Analyze Seahorse data files, Seahorse Wave desktop software|Agilent). On the day of the assay, prepare the inhibitors/substrates that are to be injected into the ports. Then, separate 3 × 108 trophozoite-stage parasites from the uninfected red blood cells (RBCs) and ring-stage parasites using a MACS® magnetic column. Check the purity of the parasites with Giemsa-stained smears. Determine the concentration of infected RBCs in the sample using a hemocytometer and dilute to approximately 5 × 107 parasites per milliliter. Treat infected RBCs with saponin to permeabilize the host cell membrane and seed approximately 5 × 106 parasites (100 μL) per well in mitochondria assay solution (MAS) buffer. Supplement MAS buffer with digitonin to permeabilize the parasite plasma membrane. Load the ports with the prepared inhibitors/substrates and run the assay using a Seahorse XFe96 analyzer. Once the assay is completed, analyze the data using the Wave desktop software. Further data processing can be done using statistical analysis software.
Background
Plasmodium species cause malaria in humans and were responsible for 247 million malaria cases and 619,000 deaths in 2021 (WHO report, 2022). The emergence of parasites resistant to frontline antimalarial drugs has contributed to the increase in malaria cases and deaths in recent years (Conrad and Rosenthal, 2019; Ippolito et al., 2021). Hence, it is important to look beyond the currently available antimalarial drugs for new treatments. The mitochondrion of Plasmodium is a validated drug target, and some common antimalarial compounds interfere with its electron transport chain (ETC) (Hikosaka et al., 2015; Ke and Mather, 2017).
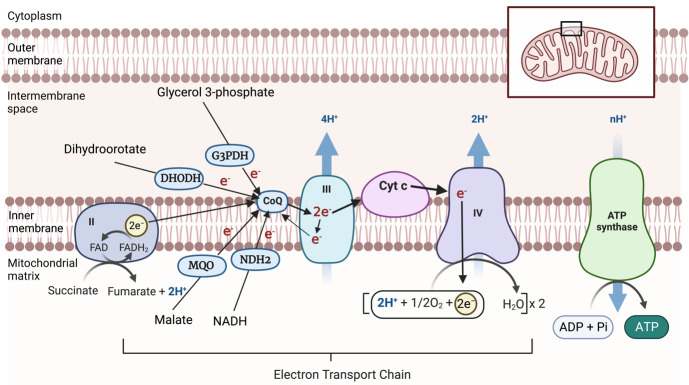
The ETC is located in the inner mitochondrial membrane of eukaryotes and facilitates the transfer of electrons through a series of redox centers to O2. Electrons are sourced from the oxidation of substrates such as malate, succinate, NADH, glycerol 3-phosphate, and dihydroorotate in the mitochondrion (Hayward and van Dooren, 2019). These electrons are donated to coenzyme Q (CoQ). Reduced CoQ docks at the cytochrome bc1 complex (also known as Complex III). Complex III facilitates the transfer of electrons to an electron carrying protein, cytochrome c (CytC). From CytC, electrons are donated to the cytochrome c oxidase complex (Complex IV), where the reduction of O2, the final electron acceptor of the ETC, occurs. The transfer of electrons through Complexes III and IV is coupled to the net movement of protons from the mitochondrial matrix to the mitochondrial intermembrane space (Figure 1). The proton motive force that is established across the inner mitochondrial membrane by the ETC is critical for the synthesis of ATP by the ATP synthase complex (Figure 1).
Figure 1. Mitochondrial electron transport chain.
Electrons from the oxidation of a range of different substrates in the mitochondrion are donated to coenzyme Q (CoQ). CoQ donates these electrons to Complex III, from where they are donated via cytochrome c (CytC) to Complex IV, where electrons ultimately reduce O2 to form H2O. Electron transport events are indicated by arrows. The passage of electrons through Complexes III and IV is coupled to the net transport of protons (H+) from the mitochondrial matrix to the intermembrane space, generating an H+ gradient that can be harnessed by ATP synthase to generate ATP. DHODH, dihydroorotate dehydrogenase; G3PDH, glycerol 3-phosphate dehydrogenase; MQO, malate: quinone oxidoreductase; NDH2, type II-NADH dehydrogenase; FADH2, Dihydroflavine-adenine dinucleotide; FAD, Flavin adenine dinucleotide. The inset indicates the position of the enlarged portion within the mitochondrion. Created with BioRender.com.
The P. falciparum ETC varies considerably from the ETC of their mammalian hosts (Hayward and van Dooren, 2019). For example, the P. falciparum ETC has a single subunit “type II” NADH dehydrogenase instead of the multi-subunit Complex I found in mammals and a malate:quinone oxidoreductase enzyme that feeds electrons from malate oxidation directly into the ETC (Figure 1; Rajaram et al., 2022). Complex III is a validated drug target in apicomplexans, and the compounds that inhibit Complex III often target either (or in rare cases both) of the CoQ binding sites in the complex. Structural differences in the CoQ binding sites of parasite Complex III compared with human Complex III likely account for the high degree of selectivity of inhibitors like atovaquone against the parasite complex (Srivastava et al., 1999; Fisher et al., 2020). Mutations in the CoQ binding sites of parasite Complex III can confer clinically relevant levels of resistance against compounds like atovaquone (Staines et al., 2018). It is therefore of interest to identify novel inhibitors of Complex III or the other components of the P. falciparum ETC, ideally ones that remain potent against parasites already resistant to existing ETC inhibitors.
Measuring cellular O2 consumption is a powerful means of assessing ETC function. We have recently developed a suite of Seahorse XFe96 flux analyzer assays to measure mitochondrial O2 consumption rates in the apicomplexan parasite Toxoplasma gondii, a species from the same phylum as Plasmodium (Hayward et al., 2022). We have now established similar assays to enable the measurement of mitochondrial O2 consumption rates in permeabilized asexual-stage Plasmodium falciparum parasites and describe them in this protocol (Graphical overview; General note 1). The assay can be used to screen compound libraries to identify ETC inhibitors in P. falciparum. The assay can aid in determining the molecular target of identified inhibitors in the ETC and the effectiveness of novel inhibitors against drug-resistant parasite strains (Hayward et al., 2023; Nguyen et al., 2023). The assay can also be used to obtain molecular insights into the functionality of the P. falciparum ETC.
