Abstract
Background
T-cell responses during chronic viral infections become exhausted, which is reflected by upregulation of inhibitory receptors (iRs) and increased interleukin 10 (IL-10). We assessed 2 iRs—PD-1 (programmed cell death protein 1) and Tim-3 (T-cell immunoglobulin and mucin domain–containing protein 3)—and IL-10 mRNAs in peripheral blood mononuclear cells (PBMCs) and their soluble analogs (sPD-1, sTim-3, and IL-10) in plasma in chronic HIV-1/hepatitis C virus (HCV) coinfection and explored the effect of HCV treatment on these markers. We also aimed to establish whether iR expression may be determined by the HCV CD8+ T-cell immunodominant epitope sequence.
Methods
Plasma and PBMCs from 31 persons with chronic HIV-1/HCV coinfection from the Swiss HIV Cohort Study were collected before and after HCV treatment. As controls, 45 persons who were HIV-1 negative with chronic HCV infection were recruited. Exhaustion markers were assessed by enzyme-linked immunosorbent assay in plasma and by quantitative reverse transcription polymerase chain reaction in PBMCs. Analysis of an HCV epitope sequence was conducted by next-generation sequencing: HLA-A*02–restricted NS31073–1081 and NS31406–1415 and HLA-A*01–restricted NS31436–1444.
Results
The study revealed higher plasma sPD-1 (P = .0235) and IL-10 (P = .002) levels and higher IL-10 mRNA in PBMCs (P = .0149) in HIV-1/HCV coinfection. A decrease in plasma sPD-1 (P = .0006), sTim-3 (P = .0136), and IL-10 (P = .0003) and Tim-3 mRNA in PBMCs (P = .0210) was observed following successful HCV treatment. Infection with the HLA-A*01–restricted NS31436–1444 ATDALMTGY prototype variant was related to higher sTim-3 levels than infection with the ATDALMTGF escape variant (P = .0326).
Conclusions
The results underscore the synergistic effect of coinfection on expression of exhaustion markers, their reduction following successful HCV treatment and imply that iR levels may operate on an epitope-specific manner.
Keywords: HCV treatment, HIV-1/HCV coinfection, sPD-1, sTim-3, T-cell exhaustion
Effector T-cell responses following primary viral infection are accompanied by orchestration of gene expression, including upregulation of inhibitory receptors, which are downregulated when the pathogen is eradicated. In chronic infections, persistent antigen load and immune response stimulation induce overexpression of inhibitory receptors, which downregulate the functional and proliferative potential of the effector cells toward exhaustion [1, 2].
T-cell exhaustion is characteristic of chronic immune-related processes, including viral infections (eg, HIV-1, hepatitis B virus, and hepatitis C virus [HCV]) and tumors [3–5]. Aggregated data suggest that immune exhaustion is a crucial factor facilitating viral persistence [2].
T-cell exhaustion in chronic HCV infection is characterized as overexpression of several inhibitory receptors—including PD-1 (programmed cell death protein 1) [6] and Tim-3 (T-cell immunoglobulin and mucin domain–containing protein 3) [7]—on total and HCV-specific CD4+ and CD8+ T cells. Blocking PD-1/Tim-3 interaction by monoclonal antibodies restored the functionality of HCV-specific T cells, manifested as higher expansion, proliferation, and effector cytokine production [7–9]. A similar approach is becoming an important therapeutic measure in oncologic treatment [5, 10]. Additionally, T-cell exhaustion is related to increased secretion of anti-inflammatory immunosuppressive interleukin 10 (IL-10) [11]. We found that successful treatment of chronic HCV monoinfection resulted in a percentage decrease of T cells with Tim-3 expression and PD-1/Tim-3 coexpression as well as in a reduction of IL-10 in plasma, indicating that T-cell exhaustion is reversible [12].
Inhibitory receptors have their corresponding analogs in a plasma-soluble form (ie, sPD-1 and sTim-3). Five alternative splicing isoforms of PD-1 encoding gene have been reported, among which the PD-1Δ3 isoform is secreted as a soluble molecule [13]. sTim-3 is an effect of enzymatic shedding from the cell surface [14]. Their levels correlated with membrane expression of their receptors on T cells; thus, they have the potential to become convenient markers approximating exhaustion [15, 16].
HIV-1/HCV coinfection is associated with higher HCV viral loads and more rapid progression of HCV-related disease, likely linked to HIV-1–induced immune disturbances (eg, HCV-specific T-cell attenuation) [9, 17]. In HIV-1/HCV coinfection, total and HCV-specific CD8+ T cells coexpressed PD-1 and Tim-3 at significantly higher frequencies than in HCV monoinfection [9]. However, antiretroviral therapy (ART) did not alter the exhaustion phenotype of CD8+ and CD4+ cells [9].
Although sPD-1 and sTim-3 have been studied in chronic HIV-1 and HCV monoinfections [14, 18, 19], they remain largely understudied in HIV-1/HCV coinfection, despite representing a unique model of 2 viruses inducing exhaustion [1, 3, 20]. Possible complex interactions, including synergistic/superseding effects on exhaustion, may be expected and eventually potentiated by HIV-1 [21]. Similarly, it is largely unknown whether treatment against chronic HCV infection may reduce exhaustion in HIV-1 coinfection, which could be due to a number of mechanisms—for example, the reduction of HCV antigen load or the elimination of viral proteins shown to inhibit immune responses [12, 22].
Although viral pathogens encode numerous immunogenic epitopes, CD8+ T cells recognize and respond only to some of them. This immunodominance is strongly determined by human leukocyte antigen (HLA) alleles [23]. Immunodominant HCV-specific CD8+ T-cell epitopes within E2, NS3, or NS5B are targeted in the majority of individuals expressing the HLA alleles [24, 25]. NS3-immunodominant CD8+ T-cell responses to genotype 1 HCV typically include NS31406–1415 and NS31073–1081, restricted by the HLA-A*02 allele, and NS31436–1444, restricted by the HLA-A*01 allele. They represent the most commonly recognized epitopes in patients infected with genotype 1, which is also determined by the highest prevalence of these 2 HLA-A alleles among Caucasians [26, 27]. Footprints of the HLA-defined T-cell response leading to escape variants, which represents an important viral strategy of immune evasion, may occur within these epitopes [28]. For example, NS31436–1444 ATDALMTGY and ATDALMTGF accounted for 93% of genotype 1 sequences and were almost equally distributed [26]. NS31436–1444 Y1444F substitution impaired epitope binding and recognition, representing an escape variant in individuals who were HLA-A*01 positive [28, 29]. Since exhaustion is driven by persistent antigen stimulation of T cells, it may be expected that epitope sequence variation, which eventually leads to abrogation of its recognition, may be associated with reduced expression of exhaustion markers. We previously demonstrated that PD-1/Tim-3 expression on T cells in chronic HCV infection may be determined by the HCV CD8+ T-cell epitope-specific sequence or its intrahost variability [30]. However, to our best knowledge, no similar data are available for HIV-1/HCV coinfection. Understanding the exact determinants of exhaustion, particularly the role of the epitope sequence context, is of high importance, since it may lead to different states of susceptibility to reinfection as well as vaccine-induced immunity.
