Abstract
A bacterial strain (strain S5) which grows aerobically with the sulfonated azo compound 4-carboxy-4′-sulfoazobenzene as the sole source of carbon and energy was isolated. This strain was obtained by continuous adaptation of “Hydrogenophaga palleronii” S1, which has the ability to grow aerobically with 4-aminobenzenesulfonate. Strain S5 probably cleaves 4-carboxy-4′-sulfoazobenzene reductively under aerobic conditions to 4-aminobenzoate and 4-aminobenzene-sulfonate, which are mineralized by previously established degradation pathways.
It is generally assumed that sulfonated azo dyes are not degraded under aerobic conditions (14). Nevertheless, there have been some reports which suggest a conversion of certain sulfonated azo dyes under aerobic conditions (3, 7, 8, 13, 15). Furthermore, certain carboxylated analogs of sulfonated azo compounds are utilized aerobically as the sole source of carbon and energy by specifically adapted bacteria (11, 12, 16, 17). However, unequivocal evidence for the productive mineralization of a sulfonated azo compound by bacteria is lacking. In the present article the first observation of the utilization of a sulfonated azo compound as the sole source of carbon and energy by a bacterial strain is reported.
Previously, a mixed bacterial culture which mineralizes sulfanilate (4-aminobenzenesulfonate) was isolated. This coculture consisted of the strains “Hydrogenophaga palleronii” S1 and Agrobacterium radiobacter S2 (4, 5). Because sulfanilate occurs as an azoaryl structural element in many azo dyes, it was of interest whether this mixed culture could adopt the ability to reduce azo bonds and release sulfanilate as growth substrate. Therefore, the model sulfonated azo compound 4-carboxy-4′-sulfoazobenzene (CSAB) was synthesized by nitro-amine condensation starting with sulfanilic acid and 4-nitrobenzoic acid (1). The precipitated CSAB was separated from the reaction mixture by filtration and purified by repeated dissolution in alkali and precipitation with acid. The identity and purity of the bright orange product were analyzed by UV-visible light spectroscopy, elementary analysis, and high-pressure liquid chromatography (HPLC). For the solid material obtained, molar extinction coefficients of 23.74 and 1.13 mM−1 cm−1 in water were determined at the wavelengths of 326 and 434 nm, respectively. The elementary analytic results were consistent with the structure of CSAB. The purity of the preparation was tested by HPLC with a reversed-phase column and a solvent gradient from 1 to 90% (vol/vol) methanol and 0.3% (vol/vol) H3PO4. A single band which showed absorbance at a wavelength of 326 nm was eluted. At 210 nm a minor contaminant (about 15% of the signal intensity of CSAB) was detected. This compound was clearly different from either 4-nitrobenzoate or sulfanilate.
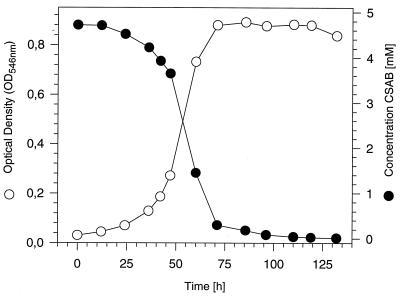
The mixed culture was grown in repeated batch cultures in a mineral medium with sulfanilate (5 mM). About every 2 weeks the culture was transferred (1:10 [vol/vol]) to fresh medium, in which the sulfanilate concentration was subsequently reduced and the CSAB concentration increased (±0.5 mM each). The color of the azo dye disappeared after 2 months. The culture was transferred to a solid mineral medium with CSAB as the sole source of carbon. From this culture was obtained strain S5, which grew aerobically with the sulfonated azo compound CSAB as its sole source of carbon and energy and with a doubling time of 9.5 h (Fig. 1). The complete disappearance of the dye was demonstrated by the loss of the orange color from the medium and by HPLC analysis, whereas CSAB was not degraded in a sterile control flask. Based on its colony morphology and the results obtained with the commercial identification system Biolog GN, this strain strongly resembled “H. palleronii” S1. Recently, it was demonstrated that, in the presence of low concentrations of biotin, cyanocobalamin, and 4-aminobenzoate, strain S1 also grows in axenic culture with sulfanilate (2). Therefore the adaptation experiment was repeated in the presence of these three substances with a pure culture of strain S1. This experiment also resulted in the isolation of a strain which grew in axenic culture with CSAB as the sole source of carbon and energy.
FIG. 1.
Aerobic growth of strain S5 with CSAB as the sole source of carbon and energy. The growth was determined photometrically (OD546), and the turnover of CSAB was measured by HPLC with a reversed-phase column and a solvent gradient consisting of H2O, methanol, and 0.3% H3PO4 with increasing concentrations of methanol (1 to 90%). An OD546 of 1 corresponded to 0.33 mg of protein ml−1.
