Abstract
Sphingomonas (Pseudomonas) paucimobilis SYK-6 is able to grow on 5,5′-dehydrodivanillic acid (DDVA), syringate, vanillate, and other dimeric model compounds of lignin as a sole carbon source. Nitrosoguanidine mutagenesis of S. paucimobilis SYK-6 was performed, and two mutants with altered DDVA degradation pathways were isolated. The mutant strain NT-1 could not degrade DDVA, but could degrade syringate, vanillate, and 2,2′,3′-trihydroxy-3-methoxy-5,5′-dicarboxybiphenyl (OH-DDVA). Strain DC-49 could slowly assimilate DDVA, but could degrade neither vanillate nor syringate, although it could degrade protocatechuate and 3-O-methylgallate. A complementing DNA fragment of strain DC-49 was isolated from the cosmid library of strain SYK-6. The minimum DNA fragment complementing DC-49 was determined to be the 1.8-kbp insert of pKEX2.0. Sequencing analysis showed an open reading frame of 1,671 bp in this fragment, and a similarity search indicated that the deduced amino acid sequence of this open reading frame had significant similarity (60%) to the formyltetrahydrofolate synthetase of Clostridium thermoaceticum.
There are many phenylmethylethers in natural aromatic compounds such as lignin. There have been many investigations of the microbial degradation of phenylmethylethers (1, 4–9, 30, 32, 34). These investigations have revealed two- or three-component enzyme systems that include terminal enzymes, such as iron-sulfur proteins, and cytochrome P-450-like enzymes (5–8). The former enzymes require NADH to carry out an O demethylation reaction (5, 7). C-O bound cleavage reactions for phenylmethylether under anaerobic conditions have also been investigated (1, 4, 9, 30, 32, 34). Berman and Frazer (4) and Stupperich and Konle (30) have noted that dl-tetrahydrofolate (THF) and ATP are essential to O demethylation reactions. However, little genetic information about O demethylation systems is available. Brunel and Davison (6) reported that the genes vanA and vanB of Pseudomonas sp. strain ATCC 19151 code for protein components of the monooxygenase system, as in the O demethylation of vanillate.
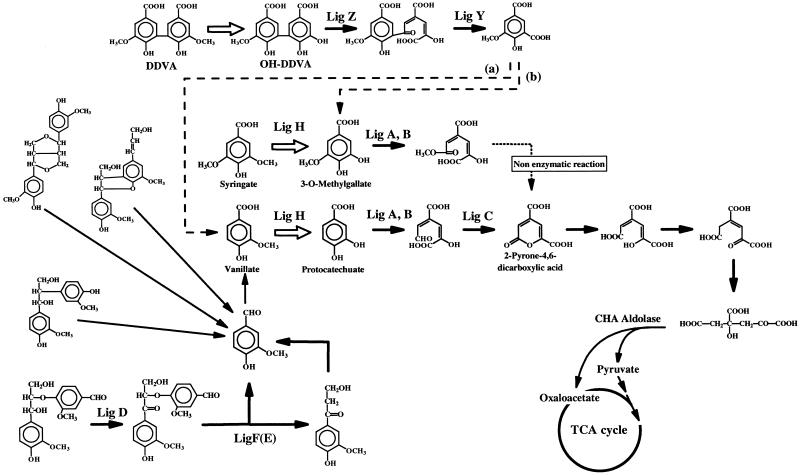
Sphingomonas (Pseudomonas) paucimobilis SYK-6, a bacterium that can grow on 5,5′-dehydrodivanillic acid (DDVA) as a sole carbon source, was isolated from pulp-bleaching wastewater in Japan. This bacterium can also grow on syringate, vanillate, and several dimeric model compounds of lignin as a sole carbon source. The metabolic pathways of DDVA and other dimeric model compounds of lignin in this bacterium, as depicted in Fig. 1, were described in our previous study (14). Several genes related to this pathway have been identified (11, 20–22, 26, 27), but the O demethylation reactions have not been investigated. DDVA, syringate, and vanillate appear to undergo O demethylation to produce the corresponding phenyldiol compounds. In this bacterium, protocatechuate and 3-O-methylgallate, from vanillate and syringate, respectively, are cleaved by protocatechuate 4,5-dioxygenase (14, 27), and 2,2′,3′-trihydroxy-3-methoxy-5,5′-dicarboxybiphenyl (OH-DDVA) is cleaved by a specific dioxygenase (OH-DDVA dioxygenase) (11). In this study, we induced mutations which resulted in alterations in the DDVA degradation pathway by using nitrosoguanidine mutagenesis and selected those mutants still possessing ring fission. The complement DNA of the new mutant was isolated with the gene library of S. paucimobilis SYK-6, which constructed in the broad-host-range cosmid vector pVK100. The minimum complement DNA (1.8 kbp) of a mutant with alterations in vanillate and syringate O demethylation was determined and sequenced.
FIG. 1.
