Abstract
Background & Aims
Nonalcoholic steatohepatitis (NASH) is a complex disease involving both genetic and environmental factors in its onset and progression. We analyzed NASH phenotypes in a genetically diverse cohort of mice, the Hybrid Mouse Diversity Panel, to identify genes contributing to disease susceptibility.
Methods
A “systems genetics” approach, involving integration of genetic, transcriptomic, and phenotypic data, was used to identify candidate genes and pathways in a mouse model of NASH. The causal role of Matrix Gla Protein (MGP) was validated using heterozygous MGP knockout (Mgp+/-) mice. The mechanistic role of MGP in transforming growth factor-beta (TGF-β) signaling was examined in the LX-2 stellate cell line by using a loss of function approach.
Results
Local cis-acting regulation of MGP was correlated with fibrosis, suggesting a causal role in NASH, and this was validated using loss of function experiments in 2 models of diet-induced NASH. Using single-cell RNA sequencing, Mgp was found to be primarily expressed in hepatic stellate cells and dendritic cells in mice. Knockdown of MGP expression in stellate LX-2 cells led to a blunted response to TGF-β stimulation. This was associated with reduced regulatory SMAD phosphorylation and TGF-β receptor ALK1 expression as well as increased expression of inhibitory SMAD6. Hepatic MGP expression was found to be significantly correlated with the severity of fibrosis in livers of patients with NASH, suggesting relevance to human disease.
Conclusions
MGP regulates liver fibrosis and TGF-β signaling in hepatic stellate cells and contributes to NASH pathogenesis.
Keywords: Gene Network Analysis, Liver Fibrosis, Nonalcoholic Steatohepatitis, Systems Genetics
Summary.
Systems genetics analyses in a mouse diversity panel identified Matrix Gla Protein (MGP) as a causal gene for nonalcoholic steatohepatitis. MGP is expressed preliminary in stellate cells and attenuated the response to TGF-β. MGP expression was also associated with fibrosis in human cohorts.
Nonalcoholic fatty liver disease (NAFLD) manifests as a spectrum of hepatic abnormalities ranging from simple steatosis and steatohepatitis (NASH) to fibrosis and cirrhosis. Although simple steatosis is benign in most cases, NASH is associated with increased overall morbidity and mortality.1 Advanced NASH can eventually progress to end-stage liver disease with increased risk of hepatocellular carcinoma. With the increasing prevalence of obesity, diabetes, and metabolic syndrome, NAFLD has rapidly become the most common form of chronic liver disease worldwide and the top non-viral cause of liver failure requiring liver transplantation.2 Despite the high prevalence of this disease, the natural history and etiology are poorly understood. NAFLD is a complex disease, involving significant genetic and environmental factors, with a heritability estimated to be 22% to 50%.3 To date, using human genome-wide association studies (GWAS), only a handful of genes for NASH have been identified and validated (review in 4), explaining only a small fraction of the genetic contribution to the disease.
We previously reported a “humanized” hyperlipidemic mouse model for NASH and have examined how it is affected by genetic background using a panel of inbred strains of mice known as the Hybrid Mouse Diversity Panel (HMDP).5 Altogether, over 900 mice (both male and female) from >100 strains were examined for hepatic fibrosis and related traits. We observed a large spectrum in hepatic fibrosis, over 100-fold, among the strains. To identify the underlying genetic factors, we have carried out dense genotyping, global transcriptomics on livers, and metabolomics on plasma from the panel. We have integrated the clinical and molecular traits using genetic mapping, correlation, and statistical modeling, an approach known as “systems genetics.”5,6
We now report the identification of matrix Gla protein (MGP), a potent inhibitor of vascular calcification and a modulator of bone morphogenesis,7 as a causal gene in this model. Although MGP has been studied extensively for its involvement in bone and vascular calcification, little is known about its function in liver and its role in NASH. The gene is expressed in various tissues, including heart, lung, kidney, skin, and arterial vessel walls8 and encodes a 10-kDa protein containing 5 residues of an uncommon amino acid, γ-carboxyglutamic acid (Gla), which are formed by a vitamin K-dependent modification of specific glutamic acid residues.9 The Gla residues confer calcium and hydroxyapatite-binding properties, which are important in its inhibitory effects on calcification. Calcium-bound MGP binds and sequesters bone morphogenetic protein 2 (BMP2) and BMP4, potent stimulators of osteochondrogenic differentiation, thereby preventing chondrogenic transformation of vascular smooth muscle cells7,10 and endothelial-mesenchymal transitions in endothelial cells.11 Additionally, MGP inhibits calcification by exerting a direct effect on elastin.12,13 In humans, mutation in the MGP gene results in Keutel syndrome, a rare disorder characterized by calcification in soft tissues.14 Homozygous MGP knockout mice are viable at birth but rapidly develop extensive calcification in the cartilage and arteries and arteriovenous malformations in multiple organs and die within 2 months from aortic rupture.15,16
To examine the role of MGP in liver fibrosis, we subjected wild-type (WT) and heterozygous MGP knockout mice to 2 diet-induced models of NASH. We found that decreased MGP expression leads to attenuation in inflammation and liver fibrosis. We show using single-cell sequencing that MGP is highly expressed in dendritic cells and stellate cells compared with other cell types in the liver. We also show that knockdown of MGP in cultured stellate cells resulted in decreased transforming growth factor-beta (TGF-β) receptor and signaling. Finally, we provide evidence that MGP variants contribute to human liver fibrosis.
Results
Systems Genetics Analysis Identified Mgp as a Candidate Gene for Hepatic Fibrosis
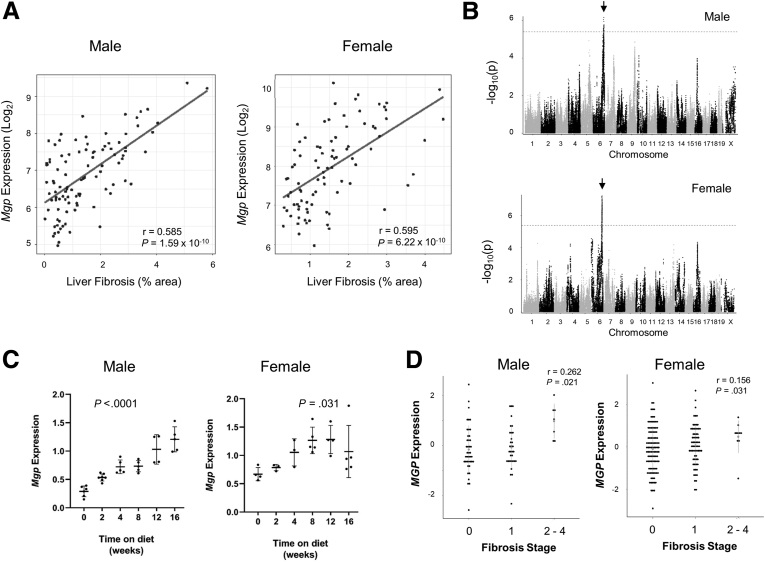
We previously reported a hyperlipidemic mouse model of NASH and examined how genetic background affects the trait in a panel of 100 strains of HMDP mice.5 Global transcriptomic profiling of livers from the HMDP panel17 revealed a strong correlation of hepatic expression of Mgp with hepatic fibrosis in both male (r = 0.585; P = 1.59 × 10-10) and female (r = 0.595; P = 6.22 × 10-10) mice (Figure 1A). Most genes highly correlated with fibrosis are likely to be downstream of fibrosis, or “reactive,” rather than causal (discussed in 18). To prioritize genes likely to be causal, we carried out expression quantitative trait locus (eQTL) analyses of all genes. Mgp exhibited significant local (cis-acting) eQTL as well as some trans-acting loci in both males and females (Figure 1B). In females, about 30% of the variance of Mgp expression was regulated locally and the local (or cis-) component exhibited correlation with fibrosis (r = 0.20; P = .05), consistent with a causal role. In males, on the other hand, only a few percent of the Mgp expression variance was explained by the local eQTL, and there was no correlation of the local component with fibrosis.
Figure 1.
Mgp expression in mouse liver. (A) Correlation of hepatic Mgp expression with hepatic fibrosis in male (top panel) and female (bottom panel) mice in the HMDP is presented. Each data point corresponds to the strain average of Mgp expression and liver fibrosis. p, P-value; r, biweight midcorrelation. (B) Manhattan plot showing the association of SNP genotype with hepatic Mgp expression in male (top panel) and female (bottom panel) mice. The genome wide significance threshold is indicated by the dotted line. Cis-eQTLs for Mgp expression on chromosome 6 are present in both male and female liver as indicated by the arrows. (C) Male C57BL/6J mice carrying human APOE∗3-Leiden and CETP transgenes were fed a “Western” high-fat, high-cholesterol diet for 0 to 16 weeks. Hepatic Mgp mRNA expression was measured by qPCR and is presented as mean ± standard error of the mean. (D) Correlation of hepatic MGP expression with fibrosis stage in patients of the KOBS study.
