Abstract
We describe the evaluation of a nested reverse transcriptase PCR (RT-PCR) procedure for the detection of small round-structured viruses (SRSVs) in molluscan shellfish and the application of this assay for the detection of SRSVs in commercially produced shellfish and in shellfish implicated in outbreaks of gastroenteritis. The range of virus strains detected and the sensitivity of detection were evaluated by using a representative panel of 21 well-characterized SRSV strains. The nested RT-PCR detected 15 of 21 SRSVs, demonstrating that the assay detects a broad range of SRSVs including strains from both genogroup I and genogroup II. Seeding experiments showed the nested RT-PCR assay to be 10 to 1,000 times more sensitive than the single-round RT-PCR assay for the detection of SRSV in shellfish. SRSV-contaminated samples were identified by nested RT-PCR for shellfish grown in polluted harvesting areas and for shellfish associated with outbreaks of gastroenteritis which were negative by a previously described single-round RT-PCR. The assay was shown to be effective for investigation of virus elimination during commercial shellfish processing procedures such as depuration and relaying and has potential applications for monitoring at-risk shellfish harvesting areas, for investigation of SRSV contamination in shellfish from producers linked to gastroenteritis outbreaks, and for the direct detection of virus in shellfish implicated in outbreaks.
The small round-structured viruses (SRSVs) are important human pathogens frequently associated with gastroenteritis following consumption of sewage-contaminated molluscan shellfish. Public health controls are hampered by the absence of methods for the detection of these viruses in shellfish, as they cannot be grown in tissue culture. Recently, genomic RNA sequences of Norwalk virus (9) and other SRSVs (4, 5, 7, 8, 10, 11, 15–18, 21) have become available and have led to the classification of SRSVs within the virus family Caliciviridae (3). The genomic characterization of a number of SRSVs has facilitated the development of highly sensitive reverse transcriptase PCR (RT-PCR) assays for the diagnosis of SRSV infection (6, 19). Several studies have demonstrated the high level of sequence diversity among SRSVs (1, 20, 23), and this has proved to be the major obstacle for the development of a diagnostic RT-PCR. Consensus primers which detect all SRSVs have not as yet been identified, but a broadly reactive primer pair which detects approximately 90% of SRSVs circulating in the United Kingdom (UK) has been described (6). The development of RT-PCR for the direct detection of SRSVs in shellfish has been further hampered by low levels of virus present in shellfish meat which may also contain potent Taq inhibitors. We (14) and others (2) have previously described the development of RT-PCR-based assays for the detection of SRSVs in molluscan shellfish. Our techniques utilize a sample extraction procedure optimized for removal of RT-PCR amplification inhibitors which largely addresses these problems (12, 14). We have described the application of these techniques to the detection of SRSVs in shellfish associated with outbreaks of human disease and in random testing of shellfish sold for consumption. By a single-round RT-PCR with broadly reactive primers followed by Southern blot hybridization with a pool of four digoxigenin-labelled SRSV-specific oligonucleotide probes, SRSVs could be detected in virtually all oyster samples associated with human disease and in a small percentage of randomly tested samples. However, positive results were frequently detectable only through the added sensitivity of Southern blot hybridization, which indicated that the RT-PCR was operating at the limits of sensitivity. This hindered attempts to confirm positives by sequencing of the amplicon and to genotype the SRSVs detected. This study describes the development and evaluation of a nested RT-PCR for SRSV which overcomes these sensitivity limitations and thus facilitates sequencing and other approaches to RT-PCR amplicon characterization. We describe the application of this method for the investigation and control of public health problems arising from consumption of molluscan shellfish.
MATERIALS AND METHODS
Viruses.