Materials and reagents
175 cm2 tissue culture flasks (Sarstedt, catalog number: 83.3912)
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) (Sigma-Aldrich, catalog number: H3375-250G)
5% (w/v) digitonin (Thermo Fisher Scientific, InvitrogenTM, catalog number: BN 20061)
Albumax II (Gibco, catalog number: 11021-045-1kg)
Atovaquone (Sigma-Aldrich, catalog number: A7986-10MG)
Cell-Tak (Corning, catalog number: 354241)
D-glucose (Sigma-Aldrich, catalog number: G7021)
D-mannitol (Sigma-Aldrich, catalog number: M9546-250G)
Dimethyl sulfoxide (DMSO) (Sigma-Aldrich, catalog number: D8418-250G)
Ethylene glycol-bis(2-amino-ethylether)-N,N,N,N’-tetra acetic acid (EGTA) (Sigma-Aldrich, catalog number: E3889-25G)
Fatty acid-free bovine serum albumin (BSA) (Sigma-Aldrich, catalog number: A3803-100G)
General plastic consumables [sterile Eppendorf tubes, sterile falcon tubes (15, 50 mL), sterile pipette tips (20, 200, and 1,000 μL), sterile serological pipettes (10, 25 mL); Sarstedt or Corning]
Gentamicin (Invitrogen, catalog number: 15710-072)
Hypoxanthine (Sigma-Aldrich, catalog number: H-9636)
L-ascorbic acid (Sigma-Aldrich, catalog number: A5960-25G)
L-malic acid (Sigma-Aldrich, catalog number: M1000-100G)
Magnesium chloride hexahydrate (MgCl2·6H2O) (Sigma-Aldrich, catalog number: M2670-100g)
Milli-Q water (fresh from any available purification system)
N,N,N,N’-tetramethyl-p-phenylenediamine (TMPD) (Sigma-Aldrich, catalog number: T7394-5G)
Potassium chloride (Sigma-Aldrich, catalog number: 7447-40-7, 500G)
Potassium phosphate monobasic (KH2PO4) (ChemSupply, catalog number: PA009-500G)
RPMI-HEPES with Glutamax (Thermo Fisher ScientificTM, catalog number: 72400120)
Sodium azide (AJAX-Finechem Univar, catalog number: AJA1222-100G)
Sodium chloride (Sigma-Aldrich, catalog number: S5886-5KG)
Sodium dihydrogen phosphate monohydrate (AnalaR, catalog number: 102454R)
Sorbitol (Sigma-Aldrich, catalog number: 50-70-4)
Sucrose (Sigma-Aldrich, catalog number: S9378-5KG)
Ascorbate stock (200 mM) (see Recipes)
Albumax II (5% w/v) (see Recipes)
Atovaquone stock (25 mM) (see Recipes)
Complete culture medium (see Recipes)
Malate stock (500 mM) (see Recipes)
MAS buffer (3× stock) (see Recipes)
MAS buffer (1× working solution) (see Recipes)
Phosphate buffered saline (10×) (see Recipes)
Saponin (0.15% w/v) (see Recipes)
Sodium azide stock (100 mM) (see Recipes)
Sodium bicarbonate (0.1 M) (see Recipes)
Sorbitol (5% w/v) (see Recipes)
TMPD stock (500 mM) (see Recipes)
Biological material
Plasmodium falciparum cells (wild type 3D7)
Recipes
-
Ascorbate stock (200 mM)
Dissolve 352 mg of L-ascorbic acid in 5 mL of 1× MAS buffer. Adjust pH to 7.4 with potassium hydroxide (KOH) and adjust final volume with 1× MAS buffer to 10 mL. Aliquot ascorbate stock (500 μL/aliquot) and store at -20 °C for long-term storage.
-
Albumax II (5% w/v)
Dissolve 25 g of Albumax II in 500 mL of RPMI medium. Filter sterilize using the Millipore Stericup Vacuum Driven Disposable Filtration System. Aliquot Albumax II stock (50 mL/aliquot) and store at -20 °C for long-term storage.
-
Atovaquone stock (25 mM)
Dissolve 10 mg of atovaquone in 1.09 mL of DMSO. Aliquot atovaquone stock (50 μL/aliquot) and store at -20 °C for long-term storage.
-
Complete culture medium
Prepare complete culture medium from RPMI 1640-HEPES with Glutamax supplemented with the cell culture media components shown below. Store complete culture medium at 4 °C and pre-warm to 37 °C before use.
Components Final concentration Volume RPMI medium - 500 mL Additional glucose (2.5 M) 10 mM 2 mL Hypoxanthine (200 mM) 480 μM 1.2 mL Gentamicin (10 mg/mL) 20 μg/mL 1 mL 5% Albumax solution 0.375% w/v 37.5 mL Heat-inactivated human serum 2.5% v/v 12.5 mL -
Malate stock (500 mM)
Dissolve 670.45 mg of L-malic acid in 5 mL of MAS buffer. Adjust pH to 7.4 with KOH and adjust the final volume with MAS buffer to 10 mL. Aliquot malate stock (1.5 mL/aliquot) and store at -20 °C for long-term storage.
-
MAS buffer (3× stock)
Weigh out the components (listed below) and dissolve in 50–75 mL of Milli-Q water by placing the solution into a water bath and/or use a magnetic stirrer. Adjust pH of the solution to 7.4 using KOH. Bring the final volume to 125 mL. Filter sterilize for long-term storage.
Components Final concentration Weight Mannitol 660 mM 15.03 g Sucrose 210 mM 8.99 g KH2PO4 30 mM 0.51 g MgCl2 15 mM 0.381 g HEPES 6 mM 0.1785 g EGTA 3 mM 0.1425 g -
MAS buffer (1× working solution)
On the day of the experiment, mix 15 mL of 3× MAS buffer with 30 mL of Milli-Q water, to get 45 mL of 1× MAS buffer. Add 90 mg of fatty acid-free BSA and 2.25 mL of 0.5 M malate.