Degradation pathway of various lignin-related compounds in S. (Pseudomonas) paucimobilis SYK-6 (14). The O demethylation steps of phenylmethylether are represented by solid arrows. The enzymes catalyzing each step are indicated on the figure as follows: Lig A, B, protocatechuate 4,5-dioxygenase small and large subunits, respectively (27); Lig C, 4-carboxy-2-hydroxymuconate-6-semialdehyde dehydrogenase (26); Lig D, arylglycerol-β-aryl ether Cα dehydrogenase (20); Lig F(E), α-keto-α-arylglycerol-β-aryl ether β-etherase isozymes (21, 22); Lig Z, OH-DDVA dioxygenase (11). OH-DDVA is converted to 5-carboxyvanillate with LigZ and LigY. Lig H, an enzyme related to the O demethylation of vanillate and syringate (this study). 5-Carboxyvanillate decarboxylase (represented by dotted arrow a) activity was detected in S. paucimobilis SYK-6 cell extract (24), and 3-O-methylgallate was detected by the metabolic experiment with 5-carboxyvanillate (represented by dotted arrow b) (14). The 2-pyrone-4,6-dicarboxylic acid metabolic pathway is represented according to findings from previous studies (18, 19, 26, 31). CHA aldolase, 4-carboxy-4-hydroxy-2-adipic acid aldolase; TCA cycle, tricarboxylic acid cycle.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1. P. paucimobilis wild-type strain SYK-6 was isolated from pulp-bleaching wastewater as described previously (14). P. paucimobilis SYK-6 has recently been reclassified as Sphingomonas paucimobilis (28, 35). Escherichia coli MV1190 and HB101 were used as host cells. A gene library constructed in the cosmid vector pVK100 (kanamycin resistant) of partially EcoRI-digested chromosomal DNA from S. paucimobilis SYK-6 has been described previously (25). Helper plasmid pRK2013 was used as described in a previous study (10). Plasmid pKT230MC was constructed from broad-host-range plasmid vector pKT230 (2) as follows. Plasmid pKT230 was digested by the restriction enzyme SacI. The obtained DNA was digested by EcoRI after the cohesive end of the SacI site had been removed by T4 DNA polymerase. The EcoRI-PvuII fragment of pUC119 (13) was then inserted into the resultant pKT230. Plasmids pKT230MC, pUC118 (13), and pUC119 were used as subcloning vehicles.
TABLE 1.
Bacteria and plasmids used in this study
| Strain or plasmid | Genotype or phenotype | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| HB101 | supE44 hsdS20 recA13 proA2 lacY1 galK2 rpsL20 xyl-5 mtl-1 ara-14 leuB6 thi-1 | Takara-Shuzo Co. |
| MV1190 | d(lac-proAB) thi supE d(sri-recA) 306::Tn10(Tetr) F′ [traD36 proAB+ lacIqZd M15] | Bio-Rad Laboratories, Inc. |
| S. paucimobilis | ||
| SYK-6 | Wild type, Nalr Smr | 14 |
| NT-1 | Mutant derived from SYK-6, deficient in DDVA O demethylation, Nalr Smr | This study |
| DC-49 | Mutant derived from SYK-6, deficient in vanillate and syringate O demethylation, Nalr Smr | This study |
| Plasmids | ||
| pUC118 | Apr | 13 |
| pUC119 | Apr | 13 |
| pRK2013 | tra+ Kmr derivative of RK2 containing ColE1 replicon | 10 |
| pVK100 | mob+ Kmr derivative of RK2 containing ColE1 replicon; S. paucimobilis SYK-6 cosmid library constructed with this plasmid | 24 |
| pKT230MC | pKT230(2) derivative + 200-bp EcoRI-PvuII fragment of pUC119 in the EcoRI-SacI site of pKT230 mob+ RSF1010/pACYC177 replicon Kmr | This study |
| pDE20 | pVK100 derivative cosmid; 20-kbp EcoRI fragment of S. paucimobilis SYK-6 chromosomal DNA inserted into EcoRI site of pVK100, Kmr Tcr | This study |
| pV9 | pDE20 derivative with 11-kbp BamHI fragment of the 20-kbp EcoRI fragment of pDE20 deleted, Kmr Tcr | This study |
| pUV9 | 9-kbp EcoRI fragment from pV9 inserted into EcoRI site of pUC119, Apr | This study |
| pUB6.5 | 6.5-kbp BamHI fragment from pV9 inserted into BamHI site of pUC119, Apr | This study |
| pUSB5.0 | 5.0-kbp SmaI-BamHI fragment from pV9 inserted into SmaI-BamHI of pUC119, Apr | This study |
| pUP4.0 | 4.0-kbp PstI fragment (blunted) from pV9 inserted into SmaI site of pUC119, Apr | This study |
| pUEX3.0 | pUSB5.0 derivative, 2.0-kbp region from BamHI side of the 5.0-kbp SmaI-BamHI fragment of pUSB5.0 deleted, Apr | This study |
| pUEX2.0 | pUSB5.0 derivative, 3.0-kbp region from BamHI side of the 5.0-kbp SmaI-BamHI fragment of pUSB5.0 deleted, Apr | This study |
| pUEX2.0u | 2.0-kbp EcoRI-XbaI fragment from pUEX2.0 inserted into EcoRI-XbaI of pUC118, Apr | This study |
| pKV9 | 9-kbp EcoRI fragment from pV9 inserted into EcoRI site of pKT230MC, Kmr | This study |
| pKB6.5 | 6.5-kbp EcoRI-XbaI fragment from pUB6.5 inserted into EcoRI-XbaI of pKT230MC, Kmr | This study |
| pKSB5.0 | 5.0-kbp EcoRI-XbaI fragment from pUSB5.0 inserted into EcoRI-XbaI of pKT230MC, Kmr | This study |
| pKP4.0 | 4.0-kbp EcoRI-XbaI fragment from pUP4.0 inserted into EcoRI-XbaI of pKT230MC, Kmr | This study |
| pKEX3.0 | 3.0-kbp EcoRI-XbaI fragment from pUEX3.0 inserted into EcoRI-XbaI of pKT230MC, Kmr | This study |
| pKEX2.0 | 2.0-kbp EcoRI-XbaI fragment from pUEX2.0 inserted into EcoRI-XbaI of pKT230MC, Kmr | This study |
Media and growth conditions.
