Abstract
Pseudomonas sp. strain ADP initiates atrazine catabolism via three enzymatic steps, encoded by atzA, -B, and -C, which yield cyanuric acid, a nitrogen source for many bacteria. In-well lysis, Southern hybridization, and plasmid transfer studies indicated that the atzA, -B, and -C genes are localized on a 96-kb self-transmissible plasmid, pADP-1, in Pseudomonas sp. strain ADP. High-performance liquid chromatography analyses showed that cyanuric acid degradation was not encoded by pADP-1. pADP-1 was transferred to Escherichia coli strains at a frequency of 4.7 × 10−2. This suggests a potential molecular mechanism for the dispersion of the atzABC genes to other soil bacteria.
Due to its widespread use over the last 30 years, for both selective and nonselective weed control (2, 39), atrazine [2-chloro-4-(ethylamino)-6-(isopropylamino)-1,3,5-triazine)] and other s-triazine derivatives have been detected in groundwater and surface water at levels exceeding the Environmental Protection Agency’s maximum contaminant level of 3 ppb (29).
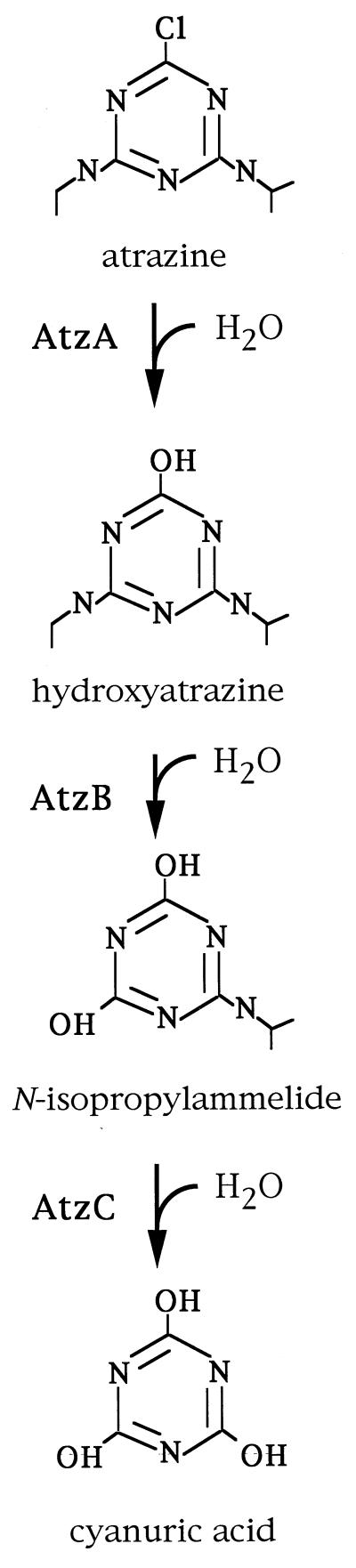
For the past 3 decades, attempts at isolating bacteria (15, 18) or fungi (28) that mineralize atrazine have been unsuccessful. In the last several years, however, a number of laboratories have independently isolated atrazine-degrading bacteria from sites that were previously exposed to atrazine (1, 4, 5, 10, 31, 32, 35). Our laboratory has studied the genes and enzymes involved in atrazine degradation by Pseudomonas sp. strain ADP, in which the first three enzymatic steps are now well defined (3, 11, 13, 37) (Fig. 1). The three genes that encode the enzymes AtzA, -B, and -C have been cloned and sequenced. The first enzyme, AtzA, catalyzes the hydrolytic dechlorination of atrazine, yielding hydroxyatrazine (11). The second enzyme, AtzB, catalyzes hydroxyatrazine deamidation, yielding N-isopropylammelide (3). The third enzyme, AtzC (or N-isopropylammelide isopropylaminohydrolase), transforms N-isopropylammelide to cyanuric acid and isopropylamine (37). An analogous catabolic pathway in Klebsiella pneumoniae 99 (25) has been reported to metabolize s-triazine compounds, but not atrazine. In that strain, the trzC, -D, and -E genes encode ammelide aminohydrolase, cyanuric acid aminohydrolase, and biuret aminohydrolase, respectively, and are located on a 113-kb plasmid (27).
