Abstract
The class II bacteriocins pediocin PA-1, from Pediococcus acidilactici, and lactococcin A, from Lactococcus lactis subsp. lactis bv. diacetylactis WM4 have a number of features in common. They are produced as precursor peptides containing similar amino-terminal leader sequences with a conserved processing site (Gly-Gly at positions −1 and −2). Translocation of both bacteriocins occurs via a dedicated secretory system. Because of the strong antilisterial activity of pediocin PA-1, its production by lactic acid bacteria strains adapted to dairy environments would considerably extend its application in the dairy industry. In this study, the lactococcin A secretory system was adapted for the expression and secretion of pediocin PA-1. A vector containing an in-frame fusion of sequences encoding the lcnA promoter, the lactococcin A leader, and the mature pediocin PA-1, was introduced into L. lactis IL1403. This strain is resistant to pediocin PA-1 and encodes a lactococcin translocation apparatus. The resulting L. lactis strains secreted a bacteriocin with an antimicrobial activity of approximately 25% of that displayed by the parental pediocin-producing P. acidilactici 347. A noncompetitive indirect enzyme-linked immunosorbent assay with pediocin PA-1-specific antibodies and amino-terminal amino acid sequencing confirmed that pediocin PA-1 was being produced by the heterologous host.
Bacteriocins of lactic acid bacteria have received considerable attention in recent years due to their potential application in the food industry as natural preservatives. Most interest has focused on lantibiotics (class I bacteriocins), e.g., nisin, and small heat-stable non-lanthionine-containing bacteriocins (class II) (22, 23). A major subgroup of class II bacteriocins (IIa) has been given the generic name of pediocin family (28) after its most extensively studied member, pediocin PA-1. Members of this class have a number of features in common, including a very strong antimicrobial activity against Listeria species (28). The food-borne pathogen Listeria monocytogenes is a major concern in the dairy industry since it can grow in a variety of dairy products at low temperature and pH (13). Although a pediocin PA-1-producing Lactobacillus plantarum strain has recently been isolated (12), this bacteriocin is generally produced by Pediococcus acidilactici strains of meat origin (3, 16, 18, 29, 31). Because of its antilisterial activity, the expression of pediocin PA-1 in strains of dairy origin would be highly desirable.
Pediocin PA-1 production, immunity, and secretion are determined by an operon containing four genes (26). The structural gene, pedA, encodes the pediocin PA-1 precursor, pedB specifies immunity, and the pedC and pedD gene products are membrane-bound proteins required for secretion of the active peptide (39). Homologs of these genes have been described for related peptides. Biosynthesis of the well-characterized class II bacteriocin, lactococcin A, produced by strains of Lactococcus lactis also involves four genes (20, 36, 40). In addition to the structural gene (lcnA) and immunity gene (lciA), there are two genes (lcnC and lcnD) whose products together form a transport system dedicated to the translocation of lactococcin through the host membrane. The LcnC protein belongs to the family of ATP-binding cassette transporter proteins (40), and LcnD acts as an accessory protein (14). These two proteins have considerable homology to PedD and PedC, respectively (39), suggesting that the latter proteins play a similar role in the transport of active pediocin. The two bacteriocins also share the double glycine-processing site found in many lactic acid bacteria class II bacteriocins, some lantibiotics, and the Escherichia coli bacteriocin, colicin V (17).
Van Belkum et al. (38) have recently investigated the role of leader sequences of the class II bacteriocins in the recognition of the precursor peptide by the dedicated translocation machinery of the host organism. By constructing hybrid genes, they demonstrated that the leader peptides of leucocin A, lactococcin A, and colicin V, which are cleaved at the Gly-Gly (positions −2 and −1) site, can direct the secretion of the nonrelated bacteriocin divergicin A. Our studies have focused on the class II bacteriocins pediocin PA-1 and lactococcin A. Since these peptides have a number of features in common, it might be expected that a pediocin PA-1 precursor could be secreted and processed by using the lactococcin A translocation machinery. L. lactis IL1403 is a plasmid-free strain that does not produce bacteriocin but contains chromosomal copies of genes analogous to lcnC and lcnD (33, 40). In addition, the natural resistance of this strain to pediocin PA-1 (8) makes it an ideal candidate for a production host to investigate the expression of pediocin PA-1 in lactococci.
This paper describes the development of an expression system geared to the production of heterologous peptides in L. lactis. Testing the system with pediocin PA-1 involved the construction of a vector containing an in-frame fusion between sequences encoding the lactococcin A leader and the structural part of mature pediocin PA-1. The hybrid genes were introduced into L. lactis IL1403, and the ability of these strains to produce and secrete pediocin PA-1 was investigated.
MATERIALS AND METHODS
Microbiological techniques, strains, and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1. Lactococcal strains were routinely grown in M17 medium (34) supplemented with 0.5% (wt/vol) glucose (GM17 medium) at 30°C without agitation. P. acidilactici was grown in MRS medium (Oxoid, Unipath Ltd., Basingstoke, United Kingdom) at 30°C without agitation. E. coli was grown in L broth (24) at 37°C on an orbital shaker. Agar plates were made by the addition of 1.5% (wt/vol) agar to broth media. Antibiotics were added as selective agents when appropriate: chloramphenicol, 5 μg ml−1 for lactococci and 15 μg ml−1 for E. coli, and ampicillin, 200 μg ml−1.