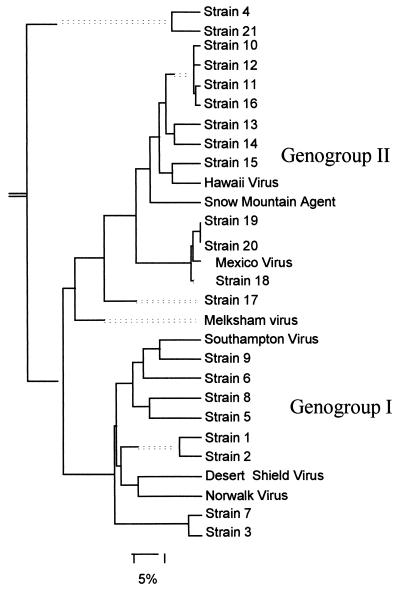
Evaluation of the range of SRSV strains amplified by the nested RT-PCR was performed with a panel of 21 fecal samples which had been shown to contain SRSV by electron microscopy and had been selected to represent the broad range of genomic diversity of SRSVs. The SRSVs had previously been characterized by sequencing of small regions of the RNA polymerase following amplification with the NI-E3 primer pair (6) and/or the SM51-31 and 52-32 primer pairs (20). The panel comprised 8 genogroup I strains and 13 genogroup II strains. Phylogenetic analysis of polymerase gene sequences from these strains and published sequences is shown in Fig. 1. Five fecal samples from the panel, two containing genogroup I SRSVs (panel strains 2 and 5) and three containing genogroup II SRSVs (panel strains 10, 15, and 18), were used in the comparison of detection sensitivities of the single and nested RT-PCR assays.
FIG. 1.
Phylogenetic analysis of strains used for evaluation of the nested RT-PCR assay. A panel of 21 fecal samples which had been demonstrated to contain SRSV by electron microscopy was used in this study. The SRSVs had previously been characterized by RT-PCR and sequencing and were selected to reflect the broad range of diversity seen among strains detected in the UK in recent years. The phylogenetic analysis was performed on a 170-bp region of the RNA polymerase with the Clustal component of the Megalign program (DNAStar). Published sequences accessed from GenBank of Snow Mountain agent (L23831), Hawaii virus (U07611), Melksham virus, Mexico virus (U22498), Southampton virus (L07418), Desert Shield virus (U04469), and Norwalk virus (M87661) are also included. Branch lengths are indicated by the unbroken lines; dotted lines are used to provide a balanced display of the phylogram and do not represent actual branch lengths.
For shellfish seeding experiments, a well-characterized stool sample shown to contain SRSVs by electron microscopy and by RT-PCR was used (panel strain 18). Sequencing of a 113-bp region of the RNA polymerase gene showed it to be a genogroup II strain most closely related to Mexico virus (10) (Fig. 1). This strain was selected because Mexico virus-like strains have been only infrequently detected in the UK in the past 3 years and thus this strain allows the exclusion of false positives in subsequent investigations of field samples derived from this positive control sample.
The fecal samples used in these experiments were prepared by making a 1:10 (wt/vol) dilution of stool in phosphate-buffered saline (PBSa; Dulbecco’s formula) followed by thorough mixing and centrifugation at 3,000 × g for 5 min. Supernatants were stored at 4°C until use. For the comparison of detection sensitivities, further dilutions from 10−1 to 10−7 were made with PBSa. For the seeding experiments, further dilutions of 1:100, 1:500, 1:1,000, 1:5,000, 1:10,000, and 1:50,000 were made with PBSa.
Shellfish.
Commercially purified oysters (Crassostrea gigas) were used in the seeding experiments. Shellfish were also obtained from commercial harvesting areas in England and Ireland subject to varying levels of pollution. Shellfish associated with a clinical outbreak in Suffolk, England, were obtained from both the implicated restaurant and the holding tank used for oyster storage prior to serving. Environmental samples were stored frozen whole at −20°C prior to use in the RT-PCR. All shellfish samples were processed from whole animals.
Shellfish processing and virus extraction and purification.
The procedure for shellfish processing has been previously described in full (12). Essentially, shellfish were shucked, homogenized, sonicated, and centrifuged, and supernatants were precipitated with polyethylene glycol. For seeding experiments and environmental samples, 50 g of shellfish flesh was processed. For shellfish associated with gastroenteritis outbreaks, up to 50 g was processed (depending on sample availability). Resuspended pellets were sonicated and centrifuged prior to further virus purification by extraction with 1,1,2-trichloro-2,2,1-trifluoroethane (Freon TF) followed by centrifugal concentration and storage at −20°C. Extracts at this stage were termed purified concentrates.