The aim of the present study was to explore PD-1, Tim-3, and IL-10 mRNA levels in peripheral blood mononuclear cells (PBMCs) as well as sPD-1, sTim-3, and IL-10 in plasma in chronic HIV-1/HCV coinfection when compared with HCV monoinfection and to assess the impact of anti-HCV treatment on immune exhaustion in chronic HIV-1/HCV coinfection. Additionally, we aimed to investigate whether the expression of exhaustion markers may be determined by an HCV CD8+ T-cell–immunodominant epitope sequence.
METHODS
Retrospective plasma and PBMC samples from 31 persons with HIV from the Swiss HIV Cohort Study (SHCS) with chronic HCV genotype 1 infection (anti-HCV+, HCV RNA+), who were receiving ART for the entire time of the study and qualified for antiviral treatment of hepatitis C, were collected before HCV treatment and at a median 12.4 months (range, 4.5–22.4) posttreatment. At baseline, the majority (n = 30, 96.8%) had a CD4+ T-cell count >200 cells/μL, and 26 (83.9%) had undetectable HIV-1 RNA, while after HCV treatment all participants had a T-cell count >200 cells/μL and undetectable HIV-1 RNA. Enrolled participants were assigned to 1 of the following treatments: pegylated interferon α (PEG-IFN) + ribavirin (n = 18), PEG-IFN + ribavirin + boceprevir (n = 1), PEG-IFN + ribavirin + faldaprevir (n = 6), PEG-IFN + ribavirin + telaprevir (n = 6), according to the standard recommendations at the time of HCV treatment in Switzerland. Effectiveness of treatment (sustained virologic response [SVR]) was assessed 6 months posttreatment, defined as undetectable HCV RNA in plasma via a polymerase chain reaction (PCR) test of sensitivity of at least 25 IU/mL (COBAS Amplicor Assay version 2.0; Roche Molecular Systems). SVR criterion was achieved in 21 of 31 (67.7%) individuals. For comparison of baseline expression of exhaustion markers, 45 participants with chronic HCV genotype 1 infection, without evidence of any other chronic viral infection, were prospectively identified from the Outpatient Clinic of the Warsaw Hospital for Infectious Diseases. The following clinical data were collected: age, sex, baseline liver fibrosis scores, HCV and HIV-1 viral loads, and CD4 cell count (when applicable). Baseline liver fibrosis scores were assessed by transient elastography [31]. HCV viral load was measured by RealTime HCV assay (Abbott Laboratories) of sensitivity 12 IU/mL or 25 IU/mL (COBAS Amplicor Assay version 2.0). HIV-1 viral load was measured by using the COBAS AmpliPrep/COBAS TaqMan HIV-1 Test version 2.0 with a limit of detection ≤20 HIV-1 RNA copies/mL in plasma. Some samples collected before 2010 were measured with former versions of the COBAS AmpliPrep/COBAS TaqMan HIV-1 Test with higher limits of detection in plasma (ie, 40 or 50 HIV-1 RNA copies/mL). Clinical and virologic characteristics of enrolled participants are presented in Table 1.
Table 1.
Clinical and Virologic Characteristics of the Study Participants
| No. or Median (Range) | ||
|---|---|---|
| HIV-1/HCV Coinfection (n = 31) | HCV Monoinfection (n = 45) | |
| Sex: male:female | 26:5 | 19:26 |
| Age, y | 46 (35–59) | 60 (28–83) |
| HCV viral load, U·mL−1 | ||
| Baseline | 1.1 × 106 (9.2 × 102–2.0 × 107) | 9.4 × 105 (1.6 × 104–1.1 × 107) |
| SVR+ | 1.4 × 106 (9.2 × 102–1.8 × 107) | … |
| SVR− | 5.0 × 105 (1.2 × 103–2.0 × 107) | … |
| End of treatment | ||
| SVR+ | 0.0a (0.0–0.0) | … |
| SVR− | 7.6 × 102 (0.0–4.7 × 105) | … |
| Follow-up | ||
| SVR+ | 0.0a (0.0–0.0) | … |
| SVR− | 6.1 × 105 (6.4 × 102–3.7 × 107) | … |
| Baseline liver fibrosis score | ||
| Minimal: F0/1 | 7 | 14 |
| Moderate: F2 | 1 | 21 |
| Severe: F3 | 2 | 10 |
| Cirrhosis: F4 | 2 | 0 |
| Not available | 19 | … |
| HIV-1 viral load, copies/mL | … | |
| Baseline | 0.0a (0.0–8.5 × 102) | |
| Follow-up | 0.0a (0.0–0.0) | |
| CD4+ cell count/μLb | … | |
| Baseline | 560 (159–1265) | |
| SVR+ | 672 (159–847) | |
| SVR− | 480 (203–1265) | |
| Follow-up | 581 (304–1567) | |
| SVR+ | 580 (304–1567) | |
| SVR− | 617.5 (315–933) | |
Ellipses (…) indicate not applicable.
Abbreviations: HCV, hepatitis C virus; SVR, sustained virologic response.
aBelow the limit of detection.
bNormal range: 500–1500 cells/μL.
Patient Consent Statement
The study was carried out according to the guidelines of the 2013 Declaration of Helsinki, and written informed consent was obtained from all participants before the enrollment. The SHCS has been approved by the ethics committee of the participating institutions: Kantonale Ethikkommission Bern (approval 21/88), Ethikkommission des Kantons St Gallen (approval 12/003), Le Comité d’Ethique des Départements des Spécialités Médicales et de Médecine Communautaire et de Premier Recours, Hôpitaux Universitaires de Genève (approval 01-142), Kantonale Ethikkommission Zürich (approval EK-793), Repubblica e Cantone Ticino—Comitato Ethico Cantonale (approval CE 813), Commission cantonale d’étique de la recherche sur l’être humain, Canton de Vaud, Lausanne (approval 131/01), and Ethikkommission beider Basel (approval 688). The study was also approved by the bioethics committee of the Medical University of Warsaw (consent KB/77/A/2015).