TABLE 1.
Bacterial strains and plasmid used in this study
| Strain | Relevant characteristics | Source or reference |
|---|---|---|
| L. lactis subsp. lactis bv. diacetylactis WM4 | Lactococcin A producer | 32 |
| L. lactis IL1403 | Plasmid-free; LcnA−, LciA−; containing lcnC and lcnD gene analogs; pediocin PA-1 resistant | 40 |
| L. lactis MG1614 | Plasmid free; LcnA−, LciA− | 15 |
| L. lactis FI8817 | IL1403(pFI2058) | This study |
| L. lactis FI9043 | IL1403(pFI2126) | This study |
| P. acidilactici 347 | Pediocin PA-1 producer | 31 |
| P. acidilactici 347-8 | P. acidilactici 347-cured derivative | NBIIIa |
| E. faecium P21 | Pediocin PA-1-sensitive indicator | NBIIIa |
| E. coli MC1022 | Plasmid free | 7 |
NBIII, Departamento de Nutrición y Bromatología III, Facultad de Veterinaria, Universidad Complutense de Madrid, Madrid, Spain.
Antimicrobial activity in cultures was assayed by a plate diffusion bioassay performed as previously described (11) with L. lactis MG1614 and Enterococcus faecium P21 as indicator organisms sensitive to lactococcin A and pediocin PA-1, respectively. Pediocin PA-1 production was quantified with a series of pure pediocin PA-1 standards ranging from 0 to 20 μg ml−1. The zones of inhibition were measured and plotted against the logarithm of their concentration to give a standard curve from which test supernatant concentrations were estimated.
Molecular techniques.
Plasmid DNA isolation was carried out as described by Dodd et al. (10). Restriction enzymes and other DNA-modifying enzymes from various sources were used as specified by the suppliers. Recombinant plasmids were recovered by transformation of E. coli as described previously by Dodd et al. (10) or by electroporation of L. lactis by the method of Holo and Nes (19) with the modifications used by Dodd et al. (10). Conditions used for PCR were as described by Horn et al. (21), and the primers were synthesized on an Applied Biosystems DNA synthesizer (model 381A). Fragments generated for the construction of vectors were amplified with Dynazyme (Flowgen) and cloned into pCRII (Invitrogen) before nucleotide sequence confirmation. For routine PCR screening of recombinant clones, AmpliTaq DNA polymerase (Perkin-Elmer) was used. The nucleotide sequences of PCR-generated fragments were confirmed on purified plasmid DNA with an Applied Biosystems DNA sequencer (model 373A) and the manufacturer’s Taq DyeDeoxy Terminator Cycle sequencing kit.
Construction of pFI2058 (containing lcnA and lciA).
Lactococcin A genes were introduced into the shuttle vector pTG262 by PCR amplification of the relevant segment of DNA from L. lactis WM4 with flanking primers plcn1 (5′-CAATCAGTAGAGTTATTAACATTTG-3′) and plcn2 (5′-GATTTAAAAAGACATTCGATTATTAT-3′) (Fig. 1a). This generated a 770-bp PCR fragment containing the lcnA and lciA genes with the upstream promoter and downstream putative transcription terminator sites (33). The fragment was cloned into pCRII, and the nucleotide sequence of the inserted DNA was confirmed. The PCR-generated lcnA and lciA genes were recovered as an EcoRI fragment and cloned into the EcoRI site of pTG262 to generate pFI2058.
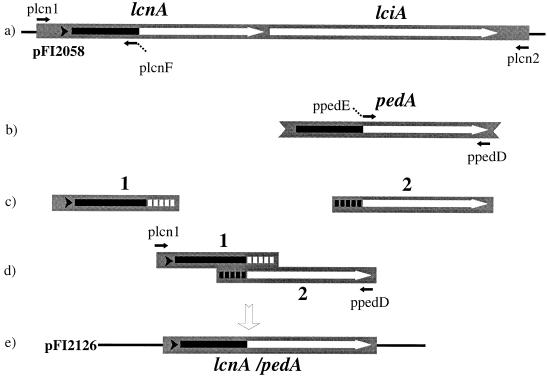
FIG. 1.
PCR strategy used for splicing the lacA and pedA genes. (a) Region of the lactococcin A operon, containing lcnA and lciA genes, cloned into pFI2058. (b) Map of the pedA gene. The thick arrows show coding regions with the amino-terminal leaders indicated in black. The small arrows above and below the maps indicate the position and direction of primers used for PCR. Nonhomologous tails on primers plcnF and ppepE are represented by dashed lines. (c) Fragments generated by the first PCR step. (d) Homologous regions in fragments 1 and 2 annealed to provide a template for the second PCR step, involving primers plcn1 and ppepD. (e) Product of the second PCR step, cloned into pFI2126. An in-frame fusion resulted in the creation of a consensus amino-terminal cleavage site (vertical arrow) between two parts of the lcnA/pedA hybrid gene.
Construction of the lcnA/pedA hybrid gene.