Extraction of viral RNA purified concentrates.
The RNA extraction procedure has been previously described in full (12). Briefly, a reaction mix of glass powder matrix and guanidine isothiocyanate (GITC) was used to extract total nucleic acid from purified shellfish concentrates. Shellfish tissue weight equivalents extracted were 7 g (neat), and a 1:3 dilution in PBSa (2.3 g), for all seeding experiments and environmental samples. GITC serves to denature cellular and nucleoprotein complexes (thereby releasing the RNA) and protects the RNA in the sample from digestion by nucleases. RNA bound to the glass powder was washed with GITC, ethanol, and acetone prior to elution in Tris buffer. RNA was then precipitated in ethanol, and RT-PCR was performed.
Oligonucleotide primers.
In the first-round SRSV RT-PCR, a broadly reactive primer combination of the three primers, G1-G2-SM31, was used. The sense primers, G1 and G2, were derived from published SRSV RNA polymerase sequences and were designed to anneal specifically with genogroup I and genogroup II strains, respectively. The antisense primer, SM31, has previously been described (20). The internal (nested) primers were a previously described primer pair, NI-E3, which amplify a 113-bp region of the RNA polymerase gene (corresponding to nucleotides 4756 to 4867 of Norwalk virus) and have been shown to detect more than 90% of strains which were circulating in the UK in 1993 and 1994 (6). Primer sequences and genomic locations are given in Table 1.
TABLE 1.
Primer sequences and locations
| Primer | Orien- tation | Target geno- group(s) | Genomic locationa | DNA sequence |
|---|---|---|---|---|
| G1 | Sense | I | 4679–4696 | TCNGAAATGGATGTTGG |
| G2 | Sense | II | 4338–4355 | AGCCNTNGAAATNATGGT |
| SM31 | Antisense | I | 4871–4853 | CGATTTCATCATCACCATA |
| II | 4607–4588 | |||
| NI | Sense | I | 4756–4776 | GAATTCCATCGCCCACTGGCT |
| II | 4492–4512 | |||
| E3 | Antisense | I | 4869–4853 | ATCTCATCATCACCATA |
| II | 4605–4588 |
Genomic locations (in nucleotides) for genogroup I strains and genogroup II strains refer to Norwalk virus and Lordsdale virus, respectively.
Reverse transcription and RT-PCR.
SRSV RT-PCR was performed by resuspending RNA pellets in 6.9 μl of sterile water, adding 20 U of RNase inhibitor (RNAsin; Promega)–1 μl of 50 mM random hexamers (PdN6; Pharmacia Biotech), and overlaying the pellets with 50 μl of mineral oil (400-5; Sigma). The mixture was heated at 70°C for 5 min, chilled on ice, and then added to 8.1 μl of reaction mix containing (final concentrations) 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 5 mMgCl2, 0.2 mM (each) deoxynucleoside triphosphate (dNTP) (Pharmacia Biotech), and 100 U of Moloney murine leukemia virus RT (fast protein liquid chromatography pure, cloned Moloney murine leukemia virus; Life Technologies). Reverse transcription was performed at room temperature for 10 min followed by incubation at 37°C for 1 h. The reaction mixture was terminated by incubation at 95°C for 5 min, the tubes were then chilled on ice, and 15 μl of RT mix was added to 35 μl of PCR mix (10 mM Tris [pH 8.3], 50 mM KCl, 1.5 mM MgCl2, 0.2 mM [each] dNTP, 20 pmol of each primer [G1-G2 and SM31], 1 U of Taq polymerase). After an initial denaturation at 94°C for 2 min, 30 amplification cycles of 95°C for 1 min, 40°C for 1 min, and 72°C for 1 min were performed followed by a final extension of 72°C for 10 min. Second-round amplification was carried out with 1 μl of the first-round amplicon and 49 μl of RT-PCR mix containing (final concentrations) 10 mM Tris-HCl (pH 8.8), 50 mM KCl, 1.5 mM MgCl2, 0.2 mM (each) dNTP, and 20 pmol of each primer (NI and E3). The cycling parameters were unchanged. RT-PCR amplicons were analyzed by electrophoresis of 20 μl of reaction mix in agarose gels (4% Nusieve 3:1; Flowgen) at 10 V/cm for 1.5 to 2 h. First-round primers G1-G2 and SM31 amplify either a 270-bp (G2-SM31) or a 190-bp (G1-SM31) region of the RNA polymerase gene, and nested primer pair NI-E3 amplifies a 113-bp region. Molecular weights were determined by comparison with a 1-kb DNA ladder (Life Technologies).