Assessment of PD-1, Tim-3, and IL-10 mRNA Levels
Total RNA was extracted from PBMCs (typically 106 cells) by a modified guanidinium thiocyanate-phenol/chloroform method with Trizol reagent (Life Technologies), followed by DNA digestion with a DNA-Free DNA Removal Kit (Life Technologies). RNA was subjected to reverse transcription with M-MLV Reverse Transcriptase (Life Technologies) and random hexamers (Life Technologies). Specific mRNA levels were assessed in triplicate by 1-step real-time PCR with a LightCycler FastStart DNA Master SYBR Green I Kit (Roche) and a set of specific primers (Supplementary Table 1) on a Light Cycler 2.0 instrument (Roche) at the following conditions: initial denaturation at 95 °C for 10 minutes and 45 cycles of amplification, each comprising denaturation (95°C, 1 minute), primer annealing (65 °C, 1 minute) and elongation (72°C, 1 minute), and DNA melting (60–95 °C, 15 minutes).
PD-1, Tim-3, and IL-10 expression was normalized to a histone 3 housekeeping gene by a ΔCt method according to the following equation:
Normalized gene expression = 2ΔCt
ΔCt = Ctreference gene – Ctanalyzed gene (where Ct is cycle threshold)
Normalized mRNA expression levels were graphically visualized on a semilogarithmic scale via Prism version 9.4.0 (GraphPad).
Assessment of IL-10, sPD-1, and sTim-3 in Plasma
IL-10, sPD-1, and sTim-3 levels were assessed with an IL-10 Human UltraSensitive ELISA Kit (sensitivity, <0.2 pg/mL), PD-1 Human ELISA Kit (sensitivity, 1.14 pg/mL), and Tim-3 Human ELISA Kit (sensitivity, 35.3 pg/mL; all by Life Technologies). Results were expressed as picograms per milliliter and visualized with Prism version 9.4.0.
HLA-A*01 and HLA-A*02 Typing
The HLA-A*01 and HLA-A*02 alleles were identified by qualitative and quantitative PCR, respectively, as described elsewhere [32, 33].
Sequencing of NS31073–1081, NS31406–1415, and NS31436–1444 HCV Epitopes
Nucleotide sequence analysis of HLA-A*02–restricted NS31073–1081 and NS31406–1415 and HLA-A*01–restricted NS31436–1444 epitopes was conducted by next-generation amplicon sequencing via the Illumina MiSeq platform as previously described [30]. Data analysis was performed with Quasirecomb version 1.2 (ETH Zurich and Swiss Institute of Bioinformatics). It uses a jumping hidden Markov model to infer quasispecies sequences and their frequencies from the next-generation sequencing data [34]. Epitope sequences were visualized with a web-based application (WebLogo version 2.8.2; University of California, https://weblogo.berkeley.edu/) [35].
Statistical Analysis
Results were expressed as median (range). A Wilcoxon matched-pair test was used to compare immune exhaustion markers before and after treatment. Furthermore, a Mann-Whitney U test was used to compare exhaustion markers between HIV-1/HCV coinfection and HCV monoinfection and among HIV-1/HCV cases harboring different variants of the epitopes. All P values were 2-tailed, and P < .05 was considered significant. The analysis was performed with Prism version 9.4.0.
RESULTS
Soluble Exhaustion Markers Were Higher in HIV-1/HCV Coinfection and Decreased Following Successful HCV Antiviral Treatment
We compared the expression of exhaustion markers between HIV-1/HCV coinfected and HCV-monoinfected cases and assessed the impact of HCV antiviral treatment on these markers in HIV-1/HCV coinfection.
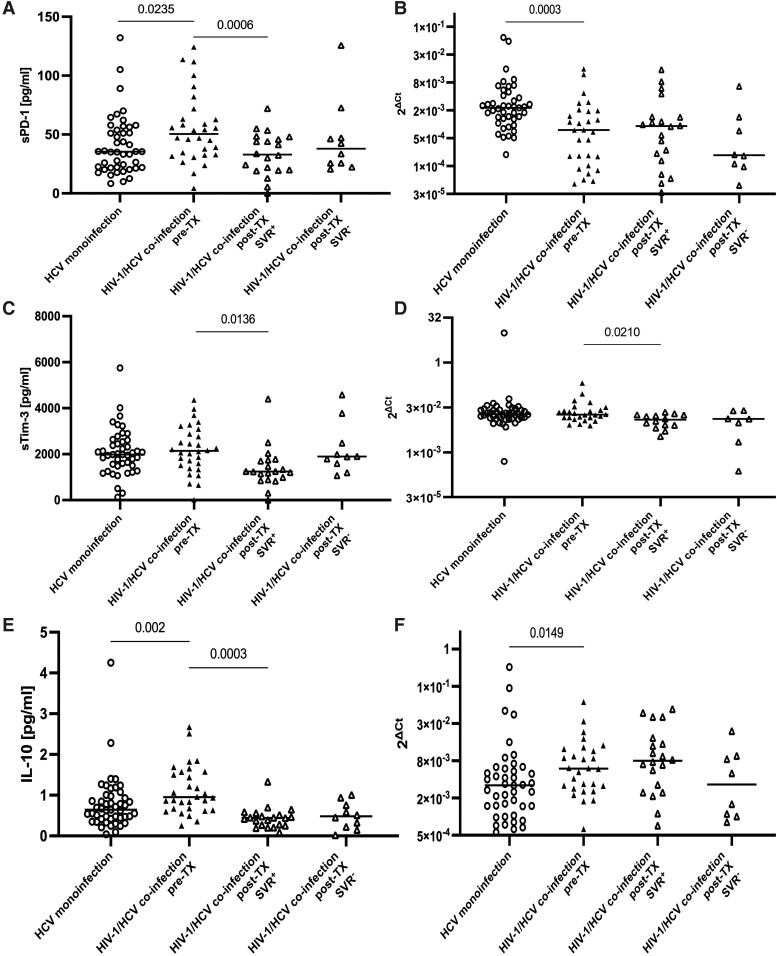
Baseline median (range) sPD-1 levels were higher in cases of coinfection than HCV monoinfection (50.4 [4.3–124.3] vs 35.5 [8.4–132.1] pg/mL, P = .0235; Figure 1A). Differences in sPD-1 levels remained significant even when 5 participants who were HIV-1 RNA+ were excluded from the analysis (47.2 [4.3–124.3] vs 35.5 [8.4–132.1] pg/mL, P = .0340).
Figure 1.
sPD-1, sTim-3, and IL-10 in plasma (A, C, E) and their mRNA levels in PBMCs (B, D, F) in HCV monoinfection (n = 43–45) and HIV-1/HCV coinfection (n = 26–30). The effect of antiviral treatment on these markers in HIV-1/HCV coinfection. SVR+, individuals who achieved sustained virologic response; SVR−, individuals who did not achieve sustained virologic response. Each symbol (triangle, circle) represents a mean of 3 replicates obtained from each participant. Horizontal lines within each population of results represent median values. Numbers above lines representing pairwise comparisons express P values. HCV, hepatitis C virus; IL-10, interleukin 10; PBMC, peripheral blood mononuclear cell; PD-1, programmed cell death protein 1; SVR, sustained virologic response; Tim-3, T-cell immunoglobulin and mucin domain–containing protein 3; TX, treatment.
