Abstract
The influenza A virus genome is segmented into eight viral RNAs (vRNA). Secondary structures of vRNA are known to be involved in the viral proliferation process. Comprehensive vRNA structures in vitro, in virio, and in cellulo have been analyzed. However, the resolution of the structure map can be improved by comparative analysis and statistical modeling. Construction of a more high-resolution and reliable RNA structure map can identify uncharacterized functional structure motifs on vRNA in virion. Here, we establish the global map of the vRNA secondary structure in virion using the combination of dimethyl sulfate (DMS)-seq and selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE)-seq with a robust statistical analysis. Our high-resolution analysis identified a stem-loop structure at nucleotide positions 39 – 60 of segment 6 and further validated the structure at nucleotide positions 87 – 130 of segment 5 that was previously predicted to form a pseudoknot structure in silico. Notably, when the cells were infected with recombinant viruses which possess the mutations to disrupt the structure, the replication and packaging of the viral genome were drastically decreased. Our results provide comprehensive and high-resolution information on the influenza A virus genome structures in virion and evidence that the functional RNA structure motifs on the influenza A virus genome are associated with appropriate replication and packaging of the viral genome.
Keywords: Influenza virus, SHAPE-seq, DMS-seq, RNA structure, Chemical probing
Graphical Abstract
1. Introduction
The influenza A virus (IAV) genome consists of eight single-stranded negative-sense RNA segments (vRNA). One copy of each segment is packaged together into a single virus particle, in which eight segments are organized in a conserved ‘7 + 1′ configuration [1], [2]. Segment reassortment is one of the driving forces for IAV evolution. For example, genetic reassortment between the human IAV and the avian/animal IAV can lead to the emergence of a new subtype of IAV, a candidate for a pandemic influenza strain. Each genome segment forms the viral ribonucleoprotein (vRNP) with the viral RNA polymerase and nucleoprotein (NP), a single-stranded RNA-binding viral protein. In a previous study, the vRNPs in viral particles were revealed to form a double-helical structure with the polymerase at one end and a short loop at the other [3]. Previous studies have examined motif sequences required for efficient genome packaging and bundling [4]. The signal sequences for efficient genome packaging and bundling were initially found to be located in the coding regions at both ends of each segment, so-called “packaging signals”. However, such signal sequences are also found in the middle of coding regions that are outside the “packaging signal” [5].
In many RNA viruses, specific regions of the viral RNA genomes act as cis-acting regulatory elements that mediate virus propagation. These cis-acting RNA elements often form highly specialized structural motifs such as stem-loops or pseudoknots [6]. In IAV, the RNA structure motifs at the promoter region located at the 5′ and 3′ termini of the vRNA and their reformation at the promoter in the transcription step are elucidated by crystal structure and cryo-EM analyses [7], [8], [9]. Other than promoter regions, the comprehensive analyses of specialized structural motifs coded on the IAV genome RNA were also carried out by RNA secondary structure prediction methods. These studies identified the conservation of stem-loop structures on the IAV genome, suggesting the requirement of RNA structures for IAV propagation [10], [11]. This is also supported by the previous findings that the mutations that disrupt predicted stem-loop and pseudoknot structures have been shown to reduce virus propagation [10], [11], [12], [13]. One of the functions of RNA structures on vRNP is hypothesized to mediate intersegment interaction networks. Gavazzi et al. discovered a direct interaction between segments 2 and 8 of an H5N2 avian IAV strain in vitro. The regions involved in the intersegment interaction contain complementary sequences and potentially form a kissing-loop complex to initiate the intersegment interaction [5]. Thus, secondary structures formed on the IAV genome may have various functions in viral propagation.
NP-free vRNA segment structures were determined by the chemical probing method using in vitro synthesized vRNA segments [14], [15], [16]. However, recent studies by cross-linking immunoprecipitation (CLIP) analyses have revealed that the secondary structures of vRNAs are partially unwound by binding NP [17], [18]. Since NP is observed to bind vRNA in sequence- and RNA structure-independent manners [17], [18], [19], the vRNA structure in virion can be unpredictable or highly diverged from the structure determined in vitro. To investigate the functional RNA secondary structure motifs, high throughput sequencing (HTS) approaches have been developed, in which the secondary structures are determined by chemical mapping and reverse transcription followed by sequencing. Dadonaite et al. revealed the secondary structures of the IAV genome in the virion using selective 2′-hydroxyl acylation analyzed by primer extension and mutational profiling (SHAPE-MaP). Their SHAPE-MaP profiles revealed that some vRNA secondary structures remain in the context of vRNP [20]. Moreover, the secondary structures of the IAV genome of H1N1pdm strain in virion and in infected cells were also identified by SHAPE-MaP and dimethyl sulfate mutational profiling with sequencing (DMS-MaPseq) [21]. Although RNA secondary structure on vRNP has been analyzed by the HTS approaches, the construction of a more high-resolution and reliable in vivo and in virio structure map is an essential step to investigate the uncharacterized functional structure motifs on vRNP.
In this study, we revealed the secondary structures of the IAV genome in the virion using multiple HTS technologies and statistical model-based approaches to create comprehensive and high-resolution information on the IAV genome structures. While it is crucial to obtain the conformational information of vRNP determined in vivo, each HTS approach may possess its own detection bias for base reactivity [22]. To overcome the problem, we applied statistical model-based approaches developed to estimate reactivity from HTS data more accurately by integrating multiple datasets with reducing technology-specific biases. Specifically, we obtained a robust RNA secondary structure map by combining two high-throughput and massive-scale sequencing techniques, DMS-seq [23], [24], [25] and SHAPE-seq [26], [27], and multiple bioinformatical tools for calculating SHAPE reactivity, BUMHMM [28] and reactIDR [29]. As a result, we identified unique structural motifs on the vRNP. Furthermore, the disruption of the structural motifs by introducing mutations inhibited the replication of a specific segment, and moreover, all segments in some cases. Taken together, we construct comprehensive and high-resolution information on the IAV genome structures in virio and show that the structural motifs found on vRNP are associated with the regulation of viral genome replication.
2. Materials and methods
2.1. Cells
MDCK (NBL-2) cells (American Type Culture Collection, Manassas, VA) were maintained in a minimal essential medium (MEM) (Sigma-Aldrich, ST. Louis, MO) containing 10% fetal bovine serum and penicillin/streptomycin (Nacalai Tesque, Kyoto, Japan). HEK293T cells (kindly provided by Dr. Y. Kawaoka) were maintained in a Dulbecco’s modified Eagle’s medium (DMEM) with high glucose concentration (Sigma-Aldrich) containing 10% fetal bovine serum and penicillin/streptomycin.
2.2. Viruses
Influenza virus A/PR/8/34 (H1N1) (PR8) was grown in the allantoic sacs of 11 days-old chick embryos at 35.5°C for 48 h. The purified virion and the vRNP from the purified virion were prepared as previously described [19]. Briefly, virion in allantoic fluid was precipitated by PEG. The precipitate was suspended in PBS(+) and centrifuged on 60% and 30% sucrose in PBS(+) at 76k × g for 90 min in an SW28 rotor (Beckman Coulter, Brea, CA) at 4 °C. The virus band formed on the 60% sucrose layer was collected and re-precipitated. The precipitate was suspended in the buffer containing 10 mM Hepes-NaOH (pH 7.4), 100 mM NaCl, 1 mM DTT, and 20% glycerol. To prepare vRNP, purified virion was treated with a disruption buffer (50 mM Hepes-NaOH [pH 7.4], 150 mM NaCl, 5 mM MgCl2, 5 mM DTT, 5% glycerol, 1% Triton X-100%, and 2% lysolecithin) at 30°C for 10 min. The sample was centrifuged on 30–60% glycerol gradients in 50 mM Hepes-NaOH (pH 7.9), 150 mM NaCl, and 1 mM at 200k × g for 3 h in an SW55 rotor (Beckman Coulter) at 4°C. Fractionation was carried out from the top of the gradient, and vRNP fractions were confirmed by SDS-PAGE followed by CBB staining. To construct the pPolI-PR8 mutant vector, an inverted PCR was performed using the pPolI-PR8 vector as a template with specific primer sets (Primers used In this study were listed in Table S1). After DpnI treatment, phosphorylation, ligation, and transformation into an Escherichia coli Mach1 (Thermo Fisher Scientific) were performed. Recombinant viruses were generated using a reverse genetics approach [30]. Viral protein expression vectors [30] and the viral RNA expression vectors derived from the PR8 strain [31] were transfected to 293 T cells. To propagate the recombinant virus, MDCK cells were infected with the recombinant virus at a multiplicity of infection (MOI) of 0.1. At 48 h post infection (hpi), the supernatants were collected, and cell debris were removed by low-speed centrifugation (3k × g, 5 min). The virus titer was determined by a plaque assay. To prepare purified virion, MDCK cells were infected with the recombinant virus at an MOI of 0.1, and the supernatant was collected at 48 hpi. After removal of cell debris by low-speed centrifugation (500 × g, 5 min) and filtration through a 0.45-µm filter, the supernatant was ultracentrifuged at 100k × g for 1.5 h using an SW28 rotor at 4°C. The pellet was suspended in PBS(-) and centrifuged on 30–60% sucrose gradients in PBS(-) at 100k × g for 1.5 h in an SW28 rotor at 4 °C. Viral bands were pooled and re-precipitated by centrifugation in PBS(-) at 120k × g for 1.5 h in an SW55 rotor (Beckman Coulter) at 4 °C. The precipitated virion was suspended in a DMS buffer (40 mM Hepes-NaOH [pH 7.4], 100 mM NaCl, and 0.5 mM MgCl2) or PBS(-) and stored at − 80 °C until use.