To ensure that the genetic backgrounds of strains S5 and S1 were identical, the genes for the 16S rRNAs were amplified by PCR with different universal primers (6) and sequenced in comparison to the corresponding gene from the type strain, H. palleronii DSM 63. It was found that the sequences from strains S1 and S5 were > 99.8% identical (there were only two discrepancies between the two sequences), but they showed only 97.7 to 97.9% identity with the 16S rRNA gene from H. palleronii DSM 63. It was therefore concluded that strain S5 was derived from strain S1 and that the strains do not belong to the species H. palleronii.
A reductive cleavage of the azo bond of CSAB would result in the formation of 4-aminobenzoate and sulfanilate. Like the parent strain, S1, strain S5 grew in the presence of sulfanilate, 4-aminobenzoate, and 4-sulfocatechol. The doubling times with these compounds were 6.2 to 6.4 h. We therefore investigated whether reductive cleavage of CSAB by strain S5 occurs. Strain S5 was grown aerobically with 5 mM CSAB, and cell extracts were prepared (10) in different buffers. These cell extracts were incubated aerobically in cuvettes containing 50 mM Tris-HCl buffer (pH 8.0), 0.5 mM CSAB, 1 mM NADH, or 1 mM NADPH and with various mixtures of possible cofactors. The enzyme activity was measured spectrophotometrically at the absorption maximum for CSAB (at a wavelength of 434 nm), but no significant decrease in absorbance was observed. Neither addition of a membrane fraction nor performing the enzyme assays under anaerobic conditions (9) improved the turnover of CSAB in the cell-free system. Furthermore, there was no significant increase in azo reductase activity when harvested cells were resuspended in the culture supernatant instead of Tris-HCl buffer.
The maximal enzyme activities observed for cell extracts were only about 30% of the activities found for intact cells. This suggested that during the disruption of the cells some important components of the azo reductase system were destroyed or some cofactors were present in only limiting quantities.
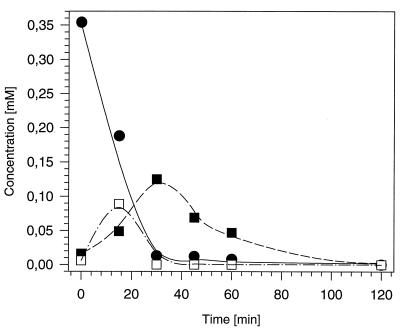
Because it was difficult to obtain reproducible enzyme activities with cell extracts, the turnover of CSAB by resting cells was investigated. Cells of strain S5 were grown with CSAB (5 mM), harvested by centrifugation, resuspended in Tris-HCl at an optical density at 546 nm (OD546) of 5.3, and incubated in a water bath shaker (140 rpm; 30°C) with 0.5 mM CSAB (Fig. 2). Thus, the transient accumulation of two metabolites in the supernatants was observed by reversed-phase HPLC (column size, 250 by 4.6 mm) (SIL 100; Grom, Herrenberg, Germany). The solvent system consisted of a solvent gradient with increasing concentrations of methanol, starting with 1% (vol/vol) methanol, 98.9% (vol/vol) water, and 0.1% H3PO4. The flow rate was 0.7 ml min−1. The metabolites formed were identified as sulfanilate and 4-sulfocatechol by comparison of their retention times and in situ UV-visible light-spectra with authentic standards. Surprisingly, the concentration of 4-sulfocatechol in the medium increased (and decreased) during the experiment more rapidly than the concentration of sulfanilate (Fig. 2). 4-Sulfocatechol also temporarily accumulated when resting cells of strain S1 were incubated with sulfanilate (4, 5). This suggested that in the resting-cell assay the initial activity of the sulfanilate-converting enzyme was higher than the activity of the 4-sulfocatechol-oxidizing enzyme protocatechuate-3,4-dioxygenase type II. Presumably, the activity of the sulfanilate-converting enzyme decreased during the experiment more rapidly than the activity of protocatechuate-3,4-dioxygenase type II. No accumulation of 4-aminobenzoate or protocatechuate was found by HPLC analysis during the experiment. In a control experiment with cells of strain S1 grown with 4-aminobenzenesulfonate, no turnover of CSAB was observed by HPLC analysis.
FIG. 2.
Conversion of CSAB (•) to sulfanilate (▪) and 4-sulfocatechol (□) by resting cells of strain S5. Strain S5 was grown in a mineral medium with CSAB as the sole source of carbon and energy, and resting cells were prepared as described in the text.
The detection of sulfanilate derived from CSAB suggested a reductive cleavage of CSAB, yielding sulfanilate as one of the reduction products. This reaction should also proceed in the absence of oxygen. Therefore, resting cells were incubated under anaerobic conditions with CSAB. Surprisingly, the rate of CSAB turnover under anaerobic conditions was <2% of the turnover rate under aerobic conditions.