RESULTS
Evaluation of the specificity and sensitivity of the nested RT-PCR assay.
A nested RT-PCR with two primer sets derived from alignments of the polymerase region of published SRSV genomic sequences was developed. The assay was evaluated for specificity and sensitivity by using a panel of 21 fecal samples which contained SRSV particles by electron microscopy. These SRSVs had previously been characterized by RT-PCR and sequencing and were selected to represent the broad range of diversity seen among genogroup I and genogroup II strains (Fig. 1). These strains were tested by both a single-round RT-PCR with primers NI-E3 and the nested RT-PCR. The results (Table 2) show that the single-round RT-PCR amplified 13 of the 21 strains while the nested RT-PCR detected 15 of the 21 strains. The nested PCR detected all but one of the strains amplified by the single-round RT-PCR and also amplified a further three strains, showing that the nested RT-PCR gave wider strain cross-reactivity than did the single-round RT-PCR.
TABLE 2.
Single-round and nested RT-PCR results for a panel of genomically representative genogroup I and II SRSVs
| Strain no. | Strain designation | Result fora:
|
||
|---|---|---|---|---|
| Geno- group | Direct NI-E3 | Nested NI-E3 | ||
| 1 | MPH/88/UK | I | Pos | |
| 2 | B291/94/UK | I | Pos | |
| 3 | Winchester/94/UK | I | ||
| 4 | 6708/90/UK | II | Pos | Pos |
| 5 | QA/92/UK | I | Wk pos | Pos |
| 6 | Haytread/89/UK | I | Wk pos | |
| 7 | Blakemore/95/UK | I | ||
| 8 | Malta/95/Malta | I | ||
| 9 | LordHarrisCt/95/UK | I | ||
| 10 | Sym Green/95/UK | II | Pos | Pos |
| 11 | Grimsby/95/UK | II | ||
| 12 | CMH/95/UK | II | Pos | Pos |
| 13 | Pilgrim/90/UK | II | Wk pos | |
| 14 | SP630/90/UK | II | Pos | Pos |
| 15 | Girlington/93/UK | II | Pos | Pos |
| 16 | Wakefield/95/UK | II | Pos | Pos |
| 17 | Hillingdon/95/UK | II | Pos | Pos |
| 18 | Stanroyd/94/UK | II | Pos | Pos |
| 19 | BHM240/93/UK | II | Pos | Pos |
| 20 | BHM132/94/UK | II | Pos | Pos |
| 21 | Creche/90/UK | II | Pos | Pos |
| Total no. | 13 | 15 | ||
A positive (Pos) reaction was defined as an amplicon band of the correct size of intensity greater than or equal to that of the amplicon obtained with the positive control sample on an ethidium bromide-stained agarose gel. A weak positive (Wk pos) reaction was a visible band of the correct size but of weaker intensity than the positive control sample. Where no amplicon band was visible, the result was negative.