2.3. DMS-seq and SHAPE-seq
The vRNA was prepared by proteinase K treatment of virion purified from the allantoic fluid at 37°C for 30 min in SDS buffer (0.25% SDS and 100 µg/ml proteinase K in PBS(-)) followed by phenol/chloroform extraction. NAI was synthesized using a previously described method [26]. One µl of DMS (Wako Pure Chemical Industries, Osaka, Japan) or 5 µl of NAI was added to the purified virion (5 µl of the purified virion from allantoic fluid or from 80 ml of cell culture supernatant of infected cells), vRNP (25 µl of vRNP fraction), or vRNA (from 5 µl of purified virion) in 100 µl of DMS buffer. The final concentrations of DMS and NAI were 136.7 mM and 50 mM, respectively. After incubation for 5 min (DMS) or 15 min (NAI) at 25°C, 10 µl of 1 M DTT was added to stop the reaction. Then, the RNA was extracted with phenol/chloroform. Sequencing libraries were prepared using a previously described method [32]. Briefly, cDNA was synthesized with random hexamers containing the Illumina adapters at their 5′- ends in 50 mM Tris-HCl (pH8.3), 75 mM KCl, 3 mM MgCl2, 10 mM DTT, 1 mM dNTP, and 10 U of mutated M-MLV reverse transcriptase (ReverTra Ace [Toyobo, Osaka, Japan]) at 25°C for 10 min and 42°C for 50 min. The ssDNA linker containing a 5′ phosphate and 3′ C3 spacer was ligated to the synthesized cDNA using 20 U of the Circligase I (Lucigen, Middleton, WI). The resultant cDNA was amplified by an adapter-based PCR using the KAPA HiFi DNA polymerase (Roche, Basel, Switzerland). Sequencing was performed using a MiSeq (Illumina, San Diego, CA) (2 × 75-bp PE) and NovaSeq6000 (Illumina) (2 × 150-bp PE). Raw reads were cleaned and trimmed with Trimmomatic v0.36 [33]. To ensure that the number of reads was almost constant for each condition before statistical analysis was performed, the raw reads were downsampled with Picard (http://broadinstitute.github.io/picard/). The cleaned reads were aligned to the A/PR/8/34 genome using bowtie2 with default parameters. We performed duplicate DMS-seq and SHAPE-seq experiments on two independent samples, and the reactivities of each nucleotide were calculated using reactIDR [29] with –DMS option and with a default setting, respectively, and BUMHMM [28] with a default setting. All calculated probabilities of modification are presented in supplemental data. Base-pairing probabilities and Shannon entropies were calculated by rf-fold in RNA framework [34], [35] (options are -g -dp -sh -nlp -md 500 -w -pk) from the probabilities of modification calculated by reactIDR. The SHAPE annotated RNA secondary structure prediction was performed by RNAstructure [36]. SHAPE intercept and SHAPE slope to predict a SHAPE annotated secondary structure were − 0.6 and 1.8, respectively. A predicted RNA structure with maximum expected accuracy was shown in this study. Computational prediction of the pseudoknot structure was performed by IPknot [37].
A 30-nt moving average of the probability of modification and Shannon entropy of each segment was calculated, and we defined the regions with both probability of modification and Shannon entropy less than the median of the segment as a structured region. Common structured regions across all three conditions that had robust probabilities of modification and entropies from DMS-seq and SHAPE-seq were identified as structured regions formed on vRNP. We predicted whether the identified structure regions could form secondary structures by RNAstructure.
To analyze the probability of modification in high-NP binding regions, NP PAR-CLIP data sets (PR8 strain) were downloaded from Sequence Read Archive (SRX3545111) and aligned to the PR8 genome using bowtie2 with default parameters. The coverage of each nucleotide of PAR-CLIP and control RNA experiments was calculated by IGV [38]. We normalized the number of coverages per nucleotide to the total number of coverages to yield a normalized coverage ratio from both PAR-CLIP and control RNA sequencing. vRNA nucleotides with fold-change > 2 were identified, and the regions were extracted. Due to the number of reads, we used only one dataset of PAR-CLIP and control RNA-seq.
2.4. RT-qPCR
Total RNA was extracted from MDCK cells infected at an MOI of 1 using the ISOGEN reagent (Nippon Gene, Tokyo, Japan). For the preparation of the vRNA in the supernatant from the infected cells, MDCK cells were infected with the virus at an MOI of 0.1, and the cells were suspended in MEM containing 0.6 µg/ml TPCK-trypsin (Sigma-Aldrich). At 48 hpi, the supernatant was collected, and cell debris was removed by low-speed centrifugation (500 × g, 5 min) and filtration through a 0.45-µm filter (EMD Millipore, Billerica, MA). The supernatant was mixed with chicken red blood cells (Japan Bio Serum, Tokyo, Japan) at 4°C for 30 min. The cells with the bound virus were pelleted by centrifugation (1k × g, 5 min) and were washed with PBS(-). The RNAs were extracted by ISOGEN reagent (Nippon Gene) from the cells with the bound virus.
For RT-qPCR, the cDNA was synthesized with the Uni12 primer using ReverTra Ace. The synthesized cDNA was mixed with the Thunderbird SYBR qPCR mix (Toyobo) and a specific primer set for each segment. The qPCR reactions were performed using a Thermal Cycler Dice Real-Time System TP800 (Takara Bio), and the relative amounts of each segment were calculated.
2.5. Western blotting
MDCK cells were infected with the recombinant virus at an MOI of 1 in MEM, and the infected cells were collected at 8 hpi. The infected cells were suspended in lysis buffer (20 mM Tris-HCl [pH 7.9], 150 mM NaCl, 1 mM EDTA, and 0.2% NP-40). Viral proteins in the cell lysate were separated by SDS-PAGE and detected by western blotting using an ImageQuant LAS 4000 (GE Healthcare, Milwaukee, WI). NA and M1 were detected using a sheep polyclonal antibody (R&D Systems, Minneapolis, MN) and a rabbit polyclonal antibody [39]. Band intensities were measured using ImageJ software [40], and standard curves were generated to semi-quantify the relative amount of viral proteins.
2.6. Mini-replicon assay
293 T cells were transfected with the expression plasmids encoding PB2, PB1, PA, and NP and vRNA expression plasmid. To avoid NP mRNA synthesis from vRNA, mutations were introduced into the pPolI-PR8-Seg5 plasmid (pPolI-PR8-Seg5stop) [41]. pPolI-PR8-Seg5stop, pPolI-PR8-Seg5stop 87mut, and pPolI-PR8-Seg5stop 87rec were used for vRNA expression plasmid to analyze the effect of mutations in segment 5 on viral RNA synthesis, and pPolI-PR8-Seg6 and pPolI-PR8-Seg6 39mut were used to analyze the effect of mutations in segment 6. At 24 h after transfection, cells were collected, and total RNA was extracted by the ISOGEN reagent (Nippon Gene). The cDNA was synthesized with the Uni12 primer using ReverTra Ace, and the amount of segment 5 or segment 6 vRNA was determined by qPCR. To determine the amount of NP mRNA for normalization, the cDNA was synthesized with the oligo(dT) primer, and the qPCR reactions were performed using a specific primer set for segment 5 (NP). The amount of vRNAs from the cells transfected with mutant plasmid was double normalized by that transfected with the wild type plasmid and the amount of NP mRNA.
3. Results
3.1. Identification of regions whose structure is altered by vRNP formation
To reveal the vRNA secondary structures that form in the presence of binding proteins, we performed DMS-seq and SHAPE-seq for the IAV genome RNA under three different conditions; the vRNA, vRNP, and virion. These methods are aimed at detecting the RNA regions that are more accessible and likely to be attacked by the reagents. Thus, we can infer single-stranded regions and flexible or loosely structured regions at a single base resolution according to the reactivity scores. We utilized both DMS-seq and SHAPE-seq to uncover the whole landscape of the secondary structures of vRNA in the presence of the viral RNA-binding proteins. The specificity of DMS and NAI to modify moieties of a single-stranded RNA is different [42]. NAI modifies the 2′-OH group in the ribose backbone, whereas DMS modifies adenine and cytosine residues. This means that the effect on nucleotide modification of RNA-binding proteins, especially NP which is known to bind to the sugar-phosphate backbone [3], differs between DMS and NAI. Thus, by comparing the results of DMS-seq and SHAPE-seq and identification of structural regions common to both DMS-seq and SHAPE-seq, the secondary structure region in the vRNP can be estimated more accurately without the bias of RNA-binding proteins.