-
Phosphate buffered saline (10×)
Components Weight NaCl 80 g KCl 2 g Na2HPO4 14.4 g KH2PO4 1.9 g Dissolve the compounds in 800 mL of Milli-Q water. Adjust pH of the solution to 7.4 using HCl. Bring the final volume to 1 L. Sterilize the solution by autoclaving for 20 min at 15 psi. Store at room temperature. When needed, mix 100 mL of 10× PBS with 900 mL of Milli-Q water to get 1× PBS.
-
Saponin (0.15% w/v)
Dissolve 0.15 g of saponin in 100 mL of PBS. Filter-sterilize the solution using a 0.2 μm filter. Store at 4 °C for long-term storage. When needed, mix 1 mL of 0.15% saponin with 2 mL of PBS to obtain a 0.05% (w/v) saponin working solution.
-
Sodium azide stock (100 mM)
Dissolve 65.1 mg of sodium azide in 10 mL of MAS buffer. Aliquot sodium azide stock (500 μL/aliquot) and store at -20 °C for long-term storage.
-
Sodium bicarbonate (0.1 M)
Dissolve 84 mg of sodium bicarbonate in approximately 8 mL of Milli-Q water. Adjust pH to 8.0 and make up the volume to 10 mL. Filter-sterilize using a 0.2 μm filter. Store at room temperature.
-
Sorbitol (5% w/v)
Dissolve 5 g of sorbitol in 100 mL of Milli-Q water. Filter-sterilize the solution using a 0.2 μm filter. Store at 4 °C for long-term storage.
-
TMPD stock (500 mM)
Dissolve 82.2 mg of TPMD in 1 mL of ethanol. Aliquot TMPD solution (50 μL/aliquot) and store at -20 °C for long-term storage.
Equipment
4 °C fridge and -20 °C freezer
Tabletop centrifuge for 15 and 50 mL tubes (Beckman Coulter, model: Allegra X-15R)
Tabletop centrifuge for microcentrifuge tubes (Thermo Scientific, model: Heraeus Pico 17)
Improved Neubauer hemocytometer (Hirschmann, model: 8100104)
Humidified 37 °C non-CO2 incubator
MACS® magnetic column (CS columns, Miltenyi Biotec, catalog number: 130-041-305) with a sterile 3-way stopclock
SuperMACS II separator (Miltenyi Biotec, catalog number: 130-044-104)
Multichannel pipettes (30–300 μL) (Gilson)
pH meter (Cyberscan pH510)
Pipettes (0.2–2, 2–20, 20–200, and 100–1,000 μL) (Gilson)
Reagent reservoirs
Seahorse XFe96 Analyzer (Agilent)
Seahorse XFe96 FluxPak (XFe96 cell culture microplate, XFe96 sensor cartridge and utility plate, XF Calibrant Solution) (Agilent, catalog number: 102416-100)
Standard inverted light microscope (Olympus CKX41), fitted with 20× objective lense
Biological safety cabinet (Safemate 1.2 vision, Edwards Group)
Water bath, 37 °C (WB7, Ratek)
Zip-lock bags
Software
Wave Desktop Software, version 2.6.3 (for running the Seahorse XFe96 assay)
GraphPad Prism 9 (for statistical analyses of the results)
Procedure
-
Parasite culturing
Maintain P. falciparum parasites in complete culture medium at 4% hematocrit and approximately 3% parasitaemia in cell culture plates and flasks (Maier and Rug, 2013).
Every second day, check the parasitaemia using Giemsa staining technique; replace the culture with fresh complete culture medium pre-warmed to 37 °C and dilute the parasite cultures to 0.2% parasitaemia. Gas the culture flask with malaria gas mixture of 1% O2, 5% CO2, and 94% N2 for 30 s; close the cap of the flask and incubate at 37 °C.
Maintain a synchronized P. falciparum culture by treating the parasites with sorbitol according to Lambros and Vanderberg (1979).
Expand the cell culture to approximately 3 × 108 trophozoite-infected RBCs and proceed with the Seahorse XFe96 assay (General note 2).
-
XF96 cell culture microplate pre-treatment (at least 1 day prior to the experiment)
Figure 2 depicts various parts of the Seahorse XFe96 sensor cartridge used in the protocol. The XFe96 analyzer uses only 96-well plates. A 24-well plate format is available for use with a Seahorse XFe24 analyzer, with a previously published protocol describing the analysis of O2 consumption rates in P. falciparum using this approach (Sakata-Kato and Wirth, 2016).
In order to immobilize Plasmodium falciparum cells on the Agilent Seahorse XFe96 plate during the experiment, coat each well with Cell-Tak, according to the manufacturer’s protocol, 1–5 days prior to the experiment. Briefly, prepare Cell-Tak in 0.1 M sodium bicarbonate at a final concentration of 22.12 μg/mL (General note 3); 2.5 mL is enough for coating one 96-well plate. Dilute Cell-Tak right before use, mix thoroughly, and dispense immediately.
Dispense 25 μL of Cell-Tak in each well of the XFe96 cell culture microplate; 25 μL is usually sufficient to cover the entire bottom of the well, but care must be taken to spread Cell-Tak throughout the well.
Incubate the plate for 20 min at room temperature. Aspirate the solution. Invert and pat the plate to remove any residual Cell-Tak solution.
Wash each well with 200 μL of Milli-Q water, twice, to reduce any trace sodium bicarbonate.
Discard the water and air-dry the wells under sterile conditions. The plate can be left in the cell culture hood for drying.
Store the XFe96 cell culture microplate in a zip-lock bag in the fridge (4 °C) for up to one week.
On the day of the experiment, warm the plate to room temperature in a biological safety cabinet before seeding cells.