Time course studies showed that hepatic Mgp expression increased with time during NASH progression, correlating to the increase in severity in NASH in these mice (Figure 1C). To explore the relevance of our findings to human NASH pathobiology, we analyzed MGP expression in RNAseq data of the Kuopio Obesity Surgery (KOBS) clinical study.19 Hepatic MGP expression was found to be significantly correlated with fibrosis stage in both female (r = 0.156; P = .036) (Figure 1D) and male patients (r = 0.262; P = .002) (Figure 1D). MGP expression was significantly increased in the livers of patients with NASH compared with non-NASH subjects (healthy or simple steatosis) (P = .021).
As liver fibrosis is influenced by many cell types in the liver, we performed single-cell sequencing to examine the expression pattern of Mgp in the liver. We isolated non-parenchymal cells (NPCs) from frozen liver samples from mice before and after the onset of liver fibrosis (week 8 and 12),5 and conducted single-cell transcriptome sequencing using the 10× Genomics Chromium Single Cell System. Clustering of the sequenced cells using uniform manifold approximation and projection (UMAP) revealed many distinct cell clusters (Figure 2A), including vascular cells, resident immune cells, and infiltrating/circulating immune cells, in addition to a small cluster of hepatocytes (Figure 2B). Mgp was found to be highly expressed in hepatic stellate cells (HSCs) and dendritic cells, relative to other cells present in the liver (Figure 2B). This is consistent with previous proteomics studies showing high expression in the HSCs in mouse livers20 and was confirmed by Western blotting showing the presence of MGP in primary HSCs (Figure 2C). As HSCs play an important role in liver fibrosis, these data suggest that Mgp may affect fibrosis through its action in HSCs.
Figure 2.
Mgp expression in HSCs. (A) UMAP plot of single-cell sequencing data from mouse hepatic nonparenchymal cells reveals distinct cell clusters corresponding to different cell types. (B) Expression levels of Mgp in each individual cell type. Each dot represents the expression levels of Mgp in the indicated cell types. The height of the bar represents total expression of the indicated cell type. B, B-lymphocytes; DCs, dendritic cells; ECs, endothelial cells; KCs, Kuffper cells; MPs, macrophages; NKs, natural killer cells; SMCs, smooth muscle cells; T, T-lymphocytes. (C). Protein lysates (n = 3 per group) from WT and Mgp+/- HSC were resolved by SDS-PAGE and Western blotting. The amount of MGP was normalized to HSC marker Desmin.
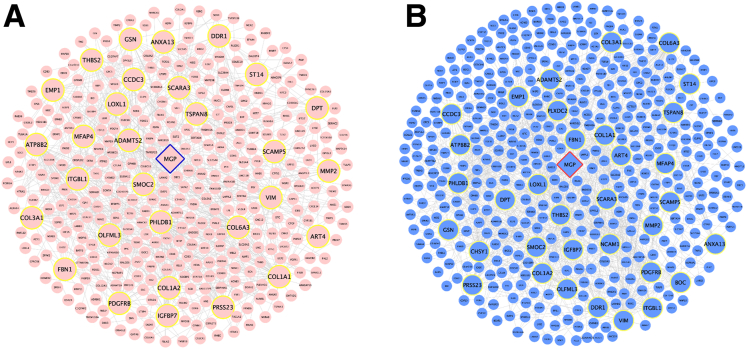
To identify potential interactions of Mgp, we performed network modeling using Mergeomics.21 Previously, we used this strategy to identify causal genes for hepatic steatosis in NAFLD.6 Consistent with eQTL analysis, this showed that Mgp is a key driver for liver fibrosis (Figure 3). Notably, Mgp associated with a number of connective tissue and matrix component genes (Col1a1, Col1a2, Col3a1, and Col6a3) and regulators of fibrosis (Tspan8, Pgdfrb, Loxl1, Smoc2, Adamts2, Mfap4, and Itgbl1).22, 23, 24, 25, 26, 27 Some of the genes in the subnetwork have been identified as potential biomarkers for liver fibrosis (Thbs2, Ddr1, and Dpt).28, 29, 30 Overall, network analysis showed that Mgp is closely connected to genes involved in fibrosis.
Figure 3.
Key driver subnetwork of Mgp. Key driver subnetwork of Mgp was derived from GWAS integration with functional genomics (A), or liver fibrosis gene expression data (B) by Mergeomics analysis.
MGP Haploinsufficiency Reduced Hepatic Fibrosis in Female but not Male Mgp+/- Mice
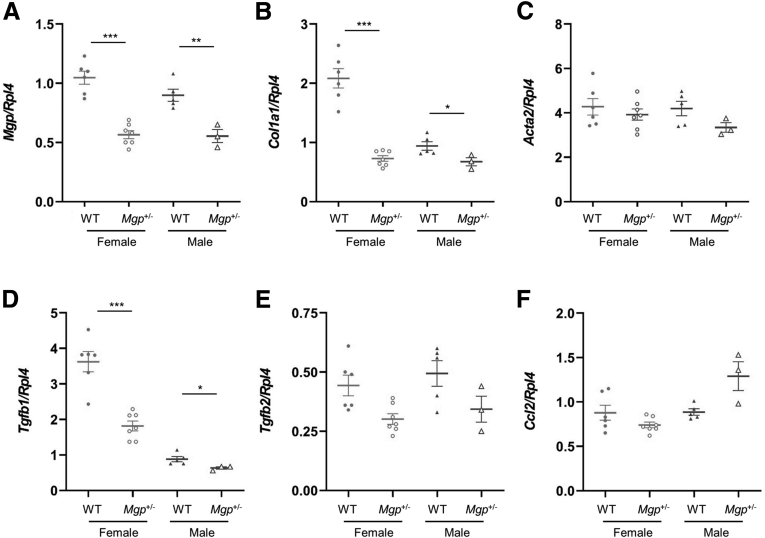
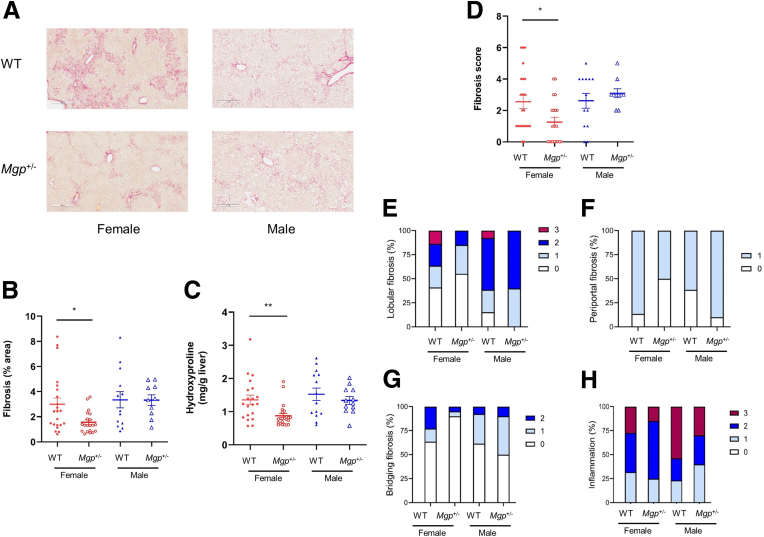
Because homozygous MGP knockout mice die within 2 months of age from aortic rupture, we performed our validation studies using heterozygous mice. We used 2 different NASH models. The first involves feeding the mice a choline-deficient, defined amino acid high fat (CDAHFD) diet containing 0.17% methionine for 12 weeks.31 Compared with the livers of WT mice, female Mgp+/- mice showed significant reduction in hepatic fibrosis with ∼54% decrease in collagen-positive stained area (Figure 4A–B), although the hydroxyproline content was not significantly different (Figure 4C). This discrepancy is likely due to a difference in sensitivity in detecting collagen by the 2 measurements. We have previously shown that computer-assisted collagen area measurements offer higher sensitivity in fibrosis assessment as it excludes the measurement of physiological collagen (from vasculature linings), whereas hydroxyproline assays do not.5 Improvements in fibrosis score, lobular fibrosis, portal fibrosis, bridging fibrosis, and inflammation were observed in female Mgp+/- mice (Figure 4D–H). In contrast, no decrease in liver fibrosis was observed in male Mgp+/- mice (Figure 4A–G). Consistent with this observed sex difference, the reduction in expression of profibrotic genes (Col1a1 and Tgfb1) was much greater in female mice than male mice (Figure 5).
Figure 4.
Assessment of liver fibrosis in mice fed the CDAHFD diet. Liver sections from control and Mgp+/- mice (n = 6–8 animals per group) were stained for collagen with picrosirius red (A). The percentage of fibrosis area in the entire liver sections was quantified by a computer algorithm (B). Liver collagen content was assessed by hydroxyproline assay (C). The fibrosis score was determined by a pathologist blinded to the study (D). Results are presented as mean ± standard error of the mean. ∗P < .05 vs control. Incidence of lobular fibrosis (E), bridging fibrosis (F), periportal fibrosis (G), and inflammation (H) are presented as a percentage of mice in each group exhibiting these pathologies. Pathologist’s scoring criteria are listed in Table 1.
Figure 5.