FIG. 1.
Pathway for atrazine catabolism to cyanuric acid in Pseudomonas sp. strain ADP.
Several studies have conclusively shown that horizontal gene transfer occurs among microorganisms in natural and laboratory environments (30). The majority of “natural” conjugal transfer experiments have been carried out in nonsterile soil and involve the use of highly promiscuous plasmids and introduced organisms (30, 42). In general, these results suggest that plasmid transfer is a prominent factor in gene flow in natural systems. Previously, horizontal gene transfer has been invoked to explain the appearance of similar tfd genes in different 2,4-dichlorophenoxyacetic acid-degrading bacterial strains isolated from different regions (16, 26). Catabolic plasmids have also been implicated in the dispersal of genes for 3-chlorobenzoate, chlorocatechol, and naphthalene biodegradation (17, 22, 34).
Recently, the PCR technique was used to demonstrate the presence of DNA that is strikingly homologous to the atzABC genes in atrazine-degrading strains obtained from geographically diverse locations (12). In this study, we report the physical linkage of the atzA, -B, and -C genes on a large plasmid, pADP-1, which is self-transmissible to gram-negative bacteria.
Instability of atrazine degradation phenotype.
Pseudomonas sp. strain ADP has an unstable atrazine-clearing phenotype during cultivation and propagation on complex laboratory growth media. This phenotypic instability, assayed with the plate-clearing procedure, was especially conspicuous in cells grown with NH4Cl in the absence of atrazine as the sole nitrogen source (9). Upon repeated subculturing, clearing-negative strains of ADP (Atr−) that had spontaneously lost the ability to degrade atrazine and hydroxyatrazine on Luria-Bertani (LB) medium (38) containing atrazine or hydroxyatrazine (500 μg/ml) were obtained. These observations suggested that the Atr− strains were lacking at least the first two enzymes in the atrazine degradation pathway. PCR analyses done with genomic DNA from the Atr− strains and the atzA, -B, and -C primers confirmed that the first three genes were missing in the Atr− strains. Moreover, Southern hybridization experiments done with radiolabeled probes specific for the atzA, -B, and -C genes and total genomic DNA isolated from wild-type Pseudomonas strain ADP (Atr+) and the Atr− strains confirmed that Atr− strains did not contain DNA homologous to the atzA, -B, and -C genes (data not shown). Phenotypic instability of biodegradation capacity has been observed in other soil bacteria (33, 40).
Transfer of atrazine degradation ability.
Based on the instability observed with the atrazine degradation phenotype in Pseudomonas sp. strain ADP, an experiment was designed to determine if the atrazine degradation genes were located on a plasmid and were self-transmissible. Mating experiments were done in the absence of a helper strain with Pseudomonas sp. strain ADP (Nalr) (Atr+) as the donor and Escherichia coli AD256 (recA56 srlC300::Tn10; Tetr) (19) as the recipient. Samples of 2 ml of an overnight culture of Pseudomonas sp. strain ADP (donor) and of E. coli AD256 (recipient) (19) were centrifuged at 10,000 × g for 1 min at 4°C, washed in a solution containing 0.85% sodium chloride and 0.01% Tween 20, and resuspended in 0.1 ml of sterile LB broth. Cell suspensions were placed on LB agar plates and incubated at 30°C overnight. Dilutions of the mating mixtures were plated on LB agar with atrazine (500 μg/ml) and tetracycline (15 μg/ml) and incubated at 37°C overnight.