-
Hydrate the sensor cartridge (1 day prior to the experiment)
The sensor cartridge consists of solid-state sensor materials, which detect changes in pH and O2 concentration during the experiment. Prepare the sensors by hydrating the sensor cartridge according to Agilent’s user guide as follows. Open the XFe96 FluxPak (cartridge + utility plate package) and place the cartridge upside down (sensor end up) next to the utility plate. Do not touch the sensors. Fill all the wells of the utility plate with 200 μL of Milli-Q water.
Place the cartridge back on the utility plate and make sure that the sensors are submerged completely. Incubate the sensor cartridge and utility plate in a 37 °C humidified incubator overnight.
On the day of the assay, remove the cartridge and place it upside down (sensor end up). Discard the water from the utility plate and fill all the wells with 200 μL of XF Calibrant pre-warmed to 37 °C.
Place the cartridge back in the utility plate and move the cartridge up and down, twice, to remove any trapped air bubbles from the sensor.
Incubate the utility plate with the sensor cartridge and calibrant solution in a 37 °C humidified incubator for at least 1 h prior to the start of the experiment.
-
Wave program template preparation
Set up Wave desktop software.
-
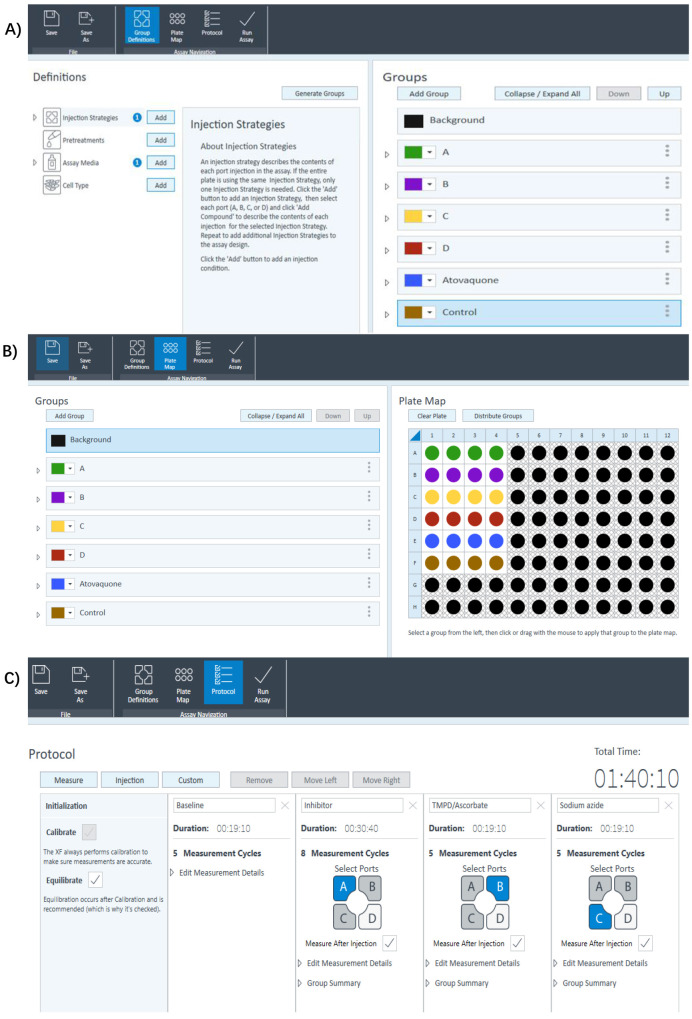
Under the group definitions tab, define the injection strategies (Figure 3A). An example is shown below.
Port A, inhibitors/atovaquone (5 μM).
Port B, TMPD (0.2 mM) and ascorbic acid (2 mM).
Port C, sodium azide (NaN3; 10 mM).
Configure the plate map by assigning individual groups to different conditions of the experiment (Figure 3B). Make sure to include background wells (measurement without cells). A–D represents different candidate P. falciparum ETC inhibitors. Atovaquone is used as the positive control. Control wells have parasites without drug treatment (i.e., solvent control injected from Port A). Background wells do not have parasites. Four technical replicates are included for each condition.
-
Under the protocol tab, click on injection and specify the timing and repeats of the mixing and measurement cycles for each injection (Figure 3C). e.g., eight cycles of measurement after inhibitor injection (five cycles of measurements otherwise). Each cycle of measurement has:
20 s of mixing,
1 min of waiting,
2.5 min of measuring.
Save this template. On the day of the experiment, open the template and click on the run assay tab to start the assay.
-
Inhibitor/substrate preparation
Prepare 45 mL of 1× MAS buffer (see Recipes). Pre-warm 1× MAS buffer, PBS, and saponin in a water bath (37 °C) until use.
-
Prepare the substrates/inhibitors/controls that will be injected in the ports of the sensor cartridge before parasite preparations (General note 4). For the model experiment, we used:
8× atovaquone/inhibitors solution. Prepare a 40 μM solution to obtain a final concentration of 5 μM following injection into the wells during the assay. To make up 40 μM stocks, add 1.4 μL of 10 mM inhibitor to 348.6 μL of MAS buffer, or add 1.1 μL of 25 mM atovaquone to 698.9 μL of MAS buffer.
9× TMPD/ascorbate solution. Prepare a solution containing 1.8 mM TMPD and 18 mM ascorbate to obtain a final concentration following injection of 0.2 mM TMPD/2 mM ascorbate. Add 6.5 μL of 500 mM TMPD and 162 μL of 200 mM ascorbate to 1,631.5 μL of MAS buffer.
10× sodium azide solution. Prepare a solution of 100 mM NaN3 to obtain a final concentration following injection of 10 mM NaN3. Add 300 μL of 1 M NaN3 to 2,700 μL of MAS buffer (General note 5).
-
Parasite preparation
Assemble the MACS® magnetic column with three-way stopcock inside the biological safety cabinet. Pre-wash the column by passing 30 mL of 100% ethanol, 30 mL of 80% ethanol, 30 mL of PBS (pre-warmed to 37 °C), and 30 mL of RPMI-HEPES with Glutamax, pre-warmed to 37 °C (General note 6).