The technique of spliced overlap extension was used in the construction of in-frame fusions of sequences encoding the lactococcin A leader (Fig. 1a) and the structural portion of mature pediocin PA-1 (Fig. 1b). This initially involved the amplification of two DNA fragments. Primers plcn1 and plcnF (5′-CCATTACCGTAGTATTTTCCTCCGTTAGCTTC-3′) were used to amplify a 170-bp fragment, encoding the lactococcal A leader and promoter (fragment 1 [Fig. 1c]). DNA from colonies of L. lactis WM4 was used as the template. The 17 nucleotides forming a tail at the 5′ end of primer plcnF (underlined) are complementary to the amino-terminal sequences of mature pediocin PA-1, i.e., after cleavage at the Gly-Gly site of the leader peptide (26). Primers ppedD (5′-ACCCCGGGATTGATGCCAGCTC-3′) and ppedE (5′-GAAGCTAACGGAGGAAAATACTACGGTAATGG-3′) were used to amplify a 180-bp fragment 2 (Fig. 1c), comprising exclusively the part of the pedA gene that encodes mature pediocin PA-1 (26). The template was provided by DNA from colonies of P. acidilactici 347. Primer ppedE was designed with a 5′ tail corresponding to sequences within the lactococcin A leader. These 19 nucleotides (underlined) are complementary to the 3′ end of fragment 1 (Fig. 1c). Fragments 1 and 2 were diluted (1/200) in distilled water, and equal quantities were mixed. This mixture was used as the template to amplify a 312-bp fragment with primers plcn1 and ppedD (Fig. 1d). The fragment was cloned into pCRII, and nucleotide sequence analysis confirmed that it was composed of sequences corresponding precisely to the in-frame fusion of the lactococcin A leader and mature pediocin PA-1 (Fig. 1e). The hybrid gene and upstream promoter region was isolated as an EcoRI fragment and cloned into pTG262 to generate pFI2126. Transformation of L. lactis IL1403 with this recombinant plasmid generated strain FI9043.
Purification and amino acid sequencing of pediocin PA-1.
The bacteriocin produced by L. lactis FI9043 was purified from a 1-liter culture grown in MRS broth at 30°C to late logarithmic phase. The procedure, involving ammonium sulfate precipitation and, successively, cation-exchange, hydrophobic interaction, and reverse-phase chromatography (PepRPC HR5/5 fast protein liquid chromatography system; Pharmacia LKB, Uppsala, Sweden) was essentially as previously described (9, 30) except that the fraction obtained after ammonium sulfate precipitation was applied to a Sephadex G-25 gel filtration column (Pharmacia) and equilibrated with 20 mM sodium phosphate buffer (pH 5.8). The fraction displaying activity was then applied to the cation-exchange column. The active fraction, obtained after hydrophobic interaction chromatography, was applied to the reverse-phase column, and the bacteriocin was eluted with a linear gradient ranging from 10 to 60% 2-propanol containing 0.1% trifluoroacetic acid. Purification steps were performed at room temperature, and the chromatographic equipment and reagents were obtained from Pharmacia and used as specified by the supplier. The microtiter plate assay system developed by Holo et al. (20) was used to quantify the bacteriocin activity during the purification process. One bacteriocin unit was defined as the reciprocal of the highest dilution causing 50% growth inhibition of the indicator organism, Enterococcus faecium P21.
The reverse-phase fraction containing the bacteriocin was desiccated by rotary evaporation and resuspended in an equivalent volume of deionized water. The concentration of pure bacteriocin was estimated by using the molar extinction coefficient of pediocin PA-1 (an absorbance at 280 nm of 3.1 corresponds to 1.0 mg ml−1). The amino-terminal sequence of the purified bacteriocin was determined by Edman degradation with an automatic sequencer (model 47A; Applied Biosystems).
Specific detection of pediocin PA-1 by ELISA.
The production of pediocin PA-1 by strains used in this study was assessed using a noncompetitive indirect enzyme-linked immunosorbent assay (NCI-ELISA), based on the method of Bubert et al. (6). Briefly, 100 μl of pure bacteriocin samples (100 μl) was serially diluted in 0.1 M sodium carbonate-bicarbonate buffer (pH 9.6) to give a range of concentrations from 0 to 2.5 μg ml−1. Samples were incubated in 96-well polystyrene microtiter plates (Maxisorp; Nunc, Roskilde, Denmark) for 3 h at 37°C. After the coated bacteriocin was washed with phosphate-buffered saline (PBS), wells were blocked for 1 h at 37°C with 0.01 M PBS (pH 7.2) containing 1% ovalbumin (OA). A 50-μl volume of antiserum, diluted 1:1,000 in PBS, was then added, and the plates were incubated for 1 h at 37°C. The antiserum contained rabbit antibodies raised against a synthetic peptide (PH2) composed of the carboxy-terminal 11 amino acids of pediocin PA-1 (25). For colorimetric reactions, horseradish peroxidase-conjugated goat anti-rabbit antibodies (Cappel Laboratories, West Chester, Pa.), diluted 1:500 in PBS, and the substrate 2,2′-azinobis(3′-ethylbenzothiazoline-6-sulfonic acid) (Sigma, St. Louis, Mo.) were used. The absorbance was read at 405 nm in a Labsystems iEMS reader with a built-in software package for data analysis (Labsystems, Helsinki, Finland). PH2 conjugated to OA by the glutaraldehyde method (OA-PH2) (5), pure nisin (Aplin and Barrett, Trowbridge, United Kingdom), pediocin PA-1 (produced by P. acidilactici 347 and purified by the same method cited above), and the protein fraction obtained from culture supernatants of P. acidilactici 347-8 (a pediocin PA-1 nonproducer) were used as controls at the equivalent concentrations.