Fifty E. coli colonies (Tetr) from the mating mixture were analyzed for atrazine degradation ability, and three colonies, designated E. coli 3A, 3B, and 3C, degraded atrazine in the plate-clearing assay (13). These colonies did not grow on plates containing nalidixic acid (20 μg/ml), indicating that they were not Pseudomonas sp. strain ADP that had acquired resistance to tetracycline. Moreover, sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis revealed that the soluble protein profiles of the atrazine-degrading E. coli strains were different from those of Pseudomonas sp. strain ADP. Mating experiments with Pseudomonas sp. strain ADP as the donor strain yielded atrazine-degrading E. coli transconjugants at a frequency of 4.7 × 10−2 per recipient. In addition the plasmid encoding atrazine degradation activity was transferable to several gram-negative soil bacteria (6).
Cell extracts were prepared from E. coli 3A, 3B, and 3C, subjected to SDS-polyacrylamide gel electrophoresis with a Mini-PROTEAN II gel apparatus (Bio-Rad), and Western blotted with anti-AtzA antibody (9). These procedures showed that AtzA was expressed in all three transconjugant strains. Cell extracts from wild-type Pseudomonas sp. strain ADP served as a positive control. Extracts from Pseudomonas sp. strain ADP (Atr−) and E. coli AD256 were used as negative controls, and these strains failed to yield any protein that reacted with the anti-AtzA antibody.
Plasmid content of bacteria.
Previously, using conventional plasmid isolation techniques, we failed to observe a plasmid that contained the atzABC genes in Pseudomonas sp. strain ADP (13). In this study, plasmid profiles were determined on horizontal agarose gels by a modified in-well lysis method (20). Gels were prepared in TBE buffer (89 mM Tris-borate–2 mM EDTA; pH 8.0) with 0.75% (wt/vol) agarose and 1% (wt/vol) SDS. Cells were grown with the appropriate antibiotics as described above. Log-phase cells (400 μl) of Pseudomonas, Sinorhizobium, or the transconjugant E. coli strains were centrifuged, washed with 0.5 M NaCl, resuspended in 50 μl of 20% (wt/vol) sucrose, and added to wells preloaded with modified lysis solution (lysozyme [1.0 mg/ml], RNase [10 μg/ml], and 20% [wt/vol] sucrose in TBE). After 10 min of incubation at room temperature, voltage was applied as follows: 5 V for 30 to 45 min, 14 V for 15 min, 40 V for 60 min, and 80 V for 7 to 8 h. Molecular weights of the Sinorhizobium plasmids (21, 36) were used to estimate the sizes of the plasmids present in the Pseudomonas and the recombinant E. coli strains. The approximate molecular masses in kilobases of the indigenous plasmids in the Sinorhizobium fredii strains were as follows: USDA 191, >455, 347, and 105; USDA 205, >455, 342, 177, 126, and 53; USDA 206, >455, 320, 99, and 85; and USDA 217, >455, 335, and 146 (21, 36).
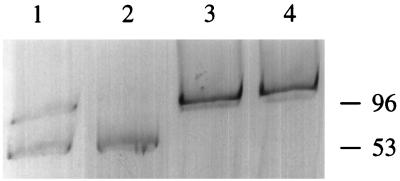
Results of the in-well lysis studies indicated that Pseudomonas sp. strain ADP (Atr+) contained at least two plasmids, pADP-1 and pADP-2, of approximately 96 and 53 kb, respectively (Fig. 2). The pADP-1 plasmid was missing in the Atr− Pseudomonas sp. ADP strains. The E. coli transconjugants (Tetr) that acquired atrazine degradation ability gained a plasmid of approximately the same size (Fig. 2). Plasmid pADP-2 did not hybridize with any of the atzA, -B, and -C gene probes.
FIG. 2.