We carried out duplicate DMS-seq and SHAPE-seq experiments. The coverages were enough to calculate the reactivities except for the 3′ end of segments (Fig. S1A). Reproducibility was evaluated by the drop-off rate of reverse transcriptase. The coefficient of determination (R2) of each duplicate experiment ranged from 0.46 to 0.94 (Fig. S1B). The ratio of bases where reverse transcriptase dropped off was skewed toward A and C in DMS treatment, as previously reported [23] (Fig. S2A). To calculate a reliable score from these samples containing relatively low R2, we utilized robust statistical analyses. The probabilities of modifications for all nucleotides were calculated from the large-scale sequencing data using reactIDR [29]. reactIDR uses the irreproducible discovery rate [43] with a hidden Markov model to discriminate between true and spurious signals from the duplicate experiment and output normalized probability that is an index of reactivity. Hereafter, we use the term “probability of modification” to indicate an index of reactivity of each nucleotide.
From these DMS-seq and SHAPE-seq data, we analyzed the regions whose structure is altered by vRNP formation. First, we compared the probabilities of modifications in high-NP binding regions identified by PAR-CLIP analysis [18] with that of the other regions (Tables S2 and S3). The definition of a high-NP binding region is described in materials and methods. The probability of modification in high-NP binding regions in vRNA was lower than that of the other regions. However, the probabilities of modification in high-NP binding regions in vRNP and virion were higher than or comparable with those of the other regions. These results suggest that the structure of the high-NP binding regions changes by forming vRNP. Next, probabilities of modification of vRNP and virion were compared to the probability of modification of vRNA by deltaSHAPE software [44]. We first performed this analysis using SHAPE-seq data. deltaSHAPE values for each nucleotide that differed significantly between vRNA and vRNP or virion are shown in Fig. 1A. Nucleotides with a deltaSHAPE value significantly less than 0 were defined as more structured in vRNP or virion, and nucleotides with a deltaSHAPE value significantly greater than 0 were defined as less structured. The formation of vRNPs changed the structure of the entire region, but there were more regions where the RNA structure was dissolved (Fig. 1A and Table 1). We focused on the terminal regions of the segments and analyzed the structural changes in these regions. The regions within 200 bases from the 5′ end were more structured by forming vRNP (Fig. 1A and Table 1). The 36 bases at the 3′ end of each segment were excluded from the calculation due to low coverage, but the regions within 200 bases from the 3′ end were more structured in the virion and less structured in vRNP. DMS-seq data is limited to A and C nucleotides, but the results of the analysis of the DMS-seq data show similar trends to those obtained from the SHAPE-seq data (Fig. 1B). These results suggest that the terminal regions of the segments in vRNP are more structured than in vRNA, especially in the virion.
Fig. 1.
vRNA secondary structure changes in vRNP and virion. Nucleotides with significantly different probabilities of modification between vRNA and vRNP or virion were detected from SHAPE-seq data (A) and DMS-seq data (B) using deltaSHAPE. Purple dots indicate the region within 200 bases from the 5′ end of each segment, and orange dots indicate the region within 200 bases from the 3′ end. The 36 bases at the 3′ end of each segment were excluded from the calculation due to low coverage. More structured in RNP or virion is defined with a deltaSHAPE value > 0, and less structured in RNP or virion is defined with a deltaSHAPE value < 0.
Table 1.
Nucleotides with different probability of modification values between vRNA and vRNP or virion.
| vRNA vs vRNP |
vRNA vs virion |
|||||
|---|---|---|---|---|---|---|
| 5′ 200 base | internal | 3′ 200 base | 5′ 200 base | internal | 3′ 200 base | |
| More structured nucleotides (deltaSHAPE>0) | 231# | 1261 | 178 | 467 | 968 | 358 |
| Less structured nucleotides (deltaSHAPE<0) | 165 | 1331 | 463 | 69 | 1733 | 254 |
Number of nucleotides determined to have a significantly different probability of modification by deltaSHAPE
3.2. Identification of secondary structures formed in the vRNP
We next analyzed the regions whose structure is not altered by vRNP formation. To identify secondary structures formed on vRNP, base-pairing probabilities and their Shannon entropies from DMS-seq and SHAPE-seq were calculated by rf-fold in RNA Framework [34], [35]. Shannon entropy which is expected to be low in uniquely predicted structural regions, and base pairing probability can be used to quantify the conformational determinants of RNA in the given region. To examine the correlation between DMS-seq data and SHAPE-seq data, ROC curves and Precision-Recall curves were generated from all three conditions (Figs. S2B and S2C). Probabilities of modification from DMS-seq can partially predict structure models constructed from SHAPE-seq data, but prediction accuracy is weak when probabilities of modification from SHAPE-seq are used to predict structure models constructed from DMS-seq. Shannon entropies from SHAPE-seq and DMS-seq were weakly correlated in all three conditions (Fig. S2D). These results suggest that DMS-seq data correlate with SHAPE-seq data and vice versa, though DMS-seq is less informative than SHAPE-seq. We examined the correlation between our SHAPE-seq data and a previous SHAPE-MaP study using the same virus strain [20]. Probabilities of modification from our SHAPE-seq were correlated with reactivities from a previous SHAPE-MaP study (Fig. S2E). These results suggest that our SHAPE-seq data correlates with that of the previous study.
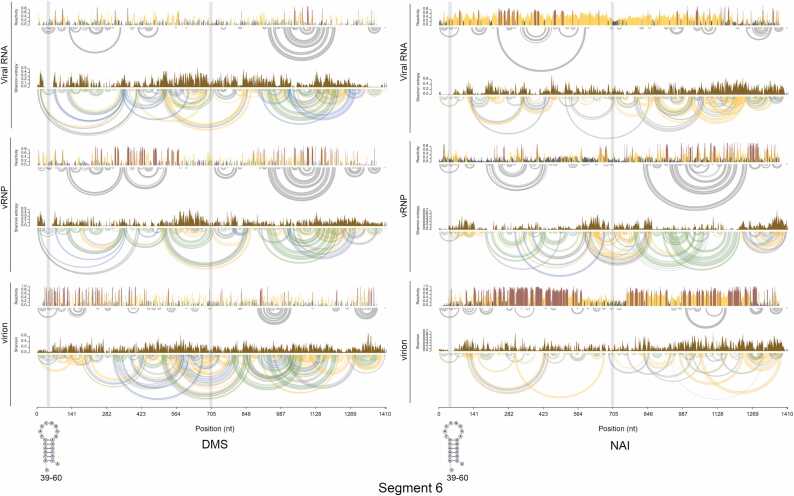
Base-pairing probability and Shannon entropy of vRNA, vRNP, and virion from DMS-seq and SHAPE-seq were shown in Fig. 2 and Fig. S3. Common structures across all three conditions that had robust probabilities of modification and entropies were identified as structured regions formed on vRNP (see Material and Methods). The identified regions are summarized in Table S4. The predicted secondary structures of the identified regions are shown in Fig. 2 and Fig. S3. Simple stem-loop and multi-branch loop structures were isolated as secondary structures formed on vRNP.
Fig. 2.
Base-pairing probability and Shannon entropy of segment 6 from DMS-seq and SHAPE-seq. Base-pairing probabilities and Shannon entropies from DMS-seq (left) and SHAPE-seq (right) were calculated from the output of reactIDR. vRNA sequence is numbered from 5′ to 3′. Predicted base pairs shown in the bar plot of base-pairing probabilities were plotted as arcs. Base-pairing probabilities of 80%, 30%, 10%, 3%, or higher are shown as green, blue, yellow, and grey arcs, respectively. The gray boxes indicate identified structured regions. Predicted RNA structures were shown at the bottom.