Hepatic gene expression of mice fed the CDAHFD diet. Expression of profibrotic genes (as indicated) in the livers from control and Mgp+/- mice (n = 3–7 animals per group) was measured by qPCR and normalized to the level of the housekeeping gene Rpl4. Relative gene expression levels are presented as mean ± standard error of the mean. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001 vs WT control.
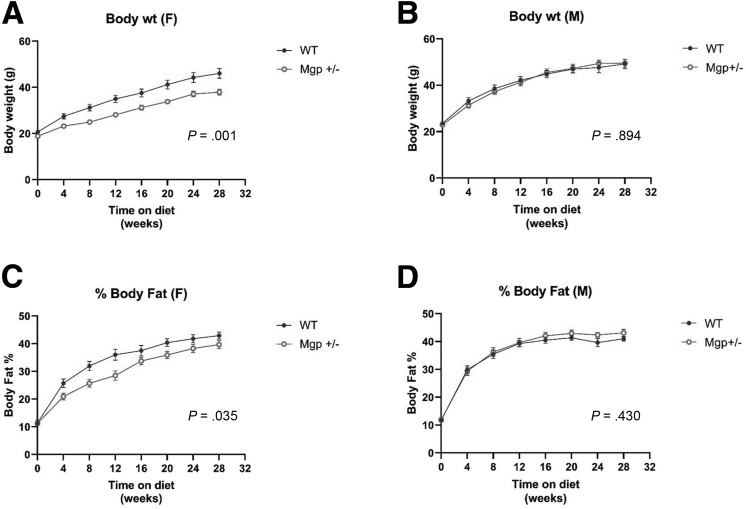
We also studied Mgp using an over-nutrition mouse model of NASH that involves feeding mice a Gubra amylin NASH (GAN) diet (40 kcal-% fat. of these 0% trans-fat and 46% saturated fatty acids by weight, 22% fructose, 10% sucrose, 2% cholesterol) for 30 weeks. Similar to the findings from the CDAHFD diet study, female Mgp+/- mice developed less liver fibrosis than WT mice, whereas no difference was observed between the 2 groups in male mice (Figure 6A–D). Lobular fibrosis, bridging fibrosis and inflammation were decreased in female Mgp+/- mice, whereas only inflammation was improved in male mice (Figure 6E–H). Hepatic expression of profibrotic genes (Col1a1, Tgfb1, Tgfb2, Ccl2, and Timp1) was significantly reduced in female Mgp+/- mice, whereas no reduction was observed in male mice (Figure 7). The levels of hepatic lipids (triglyceride, total cholesterol, cholesterol ester, free cholesterol, and phospholipids) were not significantly changed in Mgp+/- mice, suggesting that the protective effect in liver fibrosis is not due to reduction in lipid accumulation in the liver (Figure 8). Plasma levels of triglyceride, total cholesterol, unesterified cholesterol, and glucose were decreased in female Mgp+/- mice but not in male mice (Figure 9). Consistent with reduced fibrosis, alanine transaminase levels were decreased in female mice, whereas no differences were observed in male mice (Figure 9F). The female Mgp+/- mice showed decreased weight gain (Figure 10A–B) and adiposity (Figure 10C–D).
Figure 6.
Assessment of liver fibrosis in mice fed the GAN diet. Liver sections from control and Mgp+/- mice (n = 12–21 animals per group) were stained for collagen with picrosirius red (A). The percentage of fibrosis area in the entire liver sections was quantified by a computer algorithm (B). Liver collagen content was assessed by hydroxyproline assay (C). The fibrosis score was determined by a pathologist blinded to the study (D). Results are presented as mean ± standard error of the mean. ∗P < .05; ∗∗P < .01 vs control. Incidence of lobular fibrosis (E), bridging fibrosis (F), periportal fibrosis (G), and inflammation (H) are presented as a percentage of mice in each group exhibiting these pathologies. Pathologist’s scoring criteria are listed in Table 1.
Figure 7.
Hepatic gene expression of mice fed the GAN diet. Expression of profibrotic genes (as indicated) in the livers from control and Mgp+/- mice (n = 12–21 animals per group) was measured by qPCR and normalized to the level of the housekeeping gene Rpl4. Relative gene expression levels are presented as mean ± standard error of the mean. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001 vs WT control.
Figure 8.
Hepatic lipid levels in mice fed the GAN diet. Hepatic levels of triglycerides (A), phospholipids (B), total cholesterol (C), cholesterol ester (D), and unesterified cholesterol (E) are presented as mean ± standard error of the mean from 3 to 7 mice per group.
Figure 9.
Plasma metabolite and alanine transaminase levels in mice fed the GAN diet. Control and Mgp+/- mice were fasted for 4 hours. Plasma triglycerides (A), total cholesterol (B), unesterified cholesterol (C), high-density lipoprotein cholesterol (D), glucose (E), and alanine transaminase (F) were measured by colorimetric assays. Data are presented as mean ± standard error of the mean (n = 3–7 mice per group). ∗P < .05; ∗∗P < .01 vs control.
Figure 10.
Body composition of mice fed the GAN diet. Control and Mgp+/- Body weight of female (A) and male (B) mice were measured every 4 weeks. Adiposity was measured by nuclear magnetic resonance imaging in female (C) and male (D) mice. Data are presented as mean ± standard error of the mean (n = 18–22 mice per group). P values from 1-way analysis of variance analyses are presented.
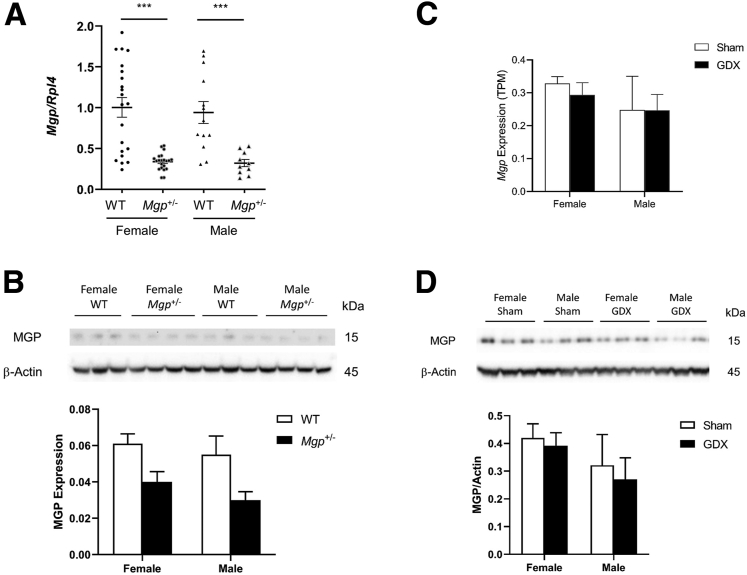
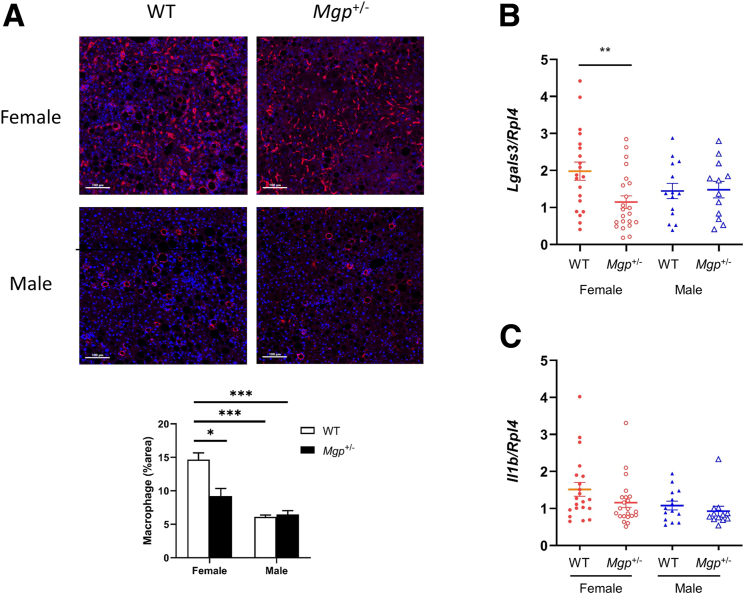
The sex difference in fibrosis response is not associated with a difference in MGP expression. No significant differences in MGP mRNA and protein expression in the liver were observed between the 2 sexes in both WT and Mgp+/- mice (Figure 11A–B). Furthermore, MGP mRNA and protein expression in the liver was not affected by gondadectomy, suggesting that sex hormones do not play a significant role in regulating MGP expression in the liver (Figure 11C–D). Consistent with the finding of reduced fibrosis in female Mgp+/- mice, a significant reduction in macrophage staining was observed in the liver (Figure 12A). Expression of galectin-3 (Lgals3), an inflammation marker, was also significantly reduced in the livers of female Mgp+/- mice (Figure 12B), whereas IL1-β expression was not different (Figure 12C). Macrophage infiltration was significantly reduced in the livers of male mice compared with female mice (Figure 12A), whereas galectin-3 and IL1-β expression did not differ (Figure 12B–C).