Baseline median (range) IL-10 levels were higher in cases of coinfection than HCV monoinfection (0.951 [0.254–2.674] vs 0.642 [0.040–4.252] pg/mL, P = .002; Figure 1E). Differences in IL-10 levels remained significant even when 5 participants who were HIV-1 RNA+ were excluded from the analysis (0.878 [0.254–2.674] vs 0.642 [0.040–4.252] pg/mL, P = .0103).
Baseline sTim-3 levels were similar (2142.2 [18.3–4354.2] vs 1985.3 [120.8–5746.8] pg/mL, P = .5270; Figure 1C).
In the SVR+ subgroup, there was a significant reduction of sPD-1 (median [range]; from 50.5 [4.3–124.3] to 33.0 [0.0–72.1] pg/mL, P = .0006), which was observed even when 3 participants who were HIV-1 RNA+ were excluded from the analysis (from 47.0 [4.3–124.3] to 41.3 [0.0–72.1] pg/mL, P = .0032). Within this subgroup, there was also a reduction of sTim-3 (from 2089.9 [18.3–4354.2] to 1243.4 [0.0–4406.0] pg/mL, P = .0136), even when 3 participants who were HIV-1 RNA+ were excluded from the analysis (from 2166.0 [18.3–4354.2] to 1238.6 [0.0–4406.0] pg/mL, P = .0305). Similarly, a reduction of IL-10 levels was observed (from 1.104 [0.254–2.674] to 0.438 [0.088–1.332] pg/mL, P = .0003), even after exclusion of 3 participants who were HIV-1 RNA+ (from 0.940 [0.254–2.674] to 0.448 [0.194–1.332] pg/mL, P = .0021).
In contrast, among SVR− cases, the changes did not reach statistical significance (from 50.4 [16.9–90.7] to 38.0 [20.6–125.6] pg/mL for sPD-1, P = .7695 [Figure 1A]; from 2481.4 [1109.9–3965.0] to 1897.3 [1068.7–4586.3] pg/mL for sTim-3, P = .4922 [Figure 1C]; from 0.852 [0.358–1.688] to 0.482 [0.018–1.006] pg/mL for IL-10, P = .0645 [Figure 1E]).
Expression of Exhaustion Markers in PBMCs
Baseline PD-1 mRNA levels in PBMCs were lower in the coinfected cases than in HCV monoinfection (P = .0003, Figure 1B), while Tim-3 mRNA levels were similar (Figure 1D). In contrast, IL-10 mRNA levels were higher in coinfected cases than in HCV monoinfection (P = .0149, Figure 1F). There was a reduction in Tim-3 mRNA levels in the SVR+ cases (P = .0210, Figure 1D), which was retained even when 3 participants who were HIV-1 RNA+ SVR+ were excluded from the analysis (P = .0195). No posttreatment changes were found in PD-1 and IL-10 mRNA levels (Figure 1B and 1F, respectively).
Epitope Sequence Polymorphisms Association With Baseline sPD-1, sTim-3, and IL-10 Levels in HIV-1/HCV Coinfection
We assessed exhaustion markers in the context of the HCV epitope sequence in cases of HIV-1/HCV coinfection. The analysis of immunodominant epitopes—HLA-A*02–restricted NS31073–1081 and NS31406–1415 and HLA-A*01–restricted NS31436–1444—was performed in 22 individuals infected with HCV 1a in whom epitope sequences were successfully inferred. Of these, 10 (45.4%) were HLA-A*02 positive and 6 (27.3%) were HLA-A*01 positive.
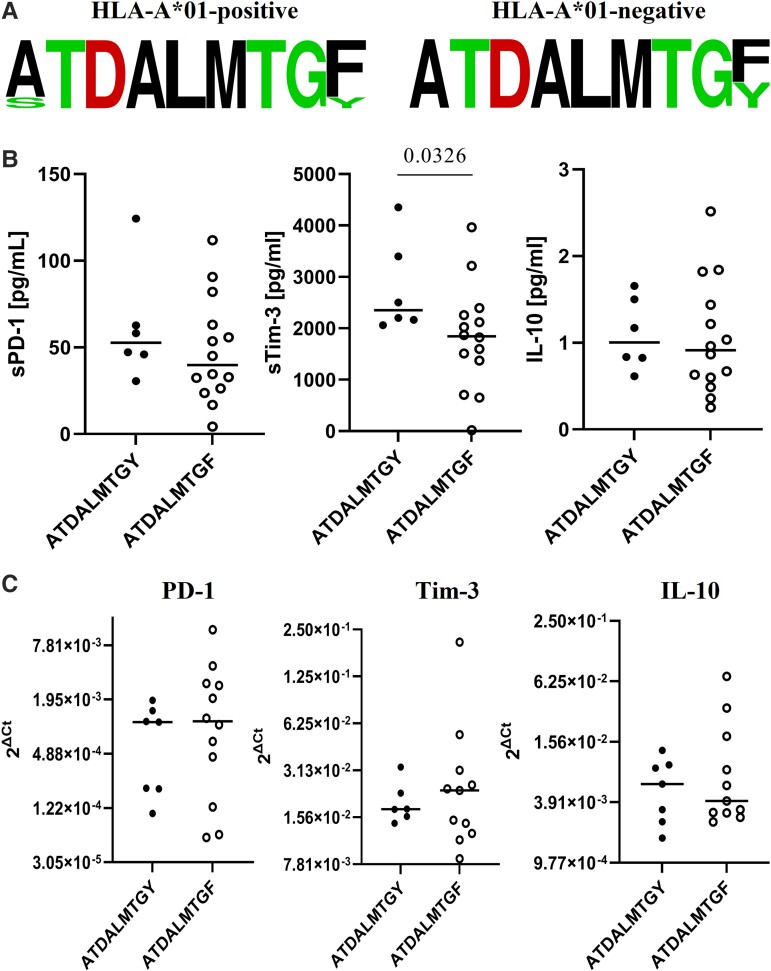
The HLA-A*01–restricted prototype ATDALMTGY sequence was present as a major variant in 1 of 6 (16.7%) of HLA-A*01+ cases, while the escape variant ATDALMTGF was present in 4 of 6 (66.7%). The remaining major sequence was STDALMTGF (1 patient, 16.6%). Intrahost variability (in all cases, 2 variants) was present in 2 HLA-A*01+ cases: major ATDALMTGF and minor ATDALMTGY in 1 and major STDALMTGF and minor ATDALMTGF in 1.