3.3. Functional RNA structure formed at nucleotide positions 39 – 60 of segment 6
To investigate whether the identified structures formed on vRNP are functional, it is confirmed whether the propagation of recombinant viruses containing mutations in the identified structures is different from the wild type virus. We constructed four recombinant viruses that contain synonymous mutations in different positions: segment 1 (Seg1 880mut), 5 (Seg5 745mut), 6 (Seg6 39mut), and 8 (Seg8 50mut), to disrupt the predicted RNA structure (Fig. 3A and S4) from SHAPE-seq data. The propagation of four recombinant viruses was measured, and that of Seg1 880mut, Seg5 745mut, and Seg8 50mut viruses were comparable to that of the wild type virus. On the other hand, the propagation of the Seg6 39mut virus was decreased to 1/5 compared with that of the wild type virus (Fig. 3B). The stem-loop structure at nucleotide positions 39 – 60 of segment 6 was confirmed by DMS-seq data (Fig. 3A). We analyzed whether NP prefers to bind to those regions by calculating fold-changes of normalized PAR-CLIP and control RNA-seq (referred as NP binding ratio). The NP binding ratio in the regions of Seg6 39mut is 0.76 and is lower than that in the entire segment 6 (median of segment 6 NP binding ratio: 0.85), while those in the regions of Seg1 880mut, Seg5 745mut, and Seg8 50mut is higher than those in the entire segments (median of NP binding ratio, segment 1: 0.84, segment 5: 0.92, and segment 8: 0.87) (Fig. S4). These results suggest that nucleotide positions 39 – 60 of segment 6 can form a secondary structure without NP. To answer the question of whether mutations in the low NP binding ratio region, independent of RNA secondary structure, affect viral propagation, a recombinant virus which has synonymous mutations at nucleotide positions 1910 – 1931 of segment 2 was generated (Seg2 1910mut virus) (Fig. S5A). This region is a low NP biding ratio region and is predicted as a non-structured region. The propagation of the Seg2 1910mut virus was comparable to that of the wild type virus (Fig. S5B), suggesting that mutations in the low NP binding ratio region do not necessarily cause the reduction of viral propagation. Suboptimal codon pairs of viral mRNA reduce mRNA stability and translation efficiency of the deoptimized gene, and IAV with maximized frequencies of CpG dinucleotides in segment 5 showed attenuation in cell culture [45]. Thus, we examined the frequency of CpG dinucleotides and codon usage of the Seg6 39mut virus. CpGs in the Seg6 39mut virus are reduced compared to that of the wild type virus, and the average codon pair score [46] of the Seg6 39mut virus is comparable to that of the wild type virus (wild type: 0.0088, Seg6 39mut: 0.0085). Hence, our results suggest that the RNA structure at nucleotide positions 39 – 60 of segment 6 is associated with the virus propagation efficiency.
Fig. 3.
Impairment of propagation and viral genome replication of recombinant virus which has mutations at nucleotide positions 39 – 60 of segment 6. (A) Predicted structure at nucleotide positions 39 – 60 of segment 6 and mutations in the Seg6 39mut virus. The predicted structures are shown with mutated positions introduced in the Seg6 39mut virus from SHAPE-seq (left) and DMS-seq (right). The red and yellow letters indicate probabilities of modification of more than 0.40 and 0.80, respectively. The gray letters indicate G and U nucleotides that were not modified by DMS. The median of CLIP/control reads is calculated from PAR-CLIP data [18]. (B) Virus propagation of the recombinant viruses. MDCK cells were infected with the recombinant virus at an MOI of 0.01, and the supernatant was collected at 24 h post infection. The virus titer was determined by a plaque assay. The crossbars indicate average values with standard deviations from four independent experiments. The circles indicate the titer of each experiment. P-values were calculated by the Dunnett’s multiple comparison test. (C) and (D) Relative vRNA amount in cells infected with the Seg6 39mut virus (C) and in the supernatant from the Seg6 39mut virus infected cells (D). The amount of each segment was determined by RT-qPCR, and the relative amount was calculated by normalization to the wild type virus. The graph indicates average values with standard deviations from three independent experiments. The circles indicate the relative amount of segments in each experiment. P-values were calculated by Welch’s t-test, and an asterisk indicates P-values less than 0.05.
To investigate the role of the RNA structure in segment 6 for viral propagation, the amounts of vRNAs in cells infected with the Seg6 39mut virus were determined. The relative amount of segment 6 vRNA in cells infected with the Seg6 39mut virus at 8 and 16 hpi was statistically decreased, while that of other segments was comparable or increased (Fig. 3C). Since viral RNA synthesis was altered in cells infected with the Seg6 39mut virus, the expression of NA encoded in segment 6 and M1 encoded in segment 7 was analyzed. At 8 hpi, M1 expression was increased while NA expression was decreased in cells infected with the Seg6 39mut virus (Fig. S6). These results suggest that the mutations in the structured region at nucleotide positions 39 – 60 of segment 6 reduce the replication of segment 6 and consequently reduce viral protein synthesis. Next, to analyze the vRNA packaging efficiency of the Seg6 39mut virus, we determined the amount of vRNAs in the virion. The amount of segment 6 in the Seg6 39mut virus was decreased than that in the wild type virus (Fig. 3D). The relative amount of each segment in the mutant viruses was also decreased, whereas it is below the statistically significant level. In conclusion, these results suggest that the packaging efficiency of segment 6 is consistently altered in Seg6 39mut virus.
3.4. Identification of a region that forms complex RNA structure in segment 5
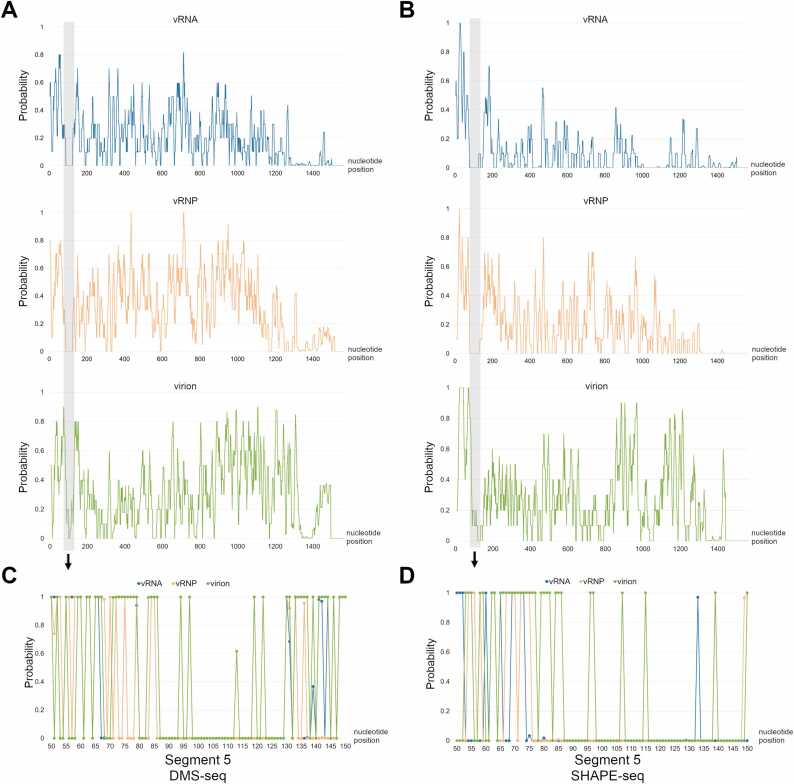
Next, to identify more candidates of functional structured regions on vRNP, the probabilities of modification calculated by BUMHMM were further analyzed. BUMHMM uses a different algorithm from reactIDR, and accounts for biological variability and biases such as coverage and sequence bias. BUMHMM outputs a probability of modification close to 0 or 1 for many nucleotides. This feature facilitates the determination of base pairing formation. The probability plot of each segment is shown in Figs. 4A, 4B, and Fig. S7. The longest less reactive region for both NAI and DMS was vRNA at nucleotide positions 87 – 130 of segment 5 (Fig. 4). This less reactive region was consistently observed in the vRNP and virion, suggesting that the secondary structure of this region is not changed by the vRNP formation. This region was not identified as a structured region from our initial criteria using base-paring probability and Shannon entropy. The Shannon entropy of this region was higher than that of the average of segment 5, suggesting several secondary structures could be predicted in this region. Furthermore, a previous study measuring codon variability in a large dataset of influenza virus genome sequences indicates that nucleotide positions 87 – 130 of segment 5 are highly conserved [47]. We tried to determine the RNA structure at nucleotide positions 87 – 130 of segment 5 by RNA structure prediction and SHAPE-seq data. We analyzed the RNA structure at nucleotide positions 87 – 130 of segment 5 only from SHAPE-seq data because DMS labeled only adenine and cytosine residues, and thus, the resolution of DMS-seq was lower than that of SHAPE-seq in the region. The SHAPE annotated RNA structures at nucleotide positions 87 – 130 of segment 5 in vRNA, vRNP, and virion were predicted by RNAstructure [36]. The predicted structure in vRNA and vRNP was stem-loop structure at nucleotide positions 98 – 130 (3′ stem-loop structure), while that in virion was stem-loop structure at nucleotide positions 87 – 115 (5′ stem-loop structure) (Fig. 5). The resolution of DMS-seq was lower than that of SHAPE-seq because only adenine and cytosine residues are labeled by DMS, but 5′ stem-loop structure was predicted from DMS-seq data of vRNA, vRNP, and virion (Fig. 5). Whole segment structure models from DMS-seq and SHAPE-seq that we constructed in Fig. S4 indicated that not only 3′ and 5′ stem-loop structures but multi-branch loop structures were formed around nucleotide positions 87 – 130 (Fig. S8). However, predicted multi-branch loop structures are not consistent with BUMHMM results, and thus, 3′ and 5′ stem-loop structures are considered the most appropriate structure around nucleotide positions 87 – 130. Interestingly, this region is categorized into the low-NP-binding regions [18] and predicted to form a pseudoknot structure in the previous studies [10], [18]. We confirmed that this region could form a pseudoknot structure by a pseudoknot structure prediction tool, IPknot. The predicted pseudoknot structure is formed by a combination of 3′ stem-loop and 5′ stem-loop structures (Fig. 6A). From these results, we speculate that nucleotide positions 87 – 130 form 3′ stem-loop, 5′ stem-loop, and pseudoknot structures, with transitions between these three structures.