RESULTS
Lactococcin A expression.
The production of lactococcin A in L. lactis IL1403 involved cloning the structural gene (lcnA) and immunity gene (lciA) of L. lactis subsp. diacetylactis WM4 into the shuttle vector pTG262, to generate pFI2058 (Fig. 1a). To determine whether the host containing homologs of the lcnC and lcnD genes could complement the equivalent processing genes, missing from pFI2058, the recombinant plasmid was introduced into this strain and a bioassay was carried out on the transformants. Supernatants from L. lactis IL1403 cells harboring pFI2058 (strain FI8817) inhibited the growth of the indicator strain L. lactis MG1614, indicating that lactococcin A was being produced (data not shown). Cells carrying the vector alone showed no inhibitory effect. The level of antimicrobial activity displayed by FI8817 was approximately 80% of that of the lactococcin A-producing parental strain, L. lactis WM4.
lcnA/pedA hybrid gene.
To determine whether pediocin PA-1 could be expressed and secreted in L. lactis IL1403, using the translocation machinery of lactococcin A, a hybrid lcnA/pedA gene was constructed (Fig. 1d). Plasmid pFI2126 contains an in-frame fusion of sequences encoding the lactococcin A leader and the mature part of pediocin PA-1 and is preceded by the promoter-active region upstream of the lcnA gene (Fig. 1e). The downstream lactococcal sequences, including the lciA gene, were not included, nor was the pediocin PA-1 immunity gene (pedB) necessary because of the natural resistance of the lactococcal host to this bacteriocin.
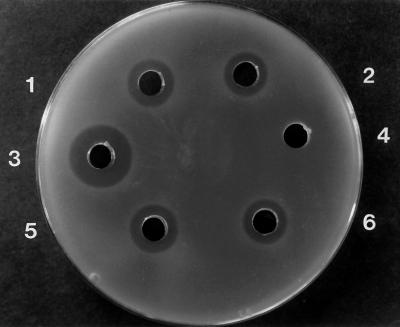
Transformation of L. lactis IL1403 with pFI2126 generated strain FI9043, which, after growth in either MRS or GM17 broth, was tested for antimicrobial activity. In plate diffusion bioassays, inhibition of the pediocin-sensitive indicator organism, E. faecium P21, was detected (Fig. 2), with cultures grown in MRS broth (final pH 4.6) displaying slightly higher antimicrobial activity than those grown in GM17 broth (final pH 5.3). The bacteriocin production level of L. lactis FI9043 was lower than that of the natural pediocin PA-1 producer, P. acidilactici 347 (Fig. 2). The zones of inhibition displayed by the L. lactis IL1403 derivatives (∼270 ng ml−1) represent approximately one-quarter of the pediocin produced by the homologous host (∼1,200 ng ml−1).
FIG. 2.
Agar diffusion bioassay for detection of bacteriocin activity against E. faecium P21. 1 and 5, L. lactis FI9043 (MRS culture); 2 and 6, L. lactis FI9043 (GM17 culture); 3, P. acidilactici 347; 4, L. lactis IL1403.
Bacteriocin purification and characterization.
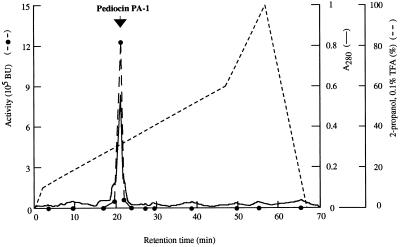
Subsequent analysis of the bacteriocin produced by L. lactis FI9043 involved purification of the active peptide with the various stages of the recovery procedure summarized in Table 2. Fractions from the first run on the reverse-phase column which showed the highest activity were collected and rechromatographed. An absorbance peak, coincident with the activity peak, was observed (Fig. 3). The final specific activity of the pure bacteriocin was approximately 106-fold higher than that in the crude culture supernatant, and the recovery was 617%.
TABLE 2.
Purification of pediocin PA-1 from L. lactis FI9043
| Purification stage | Vol (ml) | Total A280a | Total activity (BU) | Sp actb | Increase in sp act (fold) | Yield (%) |
|---|---|---|---|---|---|---|
| Culture supernatant | 1,000 | 28,500 | 202,295 | 7 | 1 | 100 |
| Fraction: | ||||||
| I. Ammonium sulfate precipitation | 100 | 1,490 | 92,494 | 62 | 8.7 | 46 |
| II. Gel filtration chromatography | 200 | 700 | 96,701 | 138 | 19.7 | 48 |
| III. Cation-exchange chromatography | 50 | 29.3 | 41,142 | 1,404 | 198 | 20 |
| IV. Hydrophobic-interaction chromatography | 10 | 12.6 | 1,316,750 | 104,090 | 14,681 | 650 |
| V. Reverse-phase chromatography | 0.7 | 0.16 | 1,248,026 | 7,849,220 | 1,107,083 | 617 |
Total A280 equals the optical density at 280 nm multiplied by the volume (in milliliters).
Specific activity is bacteriocin units (BU) per milliliter divided by the optical density at 280 nm.