Figure 11.
MGP expression in mice: effect of gonadectomy. (A) Expression of Mgp in the livers from control and Mgp+/- mice fed the GAN diet (n = 12–21 animals per group) was measured by qPCR and normalized to the level of the housekeeping gene Rpl4. Relative gene expression levels are presented as mean ± standard error of the mean. ∗∗∗P < .001 vs WT control. (B) Liver protein lysates from control and Mgp+/- mice (n = 6 per group) were resolved by SDS-PAGE and Western blotting. The amount of MGP was normalized to actin and is presented as mean + standard error of the mean. (C) Hepatic Mgp expression from gonadectamized (GDX) and sham control chow-fed C57BL/6J mice was determined by RNA sequencing. Results are presented as mean + standard error of the mean from 4 mice in each group. (D) Liver protein lysates from control and GDX mice (n = 3 per group) were resolved by SDS-PAGE and Western blotting. The amount of MGP was normalized to actin and is presented as mean + standard error of the mean.
Figure 12.
Effect of Mgp on inflammation. (A) Liver sections were stained with antibody against F4/80 (red) and DAPI (blue) and quantified by Image J software. Results is presented as the mean ± standard error of th emean from 9 random sections from 3 mice in each group. ∗P < .05; ∗∗∗P < .001 vs female WT control. (B–C) Hepatic mRNA expression of galectin-3 (B) and IL1-β was measured by qPCR and normalized to the level of the housekeeping gene Rpl4. Gene expression levels are presented as mean ± standard error of the mean. ∗∗P < .01 vs female WT control.
Mgp Knockdown Attenuates TGF-β Response in Cultured Human LX-2 Stellate Cells
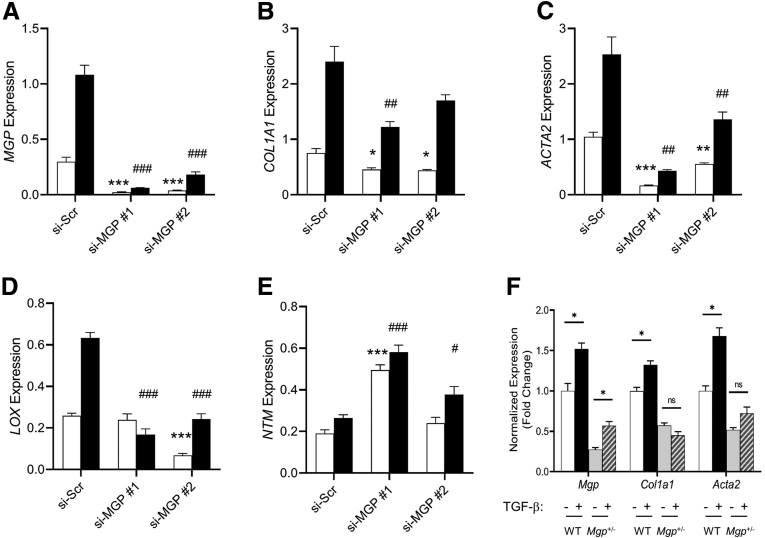
HSCs have been shown to be the major cell type contributing to the fibrotic process. Upon activation by cytokines such as TGF-β, they secrete extracellular matrix and other pro-fibrotic factors. As Mgp is highly expressed in HSC in mice, we tested whether the TGF-β-mediated fibrotic response is affected by Mgp expression in HSC. Cultured human LX-2 stellate cells were transfected with siRNA specific for MGP to knock down MGP expression. The efficiency of knockdown was >90% (Figure 13A). Treatment of cells with TGF-β1 showed that MGP expression is induced by TGF-β1, and that this induction is greatly reduced in cells with knockdown of MGP expression (Figure 13A). Knockdown of MGP expression also significantly reduced expression of COL1A1 and ACTA2 in the absence and presence of TGF-β1 (Figure 13B–C), indicating that MGP expression directly impacts the fibrogenic response to TGF-β in stellate cells. Consistent with this finding, expression of the activated HSC marker, lysyl oxidase (LOX), was also greatly suppressed in cells with MGP knockdown (Figure 13D). Conversely, expression levels of quiescent HSC marker, neurotrim (NTM), were significantly higher in cells with MGP knockdown (Figure 13E). TGF-β response was also attenuated in primary HSC from Mgp+/- mice compared with that from WT mice (Figure 13F). These data suggest that MGP is important in TGF-β-dependent activation of HSC.
Figure 13.
Effects of MGP knockdown on TGF-β response in LX-2 cells. LX-2 cells were transfected with scramble siRNA (si-Scr) or 2 different MGP siRNA (si-MGP #1 and si-MGP #2). Cells were treated with vehicle (empty bars) or TGF-β (2.5 ng/mL) (filled bars). RNA was isolated 24 hours after treatment and gene expression was determined by qPCR. Results are presented as mean ± SEM (n = 6 per group). ∗P < .05; ∗∗∗P < .001 vs untreated si-Scr control; #P < .05; ##P < .01; and ###P < .001 vs TGF-β-treated si-Scr control. (F) Primary HSC from WT and Mgp+/- mice were treated with vehicle or TGF-β (2.5 ng/mL) as indicated. RNA was isolated 24 hours after treatment and gene expression was determined by qPCR. Results are presented as mean ± standard error of the mean (n = 4 per group). ∗P < .05 vs untreated control.
Receptor-regulated SMADs (R-SMADs) are essential intracellular signaling molecules in the TGF-β superfamily signaling pathway. R-SMADs are phosphorylated by activated TGF-β receptors. This covalent modification is crucial for their heterodimerization with the common partner SMAD4 (Co-SMAD4), and translocation to the nucleus, where downstream target gene expression is activated. We examined whether MGP expression would affect this signaling pathway. As shown in (Figure 14A), TGF-β-induced phosphorylation of SMAD1, SMAD2, and SMAD3 was significantly reduced in MGP-knockdown cells.
Figure 14.
Effects of MGP knockdown on R-SMAD phosphorlyation. (A) LX-2 cells were transfected with scramble siRNA (si-Scr) or MGP siRNA (si-MGP). Cell homogenates (n = 6 per group) from control (empty bars) and TGF-β (2.5 ng/mL) treated cells (filled bars) were resolved by SDS-PAGE and Western blotting. (B) Liver homogenates (n = 6 per group) from WT and Mgp+/- mice fed on CDAHFD diet were resolved by SDS-PAGE and Western blotting. Amounts of individual phospho-SMADs were normalized to the corresponding total SMAD. ∗P < .05 vs control group of the same treatment.
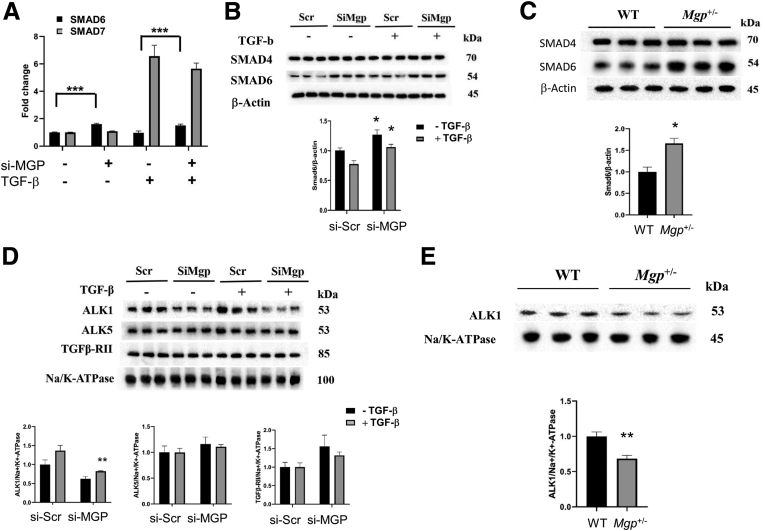
The effects of reduced Mgp expression in liver of Mgp+/- mice were similar to those observed in cultured stellate cells. Thus, decreases in phosphorylation of SMAD1, SMAD2 and SMAD3 were also observed in the livers of Mgp+/- mice fed the CDAHFD diet (Figure 14B). This finding is consistent with the observed reduction in TGF-β responsive gene expression. The TGF-β signaling pathway is subjected to negative feedback regulation by inhibitory SMADs (I-SMADs), SMAD6 and SMAD7. We examined the expression of inhibitory I-SMADs and their induction by TGF-β in LX-2 cells. Expression of SMAD6 was higher in MGP-knockdown cells, in both basal and TGF-β states whereas SMAD7 expression was not affected by MGP knockdown (Figure 15A). Protein levels of SMAD6 were significantly higher in both MGP-knockdown LX-2 cells (Figure 15B) and livers of Mgp+/- mice (Figure 15C), indicating that decreased MGP expression led to a more inhibitory potential to BMP and TGF-β simulation. At the receptor level, the canonical signaling pathway for TGF-β requires binding to a TGF-β type II receptor (Tβ-RII) that recruits a TGF-β type I receptor (TβRI) with serine/threonine kinase activity. We examined the expression of the type I receptors ALK1 and ALK5 in LX-2 cells. Knockdown of MGP led to decreased protein levels of ALK1 expression both in the basal and TGF-β stimulated cells, whereas there was no significant difference in ALK5 and TGFβ-RII expression between control and MGP-knockdown cells (Figure 15D). The expression of ALK1 was also found to be decreased in the livers of Mgp+/- mice compared with WT mice (Figure 15E). Together, these data indicate that MGP plays an important role in liver fibrosis and stellate cell activation during NASH progression.