E. coli and S. paucimobilis strains were routinely grown in Luria-Bertani (LB) medium at 37 and 28°C, respectively. When DDVA, vanillate, and other phenolic compounds were used as a carbon source, each was added to W medium (36) at a final concentration of 0.2% (wt/vol). Kanamycin (25 mg/liter), ampicillin (100 mg/liter), and nalidixic acid (25 mg/liter) were added to selective media.
Substrates, enzyme, and reagents.
DDVA, OH-DDVA, and 3-O-methylgallate were synthesized as reported previously (14). Vanillate and syringate were purchased from Tokyo Kasei Co. (Tokyo, Japan). All restriction enzymes, T4 DNA ligase, T4 DNA polymerase, and a Kilosequence kit were obtained from Takara Shuzo Co. (Kyoto, Japan). All antibiotics were purchased from Wako Pure Chemical Industries, Ltd. (Saitama, Japan).
Mutagenesis and screening.
Nitrosoguanidine mutagenesis of S. paucimobilis SYK-6 was performed as described by Miller (23). The final concentration of nitrosoguanidine was 50 μg/ml. The mutants were screened for a decrease in the ability to use DDVA as a carbon source. The capacity to degrade DDVA, OH-DDVA, syringate, 3-O-methylgallate, vanillate, and protocatechuate by these mutants was assessed as follows. S. paucimobilis SYK-6 and mutants were cultured in LB medium containing nalidixic acid (25 mg/liter). When the optical density at 550 nm (OD550) reached 0.7, the culture was centrifuged. Cells from the 10-ml culture were washed twice with 50 mM KH2PO4-NaOH buffer (pH 7.0) and resuspended in 0.5 ml of W medium. Next, 0.1 ml of this cell suspension was added to 0.4 ml of W medium containing 0.2% substrate, and this mixture was incubated at 28°C with shaking. Utilization of the substrate in the culture was observed by a decrease in UV absorption at 200 to 350 nm with a UV-2100PC spectrophotometer (Shimadzu Co., Kyoto, Japan), and the quantity of substrate was determined by high-performance liquid chromatography analysis (column, Bondasphere 5 μ C18, 3.9 by 150 mm; Millipore; mobile phase, 2% acetic acid and 10% methanol; flow rate, 0.7 ml/min; detection, 280 nm) with the Shimadzu Co. LC-6A system.
Preparation of cell extracts and enzyme assay.
S. paucimobilis SYK-6 and the mutants were cultured in LB medium containing nalidixic acid (25 mg/liter). The mutants having pVK100, pKT230MC, or their derivatives were cultured in LB medium containing nalidixic acid (25 mg/liter) and kanamycin (25 mg/liter). When OD550s reached 0.7, the cultures were centrifuged. Cells from the 100-ml cultures were washed twice with 50 ml of 50 mM KH2PO4-NaOH buffer (pH 7.0) and resuspended in 5 ml of the same buffer. The cell suspensions were subjected to sonication at 0°C to disrupt the cells, which were then centrifuged at 10,000 × g for 20 min at 4°C. The supernatants were then used as cell extracts for the enzyme reactions. The extracts were assayed by a protein assay kit (Bradford type of reagent) purchased from Bio-Rad Laboratories, Inc., Richmond, Calif.). The protocatechuate and OH-DDVA dioxygenase activities were measured as follows. One milliliter of 50 mM KH2PO4-NaOH buffer (pH 7.0) containing 1 to 3 mg of protein from the cell extract per ml of buffer was prepared in a 2-ml-volume reaction cuvette (Iijima Denshi Co., Aichi, Japan) at 30°C. Protocatechuate or OH-DDVA was added to the reaction mixture to final concentrations of 0.6 and 1.2 mM, respectively, and then the substrate-dependent oxygen consumption was examined with a galvanic cell electrode purchased from Iijima Denshi Co. The O demethylation activities of vanillate and syringate were measured as follows. The complete reaction mixture contained (in a final volume of 1 ml) 20 mM Tris-HCl buffer (pH 7.5), 3.5 mM MgCl2, 5 mM tetrahydrofolate, 5 mM ATP, 1 mM EDTA, 1 mM substrate (vanillate or syringate), and 1 to 3 mg of protein from the cell extract. After incubation for 3 h at 30°C, the reaction mixture was acidified to pH 2 with 2 M hydrochloric acid and then extracted twice with 0.5 ml of ethyl acetate. The total organic solvent was then completely evaporated, and the residue was dissolved in 0.1 ml of pyridine. A 0.02-ml portion of the pyridine solution was mixed with the same volume of N,O-bis (trimethylsilyl)-trifluoroacetamide (Tokyo Kasei Co.). After incubation for 30 min at room temperature, protocatechuate and 3-O-methylgallate in the resultant reaction mixtures were subjected to gas chromatography analysis with a GC-390 gas chromatograph with a flame ionization detector (GL Science Co., Tokyo, Japan). A CP-Sil 5CB capillary column (0.32 mm by 25 m; GL Science Co.) was used. The oven temperature program was as follows: initial, 100°C; final, 280°C; rate of increase, 5°C/min. The carrier gas was N2, with a flow rate of 10 ml/min.