FIG. 3.
Reverse-phase chromatography of pediocin PA-1. The amount applied to the column was obtained from a 1-liter culture of L. lactis FI9043. BU, bacteriocin units.
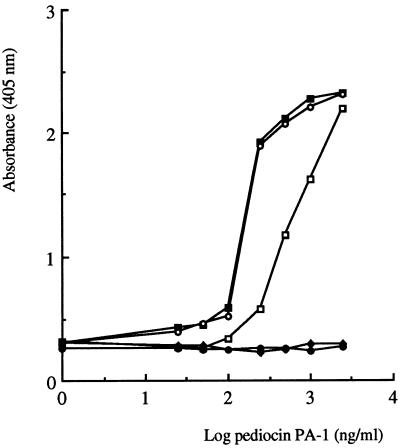
Further characterization of the bacteriocin was carried out by NCI-ELISAs with specific anti-pediocin PA-1 antibodies. L. lactis FI9043 crude culture supernatants did not cross-react with the antibodies despite exhibiting antimicrobial activity (Fig. 2). This is due to the calculated bacteriocin level in the supernatants being below the sensitivity level of the pediocin PA-1 immunoassay (25). However, a strong reactivity with the antibodies was observed when purified bacteriocin from L. lactis FI9043 was tested at concentrations greater than 500 ng ml−1, indicating that this host strain was producing pediocin PA-1 (Fig. 4). Additional data supporting this result was supplied by amino-terminal sequence analysis of the purified peptide. The first 6 residues at the amino-terminal end of the secreted peptide were KYYGNG, which is the correct sequence for the amino terminus of pediocin PA-1 and one that distinguishes it from lactococcin A. Moreover, this result established that correct processing of the hybrid precursor peptide had occurred and was consistent with the heterologous production of pediocin from L. lactis FI9043.
FIG. 4.
Standard curves for pediocin PA-1 purified from P. acidilactici 347 (▪) and L. lactis FI9043 (○), OA-PH2 (□), nisin (•), and the protein fraction obtained from culture supernatants of P. acidilactici 347-8, a P. acidilactici 347-cured derivative that does not produce pediocin PA-1 (⧫), as determined by NCI-ELISA with anti-pediocin PA-1 rabbit antibodies. Each datum point represents the average value of triplicate determinations in a single microtiter plate.
DISCUSSION
An expression system for heterologous peptides was developed in L. lactis IL1403, based on the genes and transcription signals required for lactococcin A production. In this host, the translocation functions (lcnC and lcnD) necessary for processing and secretion of lactococcins are provided by chromosomal gene analogs (33, 40). Hence, expression of the lcnA and lciA genes is the minimum requirement for production of lactococcin A.
The flexibility of the translocatory apparatus of class II bacteriocins was recently demonstrated by van Belkum et al. (38). Gene fusions were generated in which sequences encoding the leader peptides of leucocin A, lactococcin A, and colicin V were fused to divergicin A, an alternative bacteriocin that is secreted via the general secretion pathway of the cells (42). The different leader peptides were able to direct the secretion of divergicin in Leuconostoc gelidum, L. lactis, and E. coli, respectively (i.e., the homologous hosts). Furthermore, certain host-vector combinations gave rise to the production of divergicin when the leader peptides were used in heterologous hosts. The same strategy was also used for the production of colicin V from L. lactis IL1403. In this case, the E. coli gene was fused to sequences encoding the leucocin A leader peptide (38). The various components of the class II translocatory apparatus are not universally interchangeable indicating that some leader peptides are poorly recognized by heterologous ATP-binding cassette transporter proteins (35, 38). Allison et al. (2) have shown that both peptides of the two-component lactacin F complex can use the secretion machinery of Carnobacterium piscicola LV17, a strain that produces carnobacteriocins A, BM1, and B2 (1). The fact that the amino-terminal leaders of these carnobacteriocins and lactacin F peptides have the highest degree of homology among class II bacteriocins may have facilitated the secretion of lactacin F peptides in this heterologous host. In contrast, the translocatory apparatus for lactococcin A was not able to bring about secretion of leucocin A in L. lactis (35).
Pediocin PA-1 and lactococcin A are both class II bacteriocins and hence are likely candidates for expression and secretion via a heterologous translocatory apparatus. A lcnA/pedA hybrid gene was constructed by substituting the nucleotide sequences downstream of the lactococcin A Gly-Gly cleavage site with the equivalent region of the pedA gene. This gene, expressed in L. lactis FI9043, gave rise to antimicrobial activity against the pediocin-sensitive strain E. faecium P21 (Fig. 2). Confirmation that this strain was producing pediocin PA-1 came from amino-terminal sequencing of the purified product and also from immunoanalysis with antibodies which specifically recognize pediocin PA-1 (Fig. 4). This established that the lactococcin A leader peptide was capable of directing the secretion of pediocin PA-1 from L. lactis IL1403 and that correct processing of this leader peptide had occurred at the consensus cleavage site with release of mature pediocin into the growth medium.