Figure 15.
Effects of MGP knockdown on I-SMAD expression. (A) LX-2 cells were transfected with scramble siRNA (si-Scr) or MGP siRNA (si-MGP). Cells were treated with vehicle or TGF-β (2.5 ng/mL). RNA was isolated 24 hours after treatment and gene expression was determined by qPCR. SMAD6 (empty bars) and SMAD7 (filled bars) expression are presented as mean ± standard error of the mean (n = 6 per group). ∗∗∗P < .001 vs si-Scr control. (B) Protein levels of SMAD6 in LX-2 cells are presented as mean ± standard error of the mean (n = 6 per group). ∗P < .05 vs si-Scr control. (C) Protein levels of SMAD6 in liver homogenates (n = 6 per group) from WT and Mgp+/- mice fed on CDAHFD diet are presented as mean ± standard error of the mean (n = 6 per group). ∗P < .05 vs WT. (D) Protein levels of TGF-β receptor (ALK1, ALK5, and TGFβ-RII) in LX-2 cells are presented as mean ± standard error of the mean (n = 6 per group). ∗∗P < .01 vs si-Scr control. (E) Protein levels of TGF-β receptor (ALK1, ALK5, and TGFβ-RII) in liver homogenates (n = 6 per group) from WT and Mgp+/- mice fed on CDAHFD diet are presented as mean ± standard error of the mean (n = 6 per group). ∗∗P < .01 vs WT.
Discussion
Systems genetics studies of a mouse model of NASH identified Mgp as a likely causal gene, and we confirmed this in female mice using heterozygous (Mgp+/-) gene targeted mice in 2 different NASH models. In contrast to female mice, the local regulation of Mgp expression in male mice was only modestly correlated with fibrosis, and heterozygous knockout mice did not differ significantly from WT mice in liver fibrosis. We investigated the mechanism by which Mgp influences NASH using single-cell sequencing, network modeling, and studies of a human stellate cell line. In liver, we found that Mgp is expressed primarily in HSCs and in our network is associated with collagen and extracellular matrix genes. The in vitro results indicated that MGP knockdown blocks SMAD signaling and fibrotic gene activation by TGF-β. We discuss these points in turn below.
Our single-cell RNA sequencing data shows that Mgp has relatively low expression in hepatocytes but is highly expressed in HSCs and dendritic cells among liver non-parenchymal cells (Figure 2). HSCs are the central effector of hepatic fibrosis, and their activation is crucial for the extracellular collagen deposition and fibrosis in NASH. Here, we show that MGP expression is important for proper TGF-β signaling. Knockdown of MGP in cultured HSCs leads to diminished response to TGF-β (Figure 13). Expression of Mgp in HSC is induced by TGF-β (Figure 13A). We also showed that Mgp expression increases during the progression of NASH and is positively correlated with fibrosis severity in mice (Figure 1A and 1C) and humans (Figure 1D). These findings suggest that MGP plays a positive feedback role in the fibrogenic program, and induction of Mgp expression is essential for maintaining the action of TGF-β. As activation of TGF-β signaling pathways is common to fibrogenesis in various tissue injury models, our findings suggest that MGP may have a broader role in liver fibrosis process.
Our data show that hepatic Mgp expression increased during NASH progression in mice (Figure 1C). Similarly, hepatic MGP expression was shown to be significantly correlated with fibrosis severity in a human study (Figure 1D), suggesting that our findings in mice are relevant to the human disease. Fibrosis is the strongest predictor for clinical outcomes, liver-related mortality, and liver transplantation.32,33 However, although human NAFLD GWAS have identified many loci, only a handful of causal genes for fibrosis have been validated: PNPLA3, TM6SF2, MBOAT7, and HSD17B13 (reviewed in 4). As NAFLD is a complex disease determined by interactions among genetic and environmental factors, controlling for environmental variations in human is difficult. Mouse studies have the advantage that it is feasible to better control for diet and other environmental factors.
We employed two in vivo models of diet-induced NASH. The CDAHFD diet induces NASH by increasing hepatic fatty acid uptake and diminishing VLDL secretion,34 whereas the GAN diet induces obesity, insulin resistance and hyperlipidemia.35 We show that Mgp+/- mice exhibited decreased hepatic fibrosis in both models of NASH (Figures 4 and 6). Because these two models have different mechanisms of NASH induction, these data suggest that Mgp likely affects a common fibrosis pathway downstream of NASH induction. Recent bioinformatics analysis of Gene Expression Omnibus datasets of human liver and kidney fibrosis identified MGP as one of the potentially important genes with altered expression in fibrosis.28 It is interesting to note that renal MGP expression is increased in human and experimental chronic kidney disease. Renal tubulointerstitial MGP expression strongly correlates with interstitial fibrosis and interstitial inflammation.36 In addition to liver and kidneys, MGP expression was found to be increased in intestine samples from patients with Crohn’s disease and ulcerative colitis.28 These findings suggest that MGP may play a general role in tissue fibrosis, not limited to NASH and liver, and that it may be a potential pharmacological target for intervening in fibrotic diseases such as NASH and chronic kidney disease.
In both models of NASH, the protective effects on liver fibrosis were observed in female Mgp+/- mice but not in male mice. Notably, the key driver gene networks of both sexes were similar (Figure 3). Expression of MGP gene and protein in the liver was not different between male and female mice (Figure 11A), suggesting that the difference in NASH phenotypes is not due to a difference in the amount of MGP. Sex hormones did not appear to affect hepatic Mgp expression (Figure 11B). Male mice showed lower macrophage infiltration than WT female mice, despite having similar expression of Ccl2 (Figures 7C and 12A). These data suggest there may be a difference in inflammatory response between sexes. There are a number of other sex differences that could contribute a stronger effect on NASH, including lower plasma lipid and alanine transaminase levels (Figure 9), reduced adiposity in females (Figure 10), differences in diet response, and contribution by extra-hepatic tissues.
Knockdown studies in LX-2 stellate cells identified 3 mechanisms by which reduced MGP expression blunted the TGF-β signaling pathway. First, knockdown of MGP led to reduced phosphorylation of R-SMADs (SMAD 2 & 3) (Figure 14A). Second, knockdown of MGP expression increased the expression of I-SMAD (SMAD6) (Figure 15A–B). Third, ALK1 receptor expression was decreased by MGP knockdown (Figure 15D). These findings reveal an integral role of MGP in liver fibrogenesis during NASH progression through its action in HSCs. Our study does not preclude the possibility that MGP also exerts its effects on liver fibrosis through HSC-independent or extrahepatic mechanisms. Future studies using HSC-specific MGP knockout mice would be useful to assess the relative contribution of HSC- and non-HSC-dependent mechanisms on liver fibrosis.
MGP is an inhibitor of BMPs, members of the TGF-β superfamily. Previous studies in endothelial cells showed that MGP inhibits BMP-4 activity and that ALK1 expression is regulated by BMP-4 in a biphasic manner.37 Our culture cell studies show that knockdown of MGP reduced expression of ALK1 in HSCs (Figure 15D). It is possible that MGP alters ALK1 expression in HSCs through a similar mechanism. ALK1 is activated by BMP9, which has been shown to stimulate HSCs and promotes fibrosis in the liver.38 Reduction of ALK1 expression may have additional beneficial effects on fibrosis through the BMP pathway.39 Additionally, our data show that SMAD6 expression is increased in MGP knockdown cells. SMAD6 efficiently blocks BMP signaling but only weakly affects TGF-β/activin signaling.40 Hence, changes in SMAD6 expression levels could also affect ALK1 expression through its effects on BMP signaling.
In conclusion, a major challenge in studies of advanced NAFLD is the identification of genes and pathways contributing to disease progression. Thus far, only a few genes have been validated, and we now add MGP to the list. Also, our network analyses identified other strong fibrosis candidates. MGP appears to act in stellate cells by attenuating TGF-β signaling. A surprising finding was that MGP contributes to fibrosis in a sex-specific manner.
