Dear Editor,
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has caused nearly seven million deaths worldwide. Children often develop asymptomatic or mild symptoms of COVID-19.1 However, the development of life-threatening severe diseases, such as multisystem inflammatory syndrome (MIS-C), have also been reported in children.2,3 In China, children encountered a large wave of SARS-CoV-2 infection mainly caused by the BA.5.2 or BF.7 strains, after the zero-COVID policy adjustment from December 2022 to January 2023.4 In addition, those under the age of three have not been vaccinated against SARS-CoV-2. Therefore, it is imperative to understand the cross-neutralisation of SARS-CoV-2 antibodies developed in children against potentially upcoming sub-variants such as XBB, XBB.1.5, CH.1.1, EG.5, and BA.2.86.5,6
In this study, we evaluated the cross-neutralisation of serum samples from SARS-CoV-2 convalescent children collected at Shenzhen Children’s Hospital in Guangdong Province, China. The samples were tested for the neutralisation of the SARS-CoV-2 prototype (PT) and several omicron sub-variants using a pseudovirus assay (Fig. 1A).7 A total of 310 paediatric patients were divided into five groups based on vaccination dose and immunocompromised status. These included individuals who received no SARS-CoV-2 vaccination (Group 1) and one or two shots of inactivated vaccines before breakthrough infection (BTI) (Groups 2 and 3), as well as immunocompromised individuals who received either no SARS-CoV-2 vaccination before infection for various reasons (Group 4), such as haematopoietic stem cell transplantation, leukaemia, thalassaemia, or neuroblastoma, etc, or two doses of inactivated vaccines before the BTI (Group 5) as shown in Supplementary Fig. S1 and Tables S1 and S2.
Fig. 1.
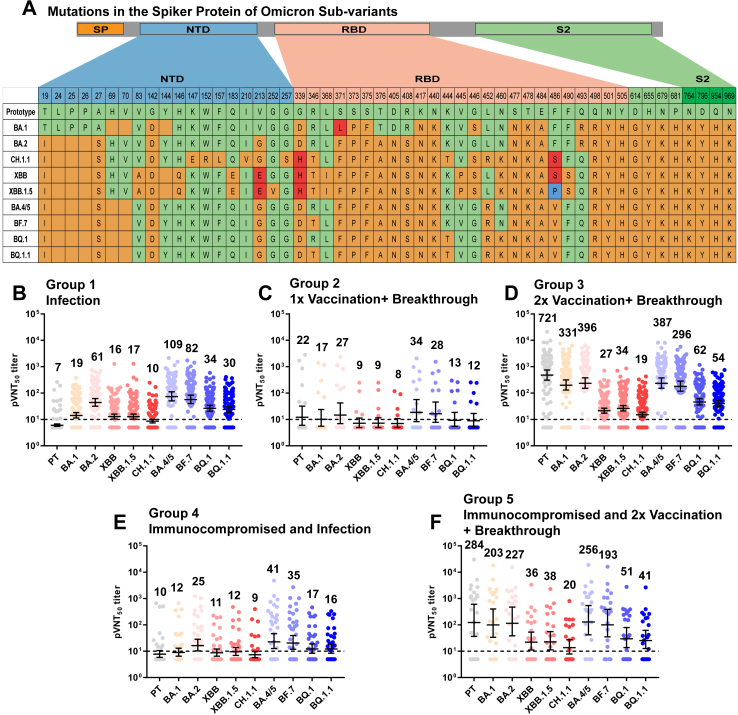
Neutralizing Antibodies against SARS-CoV-2 Variants in Convalescents’ Sera from Children. Panel A shows the mutations in the spike protein of SARS-CoV-2 PT and the omicron sub-variants analysed in this study. BA.4, and BA.5 have the same spike protein sequence; thus, they were grouped as BA.4/5. Orange indicates amino acid differences compared with PT (light green), including substitutions and deletions. The third and fourth amino acids at the same locus are highlighted in red and blue, respectively. SP, signal peptide; NTD, N-terminal domain; RBD, receptor-binding domain, S2: S2-protein subunit. Panels B–F show the 50% pseudovirus neutralisation titres (pVNT50) against SARS-CoV-2 PT and omicron BA.1, BA.2, XBB, XBB.1.5, CH.1.1, BA.4/5, BF.7, BQ.1, and BQ.1.1. Human serum was collected from convalescents of a recently passed infection wave, which was mainly caused by the omicron BA.5.2 or BF.7 sub-variant in Guangdong Province, China. Serum samples were divided into five groups according to vaccination dose and immunocompromised status. Group 1 (Infection) were individuals who received no SARS-CoV-2 vaccination before infection (n = 101). Group 2 (1x Vaccination + Breakthrough) were participants who received one dose of an inactivated virus vaccine, either CoronaVac or BBIBP-CorV (Sinopharm and Sinovac, China), before breakthrough infection (BTI) (n = 17). Group 3 (2x Vaccination + Breakthrough) were individuals vaccinated with two doses of the inactivated vaccine before the BTI (n = 134). Group 4 (Immunocompromised and Infected) were immunocompromised individuals who did not receive SARS-CoV-2 vaccination before infection (n = 36). Group 5 (Immunocompromised and 2x Vaccination + Breakthrough) were immunocompromised participants who received two doses of inactivated vaccine before BTI (n = 22). The geometric mean titre (GMT) numbers are shown at the top of each column, and the dashed line indicates the lowest detection limit (1:10). A pVNT50 value below the limit of detection was determined to be half the limit of detection.