Chikindas et al. (8) have described a similar IL1403 expression system in which the four ped determinants were cloned into a lactococcal vector. In this strain, secretion of pediocin PA-1, directed by its own pedA-encoded leader, was detected only when the ped operon was under the control of a lactococcal promoter. Under these conditions, the pediocin PA-1 yield was less than 1% of the production level by the parental Pediococcus strain. This suggests that in L. lactis, lactococcin A-directed secretion of pediocin PA-1 is more efficient than the equivalent process directed by the normal pediocin leader sequence. It was possible to increase the relative level of pediocin PA-1 production to approximately 50% when using its own dedicated PedCD translocatory machinery, by increasing the copy number of the ped operon, contained on the plasmid, in a specifically mutated lactococcal host (8).
The reduced level of pediocin PA-1 production in the L. lactis IL1403 derivative described here (∼25% of that in the parental pediococcal strain) may be attributed to the low copy number of the chromosomal lcnC and lcnD gene analogs, resulting in less efficient secretion of the bacteriocin (33). Similar observations have been presented by van Belkum et al. (37) and Holo et al. (20), who both reported a reduction in the yield of lactococcin A expressed in an IL1403 derivative. The recent analysis of the IL1403 secretion system indicated that these genes are not identical to the equivalent lactococcin A translocatory machinery (40). This may result in only partial complementation of the lcnC and lcnD genes, with less efficient processing of the bacteriocin using this secretory system. It has been reported that when the dedicated lcnC and lcnD genes were included in equivalent lactococcal expression systems, bacteriocin production was increased at least 10-fold (38, 40). The possibility that the introduction of these plasmid genes from the lactococcin A-producing strain L. lactis WM4 (33) into FI9043 has a similar effect on pediocin PA-1 production is being investigated.
Culture pH may also play a role in the reduced yield of pediocin PA-1 from the heterologous lactococcal host. In pediocin PA-1 bioassays involving L. lactis FI9043, larger inhibition zones were generated from supernatants of cultures grown in MRS broth (final pH 4.6) than from those in GM17 broth (final pH 5.3) (Fig. 2). It has been reported that processing of the prepediocin to active pediocin PA-1 by P. acidilactici strains can take place efficiently only when the final pH of the culture medium is less than or equal to 5.0 (4, 12, 43). In contrast, Ennahar et al. (12) reported that the production of pediocin AcH-1 from Lactobacillus plantarum WHE 92 was not reduced when the pH was raised to 6.0. It was suggested that the efficiency of processing of prepediocin to pediocin may differ in Lactobacillus and Pediococcus species (12). This observation has important industrial implications, since a pH of 5.0 and above is often encountered in dairy products.
Pediococci are usually associated with vegetable and meat material and are used commercially in the fermentation of vegetables and meat. Pediocin PA-1 is a bacteriocin with a broad inhibitory spectrum and is particularly effective in combating the growth of Listeria monocytogenes. However, pediococci are poorly adapted for colonizing foods in which they do not naturally reside (27) and are therefore not the ideal organisms for controlling the growth of L. monocytogenes in dairy products. In this study, we have demonstrated heterologous expression of pediocin PA-1 in L. lactis IL1403 containing a fusion of the pediocin PA-1 structural gene (devoid of the sequence encoding its natural leader peptide) behind the sequence encoding the lactococcin A leader. Expression and secretion of this bacteriocin in lactococci provides a way in which the beneficial properties of pediocin PA-1 production can be applied to the dairy industry. This approach could be extended with the aim of expressing other bacteriocins, peptides, or proteins of interest (hybrid bacteriocin molecules with a broader antimicrobial spectrum, cecrapin, yeast killer toxin) in food-grade strains. A strategy involving a dedicated secretory system could also be used to investigate vaccine delivery vehicles in mucosal environments by using lactic acid bacteria (41).
ACKNOWLEDGMENTS
This work was partially supported by grants ALI94-1026 and ALI97-0559 from the Comisión Interministerial de Ciencia y Tecnología (CICYT), Madrid, Spain, and by contract BIOT-CT94-3055 from the Commission of the European Communities. M.I.M. is a research working under the European Contract, and J.M.M. holds a fellowship from the Comunidad Autónoma de Madrid, Spain.
We are grateful to L. L. McKay (Dept. of Food Science and Nutrition, University of Minnesota) for supplying strain L. lactis WM4 and to J. Vázquez (Protein Chemistry Facility, Centro de Biología Molecular Severo Ochoa, Madrid, Spain) for performing the amino-terminal sequence analysis.