Enrich the culture for trophozoite-stage parasites by passing 3 × 108 trophozoite-infected RBCs drop-by-drop through a MACS® magnetic column placed in the magnetic field of a SuperMACS II separator (Ridgway et al., 2021). The volume of the culture is adjusted to yield a hematocrit of approximately 15% before passing through the column. Uninfected and ring-stage infected RBCs will pass through the column, while RBCs infected with mature-stage parasites (trophozoites and schizonts) will be retained.
Remove any residual uninfected red blood cells or ring-stage parasites retained in the column by passing 30–50 mL of complete culture medium pre-warmed to 37 °C through the column.
Remove the column from the magnet and elute RBCs infected with mature-stage parasites in 20–30 mL of complete culture medium.
Determine the parasite-infected RBC yield using a hemocytometer. Check the purity of the parasites in the eluate using Giemsa-stained blood smears.
Free 2 × 108–3 × 108 trophozoite-stage parasites from erythrocytes by treating infected RBCs with 1 mL of pre-warmed 0.05% (w/v) saponin in PBS at 37 °C for 3 min. Wash the cells with pre-warmed PBS three times or until the supernatant is no longer red in color (i.e., until the hemoglobin is completely removed), pelleting the parasites by centrifugation at 2,000× g for 2 min at room temperature after each wash.
The parasite yield should be approximately 2 × 108–3 × 108 cells. Adjust the parasite concentration to 5 × 107 parasites/mL in mitochondrial assay solution (MAS) buffer containing 0.002% (w/v) digitonin. Digitonin permeabilizes the parasite plasma membrane. Pre-heat digitonin solution to 62 °C (to dissolve it) before adding. Parasites start losing viability after detergent treatment and it is advisable to start the O2 consumption rate (OCR) measurements within one hour of the detergent treatment.
Seed 100 μL of the permeabilized parasites per well in the Cell-Tak-coated XFe96 cell culture plate. This results in a density of 5 × 106 cells per well. Centrifuge at 800× g for 10 min at room temperature with the centrifuge brake set to low. This step ensures the parasites adhere to the bottom of the wells. Check the plates under the microscope to ensure that a monolayer of cells has formed (see Figure 4).
Carefully add 75 μL of MAS buffer to the side of the wells at an angle, without agitating and detaching the adhered parasites. Check the plates under the microscope again to confirm that the monolayer of cells has not been disturbed.
Transfer the plate to an incubator (37 °C) at atmospheric CO2 to degas the plate of excess CO2 levels, and store until use. This step is crucial to effectively degas the plastic material of any CO2 (Thangaraj et al., 2018). Degassing adjusts the CO2 in the assay plate to (low) atmospheric CO2 levels, ensuring pH values of the assay solutions are not influenced by changes in CO2 levels during the assay.
-
OCR measurements
At least one hour prior to the experiment, turn on the XFe96 flux analyzer and the temperature controller to calibrate the instrument.
Open the prepared experiment template in Wave software and export it as a design file (.asyt) to run the assay.
Carefully load 25 μL of each inhibitor/substrate into the injection ports after the cartridge has been placed in the utility plate. For the assay we describe in this manuscript: Port A—Compounds (8× the final concentration), port B—TMPD (9× the final concentration), port C—NaN3 (10× the final concentration) port D—empty in this experiment. To load the solution into the port, insert the pipette tip to near the bottom of the port. Lift the pipette tip slightly and angle the tip alongside the wall. Slowly dispense the inhibitor/substrate into the port (General note 7).
Ensure all ports are filled, including ports of blank wells and ports of unused wells without cells. In the sample experiment, assay ports A, B, and C of all wells must be filled to enable compound/substrate dispensing. Port D, which is not used, can remain empty.
Click Run assay in the Wave software and, when prompted, load the cartridge (filled with the substrates/inhibitors to be injected) with the utility plate onto the plate holder. Make sure to remove the lid from the utility plate and place the plate in the right orientation onto the plate holder.
The sensor calibration takes approximately 25–30 min.
Once the calibration is finished, the utility plate is automatically ejected. The sensor cartridge is retained inside the XFe96 analyzer. Replace the utility plate with the cell plate containing the adhered parasites (again without the lid) and resume the run.
Once the experiment is complete, the cell culture plate is ejected. Remove the plate and the sensor cartridge and examine the injection ports to verify that all compounds were injected (General note 8).
The results file (.asyr) is automatically saved on the computer. Save the result file onto a USB drive to transfer to another computer with the analysis software.
Discard the cell culture plate with the cartridge containing the parasites and the substrates safely and appropriately (General note 9).
Figure 2. Parts of the Seahorse XFe96 sensor cartridge.
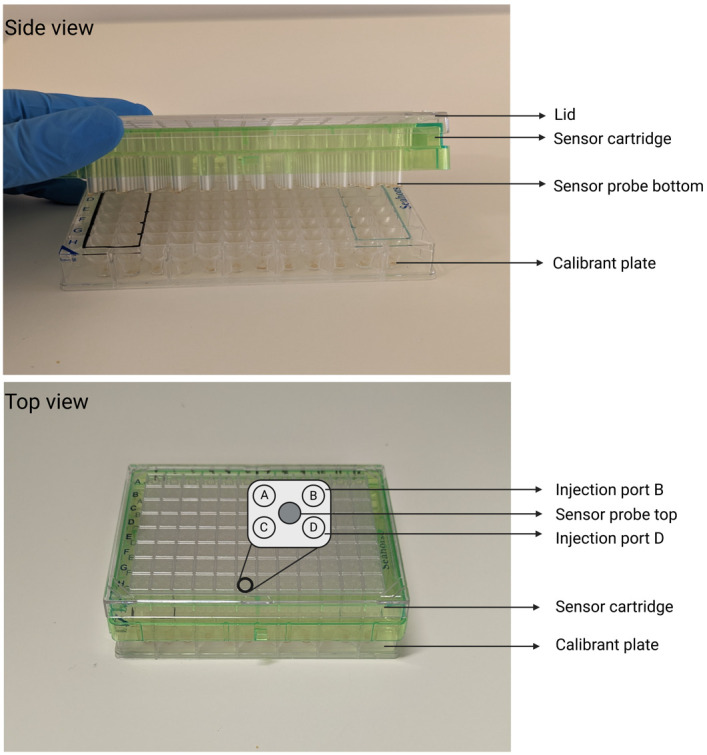
A) Side view shows the bottom of the sensor probes attached to the green sensor cartridge. B) Top view shows an example of the four injection ports above each of the 96 wells. The inset shows ports A–D and the top of the sensor probe. Created with BioRender.com.