Plasmid profiles. Lanes: 1, Pseudomonas sp. strain ADP (31), 2, Atr− strain of Pseudomonas strain ADP, 3 and 4, transconjugants 3A and 3B from the mating of ADP with E. coli strain AD256. The values to the right of the gel are mean molecular masses in kilobases determined with reference to plasmids from Sinorhizobium strains (36).
Restriction enzyme analyses of plasmid DNA from E. coli 3A, 3B, and 3C indicated that all three strains contained identical plasmid bands (data not shown). Total genomic and plasmid DNA was isolated from the E. coli clones as described previously (38) and 500 ng of DNA was used as a template in the PCRs with primers designed specifically for atzA, atzB, and atzC (12, 23). PCR analysis indicated that atzABC were present on the plasmid(s) acquired by E. coli AD256 (data not shown). Southern blotting and hybridizations were done with radiolabeled probes specific for atzA, -B, and -C genes (12, 38) and plasmid DNA from E. coli 3A, 3B, and 3C digested with HindIII. These data confirmed the PCR results indicating that the atzABC genes were located on the transferred plasmid.
In a parallel study, Topp et al. (41) characterized a number of atrazine-degrading bacteria from agricultural soil and found that all the isolates metabolized atrazine through hydroxyatrazine as an intermediate, and one plasmid of approximately 97 kb was common to all the atrazine-catabolizing bacteria. The relationship of this plasmid to pADP-1 remains to be determined.
Presence of atrazine- and cyanuric acid-metabolizing enzymes.
To determine if pADP-1 contained a gene that encoded cyanuric acid degradation, resting cells and cell extracts from Pseudomonas sp. ADP strains (Atr+ and Atr−), E. coli 3A (Atr+), and E. coli AD256 (Atr−) were tested for their abilities to catabolize atrazine and cyanuric acid. High-performance liquid chromatography analysis was done on supernatants from whole resting cells and cell extracts incubated with atrazine or cyanuric acid (200 μg/ml) with a Hewlett-Packard HP 1090 liquid chromatograph system as described previously (11). Atrazine and its metabolites were resolved with a Nova-Pak analytical C18 reverse-phase high-performance liquid chromatography column (4-μm spherical packing, 150 by 3.9 mm; Waters Corp.) and an acetonitrile gradient in water at a flow rate of 1.0 ml/min as described previously (11). Cyanuric acid was resolved with an analytical normal-phase column (Lichrosorb RP-18 column, 5-μm spherical packing, 250 by 4.6 mm; Alltech, Deerfield, Ill.) as described previously (37). Authentic atrazine and cyanuric acid were analyzed simultaneously. Cells were incubated with 200 μg of atrazine per ml and 200 μg of cyanuric acid per ml at 30°C for 12 h.
At the end of the experiment, only 16 and 1% of the atrazine with Pseudomonas sp. strain ADP (Atr+) and E. coli 3A (Atr+), respectively, were detectable. However, with Pseudomonas sp. strain ADP (Atr−) and E. coli AD256 cells, 80 and 100% of the atrazine, respectively, were recovered. Moreover, while 88 to 98% of the cyanuric acid was recovered after 12 h in cultures of E. coli 3A or E. coli AD256, with Atr+ and Atr− Pseudomonas sp. ADP strains, only 30 and 49%, respectively, of the cyanuric acid were detected at the end of the experiment. Similar results were obtained with the cell extracts. These results indicated that the transconjugant E. coli strain 3A (Atr+) acquired only the atzABC genes and not genes encoding enzymes involved in the degradation of cyanuric acid. Moreover, Pseudomonas sp. strain ADP (Atr−) that lacked atzA, -B, and -C retained the ability to degrade cyanuric acid. These results suggest that the gene(s) encoding the degradation of cyanuric acid is not located on pADP-1.