The ATDALMTGF variant predominated over the ATDALMTGY among individuals harboring the HLA-A*01 allele (4/5, 80.0%) vs those not harboring the allele (10/16, 62.5%), but the difference was not statistically significant (P = .6244, Figure 2A). Minor variants in participants who were HLA-A*01 positive included 1 ATDALMTGY and 1 ATDALMTGF.
Figure 2.
A, Distribution of dominant amino acid sequences in epitope NS31436–1444 among individuals coinfected with HIV/HCV: HLA-A*01 positive (n = 6) and HLA-A*01 negative (n = 16). Height of letters within the stack indicates the relative frequency of each amino acid at each position. Data reflecting levels of sPD-1, sTim-3, and IL-10 in plasma (B) and their mRNA levels in PBMC (C) in all individuals (A*01 positive and HLA-A*01 negative) infected with the NS31436–1444 prototype or variant epitope sequence as the dominant strain. Each symbol (circle) represents a mean of 3 replicates obtained from each participant. Horizontal lines within each population of results represent median values. Numbers above lines representing pairwise comparisons express P values. HCV, hepatitis C virus; IL-10, interleukin 10; PD-1, programmed cell death protein 1; Tim-3, T-cell immunoglobulin and mucin domain–containing protein 3.
Infection with ATDALMTGY was associated with higher baseline sTim-3 levels than infection with ATDALMTGF (2352.0 [2061.3–4354.2] vs 1844.5 [18.3–3965.0] pg/mL, respectively; P = .0326), but this difference was not present in the case of PBMC mRNA levels (Figure 2B).
The HLA-A*02–restricted NS31406–1415 epitope was the most variable among the analyzed epitopes. Variants KLVALGVNAV and KLVALGINAV are genotype 1a sequences that are reported to be dominant (74%) at a population level as well as cross-reactive (ie, eliciting a similar-magnitude ex vivo IFN-γ response of T cells) [26]; as such, these were the most prevalent in HLA-A*02+ cases, constituting 8 of 10 major variants (80.0%; n = 5 [50.0%] and 3 [30.0%], respectively). The remaining variants, KLTALGVNAV and KLVGLGVNAV, were found in only 1 patient each (10.0%). The intrahost variability of the epitope was not detected in the participants who were HLA-A*02+.
No clear pattern in exhaustion marker expression could be seen between individuals infected with KLVALGVNAV and other variants or those infected with KLVALGVNAV or KLVALGINAV and other variants (Supplementary Figure 1). Similarly, no significant differences were observed within the HLA-A*02–positive and HLA-A*02–negative subgroups. However, HLA-A*02–negative cases infected with KLVALGVNAV or KLVALGINAV variants displayed significantly higher plasma IL-10 than those infected with other variants (median [range], 1.4 [0.7–2.5] vs 0.6 [0.4–0.8]; P = .0485).
The HLA-A*02–restricted NS31073–1081 epitope CINGVCWTV sequence, representing the 1a prototype—previously shown to account for >90% of 1a variants at a population level [26]—was present as a major variant in 8 of 10 (80%) HLA-A*02+ cases. Another major sequence was CVNGVCWTV (n = 2; Supplementary Figure 2), being previously shown to be cross-reactive with CINGVCWTV [26].
Because of the limited number of observations, no meaningful statistical comparisons could be made between the exhaustion markers and the prototype CINGVCWTV vs variant CVNGVCWTV sequence among the HLA-A*02–positive cases.
Effect of Epitope Sequence Change on sPD-1, Tim-3, and IL-10 Levels in Nonresponders to Treatment
We explored whether eventual evolution (ie, change in epitope sequence) following unsuccessful treatment is related to change in the expression of exhaustion markers. The analysis was performed in 8 individuals, among whom 7 were infected with genotype 1a and 1 with 1b. Evolution occurred in 4 (50%), concerned NS31406–1415 and/or NS31436–1444 epitopes (Supplementary Figure 3), and comprised qualitative as well as quantitative changes, manifesting in reduction in complexity (the number of at least 1 epitope variant). In 2 participants (Nos. 14 and 35), evolution of NS31406–1415 and NS31436–1444 epitopes occurred in parallel. In 6 participants—3 each in whom the epitope evolution occurred and did not occur—a reduction in sTim-3 and IL-10 in plasma was observed (Supplementary Table 2). sPD-1 was reduced in 4 nonresponders: 2 in whom the epitope evolution occurred and 2 with no evolution.
DISCUSSION
The major aim of the study was to explore the effect of HIV-1 on immune exhaustion in chronic HCV infection and HCV antiviral treatment in HIV-1/HCV coinfection. To our best knowledge, this is the first study of this kind.
The study revealed higher sPD-1 and IL-10 levels in plasma and higher IL-10 mRNA in PBMCs in chronic HIV-1/HCV coinfection than in HCV monoinfection (Figure 1), which suggests more profound exhaustion, despite the fact that these patients were receiving ART.
We found opposite levels of PD-1 mRNA in PBMCs and sPD-1 in plasma between HIV-1/HCV coinfection and HCV monoinfection: while PD-1 expression levels were higher in coinfection, sPD-1 levels were higher in monoinfection (Figure 1). The reason for this observation is possibly due to the effect of alternative splicing and differential expression of the isoforms between the groups. While the mRNA levels of a full-length (flPD-1) membrane-associated variant was assessed in PBMCs, a soluble PD-1Δ3 isoform was measured in plasma. It was previously reported that there was no correlation between levels of sPD-1 and PD-1 mRNA [36].
Plasma sTim-3 as well as Tim-3 mRNA levels were similar in both groups, implying a dominant role of HCV infection that contributed to Tim-3 expression in coinfection (Figure 1). This hypothesis was supported by another study, in which sTim-3 levels were elevated in HIV-1 infection, more pronounced in HCV coinfection, and positively correlated with HCV viremia [21].
We observed a reduction in sPD-1, sTim-3, and Tim-3 mRNA levels after treatment of SVR+ cases (Figure 1), underscoring the beneficial effects of HCV antigen load elimination on reduction of exhaustion. Importantly, these effects were unlikely to be caused by HIV-1, since the statistically significant differences were retained even when patients with detectable HIV-RNA were excluded. Although we could not find a similar study that analyzed sPD-1 and/or sTim-3 in plasma in SVR+ HIV-1/HCV-coinfected cases receiving ART, reduced expression of HLA-DR and CD38 on CD4+ and CD8+ T cells and reduced proviral HIV-DNA, sCD14, LPS, 16S rDNA, and D-dimer levels were previously observed in patients with HIV/HCV coinfection who were receiving antiretroviral treatment and achieved SVR with IFN-free regimens [37].
Successful HCV treatment resulted in a reduction of IL-10 in plasma (Figure 1), similar to our previous observation in chronic HCV monoinfection [12]. This suggests that the IL-10 levels in coinfection are likely driven by chronic inflammation and immune regulatory mechanisms [11] but also by HCV proteins that were shown to directly stimulate IL-10 production [22, 38].