Figure 3. Designing a Seahorse XFe96 experimental template in the Wave software.
A) Under Group Definitions tab, fill in the injection strategies as follows. Port A: Inhibitors/atovaquone; Port B: TMPD; Port C: Sodium azide. B) Under the Plate Map tab, customize the plate map according to your group definition. Ensure background wells (wells without cells) are included. C) Under the Protocol tab, specify the timing and substrates to be injected from a specific port. Specify the number of mixing and measurement cycles as follows. After inhibitor injection 8 cycles (5 cycles for all other substrate injection); 20 s mixing; 1 min waiting; 2.5 min measuring.
Figure 4. Outline of P. falciparum in a cell monolayer.
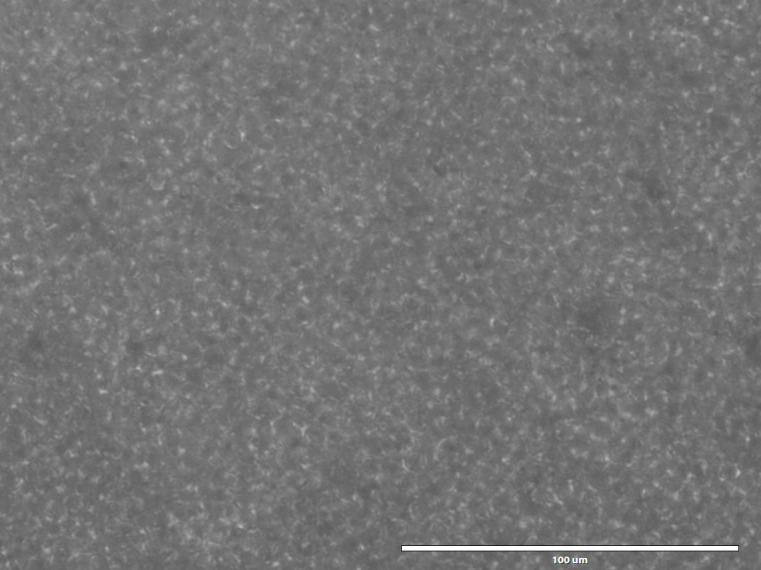
Cell-Tak-coated P. falciparum cell monolayer in XFe96 cell culture plate, after centrifuging the plate with low brake.
Data analysis
Open the results file, using Wave software 2.6.3 or another version.
Click on add view and select overview to view the results and plate map.
Make sure the background correction box is ticked. Background wells lack parasites and, once background correction box is ticked, the OCR values from these background wells will be subtracted from the experimental wells.
Occasionally, substrates from one or more ports may not dispense properly (e.g., fail to inject or inject prematurely). This will lead to errors in the data. In addition to checking the injection ports following the experiment (General note 8), analyze the data from each group on the plate map and look for instances where premature injection of substrates from some ports may have occurred. If you have a strong reason to believe that improper injection has occurred into a well, you can consider excluding the data from that particular well from the analysis by unselecting it (General note 10).
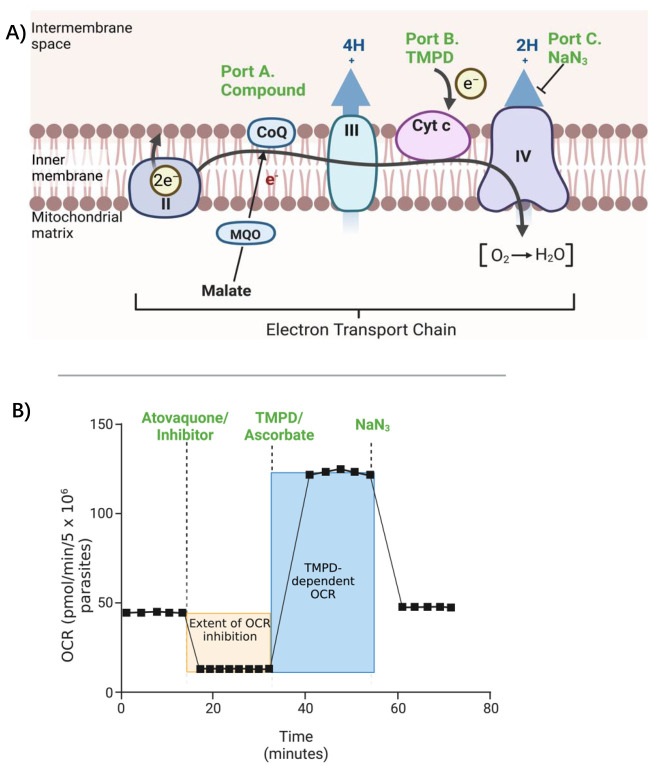
The result indicates the effect of different substrates or inhibitors on the OCR of P. falciparum over time (Figure 5A). The results can be plotted onto a graph as time (minutes) vs. OCR (pmol/min) (Figure 5B), typically as the average OCR value of the technical replicates at each time point in the plate. From these data, it is possible to calculate the extent of OCR inhibition caused by the candidate inhibitor, the extent of TMPD-dependent OCR recovery, and NaN3-dependent OCR inhibition. To ensure confidence in the data, independent biological repeats should be performed on different days.
Click on export to export the results to statistical analysis software like GraphPad Prism or Microsoft Excel to further process the data.
Figure 5. Model data based on the injection strategies performed.