In summary, bacterial growth on cyanuric acid is thought to be a relatively common phenotype in soil (7, 8, 14, 24, 25, 43). However, fewer bacteria are thought to catabolize atrazine and these have only been identified recently (1, 4, 5, 10, 31, 32, 35). The atzABC genes confer on a host bacterium the ability to metabolize atrazine to cyanuric acid. The identification of the self-transmissible plasmid pADP-1 containing the atzABC genes demonstrates a mechanism for conferring atrazine-mineralizing ability on bacteria capable of metabolizing cyanuric acid. In this context, it is important to further delineate the structure and evolution of pADP-1 and related plasmids. Such studies are in progress.
Acknowledgments
This work was supported in part by a grant from Novartis Crop Protection (formerly Ciba-Geigy Corporation) and by grant 94-34339-1122 from the U.S. Department of Agriculture-BARD program.
We thank Janis McFarland and Steven Dumford of Novartis Crop Protection for providing s-triazine compounds; William Koskinen, David Gartner, and Mark Sanders for experimental assistance; and Olga Selifonova and Jennifer Seffernick for helpful discussions.
Footnotes
Article 981250043 in the University of Minnesota Agricultural Experiment Station series.
REFERENCES
- 1.Assaf N A, Turco R F. Accelerated biodegradation of atrazine by a microbial consortium is possible in culture and soil. Biodegradation. 1994;5:29–35. doi: 10.1007/BF00695211. [DOI] [PubMed] [Google Scholar]
- 2.Belluck D A, Benjamin S L, Dawson T. Groundwater contamination by atrazine and its metabolites: risk assessment, policy and legal implications. In: Somasundaram L, Coats J R, editors. Pesticide transformation products: fate and significance in the environment. Washington, D.C: American Chemical Society; 1991. pp. 254–273. [Google Scholar]
- 3.Boundy-Mills K L, de Souza M L, Mandelbaum R T, Wackett L P, Sadowsky M J. The atzB gene of Pseudomonas sp. strain ADP encodes the second enzyme of a novel atrazine degradation pathway. Appl Environ Microbiol. 1997;63:916–923. doi: 10.1128/aem.63.3.916-923.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boundy-Mills, K. L., L. P. Wackett, and M. J. Sadowsky. Unpublished data.
- 5.Bouquard C, Ouazzani J, Promé J-C, Michel-Briand Y, Plésiat P. Dechlorination of atrazine by a Rhizobium sp. isolate. Appl Environ Microbiol. 1997;63:862–866. doi: 10.1128/aem.63.3.862-866.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Contolatis, C., and H.-G. Hur. Unpublished data.
- 7.Cook A M. Biodegradation of s-triazine xenobiotics. FEMS Microbiol Rev. 1987;46:93–116. [Google Scholar]
- 8.Cook A M, Beilstein P, Grossenbacher H, Huetter R. Ring cleavage and degradative pathway of cyanuric acid in bacteria. Biochem J. 1985;231:25–30. doi: 10.1042/bj2310025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Souza, M. L. Unpublished data.