Cloning and nucleotide sequencing.
All of the recombinant DNA methods used to construct the plasmids or to study the cloned fragments have been described previously (17). Shotgun cloning of the genes involved in O demethylation proceeded as follows. The cosmid library was introduced into the cells of S. paucimobilis DC-49 by the triparental mating method (10). Exconjugants were screened on LB medium plates containing kanamycin and nalidixic acid. Colonies growing on the plates were patched on W medium plates containing DDVA, vanillate, or syringate as a carbon source. Strains able to grow on these plates were complemented with recombinant cosmids. The various deletion derivatives (Table 1) of the pDE20 plasmid were constructed with the restriction endonucleases and exonucleases of the Kilosequence kit. Subcloning was performed with the plasmids pUC118, pUC119, and pKT230MC, and a minimum DNA fragment was determined by complementation of the degradation and assimilation abilities for vanillate and syringate of mutant DC-49. Nucleotide sequencing was performed by the dideoxy-chain termination method with an Auto Sequence kit and A. L. F. DNA Sequencer II obtained from Pharmacia Biotech (Uppsala, Sweden). The nucleotide sequence between the EcoRI and XbaI restriction sites of pUEX2.0 was determined. The nucleotide and deduced amino acid sequences were analyzed with GENETIX version 7.0 software (Software Development Co., Ltd. Tokyo, Japan), and a similarity search was carried out with the DDBJ database.
SDS-PAGE.
An overnight culture of the E. coli strain harboring the plasmids was inoculated onto fresh medium. When the OD550 of the culture reached 0.1, isopropyl-β-d-thiogalactopyranoside (IPTG) was added to create a final concentration of 1 mM. When the OD550 reached 0.8, 1 ml of the culture was centrifuged, and the cells were suspended in 50 μl of lysis buffer. Samples were boiled for 2 min, and 10 μl of each was subjected to 0.1% sodium dodecyl sulfate (SDS)–7.5% polyacrylamide gel electrophoresis (PAGE) (15). After electrophoresis, the gel was stained with Coomassie brilliant blue.
Nucleotide sequence accession number.
The nucleotide sequence data determined for this paper appear in the DDBJ, EMBL, and GenBank nucleotide sequence databases under accession no. AB006079.
RESULTS
Isolation of S. paucimobilis SYK-6 mutants altered in O demethylation.
DDVA, syringate, and vanillate appeared to undergo O demethylation, and the corresponding phenyldiol compounds are presented below. Protocatechuate and 3-O-methylgallate, from vanillate and syringate, respectively, are cleaved by protocatechuate 4,5-dioxygenase; OH-DDVA, from DDVA, is cleaved by the specific dioxygenase (OH-DDVA dioxygenase) in this bacterium, as described previously (11, 14). Thus, mutants defective in O demethylation would not be able to degrade DDVA, syringate, or vanillate, but would retain the corresponding ring fission enzymes. Two mutants of this type were isolated following nitrosoguanidine mutagenesis. The mutant strain NT-1 could not degrade DDVA but could degrade syringate and vanillate. OH-DDVA dioxygenase activity was detected in the cell extract. Strain DC-49 assimilated DDVA slowly. This strain could not degrade vanillate or syringate, but it could degrade protocatechuate and 3-O-methylgallate. Protocatechuate 4,5-dioxygenase activity was detected in the cell extract (Table 2). Strain DC-49 appeared to be able to grow on DDVA by using the 5-carboxyvanillate hydroxylation pathway, as shown in Fig. 1. The degradation and enzymatic data suggest that strain NT-1 is a mutant that is defective in the O demethylation of DDVA, and the mutation in strain DC-49 is related to the O demethylation of both vanillate and syringate.
TABLE 2.
Degradation ability and dioxygenase activity of the mutant and recombinant strains
| Strain | Degradation abilitya
|
Enzyme activityb
|
||||||
|---|---|---|---|---|---|---|---|---|
| DDVA | OH-DDVA | Syringate | 3-O-Methylgallate | Vanillate | Protocatechuate | Protocatechuate 4,5-dioxygenase | OH-DDVA dioxygenase | |
| SYK-6 | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ |
| NT-1 | − | ++ | ++ | ++ | ++ | ++ | ++ | + |
| NT-1(pDE20) | − | ++ | ++ | ++ | ++ | ++ | ++ | + |
| DC-49 | +/− | + | − | + | − | + | ++ | + |
| DC-49(pDE20) | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ |
++, more than 90% of substrate in W medium was degraded for 24 h; +, more than 50% of substrate in W medium was degraded for 48 h; +/−, more than 20% of substrate in W medium was degraded for 72 h; −, more than 95% of substrate remained in W medium after 72 h.
Substrate-dependent O2 consumption was measured for 1 min after the addition of substrate. ++, more than 30 nmol of O2 consumption per mg of protein; +, more than 10 nmol of O2 consumption per mg of protein.