Materials and Methods
Animal Studies
Generation and characterization of MGP knockout mice were described previously.15 Heterozygous MGP knockout mice in C57BL/6J background were bred with C57BL/6J mice are bred with to generate heterozygous MGP knockout mice and WT littermates as control for the experiments. Generation and analysis of the HMDP for liver fibrosis was described previously.5 Transgenic mice expressing human CETP were obtained from The Jackson Laboratory, and mice carrying the human APOE∗3-Leiden variant were kindly provided by Dr L. Havekes.17,41 These mice were interbred to create a strain carrying both transgenes in a C57BL/6J background and subsequently bred to females from about 100 common inbred strains to produce a panel of F1 heterozygous mice with C57BL/6J as a common parent. Mice (8–10 weeks old) were fed a “Western” diet (33 kcal% fat from cocoa butter and 1% cholesterol; Research Diets; catalog no. D10042101) for 16 weeks. At the end of the study, mice were euthanized after a 4-hour fast, and their livers were harvested and divided into aliquots for RNA isolation (snap-frozen) and histology (formalin-fixed and paraffin-embedded). Mgp+/– mice on a C57BL/6J background15 were previously described.42 Genotypes were confirmed by PCR. Animals were maintained on a 12-hour light–dark cycle with ad libitum access to water. Littermates were used as WT controls. All mice were fed a standard chow diet (Diet 8604, Harlan Teklad Laboratory) before switching to NASH-inducing diets. For induction of NASH, mice (8–10 weeks old) were fed a CDAHFD diet (60 kcal% fat with 0.1% methionine and no added choline; Research Diets; catalog no. A06071302) for up to 12 weeks or GAN diet (40 kcal-% fat, 46% saturated fatty acids by weight, 22% fructose, 10% sucrose, 2% cholesterol, Research Diets; catalog no. D09100310) for 30 weeks. At the end of the study, mice were fasted for 4 hours. To avoid sampling error, liver samples for histological and gene expression analyses were taken from the same lobe in each mouse. Body composition was measured by nuclear magnetic resonance (Bruker Biospin Corp). The gonadectomy and ovariectomy study was described previously in detail.43 The mice were on chow diet for 16 weeks. All animal work was approved by the University of California, Los Angeles, Animal Research Committee, the Institutional Animal Care and Use Committee.
Gene Expression and Genome-wide Association Analyses
Liver RNA was isolated from HMDP mice for global gene expression using Affymetrix HT-MG_430 PM Microarrays as described.44 eQTL in liver were mapped using the FaST-LMM algorithm with correction for population structure.45 Correlations were calculated using the bicor function from the WGCNA R package.46 High-density genotypes for inbred strains of mice were generated by the Mouse Diversity Array.47 Genome-wide association mapping of hepatic fibrosis was performed using FaST-LMM.17 Correlations between liver fibrosis and gene expression were analyzed by using the bicor package in R. Expression quantitative trait loci were mapped from TPM values by using FaST-LMM after log2-transformation. To calculate the cis-component of expression correlated with fibrosis, the expression values for each relevant gene were partitioned into groups based on the genotype of the most significant cis-eQTL with a P-value < 1 × 10-4. Medians were calculated for each resulting group, and the medians (replicated once for each individual within each group) as a whole were correlated against fibrosis.
Multiomics Integration and Key Driver Gene Analysis
We modeled fibrosis gene networks using the multi-omics HMDP data along with additional public gene expression datasets to identify pathways and predict potential “key driver” genes underlying hepatic fibrosis. We first constructed gene co-expression networks using both MEGENA48 and WGCNA46 based on fibrosis expression data across the HMDP strains and then integrated these networks with GWAS analyses of hepatic fibrosis as well as fibrosis eQTL information using the Mergeomics platform.21 We utilized the Marker Set Enrichment Analysis (MSEA) function of Mergeomics for the integration and identification of coexpression modules (groups of co-expressed genes) and biological pathways (BIOCARTA, KEGG, and REACTOME) that are enriched for hepatic fibrosis GWAS signals as well as direct fibrosis transcriptome. Subsequently, we mapped the fibrosis-associated network modules and pathways to gene regulatory Bayesian networks of liver tissues that are based on numerous genetic and gene expression datasets and our fibrosis tissues to predict potential key regulators, termed key drivers, of the hepatic fibrosis processes using the Key Driver Analysis function of Mergeomics. The GWAS network structure was derived by overlaying the GWAS SNPs mapped to genes using fibrosis liver eQTLs, whereas the transcript network was derived by overlaying fibrosis correlated genes directly onto the liver Bayesian networks.
Quantitative Assessment of Lipids and Fibrosis in the Liver
Liver lipids were extracted and quantitated as described.49 For histological examination and fibrosis quantitation, livers were fixed in 10% formalin, embedded in paraffin, sectioned at 5 μm, and stained with picrosirius red. The fibrosis score was assessed by a pathologist blinded to the study according to scoring scheme in Table 1. Slides were scanned at 20× magnification (Aperio ScanScope XT, Leica Biosystems). Using Definiens Tissue Studio (Definiens AG), we designed a fully automated image analysis algorithm that quantifies fibrosis as the percentage area of the whole tissue section, excluding normal vascular wall and liver capsular collagen. This method was previously validated by showing that the percentage of fibrosis measured strongly correlated with blinded assessments by liver pathologist and collagen content as measured by hydroxyproline assays.5 Pathological fibrosis in the entire liver cross-section was quantified and expressed as percentage area over total area of the section.
Table 1.
Scoring Criteria Used by Pathologists to Assess Fibrosis and NASH Phenotype
| 0 | 1 | 2 | 3 | 4 | |
|---|---|---|---|---|---|
| Lobular fibrosis | Absent | Focal | Multifocal | Diffuse | x |
| Bridging fibrosis | Absent | <34% | <67% | >66% | Bridging |
| Periportal fibrosis | Absent | Present | x | x | x |
| Inflammation 20X | <2 foci | 2 to 4 | >4 | x |
NASH, Nonalcoholic steatohepatitis.
Cell Culture
The human hepatic stellate cell line LX-2 cells were cultured in 2% fetal bovine serum Dulbecco’s Modified Eagle Medium supplemented with penicillin/streptomycin. For knockdown experiments, siRNA for human MGP and control scramble siRNA were obtained from Sigma. LX-2 cells were reverse transfected with siRNA of MGP or negative scramble siRNA using Lipofectamine RNAiMAX siRNA transfection reagent (Invitrogen) in a 6-well plate. Forty-eight hours later, cells were treated with TGF-β1 (2.5 ng/mL) for 24 hours.
Hepatic Stellate Cell Isolation
Primary mouse HSCs were isolated according to the method described by Zhai et al.50 NPCs from the liver were prepared using the Liver Dissociation kit (Miltenyi) and Gentle MACS tissue processor according to the manufacturer’s instructions. NPCs from 4 mice were pooled and fractionated in Optiprep by density gradient centrifugation. HSCs were collected in the fraction with density less than 1.053. Cells were then washed and cultured in Dulbecco’s Modified Eagle Medium containing 10% fetal bovine serum, 1% penicillin/streptomycin, and Glutamax.
RNA Extraction and Real-time Quantitative Polymerase Chain Reaction
Total RNA was extracted using the QIAzol (Qiagen) RNA extraction reagent according to the manufacturer’s protocol. Reverse transcription was performed using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). The real-time PCRs were performed using SYBR Green Master Mix kit according to the manufacturer’s instructions (Applied Biosystems). Quantitative PCR (qPCR) was performed using the LightCycler 480 Detection System (Roche).
Single-cell RNA-Sequencing
We previously showed that cell type heterogeneity in the liver becomes significantly detectable by qPCR analysis begins around 8 weeks in C57BL/6J mice carrying the human CETP and APOE∗3-Leiden transgenes on high-fat, high-cholesterol diet.5 Genes involved in inflammatory cell infiltration (F4/80 and MCP-1) and stellate cell activation and fibrogenesis (Col1a1) progressively increased. Liver non-parenchymal cells were isolated from C57BL/6J and F1:129X1/SvJ (resistant to diet-induced NASH) CETP and APOE∗3-Leiden transgenic mice fed on high-fat, high-cholesterol diet for 8 or 12 weeks. Mice were perfused with phosphate buffered saline, and the liver was excised and digested using the Miltenyi MACS mouse Liver Dissociation Kit to isolate the NPCs per manufacturer’s instructions. The NPC preparation was counted and then stained for viability using 7-AAD (1:20). We then sorted 7-AAD negative cells using a BD FACSAria cell sorter set with a 70-μm nozzle. An estimated 10,000 single cells were loaded onto each channel of a single-cell 3’ Chip (10X Genomics). The chip was then placed on a 10X Genomics instrument to generate single-cell gel beads in emulsion. Single-cell RNA-Seq libraries were prepared with Chromium Single Cell 3’ v2 Library and Cell Bead Kits according to the manufacturer’s instructions. Library fragment length was estimated using a TapeStation (Aligent) and library molar concentration was quantified by Qubit Fluorometric Quantitation (ThermoFisher). Libraries were then pooled and sequenced on an Illumina HiSeq 4000 (Illumina) instrument with PE100 reads and an 8-bp index read for multiplexing. Strain identity was determined post sequencing using the DMUXLET method based on known genotype information. Reads were demultiplexed and aligned to mouse genome mm10 using STAR. Digital transcript count matrices were generated using 10X Cell Ranger application. Datasets were integrated and normalized using scTransform in R with the Seurat library package. Distinct cell populations were identified by annotating the uniquely most differentially expressed genes as well as localization of known cell type markers.