Fig. 4.
Formation of the secondary structure at nucleotide positions 87–130 of segment 5 in the virion. (A) and (B) Moving average of probabilities of modification on segment 5 calculated by BUMHMM from DMS-seq (A) and SHAPE-seq (B). The gray boxes indicate low-probability regions commonly obtained by DMS-seq and SHAPE-seq. (C) and (D) Enlarged views of probabilities of modification at the region indicated by a gray box in the upper panels. vRNA sequence is numbered from 5′ to 3′.
Fig. 5.
The probabilities of modification and predicted base pairs at nucleotide positions 87 – 130 of segment 5 in vRNA, vRNP, and virion. (A-C) The probabilities of modification at nucleotide positions 87 – 130 of segment 5. The probabilities of modification of the NAI-labeled vRNA (A), vRNP (B), and virion (C) at each nucleotide position were calculated by reactIDR. The probabilities of modification at nucleotide positions 87 – 130 of segment 5 were shown, and blue lines indicate predicted base pairs. (D-F) The SHAPE annotated structure at nucleotide positions 87 – 130 of segment 5. The SHAPE annotated structure of vRNA (D), vRNP (E), and virion (F) at nucleotide positions 87 – 130 of segment 5 was shown. The red letters and yellow letters indicate probabilities of modification of more than 0.80 and 0.40, respectively, in (D) and (E), and the yellow letter indicates probabilities of modification of more than 0.20 in (F). (G-I) The DMS-seq annotated structures of vRNA (G), vRNP (H), and virion (I) at nucleotide positions 87 – 130 of segment 5 were shown. The red letters and yellow letters indicate probabilities of modification more than 0.80 and 0.40, respectively. The gray letters indicate G and U nucleotides that were not modified by DMS.
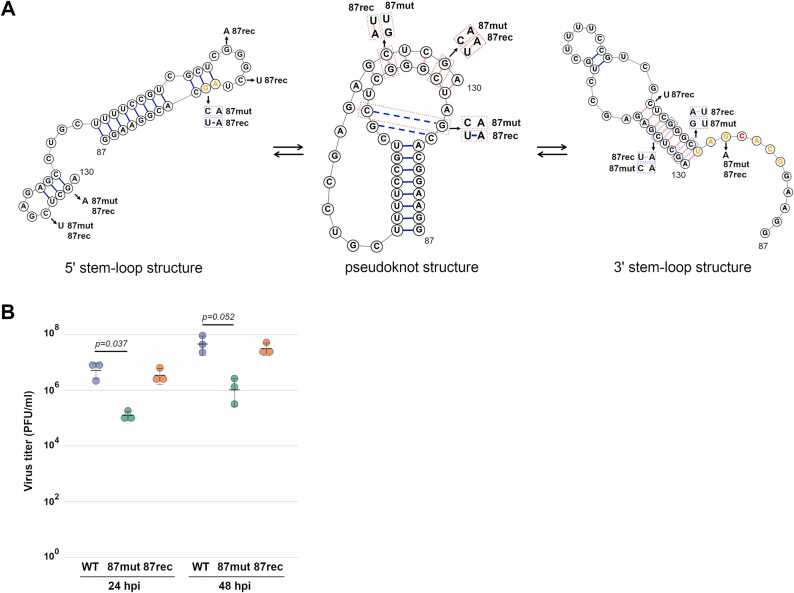
Fig. 6.
Impairment of propagation of recombinant virus which had mutations at nucleotide positions 87 – 130 of segment 5. (A) Mutations in the 87mut and 87rec viruses. Predicted structure from SHAPE-seq of the virion (5′ stem-loop structure; left) and vRNP (3′ stem-loop structure; middle) and predicted pseudoknot structure from IPknot (pseudoknot structure; right) were shown. The boxes indicate the mutated base pairs in the 87mut and 87rec viruses. Other mutations in the 87mut and 87rec viruses were also indicated. (B) Virus propagation of the 87mut and 87rec viruses. MDCK cells were infected with the wild type, 87mut, or 87rec virus at an MOI of 0.01. The supernatant was collected at indicated hours post infection (hpi), and the virus titer was determined by a plaque assay. The crossbars indicate average values with standard deviations from three independent experiments. The circles indicate the titer of each experiment. P-values were calculated by the Dunnett’s multiple comparison test.
3.5. The RNA structure at nucleotide positions 87 – 130 of segment 5 in mutant viruses
To investigate the role of the RNA structure for virus propagation, we constructed recombinant viruses where multiple synonymous mutations were introduced; one is to disrupt the 3′ stem-loop structure (referred as 87mut, hereafter), and another is to rescue the base pairs disrupted by the mutations in 87mut (referred as 87rec, hereafter) (Fig. 6A). The 87mut virus had three mutations, G96A, C126U, and G129A, that did not induce amino acid changes of NP coded on segment 5, and the 87rec virus had three additional mutations, C98A, G102A, and C105U, that were thought to revert base pairs disrupted by mutations in 87mut virus and that also did not induce amino acid changes of NP (Fig. 6A). C126U mutation in the 87mut virus does not disrupt base pairing (G-C to G-U) but reduces the thermodynamic stability of the base pair. Among the three mutations within the region, G96 is located without the loop in our predicted 3′ stem-loop structure but within the predicted pseudoknot structure. We first analyzed whether these mutations affected virus propagation. As a result, the propagation of the 87mut virus was impaired compared with that of the wild type virus, even though all three mutations did not change any amino acid residues (Fig. 6B). The propagation of the 87rec virus was comparable with that of the wild type virus (Fig. 6B). CpGs in segment 5 of the 87mut and 87rec viruses are reduced compared to that of the wild type virus, and the average codon pair scores [46] of the 87mut and 87rec viruses are comparable to that of the wild type virus (wild type: 0.0066, 87mut: 0.0081, and 87rec: 0.0076). Thus, the secondary structure changes by introducing synonymous mutations rather than suboptimal codon pairs and the frequency of CpG dinucleotides affects the replication defect. These results suggest that the RNA structure at nucleotide positions 87 – 130 of segment 5 plays an important role in virus propagation.
Next, we examined the structural differences using SHAPE-seq for the 87mut and 87rec viruses. The wild type, 87mut, and 87rec viruses were purified from the infected cell culture supernatant, and SHAPE-seq was performed. To ensure that the number of reads was comparable for each condition before statistical analysis was performed, the raw reads were downsampled. The coverages of duplicate experiments were shown in Fig. S9A, and the plots of the drop-off rate of duplicate experiments and the R2 were shown in Fig. S9B. The coverages were enough to calculate the reactivities except for the 3′ end of segments, and the R2 of each duplicate experiment ranged from 0.52 to 0.93. The probabilities of modification from duplicate experiments were calculated by reactIDR. The probabilities of modification were correlated between viruses from cell culture supernatant and allantoic fluid, wild type and 87mut viruses, and wild type and 87rec viruses (Fig. S10). The SHAPE annotated RNA structures at nucleotide positions 87 – 130 of segment 5 in the wild type, 87mut, and 87rec viruses were predicted by RNAstructure. The 3′ stem-loop structure was predicted from the probability of modification of wild type virus (Figs. 7A and 7D). In the 87mut virus, a stem-loop structure that is different from 3′ or 5′ stem-loop structure was predicted (Figs. 7B and 7E). The 5′ stem-loop structure was predicted in the 87rec virus (Figs. 7C and 7F). These results suggest that the RNA structure at nucleotide positions 87 – 130 of segment 5 is substantially reorganized and reconstituted in the 87rec virus.
Fig. 7.
Rearrangement of the RNA structure at nucleotide positions 87 – 130 of segment 5 in the 87mut. (A-C) The probabilities of modification at nucleotide positions 87 – 130 of segment 5 in the wild type, 87mut, and 87rec viruses. The probabilities of modification of the NAI-labeled wild type (A), 87mut (B), and 87rec (C) viruses at each nucleotide position were calculated by reactIDR. The probabilities of modification at nucleotide positions 87 – 130 of segment 5 were shown, and blue lines indicate predicted base pairs. Underlines indicate mutations in the 87mut and 87rec viruses. (D-F) The SHAPE annotated structure at nucleotide positions 87 – 130 of segment 5 in the wild type, 87mut, and 87rec viruses. The SHAPE annotated structures of the wild type (D), 87mut (E), and 87rec (F) viruses at nucleotide positions 87 – 130 of segment 5 were shown. Red letters and yellow letters indicate probabilities of modification of more than 0.80 and 0.40, respectively. Underlines indicate mutations in the 87mut and 87rec viruses.