REFERENCES
- 1.Ahn C, Stiles M E. Plasmid-associated bacteriocin production by a strain of Carnobacterium piscicola from meat. Appl Environ Microbiol. 1990;56:2503–2510. doi: 10.1128/aem.56.8.2503-2510.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allison G E, Worobo R W, Stiles M E, Klaenhammer T R. Heterologous expression of the lactacin F peptides by Carnobacterium piscicola LV17. Appl Environ Microbiol. 1995;61:1371–1377. doi: 10.1128/aem.61.4.1371-1377.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhunia A K, Johnson M C, Ray B. Purification, characterization and microbial spectrum of a bacteriocin produced by Pediococcus acidilactici. J Appl Bacteriol. 1988;65:261–268. doi: 10.1111/j.1365-2672.1988.tb01893.x. [DOI] [PubMed] [Google Scholar]
- 4.Biswas S R, Ray P, Johnson M C, Ray B. Influence of growth conditions on the production of a bacteriocin, pediocin AcH, by Pediococcus acidilactici H. Appl Environ Microbiol. 1991;57:1265–1267. doi: 10.1128/aem.57.4.1265-1267.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Briand J P, Muller S, Van Regenmortel M H V. Synthetic peptides as antigens: pitfalls of conjugation methods. J Immunol Methods. 1985;78:59–69. doi: 10.1016/0022-1759(85)90329-1. [DOI] [PubMed] [Google Scholar]
- 6.Bubert A, Schubert P, Köhler S, Frank R, Goebel W. Synthetic peptides derived from the Listeria monocytogenes p60 protein as antigens for the generation of polyclonal antibodies specific for secreted cell-free L. monocytogenes p60 proteins. Appl Environ Microbiol. 1994;60:3120–3127. doi: 10.1128/aem.60.9.3120-3127.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casadabad M J, Cohen S N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980;138:179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- 8.Chikindas M L, Venema K, Ledeboer A M, Venema G, Kok J. Expression of lactococcin A and pediocin PA-1 in heterologous hosts. Lett Appl Microbiol. 1995;21:183–189. doi: 10.1111/j.1472-765x.1995.tb01037.x. [DOI] [PubMed] [Google Scholar]
- 9.Cintas L M, Rodríguez J M, Fernández M F, Sletten K, Nes I F, Hernández P E, Holo H. Isolation and characterization of pediocin L50, a new bacteriocin from Pediococcus acidilactici with a broad inhibitory spectrum. Appl Environ Microbiol. 1995;61:2643–2648. doi: 10.1128/aem.61.7.2643-2648.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dodd H M, Horn N, Gasson M J. Analysis of the genetic determinant for production of the peptide antibiotic nisin. J Gen Microbiol. 1990;136:555–566. doi: 10.1099/00221287-136-3-555. [DOI] [PubMed] [Google Scholar]
- 11.Dodd H M, Horn N, Zhang H, Gasson M J. A lactococcal expression system for engineered nisins. Appl Environ Microbiol. 1992;58:3683–3693. doi: 10.1128/aem.58.11.3683-3693.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ennahar S, Aoude-Werner D, Sorokine O, van Dorsselaer A, Bringel F, Hubert J C, Hasselmann C. Production of pediocin AcH by Lactobacillus plantarum WHE 92 isolated from cheese. Appl Environ Microbiol. 1996;62:4381–4387. doi: 10.1128/aem.62.12.4381-4387.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farber J M, Peterkin P I. Listeria monocytogenes, a food-borne pathogen. Microbiol Rev. 1991;55:476–511. doi: 10.1128/mr.55.3.476-511.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franke C M, Leenhouts K J, Haandrikman A J, Kok J, Venema G, Venema K. Topology of LcnD, a protein implicated in the transport of bacteriocins from Lactococcus lactis. J Bacteriol. 1996;178:1766–1769. doi: 10.1128/jb.178.6.1766-1769.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gasson M J. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J Bacteriol. 1983;154:1–9. doi: 10.1128/jb.154.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gonzalez C F, Kunka B S. Plasmid-associated bacteriocin production and sucrose fermentation in Pediococcus acidilactici. Appl Environ Microbiol. 1987;53:2534–2538. doi: 10.1128/aem.53.10.2534-2538.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Havarstein L S, Holo H, Nes I F. The leader peptide of colicin V shares consensus sequences with leader peptide that are common among peptide bacteriocins produced by Gram-positive bacteria. Microbiology. 1994;140:2383–2389. doi: 10.1099/13500872-140-9-2383. [DOI] [PubMed] [Google Scholar]
- 18.Henderson J T, Chopko A L, Dyck van Wassenaar P D. Purification and primary structure of pediocin PA-1, produced by Pediococcus acidilactici PAC-1.0. Arch Biochem Biophys. 1992;295:5–12. doi: 10.1016/0003-9861(92)90480-k. [DOI] [PubMed] [Google Scholar]
- 19.Holo H, Nes I F. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl Environ Microbiol. 1989;55:3119–3123. doi: 10.1128/aem.55.12.3119-3123.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holo H, Nilssen Ø, Nes I F. Lactococcin A, a new bacteriocin from Lactococcus lactis subsp. cremoris: isolation and characterization of the protein and its gene. J Bacteriol. 1991;173:3879–3887. doi: 10.1128/jb.173.12.3879-3887.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horn N, Swindell S, Dodd H M, Gasson M J. Nisin biosynthesis genes are encoded by a novel conjugative transposon. Mol Gen Genet. 1991;228:129–135. doi: 10.1007/BF00282457. [DOI] [PubMed] [Google Scholar]
- 22.Jack R W, Tagg J R, Ray B. Bacteriocins of gram-positive bacteria. Microbiol Rev. 1995;59:171–200. doi: 10.1128/mr.59.2.171-200.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klaenhammer T R. Genetics of bacteriocins produced by lactic acid bacteria. FEMS Microbiol Rev. 1993;12:39–86. doi: 10.1111/j.1574-6976.1993.tb00012.x. [DOI] [PubMed] [Google Scholar]
- 24.Lennox E S. Transduction of linked genetic characters of the host bacteriophage P1. Virology. 1955;1:190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- 25.Martínez, J. M., J. M. Rodríguez, and P. E. Hernández. Unpublished data.