Cloning of genes involved in vanillate and syringate O demethylation.
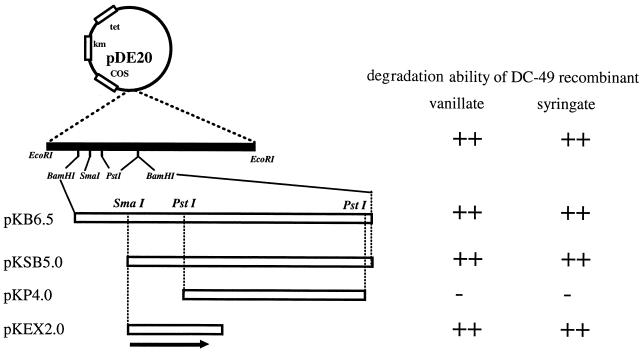
Shotgun cloning of complement DNA for strain DC-49 was conducted as described in Materials and Methods. Five exconjugants, which were selected independently, were able to grow significantly better on the W medium containing DDVA as a carbon source than did the recipient strain, DC-49. All exconjugants harbored the recombinant plasmid pDE20, which possesses 20-kbp EcoRI fragments at the EcoRI site of pVK100. Conjugation experiments with the strains DC-49 and NT-1 and the plasmid pDE20 were performed, and the degradation ability of exconjugants for DDVA, vanillate, and syringate was confirmed (Table 2). Plasmid pDE20 could complement the O demethylation of vanillate and syringate in strain DC-49. However, strain NT-1 was not complemented by pDE20 (Table 2). Subcloning experiments revealed that the 1.8-kbp region in the 6.5-kbp BamHI fragment of the pDE20 insert was able to complement the vanillate and syringate O demethylation of strain DC-49, as shown in Fig. 2.
FIG. 2.
Deletion analysis of complementation ability for vanillate and syringate degradation of strain DC-49. The plasmid construction is indicated in Table 1. The degradation ability is represented according to Table 2. The location and direction of the ligH gene are indicated by an arrow.
Sequencing of the 1.8-kbp DNA fragment of pUEX2.0.
The nucleotide sequence between the EcoRI and XbaI restriction sites of pUEX2.0 was determined. The sequence of this region had a G+C content of 65%. Computer analysis of the nucleotide sequence indicated only one open reading frame (ORF). The deduced gene product of this ORF consists of 557 amino acid residues, and the molecular weight was calculated to be 59,422. Computer analysis also showed that 90% of third bases of codons were G or C. The codon usage of this ORF was quite similar to that in other genes of S. paucimobilis SYK-6 (20–22, 26, 27). The ORF, designated here as ligH, thus appears to be a functional gene. This is supported by the finding that there is a sequence resembling a ribosome-binding site upstream from the putative ATG initiation codon (data not shown). A similarity search indicated that the deduced amino acid sequence of LigH has significant similarity (60%) to formyltetrahydrofolate synthetase of Clostridium thermoaceticum (Fig. 3) (16), Clostridium cylindrosporum (29), and Clostridium acidiurici (33). Lovell et al. (16) reported two putative ATP binding residues in formyltetrahydrofolate synthetase of C. thermoaceticum, and additional conserved regions in formyltetrahydrofolate synthetase were suggested by the PROSITE database (3) under accession no. PS00721 and PS00722. These regions were conserved in the primary structure of the ligH gene product, LigH (Fig. 3).
FIG. 3.
Amino acid alignment between LigH and formyltetrahydrofolate synthetase (FTHS) of C. thermoaceticum (16). Identical and similar amino acids are indicated by asterisks and colons, respectively. Residues proposed as putative ATP binding regions in FTHS of C. thermoaceticum are in boldface (16). Some important conserved regions in FTHSs suggested by PROSITE database (3) under accession no. PS00721 and PS00722 are boxed.
Detection of the ligH gene product.
To detect the ligH gene product in E. coli, cellular proteins were analyzed by SDS-PAGE (Fig. 4). A large amount of protein with a molecular weight of 60,000 was found in the lysate of strain MV1190(pUEX2.0u) following the addition of IPTG to the culture. The molecular weight of this protein was consistent with that of the ligH gene product deduced from the nucleotide sequence.
FIG. 4.
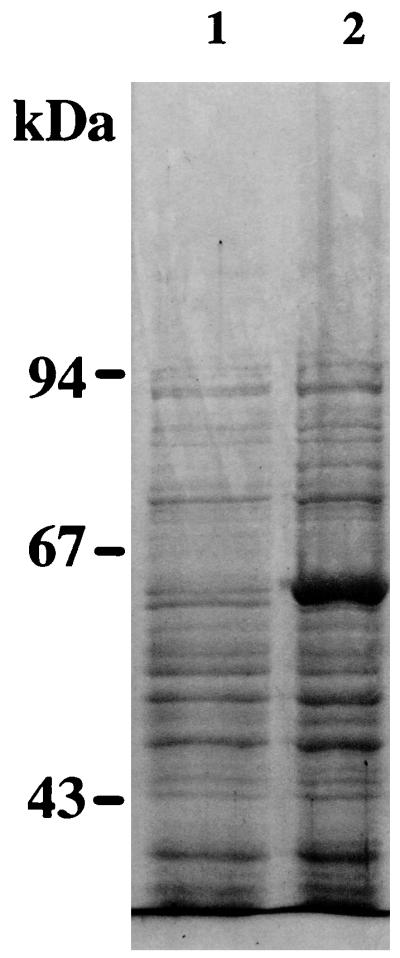
Detection of the ligH gene product. Total cellular proteins of strains grown in LB medium were electrophoresed. Lane 1, MV1190(pUC119); lane 2, MV1190(pUEX2.0u) plus IPTG.