3.6. Assessing the impairment of viral genome replication by mutations in pseudoknot structure in segment 5
To address the question of which step of virus propagation is impaired in the 87mut virus, we determined the amounts of the vRNA segments in the cells infected with the mutant viruses. The amount of the 87mut-derived vRNA was substantially decreased from the wild type virus at 8 and 16 hpi, whereas the vRNA level of the 87rec virus was comparable (Fig. 8A). These results suggest that the mutations in the structured region in segment 5 affect the replication of all segments. Furthermore, to analyze the vRNA packaging efficiency of the mutant virus, we determined the amount of vRNAs and the ratio of segments in the virion. Consequently, the amounts of vRNAs in the 87mut virus were decreased than those of the wild type virus, while those in the 87rec virus were comparable (Fig. 8B). The ratio of each segment in the mutant viruses was comparable to that in the wild type virus. These results indicate that the co-packaging efficiency of the segments is not altered in mutant viruses.
Fig. 8.
Impairment of viral genome replication in cells infected with the 87mut virus. Relative vRNA amount in cells infected with the 87mut or 87rec virus (A) and in the supernatant from the 87mut or 87rec virus infected cells (B). The amount of each segment determined by RT-qPCR is normalized by that from the condition with the wild type virus. The bar graph indicates the average of relative amounts with standard deviations from three independent experiments (shown by circles). P-values were calculated by Welch’s t-test and were adjusted by the Bonferroni correction for multiple comparisons. An asterisk indicates when the adjusted P-value is lower than 0.05. An asterisk beside 87mut at 8 hpi means adjusted P-values are lower than 0.05 for all segments.
To analyze the mechanism of reduction of viral RNA synthesis in cells infected with the 87mut and Seg6 39mut viruses, a mini-replicon assay was performed. NP expression plasmid was transfected at two concentrations because the mechanism of NP binding to vRNAs is different under low and high NP concentrations [41]. vRNA synthesis from the 87mut and 87rec vRNA templates was comparable with that from the wild type segment 5 template both under low and high NP concentrations (Fig. S11). vRNA synthesis from the Seg6 39mut vRNA template was slightly higher than that from the wild type segment 6 template. These results suggest that RNA structures in segments 5 and 6 identified in this study do not directly inhibit viral RNA synthesis.
4. Discussion
In this study, we utilized both DMS-seq and SHAPE-seq to uncover the secondary structures of vRNA with the viral proteins in the virion. The probability of modification of high-NP binding regions in the vRNP and virion tended to be higher than that of the other regions, while the opposite was observed in the vRNA (Table S3). This result indicates that the secondary structure of vRNA is likely to be dissolved by binding NP. Moreover, the terminal regions of the segments in vRNP were detected as more structured, and further in virion, compared to vRNA (Fig. 1). In vRNP, 3′ and 5′ sequences that are partially complementary anneal to form a hairpin structure. The vRNP structure formation is thought as one of the factors to make the terminal regions more stable compared to vRNA alone. Besides, intersegment interactions by the signal sequence regions at both ends of coding regions in each segment may be responsible for the low probability of modification of 5′ terminal region in virion.
Our motivation to use both DMS-seq and SHAPE-seq is to exploit their own advantages and characteristics to build up the comprehensive structure map for IAV. The different reactivity results of SHAPE-seq and DMS-seq can be explained by the accessibility of DMS and NAI to nucleotides in the RNA-protein complex. The reactivities of NAI and DMS to RNA are affected by RNA-binding proteins, and these reagents have been used for the mapping of RNA-protein complex. NAI has been shown to modify the 2′-OH group in the ribose backbone, whereas DMS modifies the base such as N1 of adenosine and N3 of cytidine. The binding of NP to the vRNA is made via the phosphodiester backbone, and potentially competes to NAI modification in vRNP or virion [48]. DMS can react with bases without the effect of steric hindrance by NP, but the apparent limitation of DMS-seq is that the reactivity is detected on only adenine and cytosine. Thus, our comprehensive structure map based on both DMS-seq and SHAPE-seq and the identification of RNA structures common to DMS-seq and SHAPE-seq are expected to complement RNA secondary structures on vRNP that were not identified in previous studies.
We have identified RNA secondary structures on vRNP, but the structures identified in the reported SHAPE-MaP [20] were not included, while SHAPE-seq probabilities obtained in this study are correlated with the SHAPE-MaP reactivities in Dadonaite et al. (Fig. S2E). This may be because our analysis uses both SHAPE-seq and DMS-seq and both probability of modification and Shannon entropy criteria to determine structure regions. Furthermore, similar structures identified in SHAPE-MaP analysis were obtained on segments 1, 3, 4, and 8, although their reliabilities are under our criteria (Fig. S3). Thus, we conclude that our structural analysis is a reliable analysis that encompasses the previously reported results and provides reliable information based on two different methods and replicates. In addition, there are additional regions predicted to form pseudoknot in the previous study, such as nucleotide positions 397 – 518 of segment 1 and nucleotide positions 804 – 867 of segment 8 [18]. We did not detect these regions as structured from our DMS-seq or SHAPE-seq results in concordance with the RNA secondary structure prediction by IPknot. Thus, the pseudoknot structures of these regions might be less stable and not detected in the virion.
Mutations in identified RNA structures did not reduce virus propagation except for mutations at nucleotide positions 39 – 60 of segment 6 (Fig. 3). The structured regions can be involved in intra- and intersegment interactions. Two complementary sequences that might form local stem-loop structures and could potentially initiate intersegment interactions by forming a kissing loop complex have been identified in vitro [5], but it is not known whether the stem-loop structures for initiating the intersegment interaction are formed in vRNP. Previous studies show that the intersegment interactions are complex and redundant [20], [49]. Due to the complexity and redundancy of the intersegment interactions, disruption of one RNA structure is not expected to affect viral propagation. The cells infected with the Seg6 39mut virus had replication and packaging defects (Fig. 3). We have revealed that C999G and A1006G mutation in segment 7 vRNA changed local RNA structures [13]. In addition, nucleotide positions 34 – 86 of segment 1 were identified as a stem-loop region by SHAPE analysis of segment 1 vRNA, and mutations of the stem-loop region reduced the packaging of segment 1 vRNA [50]. These functional RNA structures are in the packaging signal regions. The RNA structures that reduce segment packaging by mutations may function as a hotspot for intersegment interactions though not all intersegment interaction hotspots affect viral replication [49], and these hotspots may be concentrated within the packaging signal regions.
A previous in silico analysis showed that nucleotide positions 87 – 130 of segment 5 could form a pseudoknot structure [10], and CLIP analyses showed that this region was a low NP binding region in PR8 and WSN strains [17], [18]. The SHAPE-MaP analyses suggest that this region formed a different pseudoknot structure from our prediction [20] or 5′ stem-loop structure [21]. We showed a more precise structure of this region in vRNP form by using two comprehensive RNA structural sequencing with robust statistical analysis. The SHAPE annotated RNA structure at nucleotide positions 87 – 130 of segment 5 in vRNP and virion were different (Fig. 5). Moreover, the Shannon entropy of this region was higher than that of the average (Fig. S3). These results indicate that multiple modes of RNA structures may exist in this region.[39] The SHAPE annotated structure at nucleotide positions 87 – 130 of segment 5 in the 87mut virus was different from 3′ or 5′ stem-loop structure (Fig. 6, Fig. 7). In contrast, the SHAPE annotated structure of the region in the 87rec virus was predicted to 5′ stem-loop structure. This region could form both 3′ and 5′ stem-loop structures in the 87rec virus as in the wild type virus because the 3′ stem-loop and 5′ stem-loop structures could be converted via the pseudoknot structure. Taken together, it is possible that RNA structure plasticity in this region, where both 3′ and 5′ stem-loop structures can form, regulates viral genome replication. Although we have focused on the optimal RNA structure, it is possible that the suboptimal RNA structure may be important for the function. The suboptimal RNA structures at nucleotide positions 87 – 130 of segment 5 could be formed because the Shannon entropy of this region is high. If a suboptimal RNA structure of this region is functional for viral propagation, 3′ and 5′ stem-loop structures are considered to be key transition states of RNA conformation changes.
A codon variability study in a large dataset of influenza virus genome sequences can be a clue for the conservation of the structured regions. According to the analysis, it is indicated that nucleotide positions 87 – 130 of segment 5 are highly conserved but nucleotide positions 39 – 60 of segment 6 do not show significance [47]. Because the mutation rate of segment 6 encoding NA is high, complementary mutations might be introduced even if any disrupting mutation is introduced. In fact, nucleotide positions 39 – 60 of segment 6 of the WSN strain contain a mutation at the stem position, and SHAPE-MaP results for the WSN strain predicted that this region does not form a stem-loop structure though those for the PR8 strain predicted a stem-loop structure. Nucleotide positions 87 – 130 of segment 5 are well conserved in IAV strains and predicted to form a pseudoknot structure in vRNA [10], [47]. Interestingly, this region was not identified as a structural region in the recent study, where segments were scanned for local secondary structure and sequence covariance analysis [51]. Multiple structure modes in this region might trigger the weaker signals, but beyond the scope of this paper.