Western Blot Analysis
Liver homogenates and cell lysates were prepared in RIPA buffer with protease inhibitors. Western blot analysis of cell lysates was performed by SDS-PAGE using NuPAGE (Invitrogen) followed by transfer to PVDF membrane (Invitrogen). Using the appropriate antibodies, specific proteins were detected with ECL Plus chemiluminescence detection system (GE Healthcare). Chemiluminescence was detected in ChemiDoc Detection System (Bio-Rad). The expression levels of respective proteins were quantified using Quantity One software (Bio-Rad). Antibodies for SMAD1, pSMAD1/5, SMAD2/3, pSMAD2, pSMAD3, SMAD4, TGFβ-RII, and Na/K-ATPase were purchased from Cell Signaling Technology. SMAD6, ALK1, and ALK5 antibodies were purchased from Santa Cruz Biotechnology. Antibodies for MGP were purchased from Thermo Fisher.
Immunohistochemistry
Livers were dissected and subsequently fixed and stored in 4% paraformaldehyde. After dehydration, tissues were embedded in paraffin and cut into 5-μm slices. Immunohistochemical staining was performed on a Discovery XT automated stainer (Ventana Medical Systems) using rabbit anti-F4/80 antibody (1:200; cat# 70076, Cell Signaling Technology). Signal detection was performed using diaminobenzidine as a chromogen (Ventana Medical Systems). Fluorescently labeled slides were scanned on the Vectra Polaris (Akoya Biosciences) at 20× magnification using appropriate exposure times. The data from the multispectral camera were analyzed by the imaging InForm software (Akoya Biosciences). The area of F4/80 staining was analyzed using the ImageJ software.
Human NASH Study and RNA Sequencing
The KOBS study was previously described.19 The current study included 262 subjects with liver histology (55.7% without and 44.3% with different stages of fibrosis) and RNA sequencing data. Liver biopsies were obtained with ultrasonic scissors during the laparoscopic Roux-en-Y gastric bypass surgery. Overall histologic assessment of liver biopsy samples was performed by one pathologist according to standard criteria.51,52 The study protocol was approved by the Ethical Committee of Northern Savo and followed the Declaration of Helsinki. Written consent was obtained from all study subjects. Total RNA sequencing was performed for 262 liver samples from the KOBS study cohort. Briefly, RNA sequencing libraries underwent 50-nucleotide long paired-end sequencing, followed by read alignment. The gene-level count values were then normalized using a trimmed-mean of M values converted to count per million using edgeR and inverse normal transformed. Expression data were corrected for the technical and confounding factors (namely RIN, uniquely aligned reads %, 3’ bias, age, sex, and body mass index) for the subsequent analyses as described previously.19,53 Correlation of normalized gene expression data with fibrosis stage was evaluated by using partial correlation adjusted for age, body mass index, and sex.
Statistical Analysis
Differences between 2 groups were compared using the Welch t-test. P values < .05 were considered significant. Time course data were analyzed by one-way analysis of variance.
Acknowledgments
The authors thank Yonghong Meng, Zhiqiang Zhou, and Sarada Charugundla for excellent technical support. The authors are grateful to the UCLA Translational Pathology Core Laboratory (TPCL) for their outstanding service.
CRediT Authorship Contributions
Simon Hui (Conceptualization: Lead; Data curation: Lead; Formal analysis: Lead; Writing – original draft: Lead)
Lili Gong (Conceptualization: Equal; Data curation: Equal; Investigation: Equal; Writing – original draft: Equal)
Chantle Swichkow (Data curation: Equal; Investigation: Equal)
Montgomery Blencowe (Data curation: Equal; Formal analysis: Equal; Writing – original draft: Equal)
Dorota Kaminska (Data curation: Equal; Formal analysis: Equal; Writing – original draft: Equal)
Graciel Diamante (Data curation: Equal)
Calvin Pan (Data curation: Equal; Formal analysis: Equal)
Meet Dalsania (Data curation: Supporting)
Samuel French (Data curation: Equal; Formal analysis: Equal)
Clara Magyar (Data curation: Equal; Formal analysis: Equal)
Päivi Pajukanta (Funding acquisition: Equal; Resources: Equal)
Jussi Pihlajamäki (Resources: Equal; Writing – review & editing: Equal)
Kristina Boström (Resources: Lead; Writing – review & editing: Equal)
Xia Yang (Funding acquisition: Lead; Supervision: Equal)
Aldons Lusis (Conceptualization: Lead; Funding acquisition: Lead; Supervision: Lead; Writing – review & editing: Equal)
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding This work was funded by National Institutes of Health grants DK120342 and DK117850 (Aldons J. Lusis), DK117850 (Xia Yang), HG010505 and DK132775 (Päivi Pajukanta), and HL81397 and HL154548 (Kristina I. Boström).
Contributor Information
Simon T. Hui, Email: sthui@mednet.ucla.edu.
Aldons J. Lusis, Email: jlusis@mednet.ucla.edu.
References
- 1.Ratziu V., Bellentani S., Cortez-Pinto H., et al. A position statement on NAFLD/NASH based on the EASL 2009 special conference. J Hepatol. 2010;53:372–384. doi: 10.1016/j.jhep.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 2.Younossi Z., Anstee Q.M., Marietti M., et al. Global burden of NAFLD and NASH: trends, predictions, risk factors and prevention. Nat Rev Gastroenterol Hepatol. 2018;15:11–20. doi: 10.1038/nrgastro.2017.109. [DOI] [PubMed] [Google Scholar]
- 3.Sookoian S., Pirola C.J. Genetic predisposition in nonalcoholic fatty liver disease. Clin Mol Hepatol. 2017;23:1–12. doi: 10.3350/cmh.2016.0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Du X., DeForest N., Majithia A.R. Human genetics to identify therapeutic targets for NAFLD: challenges and opportunities. Front Endocrinol (Lausanne) 2021;12 doi: 10.3389/fendo.2021.777075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hui S.T., Kurt Z., Tuominen I., et al. The genetic architecture of diet-induced hepatic fibrosis in mice. Hepatology. 2018;68:2182–2196. doi: 10.1002/hep.30113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chella Krishnan K., Kurt Z., Barrere-Cain R., et al. Integration of multi-omics data from mouse diversity panel highlights mitochondrial dysfunction in non-alcoholic fatty liver disease. Cell Syst. 2018;6:103–115.e7. doi: 10.1016/j.cels.2017.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zebboudj A.F., Shin V., Bostrom K. Matrix GLA protein and BMP-2 regulate osteoinduction in calcifying vascular cells. J Cell Biochem. 2003;90:756–765. doi: 10.1002/jcb.10669. [DOI] [PubMed] [Google Scholar]
- 8.Schurgers L.J., Uitto J., Reutelingsperger C.P. Vitamin K-dependent carboxylation of matrix Gla-protein: a crucial switch to control ectopic mineralization. Trends Mol Med. 2013;19:217–226. doi: 10.1016/j.molmed.2012.12.008. [DOI] [PubMed] [Google Scholar]
- 9.Price P.A., Williamson M.K. Primary structure of bovine matrix Gla protein, a new vitamin K-dependent bone protein. J Biol Chem. 1985;260:14971–14975. [PubMed] [Google Scholar]
- 10.Wallin R., Cain D., Hutson S.M., et al. Modulation of the binding of matrix Gla protein (MGP) to bone morphogenetic protein-2 (BMP-2) Thromb Haemost. 2000;84:1039–1044. [PubMed] [Google Scholar]
- 11.Yao Y., Jumabay M., Ly A., et al. A role for the endothelium in vascular calcification. Circ Res. 2013;113(5):495–504. doi: 10.1161/CIRCRESAHA.113.301792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beazley K.E., Reckard S., Nurminsky D., et al. Two sides of MGP null arterial disease: chondrogenic lesions dependent on transglutaminase 2 and elastin fragmentation associated with induction of adipsin. J Biol Chem. 2013;288:31400–31408. doi: 10.1074/jbc.M113.495556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaartinen M.T., Murshed M., Karsenty G., et al. Osteopontin upregulation and polymerization by transglutaminase 2 in calcified arteries of Matrix Gla protein-deficient mice. J Histochem Cytochem. 2007;55:375–386. doi: 10.1369/jhc.6A7087.2006. [DOI] [PubMed] [Google Scholar]
- 14.Munroe P.B., Olgunturk R.O., Fryns J.P., et al. Mutations in the gene encoding the human matrix Gla protein cause Keutel syndrome. Nat Genet. 1999;21:142–144. doi: 10.1038/5102. [DOI] [PubMed] [Google Scholar]
- 15.Luo G., Ducy P., McKee M.D., et al. Spontaneous calcification of arteries and cartilage in mice lacking matrix GLA protein. Nature. 1997;386:78–81. doi: 10.1038/386078a0. [DOI] [PubMed] [Google Scholar]