Finally, we investigated whether exhaustion marker expression in chronic HIV-1/HCV coinfection may be determined by the autologous HCV CD8+ T-cell–immunodominant epitope sequence. Interestingly, the coinfected cases displayed lower variability of epitopes when compared with our earlier study of chronic HCV monoinfection, which employed an identical method of sequencing and epitope reconstruction [30]. For example, while the NS31406–1415 epitope was the most variable among all analyzed epitopes, only 7 dominant variants were present in coinfection as opposed to 17 in HCV monoinfection [30]. Similarly, the NS31073–1081 epitope was less variable in coinfected cases (3 vs 8 variants). Furthermore, intrahost variability of these epitopes was more common in HCV-monoinfected cases. This may imply that HIV-1 coinfection negatively affects HCV epitope variability, possibly due to diminished immunologic pressure [39]. A lower degree of HCV quasispecies variability has been documented in immune-compromised hosts, such as those undergoing hemodialysis or immune-suppressive treatment [40, 41]. Despite the fact that coinfected cases were receiving ART—which should result in the enhancement of immunocompetent T cells and HLA class I–driven immunologic pressure [42]—the increased expression of exhaustion markers shown in coinfection could have been related to the worse quality of these responses and contributed to the observed phenomenon.
Importantly, the prototype NS31436–1444 ATDALMTGY was related to significantly higher sTim-3 levels in plasma than escape ATDALMTGF, irrespective of the presence of the HLA-A*01 allele, suggesting higher exhaustion (Figure 2). Thus, immune escape seems to diminish exhaustion, possibly because it may reduce or even abrogate overstimulation with cognate antigen. A similar phenomenon was observed in our previous study on HCV monoinfection—ATDALMTGY was related to higher percentages of Tim-3+ as well as PD-1+Tim-3+CD8+ T cells [30]. Furthermore, Bengsch et al found that the exhausted phenotype of CD8+ T cells, manifested by coexpression of inhibitory receptors, was associated with an absence of sequence variations within the corresponding viral epitopes, indicating ongoing antigen triggering [43]. The epitope sequence context seems to play a role in shaping the plasma levels of exhaustion markers. However, the clinical significance of this association remains to be determined.
Administration of anti-HCV treatment may change the repertoire of the viral quasispecies because of the new selective drug-related pressure [44]. Unsuccessful therapy may be related to evolution of the viral epitope variants and consequently change in the levels of exhaustion markers. Observation of the dynamics of the analyzed HCV epitopes in cases that failed to respond to treatment revealed qualitative or quantitative changes in NS31406–1415 and/or NS31436–1444, which could have been due to drug-related pressure on the viral population (Supplementary Figure 3). Since IFN is an immunomodulatory drug that has a direct inhibitory effect on lymphocytes [45, 46], the question of whether IFN-based therapy itself mediated this evolution or was the outgrowth of variants escaping the immune system could not be elucidated. However, the observed sequence evolution did not seem to be associated with a specific pattern of the change in exhaustion markers, since participants in whom changes did or did not occur experienced a similar reduction in the levels of these markers (Supplementary Table 2).
Despite being the first of its kind, our study has shortcomings. First, it employed simple plasma-soluble measures of immune exhaustion, which may not exactly reflect the cellular mechanisms/expression of the inhibitory receptors, making the analysis of a relatively low resolution. Their clinical use might be limited, as well as their use in the description of the overall picture of immune exhaustion. Thus, validation of these results by flow cytometry on a prospective cohort may offer a future direction of the study.
Because of the limited number of epitope sequence observations and thus the limited ability to perform meaningful statistical comparisons, some associations between the expression of exhaustion markers and epitope variants could have been overlooked. Thus, further analysis of this aspect on a larger cohort would be warranted.
Furthermore, the median age of the HCV-monoinfected control cohort was higher than that of the coinfected cohort, which may raise concerns that age was related to a significant increase in exhaustion marker expression and could have resulted in an absence of statistically significant differences in sTim-3 levels.
Finally, the treatment scheme was based on IFN with or without a specific protease inhibitor, which is no longer a recommended standard and patients are not treated with such regimens anymore. This aspect would also require verification with newer treatment strategies based on direct-acting antivirals. Nevertheless, we were able to compare SVR+ and SVR− groups in terms of change in exhaustion markers and track epitope evolution in nonresponders to treatment, which would be impossible in case of IFN-free therapeutic schemes, characterized by an almost 100% success rate [47].
CONCLUSIONS
In summary, we provided evidence that HIV-1/HCV coinfection is associated with higher sPD-1 and IL-10 levels in plasma and higher IL-10 mRNA in PBMCs than in HCV monoinfection, suggesting that coinfection has an additive effect on immune exhaustion. A reduction of sPD-1, sTim-3, and IL-10 levels in plasma and Tim-3 mRNA levels in PBMCs following HCV-oriented treatment was observed, which underscores the beneficial effects of such treatment on immune homeostasis in HIV-1/HCV coinfection.
Individuals infected with the HCV prototype NS31436–1444 ATDALMTGY sequence displayed significantly higher sTim-3 levels than those infected with ATDALMTGF. These results imply that an inhibitory receptor level may be associated with the autologous viral epitope sequence context.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Contributor Information
Kamila Caraballo Cortés, Department of Immunopathology of Infectious and Parasitic Diseases, Medical University of Warsaw, Warsaw, Poland.
Sylwia Osuch, Department of Immunopathology of Infectious and Parasitic Diseases, Medical University of Warsaw, Warsaw, Poland.
Karol Perlejewski, Department of Immunopathology of Infectious and Parasitic Diseases, Medical University of Warsaw, Warsaw, Poland.
Marek Radkowski, Department of Immunopathology of Infectious and Parasitic Diseases, Medical University of Warsaw, Warsaw, Poland.
Maciej Janiak, Department of Immunopathology of Infectious and Parasitic Diseases, Medical University of Warsaw, Warsaw, Poland.
Hanna Berak, Outpatient Clinic, Warsaw Hospital for Infectious Diseases, Warsaw, Poland.
Andri Rauch, Department of Infectious Diseases, Bern University Hospital, University of Bern, Bern, Switzerland.
Jan S Fehr, Department of Public and Global Health, Epidemiology, Biostatistics and Prevention Institute, University of Zurich, Zurich, Switzerland.
Matthias Hoffmann, Division of Infectious Diseases, Cantonal Hospital Olten, Olten, Switzerland.
Huldrych F Günthard, Department of Infectious Diseases and Hospital Epidemiology, University Hospital Zurich, University of Zurich, Zurich, Switzerland; Institute of Medical Virology, University of Zurich, Zurich, Switzerland.