Detection of O demethylation activity.
The primary structure of LigH suggested that its enzymatic reaction may require THF and ATP. The enzymatic activity of O demethylation in strain SYK-6, DC-49, and DC-49’s recombinant strain were assayed (Table 3). The O demethylation activity of vanillate and syringate was detected in the cell extracts of strain SYK-6 and DC-49 harboring plasmid pKEX2.0 when THF was added to the reaction mixture, but ATP was not required in the O demethylation reaction. It was demonstrated that the complete O demethylation reaction of vanillate and syringate was dependent on THF in S. paucimobilis SYK-6. In addition, the requirement of the ligH gene for the O demethylation reaction of vanillate and syringate was confirmed (Table 3).
TABLE 3.
Enzymatic activity of O demethylation of cell extracts
| Strain | Substrate |
O-Demethylation activitya (nmol of protocatechuate or 3-O-methylgallate/60 min/ mg of protein)
|
||
|---|---|---|---|---|
| Completeb | −THF | −ATP | ||
| DC-49 | Vanillate | <0.3 | 0 | <0.3 |
| Syringate | <0.3 | 0 | <0.3 | |
| DC-49(pKEX2.0) | Vanillate | 11.0 ± 1.3 | <0.3 | 8.0 ± 1.6 |
| Syringate | 9.3 ± 1.5 | <0.3 | 8.0 ± 1.7 | |
| SYK-6 | Vanillate | 12.5 ± 1.5 | <0.3 | 11.3 ± 1.5 |
| Syringate | 8.8 ± 2.3 | <0.3 | 7.8 ± 1.8 | |
Each value is the mean ± standard deviation of duplicate reactions.
Complete reaction mixture was indicated in Materials and Methods.
DISCUSSION
The gene involved in the O demethylation of a key intermediate in the biodegradation of lignin, vanillate, and syringate was cloned and sequenced. We were able to isolate two types of O demethylation-altering mutants of S. paucimobilis SYK-6. The mutant strain NT-1 could not degrade DDVA, but could degrade syringate, vanillate, and OH-DDVA. The mutant strain DC-49 could not degrade vanillate and syringate, whereas it could degrade protocatechuate and 3-O-methylgallate. The phenotypes of these mutants were very stable, and the complement DNA, pDE20, of strain DC-49 was isolated from the cosmid library of strain SYK-6. Subcloning and complementation analysis revealed minimum plasmid pKEX2.0 (Fig. 2). The only ORF (designated ligH) found in the 1.8-kbp insert of plasmid pKEX2.0 had a coding capacity of 557 amino acids. To detect the ligH gene product in E. coli, cellular proteins were analyzed by SDS-PAGE (Fig. 4). A large amount of protein with a molecular weight of 60,000 was found in the lysate of strain MV1190 (pUEX2.0u) after the addition of IPTG to the culture. The molecular weight of this protein was consistent with that of the ligH gene product deduced from the nucleotide sequence.
It is of interest that only the ligH gene could complement both vanillate and syringate O demethylation of strain DC-49, but it was not complemented in NT-1 by pDE20 (Table 2). Thus, it appears that S. paucimobilis SYK-6 has two systems for the O demethylation of phenylmethylether and that the O demethylation of both vanillate and syringate is catalyzed by same enzyme. This is also seen in the protocatechuate 4,5-dioxygenase encoded by ligAB of this bacterium, which oxidized both protocatechuate and 3-O-methylgallate (14), but not OH-DDVA. S. paucimobilis SYK-6 has the OH-DDVA-specific ring fission enzyme encoded by ligZ as described previously (11). To investigate another O demethylation (DDVA-specific) system, further studies including the determination of the gene or genes complementing mutant NT-1 are presently being conducted in our laboratory.
A similarity search revealed that the deduced amino acid sequence of the LigH that complemented the mutation related to the O demethylation ability of vanillate and syringate of strain DC-49 showed significant similarity (60%) to formyltetrahydrofolate synthetase from C. thermoaceticum (Fig. 4) (16), C. cylindrosporum (29), and C. acidiurici (33). It was reported that the formyltetrahydrofolate synthetase reaction required ATP (12). The primary structure of LigH suggested that its enzymatic reaction may require THF and ATP. In fact, the THF-dependent O demethylation activity of vanillate and syringate was detected in the cell extract of S. paucimobilis SYK-6 and DC-49 conjugated with plasmid pKEX2.0, which possessed the ligH gene (Table 3). However, ATP was not required for the reaction, whereas putative ATP binding regions were conserved in the primary structure of LigH (Fig. 3). Unfortunately, at this time, we cannot explain the functions of conserved regions, including the putative ATP binding regions of LigH. Berman and Frazer (4) and Stupperich and Konle (30) reported the existence of a THF- and ATP-dependent O demethylation system in anaerobes. S. paucimobilis SYK-6 was able to O demethylate three phenylmethylethers (DDVA, vanillate, and syringate) under aerobic conditions. It has been reported that aerobic bacteria use hydroxylating enzymes to cleave O-methylether linkages, and the enzymes require NADH (5, 7). However, neither NADH nor ATP was required for the O demethylation reaction of vanillate and syringate in S. paucimobilis SYK-6 (Table 3). Therefore, the O demethylation reaction in S. paucimobilis would appear to be different from those previously reported for the anaerobic and aerobic O demethylation systems.