- 16.Yao Y., Yao J., Radparvar M., et al. Reducing Jagged 1 and 2 levels prevents cerebral arteriovenous malformations in matrix Gla protein deficiency. Proc Natl Acad Sci U S A. 2013;110:19071–19076. doi: 10.1073/pnas.1310905110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bennett B.J., Davis R.C., Civelek M., et al. Genetic architecture of atherosclerosis in mice: a systems genetics analysis of common inbred strains. PLoS Genet. 2015;11 doi: 10.1371/journal.pgen.1005711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hasin Y., Seldin M., Lusis A. Multi-omics approaches to disease. Genome Biol. 2017;18:83. doi: 10.1186/s13059-017-1215-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mannisto V., Kaminska D., Kakela P., et al. Protein phosphatase 1 regulatory subunit 3B genotype at rs4240624 has a major effect on gallbladder bile composition. Hepatol Commun. 2021;5:244–257. doi: 10.1002/hep4.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Azimifar S.B., Nagaraj N., Cox J., et al. Cell-type-resolved quantitative proteomics of murine liver. Cell Metab. 2014;20:1076–1087. doi: 10.1016/j.cmet.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 21.Shu L., Zhao Y., Kurt Z., et al. Mergeomics: multidimensional data integration to identify pathogenic perturbations to biological systems. BMC Genomics. 2016;17:874. doi: 10.1186/s12864-016-3198-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kesteloot F., Desmouliere A., Leclercq I., et al. ADAM metallopeptidase with thrombospondin type 1 motif 2 inactivation reduces the extent and stability of carbon tetrachloride-induced hepatic fibrosis in mice. Hepatology. 2007;46:1620–1631. doi: 10.1002/hep.21868. [DOI] [PubMed] [Google Scholar]
- 23.Luo L., Wang C.C., Song X.P., et al. Suppression of SMOC2 reduces bleomycin (BLM)-induced pulmonary fibrosis by inhibition of TGF-beta1/SMADs pathway. Biomed Pharmacother. 2018;105:841–847. doi: 10.1016/j.biopha.2018.03.058. [DOI] [PubMed] [Google Scholar]
- 24.Pan Z., Yang K., Wang H., et al. MFAP4 deficiency alleviates renal fibrosis through inhibition of NF-kappaB and TGF-beta/Smad signaling pathways. FASEB J. 2020;34:14250–14263. doi: 10.1096/fj.202001026R. [DOI] [PubMed] [Google Scholar]
- 25.Wang M., Gong Q., Zhang J., et al. Characterization of gene expression profiles in HBV-related liver fibrosis patients and identification of ITGBL1 as a key regulator of fibrogenesis. Sci Rep. 2017;7 doi: 10.1038/srep43446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang A., Yan X., Xu H., et al. Selective depletion of hepatic stellate cells-specific LOXL1 alleviates liver fibrosis. FASEB J. 2021;35 doi: 10.1096/fj.202100374R. [DOI] [PubMed] [Google Scholar]
- 27.Zhuang L., Ge X., Hu X., et al. miR-543 regulates high glucose-induced fibrosis and autophagy in diabetic nephropathy by targeting TSPAN8. BMC Nephrol. 2022;23:89. doi: 10.1186/s12882-022-02716-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jerala M., Hauptman N., Kojc N., et al. Expression of fibrosis-related genes in liver and kidney fibrosis in comparison to inflammatory bowel diseases. Cells. 2022;11:314. doi: 10.3390/cells11030314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kozumi K., Kodama T., Murai H., et al. Transcriptomics identify thrombospondin-2 as a biomarker for NASH and advanced liver fibrosis. Hepatology. 2021;74:2452–2466. doi: 10.1002/hep.31995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y., Zhang Y., Liang H., et al. Serum N-terminal DDR1: a novel diagnostic marker of liver fibrosis severity. J Clin Transl Hepatol. 2021;9:702–710. doi: 10.14218/JCTH.2021.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsumoto M., Hada N., Sakamaki Y., et al. An improved mouse model that rapidly develops fibrosis in non-alcoholic steatohepatitis. Int J Exp Pathol. 2013;94:93–103. doi: 10.1111/iep.12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Angulo P., Kleiner D.E., Dam-Larsen S., Adams L.A., et al. Liver fibrosis, but no other histologic features, is associated with long-term outcomes of patients with nonalcoholic fatty liver disease. Gastroenterology. 2015;149:389–397.e10. doi: 10.1053/j.gastro.2015.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hagstrom H., Nasr P., Ekstedt M., et al. Fibrosis stage but not NASH predicts mortality and time to development of severe liver disease in biopsy-proven NAFLD. J Hepatol. 2017;67:1265–1273. doi: 10.1016/j.jhep.2017.07.027. [DOI] [PubMed] [Google Scholar]
- 34.Rinella M.E., Elias M.S., Smolak R.R., et al. Mechanisms of hepatic steatosis in mice fed a lipogenic methionine choline-deficient diet. J Lipid Res. 2008;49:1068–1076. doi: 10.1194/jlr.M800042-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boland M.L., Oro D., Tolbol K.S., et al. Towards a standard diet-induced and biopsy-confirmed mouse model of non-alcoholic steatohepatitis: impact of dietary fat source. World J Gastroenterol. 2019;25:4904–4920. doi: 10.3748/wjg.v25.i33.4904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miyata K.N., Nast C.C., Dai T., et al. Renal matrix Gla protein expression increases progressively with CKD and predicts renal outcome. Exp Mol Pathol. 2018;105:120–129. doi: 10.1016/j.yexmp.2018.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yao Y., Zebboudj A.F., Shao E., et al. Regulation of bone morphogenetic protein-4 by matrix GLA protein in vascular endothelial cells involves activin-like kinase receptor 1. J Biol Chem. 2006;281:33921–33930. doi: 10.1074/jbc.M604239200. [DOI] [PubMed] [Google Scholar]
- 38.David L., Mallet C., Mazerbourg S., et al. Identification of BMP9 and BMP10 as functional activators of the orphan activin receptor-like kinase 1 (ALK1) in endothelial cells. Blood. 2007;109:1953–1961. doi: 10.1182/blood-2006-07-034124. [DOI] [PubMed] [Google Scholar]
- 39.Li P., Li Y., Zhu L., et al. Targeting secreted cytokine BMP9 gates the attenuation of hepatic fibrosis. Biochim Biophys Acta Mol Basis Dis. 2018;1864:709–720. doi: 10.1016/j.bbadis.2017.12.008. [DOI] [PubMed] [Google Scholar]
- 40.Hata A., Lagna G., Massague J., et al. Smad6 inhibits BMP/Smad1 signaling by specifically competing with the Smad4 tumor suppressor. Genes Dev. 1998;12:186–197. doi: 10.1101/gad.12.2.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van den Maagdenberg A.M., Hofker M.H., Krimpenfort P.J., et al. Transgenic mice carrying the apolipoprotein E3-Leiden gene exhibit hyperlipoproteinemia. J Biol Chem. 1993;268:10540–10545. [PubMed] [Google Scholar]
- 42.Yao Y., Jumabay M., Wang A., et al. Matrix Gla protein deficiency causes arteriovenous malformations in mice. J Clin Invest. 2011;121:2993–3004. doi: 10.1172/JCI57567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parks B.W., Sallam T., Mehrabian M., et al. Genetic architecture of insulin resistance in the mouse. Cell Metab. 2015;21:334–347. doi: 10.1016/j.cmet.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bennett B.J., Farber C.R., Orozco L., et al. A high-resolution association mapping panel for the dissection of complex traits in mice. Genome Res. 2010;20:281–290. doi: 10.1101/gr.099234.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lippert C., Listgarten J., Liu Y., et al. FaST linear mixed models for genome-wide association studies. Nat Methods. 2011;8:833–835. doi: 10.1038/nmeth.1681. [DOI] [PubMed] [Google Scholar]
- 46.Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rau C.D., Parks B., Wang Y., et al. High-density genotypes of inbred mouse strains: improved power and precision of association mapping. G3 (Bethesda) 2015;5:2021–2026. doi: 10.1534/g3.115.020784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Song W.M., Zhang B. Multiscale embedded gene co-expression network analysis. PLoS Comput Biol. 2015;11 doi: 10.1371/journal.pcbi.1004574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hui S.T., Parks B.W., Org E., et al. The genetic architecture of NAFLD among inbred strains of mice. Elife. 2015;4 doi: 10.7554/eLife.05607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhai X., Wang W., Dou D., et al. A novel technique to prepare a single cell suspension of isolated quiescent human hepatic stellate cells. Sci Rep. 2019;9 doi: 10.1038/s41598-019-49287-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brunt E.M., Janney C.G., Di Bisceglie A.M., et al. Nonalcoholic steatohepatitis: a proposal for grading and staging the histological lesions. Am J Gastroenterol. 1999;94:2467–2474. doi: 10.1111/j.1572-0241.1999.01377.x. [DOI] [PubMed] [Google Scholar]
- 52.Kleiner D.E., Brunt E.M., Van Natta M., et al. Nonalcoholic Steatohepatitis Clinical Research Network Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41:1313–1321. doi: 10.1002/hep.20701. [DOI] [PubMed] [Google Scholar]
- 53.de Mello V.D., Sehgal R., Mannisto V., et al. Serum aromatic and branched-chain amino acids associated with NASH demonstrate divergent associations with serum lipids. Liver Int. 2021;41:754–763. doi: 10.1111/liv.14743. [DOI] [PMC free article] [PubMed] [Google Scholar]