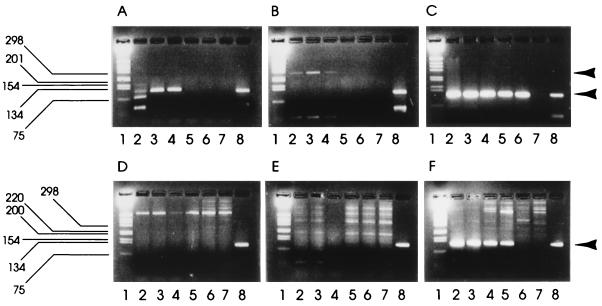
The single-round and nested RT-PCRs were further compared for sensitivity of SRSV detection. Five fecal samples containing SRSVs (genogroup I strains 2 and 5 and genogroup II strains 10, 15, and 18) were titrated in PBSa dilutions from 10−1 to 10−7, and the diluted fecal extracts were tested by both the direct and the nested procedures (Table 3). The titer was taken as the highest dilution in which a specific amplicon was detected in an ethidium bromide-stained agarose gel. By single-round RT-PCR, both genogroup I strains were detectable to a dilution of only 10−1, whereas by nested RT-PCR, the samples were positive at dilutions of 10−4 and 10−2. All three samples containing genogroup II strains were positive at a dilution of 10−2 by single-round RT-PCR but could be detected at dilutions from 10−4 to 10−6 by nested RT-PCR. Thus, significant increases in titer were demonstrated by the nested procedure for all five strains, demonstrating that the nested RT-PCR gives a higher sensitivity of detection for both genogroup I and genogroup II strains in fecal samples. Previous studies (12, 13) have shown the potent inhibitory potential of molluscan shellfish extracts for RT and/or Taq polymerase enzymes used in the RT-PCR assay. The sensitivities of the single-round and nested RT-PCRs were compared for the detection of SRSV in the presence of shellfish meat. Commercially produced oysters (C. gigas) were processed as previously described (12) to the purified shellfish concentrate stage. Seeding experiments were then performed by adding 50 μl of SRSV (Mexico strain) fecal extract dilution to 350 μl of either shellfish concentrate (equivalent to 7 g of shellfish meat) or PBSa prior to nucleic acid extraction as previously described (12). Experiments were conducted with SRSV fecal extract dilutions (in PBSa) of 1:100, 1:500, 1:1,000, 1:5,000, 1:10,000, and 1:50,000. Sensitivities were compared for both nested and single-round RT-PCRs (Fig. 2). A positive result was taken as the presence of a specific amplicon detected in an ethidium bromide-stained agarose gel. When SRSV extracts were seeded into PBSa, the first round of the nested RT-PCR (G2-SM31) and the single-round (NI-E3) RT-PCR gave comparable results with amplicon bands visible to a 1:1,000 dilution of fecal extract (Fig. 2, gels A and B). The second round of the nested RT-PCR showed amplicon bands visible to a 1:10,000 dilution of fecal extract (Fig. 2, gel C), confirming the increased sensitivity of the nested PCR previously observed. By contrast, when SRSV extracts were seeded into shellfish concentrates neither the first round of the nested RT-PCR (G2-SM31) nor the single-round (NI-E3) RT-PCR gave positive amplicon bands (Fig. 2, gels D and E). This confirms previous observations showing the inhibitory nature of shellfish for RT-PCR (12). However, the second round of the nested RT-PCR gave amplicon bands at up to a 1:5,000 dilution of SRSV fecal extract seeded into shellfish concentrates (Fig. 2, gel F). The nested RT-PCR therefore achieved at least a 50-fold improvement in sensitivity over that obtained with a single-round PCR when used to detect SRSV in shellfish extracts. This improvement was only twofold less than the optimum observed when SRSV was seeded into PBSa rather than shellfish extracts. This data suggests that the nested RT-PCR not only is much more sensitive than the single-round RT-PCR for detecting SRSVs in shellfish extracts but also overcomes difficulties with the residual RT-PCR inhibitors left after the standard shellfish processing procedure.
TABLE 3.
Endpoint titration of five fecal samples containing SRSVs and tested by the single-round and nested RT-PCRs
| Strain no. | Genogroup | RT-PCR titer (10−n)a
|
|
|---|---|---|---|
| Single round | Nested | ||
| 2 | I | −1 | −4 |
| 5 | I | −1 | −2 |
| 10 | II | −2 | −5 |
| 15 | II | −2 | −6 |
| 18 | II | −2 | −5 |
Titer, highest dilution of fecal sample which gave an amplicon of the correct size visible in an ethidium bromide-stained agarose gel following electrophoresis.
FIG. 2.