We observed that serum samples from the 2x Vaccination + Breakthrough group (Group 3) induced higher titres of neutralising antibodies (NAb) against PT and all omicron sub-variants tested compared to the samples collected from the children who were un-vaccinated (Group 1) and the 1x Vaccination + Breakthrough group (Group 2, Fig. 1, Supplementary Figs. S2 A, C and S3). However, the neutralising titres of Group 1 were slightly higher than those of Group 2 which may be related to the limited sample size. Similarly, serum samples from the Immunocompromised and 2x Vaccination + Breakthrough group (Group 5) displayed more efficient neutralisation than samples collected from the un-vaccinated immunocompromised children (Group 4, Supplementary Figs. S2B and S3). These results indicate that the two doses of the vaccine can contribute to higher NAb titres in children with and without immunocompromised diseases. Further research is needed to determine whether vaccination alone can induce comparable levels of cross-NAbs in immunocompromised children.
In the un-vaccinated group, the NAb titres against BA.4/5 and BF.7 were similar to the highest among all variants detected. The NAb titres against PT and CH.1.1 were the lowest (16.3 and 10.8 fold lower than BA.4/5), even lower than XBB and XBB.1.5 (6.7 and 6.6 fold lower than BA.4/5) (Supplementary Table S3).
In all BTI groups, the NAb titres against PT, BA.1, BA.2, BA.4/5, and BF.7 were higher, and those against CH.1.1, XBB, and XBB.1.5, which showed the most reduced titres (2.4 to 37.5 fold lower than PT), were lower than those against BQ.1 and BQ.1.1 (Supplementary Table S3). The seropositivity against XBB, XBB.1.5 and CH.1.1 was the lowest among all strains (Supplementary Fig. S4 B, C, E). The specific mutation sites in CH.1.1 XBB and XBB.1.5 compared with PT and BA.4/5 or BF.7, such as V213E, G339H, and F486S/P, may play important roles in immune escape.8, 9, 10
Next, we sub-grouped Group 1 and Group 3 individuals by age to test whether the development of the immune system in children could contribute to SARS-CoV-2 NAb induction. In Group 1, children in the 0–1 years, 1–3 years and 3–6 years sub-groups showed similar NAb titres. The NAb titres were slightly higher in the 6–17 year-old group with no significant differences owing to the small sample size (p > 0.05, Supplementary Fig. S5). Furthermore, in Group 3, children who were 6–17 years of age had slightly higher NAb titres (not significant, p > 0.05) than those in the 3–6 years sub-group (Supplementary Fig. S6). These results suggest that natural omicron infections in infants can result in cross-NAbs at levels similar to those observed in preschool children.
In summary, our results suggest that children administered two doses of inactivated vaccines can more efficiently neutralise SARS-CoV-2 PT and omicron sub-variants. Importantly, cross-NAbs can also be induced in immunocompromised children. CH.1.1, XBB, XBB.1.5, and their sub-lineages exhibited the highest immune escape potential and are major threats to future reinfections. Taken together, these findings provide an assessment of the NAb titre in children and will help evaluate the potential risk of future reinfection and inform future vaccination regimens for children.