We concluded that the formyltetrahydrofolate synthetase-like protein LigH must be essential to the O demethylation system of vanillate and syringate in S. paucimobilis SYK-6. Additional biochemical experiments will obviously be needed to obtain a better understanding of the mode of action for LigH.
ACKNOWLEDGMENTS
We thank Masao Fukuda and Eiji Masai (Department of Bioengineering, Nagaoka University of Technology, Niigata, Japan) for invaluable discussions throughout this study.
REFERENCES
- 1.Bache R, Pfennig N. Selective isolation of Acetobacterium woodii on methoxylated aromatic acids and determination of growth yields. Arch Microbiol. 1981;130:255–261. [Google Scholar]
- 2.Bagdasarian M, Lurz R, Rucker B, Franklin F C H, Bagdasarian M M, Frey J, Timmis K N. Specific purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene. 1981;16:237–247. doi: 10.1016/0378-1119(81)90080-9. [DOI] [PubMed] [Google Scholar]
- 3.Bairoch A, Bucher P. PROSITE: recent developments. Nucleic Acids Res. 1994;22:3583–3589. [PMC free article] [PubMed] [Google Scholar]
- 4.Berman M H, Frazer A C. Importance of tetrahydrofolate and ATP in the anaerobic O-demethylation reaction for phenylmethylethers. Appl Environ Microbiol. 1992;58:925–931. doi: 10.1128/aem.58.3.925-931.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernhardt F-H, Bill E, Trautwein A X, Twilfer H. 4-Methoxybenzoate monooxygenase from Pseudomonas putida: isolation, biochemical properties, substrate specificity and reaction mechanisms of the enzyme components. Methods Enzymol. 1988;161:281–294. doi: 10.1016/0076-6879(88)61031-7. [DOI] [PubMed] [Google Scholar]
- 6.Brunel F, Davison J. Cloning and sequencing of Pseudomonas genes encoding vanillate demethylase. J Bacteriol. 1988;170:4924–4930. doi: 10.1128/jb.170.10.4924-4930.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buswell J A, Ribbons D W. Vanillate O-demethylase from Pseudomonas species. Methods Enzymol. 1988;161:294–301. doi: 10.1016/0076-6879(88)61032-9. [DOI] [PubMed] [Google Scholar]
- 8.Dardas A, Gal D, Barrelle M, Sauret-Ignazi G, Sterjiades R, Pelmont J. The demethylation of guaiacol by a new bacterial cytochrome P-450. Arch Biochem Biophys. 1985;236:585–592. doi: 10.1016/0003-9861(85)90662-9. [DOI] [PubMed] [Google Scholar]
- 9.DeWeerd K A, Saxena A, Nagle D P, Jr, Suflita J M. Metabolism of the 18O-methoxy substituent of 3-methoxybenzoic acid and other unlabeled methoxybenzoic acid by anaerobic bacteria. Appl Environ Microbiol. 1988;54:1237–1242. doi: 10.1128/aem.54.5.1237-1242.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ditta G, Stanfield S, Corbin D, Helinski D R. Broad host range DNA cloning system for Gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci USA. 1980;77:7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Egashira T, Peng X, Kimbara K, Hanashiro K, Masai E, Katayama Y, Kawai S, Seto M, Takahashi C, Fukuda M, Yano K. Program of the 5th International Symposium on Pseudomonas: Biotechnology and Molecular Biology. 1995. Cloning and characterization of the gene encoding novel ring cleavage enzyme for lignin related biphenyl in Pseudomonas (Sphingomonas) paucimobilis SYK-6; p. 78. [Google Scholar]
- 12.Elliot I J, Ljungdahl L G. Chemical modification of cysteine and tyrosine residues in formyltetrahydrofolate synthetase from Clostridium thermoaceticum. Arch Biochem Biophys. 1982;215:245–252. doi: 10.1016/0003-9861(82)90301-0. [DOI] [PubMed] [Google Scholar]
- 13.Henikoff S. Unidirectional digestion with exonuclease creates targeted breakpoints for DNA sequencing. Gene. 1984;28:351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- 14.Katayama Y, Nishikawa S, Murayama A, Yamasaki M, Morohoshi N, Haraguchi T. The metabolism of biphenyl structure in lignin of the soil bacterium (Pseudomonas paucimobilis SYK-6) FEBS Lett. 1988;233:129–133. [Google Scholar]
- 15.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 16.Lovell C R, Prazybyla A, Ljungdahl L G. Primary structure of thermostable formyltetrahydrofolate synthetase from Clostridium thermoaceticum. Biochemistry. 1990;29:5687–5694. doi: 10.1021/bi00476a007. [DOI] [PubMed] [Google Scholar]
- 17.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 18.Maruyama K. Purification and properties of 2-pyrone-4,6-dicarboxylate hydrolase. J Biochem. 1983;93:557–565. doi: 10.1093/oxfordjournals.jbchem.a134210. [DOI] [PubMed] [Google Scholar]