Comparison of sensitivities of single-round and nested PCRs in the presence and absence of shellfish meat. A fecal sample containing SRSV was diluted in PBSa (gels A to C) and in purified shellfish concentrates (gels D to F) prior to extraction and RT-PCR. In each gel, lanes are as follows: 1, 1-kb molecular size marker; 2, 1:100 dilution of fecal sample; 3, 1:500 dilution; 4, 1:1,000 dilution; 5, 1:5,000 dilution; 6, 1:10,000 dilution; 7, 1:50,000 dilution; 8, positive control. Gels A and D show products of a single-round RT-PCR; gels B and E show single-round products from the nested RT-PCR; gels C and F show the nested RT-PCR second-round amplicons. Arrowheads denote positions of first-round and second (or single)-round PCR products.
Application of the nested RT-PCR for the detection of SRSV in field samples.
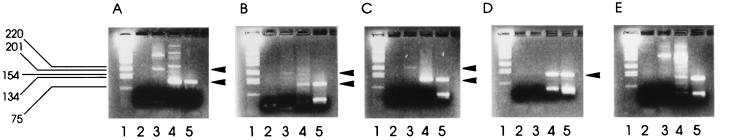
Previous studies (14) have shown the difficulty often experienced in detecting SRSV-specific RT-PCR amplicons in environmental and outbreak shellfish samples. RT-PCR amplicons were frequently visible only through the added sensitivity of Southern blot hybridization and were of insufficient quantity for further characterization of the detected SRSV strain(s) by sequencing. The nested RT-PCR was further evaluated by using environmental and outbreak shellfish samples previously found to be only weakly positive by single-round RT-PCR in order to establish its performance with field material. Figure 3 shows the analysis by both single-round and nested RT-PCRs of three shellfish samples from polluted harvesting areas and two shellfish samples associated with food poisoning outbreaks. SRSV-specific amplicons were not visible by gel electrophoresis in any sample following single-round RT-PCR. However, all samples gave positive bands by nested RT-PCR. This shows that, for all the field material tested, the nested RT-PCR was more sensitive or less susceptible to inhibition than the single-round RT-PCR. Moreover, nested RT-PCR amplicon bands were suitable for further characterization.
FIG. 3.
Application of nested PCR to field samples. Gels A to C show RT-PCR results for shellfish samples from three contaminated harvesting areas; gels D and E show results for samples implicated in two outbreaks of gastroenteritis. Lanes are as follows: 1, molecular weight markers; 2, single-round RT-PCR products; 3, nested RT-PCR first-round products; 4, nested RT-PCR second-round products; 5, nested RT-PCR-positive control. Arrowheads denote positions of first-round and second (or single)-round PCR products.
Detection of SRSVs during commercial shellfish production.
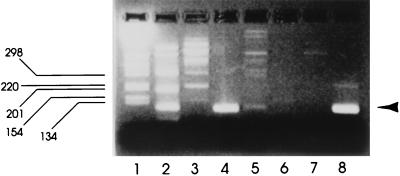
The nested RT-PCR procedure was applied to detection of SRSVs in commercially produced shellfish. Applications for commercial production included evaluation of the effectiveness of the RT-PCR for detection of virus contamination in end-product shellfish sold to the consumer and of the effectiveness of commercial shellfish purification for removal of SRSVs. These studies were performed with oysters (C. gigas) purified in a commercial processing plant associated with outbreaks of infectious disease during February and March 1996. End-product monitoring of oysters from this particular plant during this period proved valuable for demonstrating batches contaminated with SRSVs. Figure 4 shows SRSVs detected by RT-PCR in three of four samples tested during March 1996 but not in samples taken during April and June. These findings were consistent with epidemiological data showing that shellfish-associated outbreaks of infectious disease occur predominantly during the winter months (22). Further monitoring of shellfish for SRSVs before and after purification during this period clearly showed that, although the SRSV titers were reduced during purification (as judged by RT-PCR band intensity), virus was not always completely cleared by this commercial processing (Fig. 5).
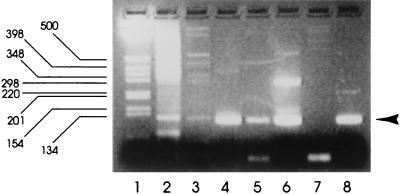
FIG. 4.