Contributors
J.D., G.F.G., and X.Z. conceptualised and supervised the study. X.Z. and W.P. drafted the manuscript. W.P. performed data analysis and created figures with the assistance of X.M. and K.T. H.W., X.M., and K.T. were responsible for sample collection, patient information collation, and evaluation. W.P. wrote the methodology. W.P., M.C., and Y.Z. performed pseudovirus neutralisation assays. J.D., G.F.G., and X.Z. were responsible for providing the resources. All the authors reviewed and edited the manuscript. All authors had full access to all data in the study and had the final responsibility for the decision to submit the manuscript for publication.
Declaration of interests
All authors declare no competing interests.
Acknowledgements
This work was supported by the Shenzhen Fund for Guangdong Provincial Highlevel Clinical Key Specialties (No. SZGSP012), and the Guangdong Engineering Technology Research Center for the accurate diagnosis of infection in children (Platform funding, 2021B295). Xin Zhao was supported by the National Science Foundation of China (82222040 and 82072289), Youth Innovation Promotion Association of the Chinese Academy of Sciences (2020092), and Beijing Nova Program of Science and Technology (20220484181). This study was supported by grants from the China Postdoctoral Science Foundation (grant numbers: 2022M723343 and 2023T160673). We thank Kefang Liu, Rong Zhang, and Pengyue Gao for their help with pseudovirus neutralisation assays. The authors thank Dr. Wei Zhang of the Institute of Microbiology, CAS, for technical guidance. We sincerely thank all the serum donors for providing blood samples and all those who assisted in completing the project.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.lanwpc.2023.100939.
Contributor Information
Xin Zhao, Email: zhaoxin@im.ac.cn.
George F. Gao, Email: gaof@im.ac.cn.
Jikui Deng, Email: szsetyydeng@sina.com.
Appendix ASupplementary data
References
- 1.Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China summary of a report of 72 314 cases from the Chinese center for disease control and prevention. JAMA. 2020;323:1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 2.Feldstein L., Rose E., Horwitz S., et al. Multisystem inflammatory syndrome in U.S. children and adolescents. N Engl J Med. 2020;383:334–346. doi: 10.1056/NEJMoa2021680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lopez L., Burgner D., Glover C., et al. Corrigendum to "lower risk of multi-system inflammatory syndrome in children (MIS-C) with the omicron variant" [The Lancet Regional Health - Western Pacific 27 (2022) 100604] Lancet Reg Health West Pac. 2023;35 doi: 10.1016/j.lanwpc.2022.100604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.COVID-19 clinical and surveillance data, 2022 to Jan 23, 2023. https://en.chinacdc.cn/news/latest/202301/t20230126_263523.html China.
- 5.Jiang X., Zhu K., Wang X., et al. Omicron BQ.1 and BQ.1.1 escape neutralisation by omicron subvariant breakthrough infection. Lancet Infect Dis. 2023;23:28–30. doi: 10.1016/S1473-3099(22)00805-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen X., Xu Y., Xie Y., et al. Protective effect of plasma neutralization from prior SARS-CoV-2 Omicron infection against BA.5 subvariant symptomatic reinfection. Lancet Reg Health West Pac. 2023;33 doi: 10.1016/j.lanwpc.2023.100758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao X., Zhang R., Qiao S., et al. Omicron SARS-CoV-2 neutralization from inactivated and ZF2001 vaccines. N Engl J Med. 2022;387(3):277–280. doi: 10.1056/NEJMc2206900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qu P., Faraone J.N., Evans J.P., et al. Enhanced evasion of neutralizing antibody response by Omicron XBB.1.5, CH.1.1, and CA.3.1 variants. Cell Rep. 2023;42(5) doi: 10.1016/j.celrep.2023.112443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Q., Iketani S., Li Z., et al. Alarming antibody evasion properties of rising SARS-CoV-2 BQ and XBB subvariants. Cell. 2023;186(2):279–286.e8. doi: 10.1016/j.cell.2022.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao Y., Jian F., Wang J., et al. Imprinted SARS-CoV-2 humoral immunity induces convergent Omicron RBD evolution. Nature. 2023;614(7948):521–529. doi: 10.1038/s41586-022-05644-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.