- 10.de Souza M L, Newcombe D, Alvey S, Crowley D E, Hay A, Sadowsky M J, Wackett L P. Molecular basis of a bacterial consortium: interspecies catabolism of atrazine. Appl Environ Microbiol. 1998;64:178–184. doi: 10.1128/aem.64.1.178-184.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Souza M L, Sadowsky M J, Wackett L P. Atrazine chlorohydrolase from Pseudomonas sp. strain ADP: gene sequence, enzyme purification, and protein characterization. J Bacteriol. 1996;178:4894–4900. doi: 10.1128/jb.178.16.4894-4900.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Souza M L, Seffernick J, Martinez B, Sadowsky M J, Wackett L P. The atrazine catabolism genes atzABC are widespread and highly conserved. J Bacteriol. 1998;180:1951–1954. doi: 10.1128/jb.180.7.1951-1954.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Souza M L, Wackett L P, Boundy-Mills K L, Mandelbaum R T, Sadowsky M J. Cloning, characterization, and expression of a gene region from Pseudomonas sp. strain ADP involved in the dechlorination of atrazine. Appl Environ Microbiol. 1995;61:3373–3378. doi: 10.1128/aem.61.9.3373-3378.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Erickson E L, Lee K H. Degradation of atrazine and related s-triazines. Crit Rev Environ Contam. 1989;19:1–13. [Google Scholar]
- 15.Fernandez-Quintanilla C, Cole M A, Slife F W. European Weed Research Society Symposium on Theory and Practice of the Use of Soil Applied Herbicides. Paris, France: Columa; 1981. Microbial and chemical degradation of atrazine in solution; pp. 301–308. [Google Scholar]
- 16.Fulthorpe R R, McGowan C, Maltseva O V, Holben W E, Tiedje J M. 2,4-Dichlorophenoxyacetic acid-degrading bacteria contain mosaics of catabolic genes. Appl Environ Microbiol. 1995;61:3274–3281. doi: 10.1128/aem.61.9.3274-3281.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fulthorpe R R, Wyndham R C. Transfer and expression of the catabolic plasmid pBRC60 in wild bacterial recipients in a freshwater ecosystem. Appl Environ Microbiol. 1991;57:1546–1553. doi: 10.1128/aem.57.5.1546-1553.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geller A. Studies on the degradation of atrazine by bacterial communities enriched from various biotypes. Arch Environ Contam Toxicol. 1980;9:289–305. doi: 10.1007/BF01057409. [DOI] [PubMed] [Google Scholar]
- 19.Ghai J, Das A. The virD operon of Agrobacterium tumefaciens Ti plasmid encodes a DNA-relaxing enzyme. Proc Natl Acad Sci USA. 1989;83:3109–3313. doi: 10.1073/pnas.86.9.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hashem F M, Kuykendall D. Plasmid DNA content of several agronomically important Rhizobium species that nodulate alfalfa, berseem, clover or leucaena. In: Graham P H, Sadowsky M J, Vance C P, editors. Symbiotic nitrogen fixation. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. pp. 181–188. [Google Scholar]
- 21.Heron D S, Pueppke S G. Mode of infection, nodulation specificity, and indigenous plasmids of 11 fast-growing Rhizobium japonicum strains. J Bacteriol. 1984;160:1061–1066. doi: 10.1128/jb.160.3.1061-1066.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herrick J B, Stuart-Keil K G, Ghiorse W C, Madsen E L. Natural horizontal transfer of a naphthalene dioxygenase gene between bacteria native to a coal tar-contaminated field site. Appl Environ Microbiol. 1997;63:2330–2337. doi: 10.1128/aem.63.6.2330-2337.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Innis M A, Gelfand D H, Sninsky J J, White T J. PCR protocols: a guide to methods and applications. New York, N.Y: Academic Press, Inc.; 1990. [Google Scholar]