- 19.Maruyama K. Purification and properties of 4-hydroxy-4-methyl-2-oxoglutalate aldolase from Pseudomonas ochraceae grown on phthalate. J Biochem. 1990;108:327–333. doi: 10.1093/oxfordjournals.jbchem.a123201. [DOI] [PubMed] [Google Scholar]
- 20.Masai E, Kubota S, Katayama Y, Kawai S, Yamasaki M, Morohoshi N. Characterization of the Cα-dehydrogenase gene involved in the cleavage of β-aryl ether by Pseudomonas paucimobilis. Biotechnol Biophys Biochem. 1993;57:1655–1659. doi: 10.1271/bbb.57.1655. [DOI] [PubMed] [Google Scholar]
- 21.Masai E, Katayama Y, Kawai S, Nishikawa S, Yamasaki M, Morohoshi N. Cloning and sequencing of the gene for Pseudomonas paucimobilis enzyme that cleaves β-aryl ether. J Bacteriol. 1991;173:7950–7955. doi: 10.1128/jb.173.24.7950-7955.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Masai E, Katayama Y, Kubota S, Kawai S, Yamasaki M, Morohoshi N. A bacterial enzyme degrading the model lignin compound beta-etherase is a member of the glutathione-S-transferase superfamily. FEBS Lett. 1993;323:135–140. doi: 10.1016/0014-5793(93)81465-c. [DOI] [PubMed] [Google Scholar]
- 23.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. pp. 125–129. [Google Scholar]
- 24.Nishikawa, S., and Y. Katayama. 1991. Unpublished data.
- 25.Nishikawa S, Katayama Y, Yamasaki M, Morohoshi N, Haraguchi T. In vitro packaging and conjugal transfer of lignin model compounds’ degradable Pseudomonas paucimobilis SYK-6 chromosomal DNA. Mokuzai Gakkaishi. 1988;34:1021–1025. [Google Scholar]
- 26.Nishikawa S, Katayama Y, Yamasaki M, Morohoshi N, Haraguchi T. Proceedings of the 6th International Symposium on Wood and Pulping Chemistry. 2nd ed. 1991. Cloning of the genes involved in protocatechuate meta cleavage pathway; pp. 311–313. [Google Scholar]
- 27.Noda Y, Nishikawa S, Shiozuka K, Kadokura H, Nakajima H, Yoda K, Katayama Y, Morohoshi N, Haraguchi T, Yamasaki M. Molecular cloning of the protocatechuate 4,5-dioxygenase genes of Pseudomonas paucimobilis. J Bacteriol. 1990;172:2704–2709. doi: 10.1128/jb.172.5.2704-2709.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oyaizu, H. (The University of Tokyo). 1995. Personal communication.
- 29.Rankin C A, Haslam G C, Himes R H. Sequence and expression of the gene for N10-formyltetrahydrofolate synthetase from Clostridium cylindrosporum. Protein Sci. 1993;2:197–205. doi: 10.1002/pro.5560020208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stupperich E, Konle R. Corrinoid-dependent methyl transfer reactions are involved in methanol and 3,4-dimethoxybenzoate metabolism by Sporomusa ovata. Appl Environ Microbiol. 1993;59:3110–3116. doi: 10.1128/aem.59.9.3110-3116.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sze I S-Y, Dagley S. Degradation of substituted mandelic acids by meta fission reactions. J Bacteriol. 1987;169:3833–3835. doi: 10.1128/jb.169.8.3833-3835.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taylor B F. Aerobic and anaerobic catabolism of vanillic acid and some other methoxy-aromatic compounds by Pseudomonas sp. strain PN-1. Appl Environ Microbiol. 1982;46:1286–1292. doi: 10.1128/aem.46.6.1286-1292.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whitehead T R, Rabinowitz J C. Nucleotide sequence of the Clostridium acidiurici (“Clostridium acidi-urici”) gene for 10-formyltetrahydrofolate synthetase shows extensive amino acid homology with the trifunctional enzyme C1-tetrahydrofolate synthase from Saccharomyces cerevisiae. J Bacteriol. 1988;170:3255–3261. doi: 10.1128/jb.170.7.3255-3261.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu Z, Daniel S L, Drake H L. Characterization of CO-dependent O-demethylating enzyme system from acetogen Clostridium thermoaceticum. J Bacteriol. 1988;170:5747–5750. doi: 10.1128/jb.170.12.5747-5750.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yabuuchi E, Yano I, Oyaizu H, Hashimoto Y, Ezaki T, Yamamoto H. Proposals of Sphingomonas paucimobilis gen. nov. and comb. nov., Sphingomonas parapaucimobilis sp. nov., Sphingomonas yanoikuyae sp. nov., Sphingomonas adhaesiva sp. nov., Sphingomonas capsulata sp. comb. nov., and two genospecies of genus Sphingomonas. Microbiol Immunol. 1990;34:99–119. doi: 10.1111/j.1348-0421.1990.tb00996.x. [DOI] [PubMed] [Google Scholar]
- 36.Yano K, Nishi T. pKJ1, A naturally occurring conjugative plasmid coding for toluene degradation and resistance to streptomycin and sulfonamides. J Bacteriol. 1980;143:552–560. doi: 10.1128/jb.143.2.552-560.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]