A) Schematic depicting the effects of the inhibitors and substrates on the mitochondrial electron transport chain. B) Initially, malate-dependent O2 consumption rate (OCR) is measured to establish the basal OCR. The compound of interest, either a putative inhibitor or atovaquone (positive control), is then injected into the wells to measure their effect on the OCR. The OCR will be diminished if the inhibitor affects the mitochondrial electron transport chain (ETC). Following several measurements, TMPD is injected into the wells. TMPD donates electrons to cytochrome c and thereby bypasses Complex III. If TMPD injection restores the OCR, it implies that the test compound is inhibiting OCR upstream of cytochrome c. After a few measurements, the Complex IV inhibitor sodium azide (NaN3) is injected into the wells. If NaN3 injection inhibits the OCR, it confirms that the TMPD-dependent OCR recovery is dependent on Complex IV activity. Created with BioRender.com.
Further uses of the Seahorse XFe96 assay
In the experiment described in this manuscript, we test candidate ETC inhibitors for their ability to inhibit mitochondrial OCR in P. falciparum, and whether these inhibitors target up- or downstream of cytochrome c in the ETC. The Seahorse XFe96 assay is a versatile one and can be used in a range of other experiments. For example, testing the extent of mitochondrial OCR inhibition at a range of compound concentrations enables determination of the potency of those inhibitors on the ETC [e.g., enables calculation of the half maximal effective concentration (EC50) of the compound on OCR (Hayward et al., 2023)], and the extent to which this changes in drug-resistant parasites. The Seahorse XFe96 assay can also be used for screening large libraries for putative ETC inhibitors, as we have done previously for similar assays in the related parasite Toxoplasma gondii (Hayward et al., 2023). The presence of four injection ports on the 96-well plate enables several hundred compounds to be screened using a single Seahorse XFe96 assay. The Seahorse XFe96 assay we describe here also has uses beyond compound screening strategies. For example, it can be used to measure ETC impairment in mutant parasite strains defective in aspects of mitochondrial biology, or to gain insights into fitness costs associated with mutations conferring resistance to ETC inhibitors.
Validation of protocol
-
Jenni, A. Hayward, F. Victor Makota, Daniela Cihalova, Rachel A. Leonard, Esther Rajendran, Soraya M. Zwahlen, Laura Shuttleworth, Ursula Wiedemann, Christina Spry, Kevin J. Saliba, Alexander G. Maier, Giel G. van Dooren: A screen of drug-like molecules identifies chemically diverse electron transport chain inhibitors in apicomplexan parasites, PLOS Pathogens 19(7): e10111517.
Figure 6. Most of the candidate ETC inhibitors target the ETC upstream of cytochrome c in P. falciparum parasites.
Table 3. Inhibitory activities of MMV Pathogen Box compounds against O2 consumption rate in T. gondii and P. falciparum.
William Nguyen, Madeline G. Dans, Iain Currie, Jon Kyle Awalt, Brodie L. Bailey, Chris Lumb, Anna Ngo, Paola Favuzza, SaiShyam Ramesh, Jocelyn Pennington, Kate E. Jarman, Alexander G. Maier, Giel G. van Dooren, Tony Papenfuss, Sergio Wittlin, Jake Baum, Delphine Baud, Stephen Brand, Paul F. Jackson, Alan F. Cowman, and Brad E. Sleebs: 7-N-Substituted-3-Oxadiazole Quinolones with Potent Antimalarial Activity Target the Cytochrome bc1 Complex; ACS Infectious Diseases. doi: https://doi.org/10.1021/acsinfecdis.2c00607
Figure 5. P. falciparum oxygen consumption rate (OCR) assay.
General notes and troubleshooting
General notes
The current protocol to measure mitochondrial O2 consumption rates in P. falciparum has key differences from the ones we established in T. gondii. P. falciparum preparation and permeabilization requires numerous steps that are detailed in section F. P. falciparum parasites are particularly sensitive to the depletion of carbon sources, which means that, unlike in T. gondii, we need to maintain Plasmodium in a carbon source at all times. In contrast to a previous study using a Seahorse XFe24 analyzer approach to measure O2 consumption in P. falciparum (Sakata-Kato and Wirth, 2016), we find that permeabilization of the parasite plasma membrane with low concentrations of digitonin is critical for enabling the robust measurement of mitochondrial O2 consumption rates in the parasite.
Make sure the parasites are synchronized and the majority is at the late-trophozoite stage on the day of the experiment.
Cell-Tak concentration varies from batch to batch and the final dilution may need to be adjusted depending on the batch. For the model experiment, we used Cell-Tak Lot # 0055009 (1.58 mg/mL) and diluted 35 μL of Cell-Tak in 2.5 mL of 0.1 M sodium bicarbonate solution (final concentration 22.1 μg/mL).
The time from initial parasite preparation to compound injection from the ports is substantial. To best maintain parasite viability, prepare substrate and inhibitor compounds (section E) before commencing parasite preparation (section F).
The substrates from port A are injected first, followed by substrates in port B, C, and D, according to the constant concentration protocol described in the manufacturer’s instructions. Once injected, the added MAS buffer volume in the well dilutes the substrate. To account for the dilution, the concentration of the substrates being injected increases proportionally with each injection. E.g., 8× the desired final concentration for compounds injected from port A, 9× the concentration for port B, 10× the concentration for port C, and 11× for port D.
Sterilizing the column with 25 mL of 100% and 80% ethanol is necessary if the magnetic columns are reused. If a new column is used, the ethanol washes are not required.
Make sure to dispense the correct substrate or inhibitor into the appropriate port, as it is very difficult to completely remove the sample if a wrong compound is dispensed into a port. It will also make the data unreliable if traces of different compounds are mixed. If a wrong compound is dispensed into a port, exclude those wells from the analysis altogether.
One way to verify whether all the ports were injected during the course of the assay is to take a photo of the cartridge before and after the experiment and then checking if all the ports have been discharged during the assay.