- 24.Jessee J A, Benoit R E, Hendricks A C, Allen G C, Neal J L. Anaerobic degradation of cyanuric acid, cysteine, and atrazine by a facultative anaerobic bacterium. Appl Environ Microbiol. 1983;45:97–102. doi: 10.1128/aem.45.1.97-102.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jutzi K, Cook A M, Hutter R. The degradative pathway of the s-triazine melamine. Biochem J. 1982;208:679–684. doi: 10.1042/bj2080679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ka J O, Holben W E, Tiedje J M. Genetic and phenotypic diversity of 2,4-dichlorophenoxyacetic acid (2,4-D)-degrading bacteria isolated from 2,4-D-treated field soils. Appl Environ Microbiol. 1994;60:1106–1115. doi: 10.1128/aem.60.4.1106-1115.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karns J S, Eaton R W. Abstracts of the 91st General Meeting of the American Society for Microbiology 1991. Washington, D.C: American Society for Microbiology; 1991. Genes specifying -triazine metabolism are plasmid-encoded in Klebsiella pneumoniae strain 99, abstr. Q-167; p. 304. [Google Scholar]
- 28.Kaufman D D, Blake J. Degradation of atrazine by soil fungi. Soil Biol Biochem. 1970;2:73–80. [Google Scholar]
- 29.Kello, D. 1989. WHO drinking water quality guidelines for selected herbicides. Food Addit. Contam. 6(Suppl.):S79–S85. [DOI] [PubMed]
- 30.Levy S B, Miller R V. Gene transfer in the environment. New York, N.Y: McGraw-Hill Publishing Co.; 1989. [Google Scholar]
- 31.Mandelbaum R T, Allan D L, Wackett L P. Isolation and characterization of a Pseudomonas sp. that mineralizes the s-triazine herbicide atrazine. Appl Environ Microbiol. 1995;61:1451–1457. doi: 10.1128/aem.61.4.1451-1457.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moscinski J K, Jayachandran K, Moorman T B. Abstracts of the 96th General Meeting of the American Society for Microbiology 1996. Washington, D.C: American Society for Microbiology; 1996. Mineralization of the herbicide atrazine by Agrobacterium radiobacter, abstr. Q-414; p. 458. [Google Scholar]
- 33.Nakatsu C, Ng J, Singh R, Strais N, Wyndham C. Chlorobenzoate catabolic transposon Tn5271 is a composite class I element with flanking class II insertion sequences. Proc Natl Acad Sci USA. 1991;88:8321–8316. doi: 10.1073/pnas.88.19.8312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ogawa N, Miyashita K. Recombination of a 3-chlorobenzoate catabolic plasmid from Alcaligenes eutrophus NH9 mediated by direct repeat elements. Appl Environ Microbiol. 1995;61:3788–3795. doi: 10.1128/aem.61.11.3788-3795.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Radosevich M, Traina S J, Hao Y-L, Tuovinen O H. Degradation and mineralization of atrazine by a soil bacterial isolate. Appl Environ Microbiol. 1995;61:297–302. doi: 10.1128/aem.61.1.297-302.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sadowsky M J, Bohlool B B. Possible involvement of a megaplasmid in nodulation of soybeans by fast-growing rhizobia from China. Appl Environ Microbiol. 1983;46:906–911. doi: 10.1128/aem.46.4.906-911.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sadowsky M J, Tong Z, de Souza M, Wackett L P. AtzC is a new member of the amidohydrolase protein superfamily and is homologous to other atrazine-metabolizing enzymes. J Bacteriol. 1998;180:152–158. doi: 10.1128/jb.180.1.152-158.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 39.Seiler A, Brenneisen P, Green D H. Benefits and risks of plant protection products. Possibilities of protecting drinking water: case atrazine. Water Supply. 1992;10:31–42. [Google Scholar]
- 40.Tomasek P H, Frantz B, Sangodkar U M X, Haugland R A, Chakrabarty A M. Characterization and nucleotide sequence determination of a repeat element isolated from a 2,4,5-T degrading strain of Pseudomonas cepacia. Gene. 1989;76:227–238. doi: 10.1016/0378-1119(89)90163-7. [DOI] [PubMed] [Google Scholar]
- 41.Topp E, Tessier L, Lewis M. Abstracts of the 97th General Meeting of the American Society for Microbiology. Washington, D.C: American Society for Microbiology; 1997. Characterization of atrazine-degrading bacteria isolated from agricultural soil, abstr. Q-407; p. 523. [Google Scholar]
- 42.van Elsas J D, Trevors J T, Starodub M E. Plasmid transfer in soil and rhizosphere. In: Klingmuller W, editor. Risk assessment for deliberate releases. Berlin, Germany: Springer-Verlag; 1988. pp. 89–99. [Google Scholar]
- 43.Wolf D C, Martin J P. Microbial decomposition of ring-14C atrazine, cyanuric acid and 2-chloro-4,6-diamino-s-triazine. J Environ Qual. 1975;4:134–139. [Google Scholar]