- 26.Marugg J D, Gonzalez C F, Kunka B S, Ledeboer A M, Pucci M J, Toonen M Y, Walker S A, Zoetmulder L C M, Vandenbergh P A. Cloning, expression, and nucleotide sequence of genes involved in production of pediocin PA-1, a bacteriocin from Pediococcus acidilactici PAC1.0. Appl Environ Microbiol. 1992;58:2360–2367. doi: 10.1128/aem.58.8.2360-2367.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mundt J O, Beattie W G, Wieland F R. Pediococci residing in plants. J Bacteriol. 1969;98:938–942. doi: 10.1128/jb.98.3.938-942.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nes I F, Diep D B, Havarstein L S, Brurberg M B, Eijsink V, Holo H. Biosynthesis of bacteriocins in lactic acid bacteria. Antonie Leeuwenhoek Int J Gen Mol Microbiol. 1996;70:113–128. doi: 10.1007/BF00395929. [DOI] [PubMed] [Google Scholar]
- 29.Nieto Lozano J C, Nissen-Meyer J, Sletten K, Pelaez C, Nes I F. Purification and amino acid sequence of a bacteriocin produced by Pediococcus acidilactici. J Gen Microbiol. 1992;138:1985–1990. doi: 10.1099/00221287-138-9-1985. [DOI] [PubMed] [Google Scholar]
- 30.Nissen-Meyer J, Holo H, Håvarstain L S, Sletten K, Nes I F. A novel lactococcal bacteriocin whose activity depends on the complementary action of two peptides. J Bacteriol. 1992;174:5686–5692. doi: 10.1128/jb.174.17.5686-5692.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodríguez J M, Cintas L M, Casaus P, Martínez M I, Suárez A, Hernández P E. Detection of pediocin PA-1-producing pediococci by rapid molecular biology techniques. Food Microbiol. 1997;14:363–371. [Google Scholar]
- 32.Scherwitz-Harmon K M, McKay L L. Restriction enzyme analysis of lactose and bacteriocin plasmids from Streptococcus lactis subsp. diacetylactis strain WM4 and cloning of BclI fragments coding for bacteriocin production. Appl Environ Microbiol. 1987;53:1171–1174. doi: 10.1128/aem.53.5.1171-1174.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stoddard G W, Petzel J P, van Belkum M J, Kok J, McKay L L. Molecular analyses of the lactococcin A gene cluster from Lactococcus lactis subsp. lactis biovar diacetylactis WM4. Appl Environ Microbiol. 1992;58:1952–1961. doi: 10.1128/aem.58.6.1952-1961.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Terzaghi B E, Sandine W E. Improved medium for lactic streptococci and their bacteriophages. Appl Environ Microbiol. 1975;29:807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Belkum M J, Stiles M E. Molecular characterization of genes involved in the production of the bacteriocin leucocin A from Leuconostoc gelidum. Appl Environ Microbiol. 1995;61:3573–3579. doi: 10.1128/aem.61.10.3573-3579.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Belkum M J, Hayema B J, Geis A, Kok J, Venema G. Cloning of two bacteriocin genes from a lactococcal bacteriocin plasmid. Appl Environ Microbiol. 1989;55:1187–1191. doi: 10.1128/aem.55.5.1187-1191.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Belkum M J, Hayema B J, Jeeninga R E, Kok J, Venema G. Organization and nucleotide sequence of two lactococcal bacteriocin operons. Appl Environ Microbiol. 1991;57:492–498. doi: 10.1128/aem.57.2.492-498.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Belkum M J, Worobo R W, Stiles M E. Double-glycine-type leader peptides direct secretion of bacteriocins by ABC transporters: colicin V secretion in Lactococcus lactis. Mol Microbiol. 1997;23:1293–1301. doi: 10.1046/j.1365-2958.1997.3111677.x. [DOI] [PubMed] [Google Scholar]
- 39.Venema K, Kok J, Marugg J D, Toonen M Y, Ledeboer A M, Venema G, Chikindas M L. Functional analysis of the pediocin operon of Pediococcus acidilactici PAC1.0: PedB is the immunity protein and PedD is the precursor processing enzyme. Mol Microbiol. 1995;17:515–522. doi: 10.1111/j.1365-2958.1995.mmi_17030515.x. [DOI] [PubMed] [Google Scholar]
- 40.Venema K, Dost M H R, Beun P A H, Haandrikman A J, Venema G, Kok J. The genes for secretion and maturation of lactococcins are located on the chromosome of Lactococcus lactis IL1403. Appl Environ Microbiol. 1996;62:1689–1692. doi: 10.1128/aem.62.5.1689-1692.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wells J M, Robinson K, Chamberlain L M, Schofield K M, Le Page R W F. Lactic acid bacteria as vaccine delivery vehicles. Antonie Leeuwenhoek. 1996;70:317–330. doi: 10.1007/BF00395939. [DOI] [PubMed] [Google Scholar]
- 42.Worobo R W, van Belkum M J, Sailer M, Roy K L, Vederas J C, Stiles M E. A signal peptide secretion-dependent bacteriocin from Carnobacterium divergens. J Bacteriol. 1995;177:3143–3149. doi: 10.1128/jb.177.11.3143-3149.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang R, Ray B. Factors influencing production of bacteriocins by lactic acid bacteria. Food Microbiol. 1994;11:281–291. [Google Scholar]