Karin J Metzner, Department of Infectious Diseases and Hospital Epidemiology, University Hospital Zurich, University of Zurich, Zurich, Switzerland; Institute of Medical Virology, University of Zurich, Zurich, Switzerland.
Notes
Acknowledgments. We thank the participants in the SHCS; the physicians and study nurses, for excellent patient care; A. Scherrer, A. Traytel, S. Wild, and K. Kusejko from the SHCS Data Centre for data management; and D. Perraudin and M. Amstad for administrative assistance. We thank the members of the SHCS.
Author contributions. Conceptualization: K. C. C., K. J. M. Sample/data acquisition: A. R., J. S. F., M. H., H. F. G., K. J. M. Data curation: S. O., H. B., K. P. Data analysis: S. O., K. P., M. J., H. B., A. R., J. S. F., M. H., H. F. G., K. J. M. Funding acquisition: K. C. C., K. J. M. Investigation: K. C. C., S. O., M. J. Methodology: K. C. C., S. O., K. P., H. B., K. J. M. Software: K. P. Supervision: K. C. C., M. R., K. J. M. Manuscript review: K. C. C., SO, K. P., M. R. M. J., H. B., A. R., J. S. F., M. H., H. F. G., K. J. M.
Members of the SHCS. K. Aebi-Popp, A. Anagnostopoulos, M. Battegay, E. Bernasconi, J. Böni, D. L. Braun, H. C. Bucher, A. Calmy, M. Cavassini, A. Ciuffi, G. Dollenmaier, M. Egger, L. Elzi, J. Fehr, J. Fellay, H. Furrer, C. A. Fux, H. F. Günthard (president of the SHCS), D. Haerry (deputy of Positive Council), B. Hasse, H. H. Hirsch, M. Hoffmann, I. Hösli, M. Huber, C. R. Kahlert (chair of the Mother and Child Substudy), L. Kaiser, O. Keiser, T. Klimkait, R. D. Kouyos, H. Kovari, K. Kusejko (head of the data center), B. Ledergerber, G. Martinetti, B. Martinez de Tejada, C. Marzolini, K. J. Metzner, N. Müller, D. Nicca, P. Paioni, G. Pantaleo, M. Perreau, A. Rauch (chair of the scientific board), C. Rudin, P. Schmid, R. Speck, M. Stöckle (chair of the clinical and laboratory committee), P. Tarr, A. Trkola, P. Vernazza, G. Wandeler, R. Weber, S. Yerly.
Financial support. This work was supported by the National Science Center, Poland (grant UMO-2015/19/D/NZ6/01303); and the Swiss National Science Foundation (grants 177499, 179571, and 201369) within the framework of the SHCS (project 780).
References
- 1. Kahan SM, Wherry EJ, Zajac AJ. T cell exhaustion during persistent viral infections. Virology 2015; 479–480:180–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wherry EJ, Kurachi M. Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol 2015; 15:486–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Trautmann L, Janbazian L, Chomont N, et al. Upregulation of PD-1 expression on HIV-specific CD8+ T cells leads to reversible immune dysfunction. Nat Med 2006; 12:1198–202. [DOI] [PubMed] [Google Scholar]
- 4. Wieland D, Hofmann M, Thimme R. Overcoming CD8+ T-cell exhaustion in viral hepatitis: lessons from the mouse model and clinical perspectives. Dig Dis 2017; 35:334–8. [DOI] [PubMed] [Google Scholar]
- 5. Zhang J, Lei F, Tan H. The development of CD8 T-cell exhaustion heterogeneity and the therapeutic potentials in cancer. Front Immunol 2023; 14:1166128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nakamoto N, Kaplan DE, Coleclough J, et al. Functional restoration of HCV-specific CD8 T cells by PD-1 blockade is defined by PD-1 expression and compartmentalization. Gastroenterology 2008; 134:1927–37.e1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Golden-Mason L, Palmer BE, Kassam N, et al. Negative immune regulator Tim-3 is overexpressed on T cells in hepatitis C virus infection and its blockade rescues dysfunctional CD4+ and CD8+ T cells. J Virol 2009; 83:9122–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Urbani S, Amadei B, Tola D, et al. Restoration of HCV-specific T cell functions by PD-1/PD-L1 blockade in HCV infection: effect of viremia levels and antiviral treatment. J Hepatol 2008; 48:548–58. [DOI] [PubMed] [Google Scholar]
- 9. Vali B, Jones RB, Sakhdari A, et al. HCV-specific T cells in HCV/HIV co-infection show elevated frequencies of dual Tim-3/PD-1 expression that correlate with liver disease progression. Eur J Immunol 2010; 40:2493–505. [DOI] [PubMed] [Google Scholar]
- 10. Postow MA, Callahan MK, Wolchok JD. Immune checkpoint blockade in cancer therapy. J Clin Oncol 2015; 33:1974–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Richter K, Perriard G, Behrendt R, et al. Macrophage and T cell produced IL-10 promotes viral chronicity. PLoS Pathog 2013; 9:e1003735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Osuch S, Laskus T, Berak H, et al. Decrease of T-cells exhaustion markers programmed cell death-1 and T-cell immunoglobulin and mucin domain-containing protein 3 and plasma IL-10 levels after successful treatment of chronic hepatitis C. Sci Rep 2020; 10:16060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nielsen C, Ohm-Laursen L, Barington T, Husby S, Lillevang ST. Alternative splice variants of the human PD-1 gene. Cell Immunol 2005; 235:109–16. [DOI] [PubMed] [Google Scholar]
- 14. Clayton KL, Douglas-Vail MB, Nur-ur Rahman AK, et al. Soluble T cell immunoglobulin mucin domain 3 is shed from CD8+ T cells by the sheddase ADAM10, is increased in plasma during untreated HIV infection, and correlates with HIV disease progression. J Virol 2015; 89:3723–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zilber E, Martin GE, Willberg CB, et al. Soluble plasma programmed death 1 (PD-1) and Tim-3 in primary HIV infection. Aids 2019; 33:1253–6. [DOI] [PubMed] [Google Scholar]
- 16. Chiu CY, Schou MD, McMahon JH, et al. Soluble immune checkpoints as correlates for HIV persistence and T cell function in people with HIV on antiretroviral therapy. Front Immunol 2023; 14:1123342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Operskalski EA, Kovacs A. HIV/HCV co-infection: pathogenesis, clinical complications, treatment, and new therapeutic technologies. Curr HIV/AIDS Rep 2011; 8:12–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang D, Zhou D, Du Q, et al. Aberrant production of soluble inducible T-cell co-stimulator (sICOS) and soluble programmed cell death protein 1 (sPD-1) in patients with chronic hepatitis C. Mol Med Rep 2013; 7:1197–202. [DOI] [PubMed] [Google Scholar]