A further indication of a reductive cleavage of CSAB into sulfanilate and 4-aminobenzoate was obtained by growing strain S5 with CSAB or a complex medium (HPG medium) (4). When the cells were grown in a mineral medium with CSAB and the turnover of the substrates was analyzed by HPLC, it was found that resting cells converted CSAB, 4-aminobenzoate, or 4-aminobenzenesulfonate with specific activities of 0.012, 0.026, and 0.011 μmol min−1 mg of protein−1, respectively. In contrast, after growth of the cells in HPG medium, these activities were only 0.007, 0.010, and 0.003 μmol min−1 mg of protein−1, respectively. Incubation of resting cells with CSAB and different potential inhibitors of ring cleavage dioxygenases showed that the turnover of CSAB was almost completely inhibited by the addition of 8-hydroxyquinoline or 2,2′-bipyridyl (1 mM each). The presence of 4-nitrocatechol (0.25 mM) also resulted in a pronounced reduction of the rate of CSAB turnover (6% of the rate in the absence of the inhibitor). In this system as well the formation of 4-sulfocatechol was observed.
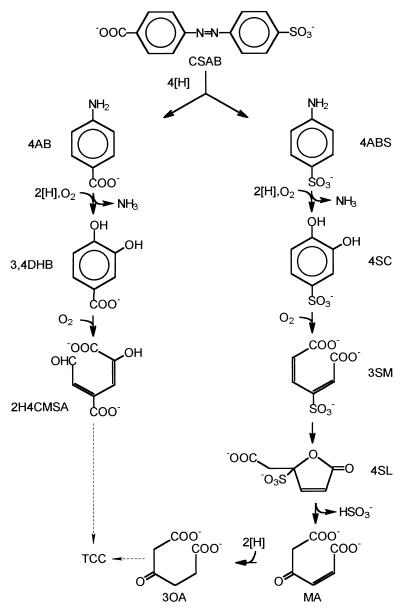
The degradation of sulfanilate and 4-aminobenzoate by strain S1 has been previously studied (5). The proposed degradation pathway for CSAB and its reduction products is shown in Fig. 3.
FIG. 3.
Proposed pathway for the degradation of CSAB by strain S5. 4AB, 4-aminobenzoate; 4ABS, 4-aminobenzenesulfonate (sulfanilate); 3,4DHB, 3,4-dihydroxybenzoate (protocatechuate); 4SC, 4-sulfocatechol; 2H4CMSA, 2-hydroxy-4-carboxymuconic semialdehyde; 3SM, 3-sulfomuconate; 4SL, 4-carboxymethyl-4-sulfobut-2-en-4-olide (4-sulfolactone); MA, maleylacetate; 3OA, 3-oxoadipate; TCC, tricarboxylic acid cycle.
To obtain some information about the substrate specificity, resting cells were incubated with CSAB, 4,4′-dicarboxyazobenzene (DCAB), 4-hydroxy-4′-sulfoazobenzene, methyl orange [4-(N,N-dimethyl)-4′-sulfoazobenzene; color index (C.I.) 13025], orange II {4-[(2-hydroxy-1-naphthalenyl)azo]-benzenesulfonic acid; C.I. 15510}, or sunset yellow FCF {6-hydroxy-5-[(4-sulfophenyl)azo]-2-naphthol-6-sulfonic acid; FD&C no. 6; C.I. 15985}. Of these compounds, only CSAB and DCAB were converted by resting cells. DCAB was also utilized by strain S5 as the sole source of carbon and energy. Furthermore, no growth of strain S5 was found with acid black 24 and 52, acid blue 113, acid red 1, amaranth, direct red 81, direct yellow 4 and 50, mordant yellow 3, and naphthol blue black.
The results presented in this study suggest that bacterial cultures with the ability to aerobically degrade simple sulfonated azo dyes may be obtained after preadaptation to sulfonated aminoaromatics and/or when reductive cleavage of the azo bond gives rise to an aerobically assimilable aminoaromatic structure, like 4-aminobenzoate. This selection scheme circumvents the problems observed during attempts to adapt bacteria with the ability to degrade carboxylated azo compounds for the degradation of sulfonated azo compounds (12). The ability of strain S5 to mineralize CSAB suggests that it is possible to degrade sulfonated azo dyes under aerobic conditions if biological systems which can grow and can mineralize the reduction products are available.
Nucleotide sequence accession number.
The nucleotide sequences for the 16S rRNAs from strains S5 and S1 have been deposited in the GenBank data library under accession no. AF019037 and AF019073, respectively.