Hutchinson et al. previously reported that propagation of a virus with mutations in conserved NP codons 464–466 (F464-L466 virus) reduced to 1/10 compared to the wild type virus [52]. NP codons 464–466 correspond to nucleotide positions 123 – 131 of segment 5 vRNA and are located at the stem site of the 3′ stem-loop structure of nucleotide positions 87 – 130. The mutations in the F464-L466 virus may disrupt the 3′ stem-loop structure, resulting in reduced viral propagation. Packaging of not only segment 5 vRNA but segment 3 vRNA was observed to be defective in F464-L466 virus [52], while the replication ratio of segment 3 vRNA was just slightly decreased but not statistically significant in 87mut virus at 8 hpi (Fig. 8B). Hence, further mutation analysis might be required to reveal the full potential of the RNA secondary structure at nucleotide positions 87 – 130 of segment 5 whether it interacts with segment 3 to regulate replication and packaging.
5. Conclusion
Overall, our study presents a reliable global secondary structure view of the IAV genome in the virion by applying statistical model-based approaches. We discovered functional structures on the vRNP that are associated with appropriate replication and packaging of the viral genome. These findings will help us to understand the molecular mechanisms by which RNA structures on the IAV genome regulate IAV replication. Antisense oligonucleotides targeting IAV genome RNA structures identified by in vitro studies have been developed [53]. Moreover, a recent report suggests that the unwinding of RNA structures in the IAV and SARS-CoV2 genome by antisense LNA decreased virus titer both in cultured cells and model animals [50]. It is expected that the structure motifs we discovered are promising targets for a new class of anti-influenza drugs that unwind a functional RNA structure motif required for the efficient propagation of IAV.
CRediT authorship contribution statement
Naoki Takizawa: Conceptualization, Methodology, Data curation, Writing – original draft, Writing – review & editing, Visualization, Funding acquisition. Risa Karakida Kawaguchi: Formal analysis, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank Dr. Yoshihiro Kawaoka (University of Tokyo) for kindly providing plasmids for the reverse genetics system and HEK293T cells, Dr. Fumitaka Momose (Kitasato University) for kindly providing plasmids, and Ms. Yukiko Iwata for technical support of experiments. This work was supported by JSPS KAKENHI Grant Number 25871077, 15K21607, and 19K07598 to N.T., Japan Program for Infectious Diseases Research and Infrastructure from AMED Grant Number JP20wm0325008 to N.T., Takeda Science Foundation, GSK Japan Research Grant 2016, and the Waksman Foundation of Japan to N.T.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.csbj.2023.10.036.
Appendix A. Supplementary material
Supplementary material
.
Supplementary material
.
Data availability
The sequence data have been deposited in DDBJ Sequence Read Archive. DRA Accession numbers are DRA014362 (https://ddbj.nig.ac.jp/resource/sra-submission/DRA014362), DRA009494 (https://ddbj.nig.ac.jp/resource/sra-submission/DRA009494), and DRA015929 (https://ddbj.nig.ac.jp/resource/sra-submission/DRA015929). The following sample DRR accession numbers can be used to find individual sample sequence data using the DDBJ search, https://ddbj.nig.ac.jp/search: DRR205184–205195, DRR205198, DRR205199, DRR383848-DRR383859, and DRR452917–452920.
References
- 1.Noda T., Murakami S., Nakatsu S., Imai H., Muramoto Y., Shindo K., et al. Importance of the 1+7 configuration of ribonucleoprotein complexes for influenza A virus genome packaging. Nat Commun. 2018;9:1–10. doi: 10.1038/s41467-017-02517-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Noda T., Sagara H., Yen A., Takada A., Kida H., Cheng R.H., et al. Architecture of ribonucleoprotein complexes in influenza A virus particles. Nature. 2006;439:490–492. doi: 10.1038/nature04378. [DOI] [PubMed] [Google Scholar]
- 3.Arranz R., Coloma R., Chichón F.J., Conesa J.J., Carrascosa J.L., Valpuesta J.M., et al. The structure of native influenza virion ribonucleoproteins. Science. 2012;338:1634–1637. doi: 10.1126/science.1228172. [DOI] [PubMed] [Google Scholar]
- 4.Gerber M., Isel C., Moules V., Marquet R. Selective packaging of the influenza A genome and consequences for genetic reassortment. Trends Microbiol. 2014;22:446–455. doi: 10.1016/j.tim.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 5.Gavazzi C., Yver M., Isel C., Smyth R.P., Rosa-Calatrava M., Lina B., et al. A functional sequence-specific interaction between influenza A virus genomic RNA segments. Proc Natl Acad Sci USA. 2013;110:16604–16609. doi: 10.1073/pnas.1314419110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rausch J.W., Sztuba-Solinska J., Le Grice S.F.J. Probing the structures of viral RNA regulatory elements with SHAPE and related methodologies. Front Microbiol. 2018;8:1–15. doi: 10.3389/fmicb.2017.02634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reich S., Guilligay D., Pflug A., Malet H., Berger I., Crépin T., et al. Structural insight into cap-snatching and RNA synthesis by influenza polymerase. Nature. 2014 doi: 10.1038/nature14009. [DOI] [PubMed] [Google Scholar]
- 8.Pflug A., Guilligay D., Reich S., Cusack S. Structure of influenza A polymerase bound to the viral RNA promoter. Nature. 2014;516:355–360. doi: 10.1038/nature14008. [DOI] [PubMed] [Google Scholar]
- 9.Wandzik J.M., Kouba T., Karuppasamy M., Pflug A., Drncova P., Provaznik J., et al. A structure-based model for the complete transcription cycle of influenza polymerase. Cell. 2020;181:877–893. doi: 10.1016/j.cell.2020.03.061. e21. [DOI] [PubMed] [Google Scholar]
- 10.Gultyaev A.P., Tsyganov-Bodounov A., Spronken M.I.J., Van Der Kooij S., Fouchier R.A.M., Olsthoorn R.C.L. RNA structural constraints in the evolution of the influenza A virus genome NP segment. RNA Biol. 2014;11:942–952. doi: 10.4161/rna.29730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kobayashi Y., Dadonaite B., Doremalen N.Van, Barclay W.S., Pybus O.G. Computational and molecular analysis of conserved influenza A virus RNA secondary structures involved in infectious virion production. RNA Biol. 2016;13:883–894. doi: 10.1080/15476286.2016.1208331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gultyaev A.P., Spronken M.I., Richard M., Schrauwen E.J.A., Olsthoorn R.C.L., Fouchier R.A.M. Subtype-specific structural constraints in the evolution of influenza A virus hemagglutinin genes. Sci Rep. 2016;6:1–15. doi: 10.1038/srep38892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takizawa N., Ogura Y., Fujita Y., Noda T., Shigematsu H., Hayashi T., et al. Local structural changes of the influenza A virus ribonucleoprotein complex by single mutations in the specific residues involved in efficient genome packaging. Virology. 2019;531:126–140. doi: 10.1016/j.virol.2019.03.004. [DOI] [PubMed] [Google Scholar]
- 14.Ruszkowska A., Lenartowicz E., Moss W.N., Kierzek R., Kierzek E. Secondary structure model of the naked segment 7 influenza A virus genomic RNA. Biochem J. 2016;473:4327–4348. doi: 10.1042/BCJ20160651. [DOI] [PubMed] [Google Scholar]
- 15.Lenartowicz E., Kesy J., Ruszkowska A., Soszynska-Jozwiak M., Michalak P., Moss W.N., et al. Self-folding of naked segment 8 genomic RNA of influenza a virus. PLoS One. 2016;11:1–21. doi: 10.1371/journal.pone.0148281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Michalak P., Soszynska-Jozwiak M., Biala E., Moss W.N., Kesy J., Szutkowska B., et al. Secondary structure of the segment 5 genomic RNA of influenza A virus and its application for designing antisense oligonucleotides. Sci Rep. 2019;9:1–16. doi: 10.1038/s41598-019-40443-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee N., Le Sage V., Nanni A.V., Snyder D.J., Cooper V.S., Lakdawala S.S. Genome-wide analysis of influenza viral RNA and nucleoprotein association. Nucleic Acids Res. 2017;45:8968–8977. doi: 10.1093/nar/gkx584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams G.D., Townsend D., Wylie K.M., Kim P.J., Amarasinghe G.K., Kutluay S.B., et al. Nucleotide resolution mapping of influenza A virus nucleoprotein-RNA interactions reveals RNA features required for replication. Nat Commun. 2018;9:465. doi: 10.1038/s41467-018-02886-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamanaka K., Ishihama A., Nagata K. Reconstitution of influenza virus RNA-nucleoprotein complexes structurally resembling native viral ribonucleoprotein cores. J Biol Chem. 1990;265:11151–11155. [PubMed] [Google Scholar]