- 19. Sperk M, Zhang W, Nowak P, Neogi U. Plasma soluble factor following two decades prolonged suppressive antiretroviral therapy in HIV-1-positive males: a cross-sectional study. Medicine (Baltimore) 2018; 97:e9759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Urbani S, Amadei B, Tola D, et al. PD-1 expression in acute hepatitis C virus (HCV) infection is associated with HCV-specific CD8 exhaustion. J Virol 2006; 80:11398–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hoel H, Ueland T, Hove-Skovsgaard M, et al. Soluble T-cell immunoglobulin mucin domain-3 is associated with hepatitis C virus coinfection and low-grade inflammation during chronic human immunodeficiency virus infection. Open Forum Infect Dis 2020; 7:ofaa033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Abdullah MAF, McWhirter SM, Suo Z. Modulation of kinase activities in vitro by hepatitis C virus protease NS3/NS4A mediated-cleavage of key immune modulator kinases. Cells 2023; 12:406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ritmahan W, Kesmir C, Vroomans RMA. Revealing factors determining immunodominant responses against dominant epitopes. Immunogenetics 2020; 72:109–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim AY, Kuntzen T, Timm J, et al. Spontaneous control of HCV is associated with expression of HLA-B*57 and preservation of targeted epitopes. Gastroenterology 2011; 140:686–696.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fitzmaurice K, Petrovic D, Ramamurthy N, et al. Molecular footprints reveal the impact of the protective HLA-A*03 allele in hepatitis C virus infection. Gut 2011; 60:1563–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kelly C, Swadling L, Brown A, et al. Cross-reactivity of hepatitis C virus specific vaccine-induced T cells at immunodominant epitopes. Eur J Immunol 2015; 45:309–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Timm J, Li B, Daniels MG, et al. Human leukocyte antigen-associated sequence polymorphisms in hepatitis C virus reveal reproducible immune responses and constraints on viral evolution. Hepatology 2007; 46:339–49. [DOI] [PubMed] [Google Scholar]
- 28. Nitschke K, Luxenburger H, Kiraithe MM, Thimme R, Neumann-Haefelin C. CD8+ T-cell responses in hepatitis B and C: the (HLA-) A, B, and C of hepatitis B and C. Dig Dis 2016; 34:396–409. [DOI] [PubMed] [Google Scholar]
- 29. Rauch A, James I, Pfafferott K, et al. Divergent adaptation of hepatitis C virus genotypes 1 and 3 to human leukocyte antigen-restricted immune pressure. Hepatology 2009; 50:1017–29. [DOI] [PubMed] [Google Scholar]
- 30. Osuch S, Laskus T, Perlejewski K, et al. CD8(+) T-cell exhaustion phenotype in chronic hepatitis C virus infection is associated with epitope sequence variation. Front immunol 2022; 13:832206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Foucher J, Chanteloup E, Vergniol J, et al. Diagnosis of cirrhosis by transient elastography (FibroScan): a prospective study. Gut 2006; 55:403–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ferrando-Martinez S, Leal M, Gonzalez-Escribano MF, Vega Y, Ruiz-Mateos E. Simplified sequence-specific oligonucleotide-based polymerase chain reaction protocol to characterize human major histocompatibility complex A*02 and A*24 specificities. Hum Immunol 2011; 72:869–71. [DOI] [PubMed] [Google Scholar]
- 33. Kasuga I, Paré PD, Sandford AJ. Specific genotyping of human leukocyte antigen-A*01 by polymerase chain reaction using allele group-specific primers. Genet Mol Biol 2006; 29:203–6. [Google Scholar]
- 34. Topfer A, Zagordi O, Prabhakaran S, Roth V, Halperin E, Beerenwinkel N. Probabilistic inference of viral quasispecies subject to recombination. J Computat Biol 2013; 20:113–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res 2004; 14:1188–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Derigs M, Heers H, Lingelbach S, Hofmann R, Hanze J. Soluble PD-L1 in blood correlates positively with neutrophil and negatively with lymphocyte mRNA markers and implies adverse sepsis outcome. Immunol Res 2022; 70:698–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lopez-Cortes LF, Trujillo-Rodriguez M, Baez-Palomo A, et al. Eradication of hepatitis C virus (HCV) reduces immune activation, microbial translocation, and the HIV DNA level in HIV/HCV-coinfected patients. J Infect Dis 2018; 218:624–32. [DOI] [PubMed] [Google Scholar]
- 38. Barrett L, Gallant M, Howley C, et al. Enhanced IL-10 production in response to hepatitis C virus proteins by peripheral blood mononuclear cells from human immunodeficiency virus-monoinfected individuals. BMC Immunol 2008; 9:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shuhart MC, Sullivan DG, Bekele K, et al. HIV Infection and antiretroviral therapy: effect on hepatitis C virus quasispecies variability. J Infect Dis 2006; 193:1211–8. [DOI] [PubMed] [Google Scholar]
- 40. Gretch DR, Polyak SJ, Wilson JJ, Carithers RL, Jr., Perkins JD, Corey L. Tracking hepatitis C virus quasispecies major and minor variants in symptomatic and asymptomatic liver transplant recipients. J Virol 1996; 70:7622–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Odeberg J, Yun Z, Sonnerborg A, Bjoro K, Uhlen M, Lundeberg J. Variation of hepatitis C virus hypervariable region 1 in immunocompromised patients. J Infect Dis 1997; 175:938–43. [DOI] [PubMed] [Google Scholar]
- 42. Knapp DJ, Brumme ZL, Huang SY, et al. Increasingly successful highly active antiretroviral therapy delays the emergence of new HLA class I-associated escape mutations in HIV-1. Clin Infect Dis 2012; 54:1652–9. [DOI] [PubMed] [Google Scholar]
- 43. Bengsch B, Seigel B, Ruhl M, et al. Coexpression of PD-1, 2B4, CD160 and KLRG1 on exhausted HCV-specific CD8+ T cells is linked to antigen recognition and T cell differentiation. PLoS Pathog 2010; 6:e1000947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sherman KE, Rouster SD, Stanford S, et al. Hepatitis C virus (HCV) quasispecies complexity and selection in HCV/HIV-coinfected subjects treated with interferon-based regimens. J Infect Dis 2010; 201:712–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gonzalez-Navajas JM, Lee J, David M, Raz E. Immunomodulatory functions of type I interferons. Nat Rev Immunol 2012; 12:125–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. McNab F, Mayer-Barber K, Sher A, Wack A, O'Garra A. Type I interferons in infectious disease. Nat Rev Immunol 2015; 15:87–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pecoraro V, Banzi R, Cariani E, et al. New direct-acting antivirals for the treatment of patients with hepatitis C virus infection: a systematic review of randomized controlled trials. J Clin Exp Hepatol 2019; 9:522–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.