REFERENCES
- 1.Bayer O, Meerwein H, Ziegler K. Stickstoffverbindungen I. In: Müller E, editor. Methoden der organischen Chemie (Houben-Weyl) 4th ed. Stuttgart, Germany: Georg Thieme Verlag; 1965. pp. 339–341. [Google Scholar]
- 2.Dangmann E, Stolz A, Kuhm A E, Hammer A, Feigel B, Noisommit-Rizzi N, Rizzi M, Reuß M, Knackmuss H-J. Degradation of 4-aminobenzenesulfonate by a two-species bacterial coculture. Physiological interactions between Hydrogenophaga palleronii S1 and Agrobacterium radiobacter S2. Biodegradation. 1996;7:223–229. doi: 10.1007/BF00058181. [DOI] [PubMed] [Google Scholar]
- 3.Dykes G A, Timm R G, von Holy A. Azoreductase activity in bacteria associated with the greening of instant chocolate puddings. Appl Environ Microbiol. 1994;60:3027–3029. doi: 10.1128/aem.60.8.3027-3029.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feigel B J, Knackmuss H-J. Bacterial catabolism of sulfanilic acid via catechol-4-sulfonate. FEMS Microbiol Lett. 1988;55:113–118. [Google Scholar]
- 5.Feigel B J, Knackmuss H-J. Syntrophic interactions during degradation of 4-aminobenzenesulfonic acid by a two-species bacterial culture. Arch Microbiol. 1993;159:124–130. doi: 10.1007/BF00250271. [DOI] [PubMed] [Google Scholar]
- 6.Gerhardt P, Murray R G E, Wood W A, Krieg N R, editors. Methods for general and molecular bacteriology. Washington, D.C: American Society for Microbiology; 1994. p. 691. [Google Scholar]
- 7.Heiss G S, Gowan B, Dabbs E R. Cloning of DNA from a Rhodococcus strain conferring the ability to decolorize sulfonated azo dyes. FEMS Microbiol Lett. 1992;99:221–226. doi: 10.1016/0378-1097(92)90030-r. [DOI] [PubMed] [Google Scholar]
- 8.Jiang H, Bishop P L. Aerobic biodegradation of azo dyes in biofilms. Water Sci Technol. 1994;29:525–530. [Google Scholar]
- 9.Kudlich M, Keck A, Klein J, Stolz A. Localization of the enzyme system involved in anaerobic reduction of azo dyes by Sphingomonas sp. strain BN6 and effect of artificial redox mediators on the rate of azo dye reduction. Appl Environ Microbiol. 1997;63:3691–3694. doi: 10.1128/aem.63.9.3691-3694.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuhm A E, Stolz A, Ngai K-L, Knackmuss H-J. Purification and characterization of a 1,2-dihydroxynaphthalene dioxygenase from a bacterium that degrades naphthalenesulfonic acids. J Bacteriol. 1991;173:3795–3802. doi: 10.1128/jb.173.12.3795-3802.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kulla H G. Aerobic bacterial degradation of azo dyes. In: Leisinger T, Cook A M, Nüesch J, Hütter R, editors. Microbial degradation of xenobiotics and recalcitrant compounds. London, England: Academic Press; 1981. pp. 387–399. [Google Scholar]
- 12.Kulla H G, Klausener F, Meyer U, Lüdeke B, Leisinger T. Interference of aromatic sulfo groups in the microbial degradation of the azo dyes orange I and orange II. Arch Microbiol. 1983;135:1–7. [Google Scholar]
- 13.Ogawa T, Yatome C, Idaka E, Kamiya H. Biodegradation of azoic acid dyes by continuous cultivation of Pseudomonas cepacia 13NA. J Soc Dyers Colourists. 1986;102:12–14. [Google Scholar]
- 14.Pagga U, Brown D. The degradation of dyestuffs: part II—behaviour of dyestuffs in aerobic biodegradation tests. Chemosphere. 1986;15:479–491. [Google Scholar]
- 15.Shaul G M, Holdsworth T J, Dempsey C R, Dostal K A. Fate of water-soluble azo dyes in the activated sludge process. Chemosphere. 1991;22:107–119. [Google Scholar]
- 16.Zimmermann T, Gasser F, Kulla H G, Leisinger T. Comparison of two azoreductases acquired during adaptation to growth on azo dyes. Arch Microbiol. 1984;138:37–43. doi: 10.1007/BF00425404. [DOI] [PubMed] [Google Scholar]
- 17.Zimmermann T, Kulla H G, Leisinger T. Properties of purified orange II azoreductase, the enzyme initiating azo dye degradation by Pseudomonas KF46. Eur J Biochem. 1982;129:197–203. doi: 10.1111/j.1432-1033.1982.tb07040.x. [DOI] [PubMed] [Google Scholar]