- 20.Dadonaite B., Gilbertson B., Knight M.L., Trifkovic S., Rockman S., Laederach A., et al. The structure of the influenza A virus genome. Nat Microbiol. 2019;4:1781–1789. doi: 10.1038/s41564-019-0513-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mirska B., Woźniak T., Lorent D., Ruszkowska A., Peterson J.M., Moss W.N., et al. In vivo secondary structural analysis of Influenza A virus genomic RNA. Cell Mol Life Sci. 2023:80. doi: 10.1007/s00018-023-04764-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sexton A.N., Wang P.Y., Rutenberg-Schoenberg M., Simon M.D. Interpreting reverse transcriptase termination and mutation events for greater insight into the chemical probing of RNA. Biochemistry. 2017;56:4713–4721. doi: 10.1021/acs.biochem.7b00323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rouskin S., Zubradt M., Washietl S., Kellis M., Weissman J.S. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature. 2014;505:701–705. doi: 10.1038/nature12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ding Y., Tang Y., Kwok C.K., Zhang Y., Bevilacqua P.C., Assmann S.M. In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature. 2014;505:696–700. doi: 10.1038/nature12756. [DOI] [PubMed] [Google Scholar]
- 25.Wan Y., Qu K., Zhang Q.C., Flynn R. a, Manor O., Ouyang Z., et al. Landscape and variation of RNA secondary structure across the human transcriptome. Nature. 2014;505:706–709. doi: 10.1038/nature12946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spitale R.C., Crisalli P., Flynn R. a, Torre E. a, Kool E.T., Chang H.Y. RNA SHAPE analysis in living cells. Nat Chem Biol. 2013;9:18–20. doi: 10.1038/nchembio.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lucks J.B., Mortimer S.A., Trapnell C., Luo S., Aviran S., Schroth G.P., et al. Multiplexed RNA structure characterization with selective 2′-hydroxyl acylation analyzed by primer extension sequencing (SHAPE-Seq) Proc Natl Acad Sci. 2011;108:11063–11068. doi: 10.1073/pnas.1106501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Selega A., Sirocchi C., Iosub I., Granneman S., Sanguinetti G. Robust statistical modeling improves sensitivity of high-throughput RNA structure probing experiments. Nat Methods. 2017;14:83–89. doi: 10.1038/nmeth.4068. [DOI] [PubMed] [Google Scholar]
- 29.Kawaguchi R., Kiryu H., Iwakiri J., Sese J. reactIDR: evaluation of the statistical reproducibility of high-throughput structural analyses towards a robust RNA structure prediction. BMC Bioinforma. 2019;20:130. doi: 10.1186/s12859-019-2645-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neumann G., Watanabe T., Ito H., Watanabe S., Goto H., Gao P., et al. Generation of influenza A viruses entirely from cloned cDNAs. Proc Natl Acad Sci. 1999;96:9345–9350. doi: 10.1073/pnas.96.16.9345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohkura T., Momose F., Ichikawa R., Takeuchi K., Morikawa Y. Influenza A virus hemagglutinin and neuraminidase mutually accelerate their apical targeting through clustering of lipid rafts. J Virol. 2014;88:10039–10055. doi: 10.1128/JVI.00586-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ding Y., Kwok C.K., Tang Y., Bevilacqua P.C., Assmann S.M. Genome-wide profiling of in vivo RNA structure at single-nucleotide resolution using structure-seq. Nat Protoc. 2015;10:1050–1066. doi: 10.1038/nprot.2015.064. [DOI] [PubMed] [Google Scholar]
- 33.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Incarnato D., Neri F., Anselmi F., Oliviero S. RNA structure framework: automated transcriptome-wide reconstruction of RNA secondary structures from high-throughput structure probing data. Bioinformatics. 2016;32:459–461. doi: 10.1093/bioinformatics/btv571. [DOI] [PubMed] [Google Scholar]
- 35.Incarnato D., Morandi E., Simon L.M., Oliviero S. RNA framework: an all-in-one toolkit for the analysis of RNA structures and post-transcriptional modifications. Nucleic Acids Res. 2018;46 doi: 10.1093/nar/gky486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reuter J.S., Mathews D.H. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinforma. 2010:11. doi: 10.1186/1471-2105-11-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sato K., Kato Y., Hamada M., Akutsu T., Asai K. IPknot: fast and accurate prediction of RNA secondary structures with pseudoknots using integer programming. Bioinformatics. 2011;27:i85–i93. doi: 10.1093/bioinformatics/btr215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takizawa N., Watanabe K., Nouno K., Kobayashi N., Nagata K. Association of functional influenza viral proteins and RNAs with nuclear chromatin and sub-chromatin structure. Microbes Infect. 2006;8:823–833. doi: 10.1016/j.micinf.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 40.Schneider C. a, Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Turrell L., Lyall J.W., Tiley L.S., Fodor E., Vreede F.T. The role and assembly mechanism of nucleoprotein in influenza A virus ribonucleoprotein complexes. Nat Commun. 2013;4:1591. doi: 10.1038/ncomms2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mitchell D., Assmann S.M., Bevilacqua P.C. Probing RNA structure in vivo. Curr Opin Struct Biol. 2019;59:151–158. doi: 10.1016/j.sbi.2019.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Q., Brown J.B., Huang H., Bickel P.J. Measuring reproducibility of high-throughput experiments. Ann Appl Stat. 2011;5:1752–1779. doi: 10.1214/11-AOAS466. [DOI] [Google Scholar]
- 44.Smola M.J., Calabrese J.M., Weeks K.M. Detection of RNA-protein interactions in living cells with SHAPE. Biochemistry. 2015;54:6867–6875. doi: 10.1021/acs.biochem.5b00977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gaunt E., Wise H.M., Zhang H., Lee L.N., Atkinson N.J., Nicol M.Q., et al. Elevation of CpG frequencies in influenza a genome attenuates pathogenicity but enhances host response to infection. Elife. 2016;5:1–19. doi: 10.7554/eLife.12735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Coleman J.R., Papamichail D., Skiena S., Futcher B., Wimmer E., Mueller S. Virus attenuation by genome-scale changes in codon pair bias. Science. 2008;320:1784–1787. doi: 10.1126/science.1155761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gog J.R., Afonso E.D.S., Dalton R.M., Leclercq I., Tiley L., Elton D., et al. Codon conservation in the influenza A virus genome defines RNA packaging signals. Nucleic Acids Res. 2007;35:1897–1907. doi: 10.1093/nar/gkm087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ye Q., Krug R.M., Tao Y.J. The mechanism by which influenza A virus nucleoprotein forms oligomers and binds RNA. Nature. 2006;444:1078–1082. doi: 10.1038/nature05379. [DOI] [PubMed] [Google Scholar]
- 49.Le Sage V., Kanarek J.P., Snyder D.J., Cooper V.S., Lakdawala S.S., Lee N. Mapping of influenza virus RNA-RNA interactions reveals a flexible network. Cell Rep. 2020;31 doi: 10.1016/j.celrep.2020.107823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hagey R.J., Elazar M., Pham E.A., Tian S., Ben-Avi L., Bernardin-Souibgui C., et al. Programmable antivirals targeting critical conserved viral RNA secondary structures from influenza A virus and SARS-CoV-2. Nat Med. 2022 doi: 10.1038/s41591-022-01908-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peterson J.M., Leary C.A.O., Moss W.N. In silico analysis of local RNA secondary structure in influenza virus A, B and C finds evidence of widespread ordered stability but little evidence of significant covariation. Sci Rep. 2022:1–10. doi: 10.1038/s41598-021-03767-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hutchinson E.C., Wise H.M., Kudryavtseva K., Curran M.D., Digard P. Characterisation of influenza A viruses with mutations in segment 5 packaging signals. Vaccine. 2009;27:6270–6275. doi: 10.1016/j.vaccine.2009.05.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Szczesniak I., Baliga-Gil A., Jarmolowicz A., Soszynska-Jozwiak M., Kierzek E. Structural and functional RNA Motifs of SARS-CoV-2 and influenza A virus as a target of viral inhibitors. Int J Mol Sci. 2023;24:1232. doi: 10.3390/ijms24021232. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Data Availability Statement
The sequence data have been deposited in DDBJ Sequence Read Archive. DRA Accession numbers are DRA014362 (https://ddbj.nig.ac.jp/resource/sra-submission/DRA014362), DRA009494 (https://ddbj.nig.ac.jp/resource/sra-submission/DRA009494), and DRA015929 (https://ddbj.nig.ac.jp/resource/sra-submission/DRA015929). The following sample DRR accession numbers can be used to find individual sample sequence data using the DDBJ search, https://ddbj.nig.ac.jp/search: DRR205184–205195, DRR205198, DRR205199, DRR383848-DRR383859, and DRR452917–452920.