The Seahorse assay plates contain reagents including TMPD, sodium azide, and potentially some of the candidate inhibitors that are toxic to humans and/or the environment, and also contain potentially infectious P. falciparum parasites. Hence, following the experiment, the parasites and the substrates should be discarded in accordance with local policies for disposing hazardous materials.
Improper injections are not common. To minimize improper injections, care should be taken not to damage the port with the pipette tip when dispensing the substrate into the port. Also, the plate should be handled gently after dispensing the substrates into the ports. Finally, including four technical replicates per experimental condition can aid in ensuring sufficient confidence in the data in instances where improper compound injection occurs, and particular wells are excluded from the analysis.
Acknowledgments
We are thankful to the Australian Red Cross for providing human RBCs and serum. This work was supported by the Research School of Biology (RSB) International Ph.D. Scholarship to S.R., an Australian National Health and Medical Research Council Ideas grant (GNT1182369) to G.v.D and A.G.M, and a Research School of Biology Innovation grant to G.v.D., A.G.M., E.R. and D.C.
Graphical overview created with BioRender.com.
Competing interests
The authors declare that they have no competing interests.
Ethical considerations
The use of human erythrocytes was approved as part of the Human Ethics committee protocol HEC 2017/351 of the Australian National University.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
Q&A
Post your question about this protocol in Q&A and get help from the authors of the protocol and some of its users.
References
- 1.Conrad M. D. and Rosenthal P. J.(2019). Antimalarial drug resistance in Africa: the calm before the storm? Lancet Infect. Dis. 19(10): e338-e351. [DOI] [PubMed] [Google Scholar]
- 2.Fisher N., Meunier B. and Biagini G.A., (2020). The cytochrome bc1 complex as an antipathogenic target. FEBS Letters 594(18): 2935-2952. [DOI] [PubMed] [Google Scholar]
- 3.Hayward J. A. and van Dooren G. G.(2019). Same same, but different: Uncovering unique features of the mitochondrial respiratory chain of apicomplexans. Mol. Biochem. Parasitol. 232: 111204. [DOI] [PubMed] [Google Scholar]
- 4.Hayward J. A., Makota F. V., Cihalova D., Leonard R.A., Rajendran E., Zwahlen S.M., Shuttleworth L., Wiedemann U., Spry C., Saliba K. J., Maier A. G., et al.(2023). A screen of drug-like molecules identifies chemically diverse electron transport chain inhibitors in apicomplexan parasites. PLOS Pathog. 19(7): e1011517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hayward J. A., Rajendran E., Makota F. V., Bassett B. J., Devoy M., Neeman T. and van Dooren G.G.(2022). Real-time analysis of mitochondrial electron transport chain function in Toxoplasma gondii parasites using a Seahorse XFe96 extracellular flux analyzer. Bio Protoc. 12(1): e4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hikosaka K., Komatsuya K., Suzuki S. and Kita K.(2015). Mitochondria of malaria parasites as a drug target. An Overview of Tropical Diseases. [Google Scholar]
- 7.Ippolito M. M., Moser K. A., Kabuya J. B. B., Cunningham C. and Juliano J. J.(2021). Antimalarial drug resistance and implications for the WHO global technical strategy. Curr. Epidemiol. Rep. 8(2): 46-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ke H. and Mather M. W.(2017). +Targeting Mitochondrial Functions as Antimalarial Regime, What Is Next? Curr. Clin. Microbiol. Rep. 4(4): 175-191. [Google Scholar]
- 9.Lambros C. and Vanderberg J. P.(1979). Synchronization of Plasmodium falciparum Erythrocytic Stages in Culture. J. Parasitol. 65(3): 418-420. [PubMed] [Google Scholar]
- 10.Maier A. G. and Rug M.(2013). In vitro Culturing Plasmodium falciparum Erythrocytic Stages. Methods Mol. Biol. 923: 3-15. [DOI] [PubMed] [Google Scholar]
- 11.Nguyen W., Dans M. G., Currie I., Awalt J. K., Bailey B. L., Lumb C., Ngo A., Favuzza P., Palandri J., Ramesh S., et al.(2023). 7-N-Substituted-3-oxadiazole Quinolones with Potent Antimalarial Activity Target the Cytochrome bc1 Complex. ACS Infect. Dis. 9(3): 668-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rajaram K., Tewari S. G., Wallqvist A. and Prigge S. T.(2022). Metabolic changes accompanying the loss of fumarate hydratase and malate–quinone oxidoreductase in the asexual blood stage of Plasmodium falciparum. J. Biol. Chem. 298(5): 101897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ridgway M., Cihalova D. and Maier A.(2021). Sex-specific Separation of Plasmodium falciparum Gametocyte Populations. Bio Protoc 11(11): e4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sakata-Kato T. and Wirth D. F.(2016). A Novel Methodology for Bioenergetic Analysis of Plasmodium falciparum Reveals a Glucose-Regulated Metabolic Shift and Enables Mode of Action Analyses of Mitochondrial Inhibitors. ACS Infect. Dis. 2(12): 903-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Srivastava I. K., Morrisey J. M., Darrouzet E., Daldal F. and Vaidya A. B.(1999). Resistance mutations reveal the atovaquone-binding domain of cytochrome b in malaria parasites. Mol. Microbiol. 33(4): 704-711. [DOI] [PubMed] [Google Scholar]
- 16.Staines H. M., Burrow R., Teo B. Y., Chis Ster I., Kremsner P. G. and Krishna S.(2018). Clinical implications of Plasmodium resistance to atovaquone/proguanil: a systematic review and meta-analysis. J. Antimicrob. Chemother. 73(3): 581-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thangaraj A., Periyasamy P., Liao K., Bendi V. S., Callen S., Pendyala G. and Buch S.(2018). HIV-1 TAT-mediated microglial activation: role of mitochondrial dysfunction and defective mitophagy. Autophagy 14(9): 1596-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.World Health Organization(2022). World malaria report 2022. World Health Organization(8 Dec 2022, ISBN: 978 92 4 0066489 8). [Google Scholar]