Application of nested RT-PCR for detecting SRSV in oysters sold for consumption. Products from a commercial shellfish supplier associated with gastroenteritis incidents were tested for SRSVs. Lanes: 1, 1-kb molecular size marker; 2, result for shellfish dated 8 March 1996; 3, result for shellfish dated 14 March 1996; 4, result for shellfish dated 22 March 1996; 5, result for shellfish dated 29 March 1996; 6, result for shellfish dated 26 April 1996; 7, result for shellfish dated 7 June 1996; 8, nested RT-PCR-positive control. The arrowhead denotes the position of the nested PCR product.
FIG. 5.
Effect of shellfish purification on SRSV content. Shellfish were tested by RT-PCR before and after commercial purification. Lanes: 1, 1-kb molecular size marker; 2, batch dated 29 March 1996 before purification; 3, that batch after purification; 4, batch dated 22 February 1996 before purification; 5, that batch after purification; 6, batch dated 14 March 1996 before purification; 7, that batch after purification; 8, nested RT-PCR-positive control. The arrowhead denotes the position of the nested PCR product.
DISCUSSION
This report describes the successful development of a nested RT-PCR for detection of SRSVs in molluscan shellfish.
The procedure was first evaluated by using a panel of well-characterized strains which represented the diversity of SRSVs detected in the last 9 years in the UK and consequently those that might be anticipated to be detected in shellfish. This showed that the nested RT-PCR assay detected a broader range of SRSVs than did the single-round RT-PCR previously described (6). The nested RT-PCR detected 3 of 8 genogroup I strains and 12 of 13 genogroup II strains, and although ideally the RT-PCR would detect all strains, a catchall primer pair has not, to date, been identified due to the extensive genomic diversity among SRSVs. Recent data have shown that genogroup II strains have predominated in recent years (8), and the nested RT-PCR assay detects the vast majority of these strains.
Comparisons of sensitivities of detection between the single-round and nested RT-PCRs were performed with five fecal samples containing genomically distinct SRSVs. Again, due to the genomic diversity among SRSVs, the primer pairs used may contain mismatches with certain strains which may decrease the sensitivity of detection for these strains. Thus, it is necessary to determine the sensitivity of the assay by using a range of strains. This investigation indicated that the nested RT-PCR, as with the single-round RT-PCR, had differing sensitivities for genogroup I and genogroup II strains. However, there was a significant increase in the sensitivity of detection (101- to 104-fold) for all strains tested. Seeding experiments demonstrated that the nested RT-PCR was also significantly more sensitive for SRSV detection in processed shellfish extracts and overcame the residual PCR inhibition frequently associated with such extracts. Application to a panel of shellfish samples from polluted harvesting areas and associated with outbreaks of infectious disease clearly demonstrated the value of the nested RT-PCR for field samples. The nested RT-PCR assay again showed a greater sensitivity and a decreased susceptibility to inhibitory substances in shellfish meats, thus overcoming the limitations previously experienced with a single-round RT-PCR (14).
This procedure was also applied to monitoring aspects of commercial shellfish production. The nested RT-PCR proved capable of detecting SRSVs in processed shellfish sold for consumption from a commercial supplier associated with incidents of gastroenteritis due to oyster consumption. Further monitoring showed that, although the commercial purification routinely applied to these oysters appeared to reduce virus content, SRSVs were not reliably eliminated. These findings concur well with the available epidemiological evidence on oyster contamination with SRSVs. The nested RT-PCR should prove valuable for further studies on the behavior of SRSVs during commercial processes such as purification and relaying. In addition, the procedure has applications for monitoring shellfish harvesting areas at risk of contamination with SRSVs, for investigation of SRSV contamination in the products of shellfish producers associated with outbreaks, and for direct investigation of shellfish causing illness. The assay may also have potential applications in other areas of environmental monitoring, including the detection of SRSVs in sewage, waters, and foodstuffs other than shellfish.
REFERENCES
- 1.Ando T, Mulders M N, Lewis D C, Estes M K, Monroe S S, Glass R I. Comparison of the polymerase region of small round structured virus strains previously classified in three antigenic types by solid-phase immune electron microscopy. Arch Virol. 1994;135:217–226. doi: 10.1007/BF01309781. [DOI] [PubMed] [Google Scholar]
- 2.Atmar R L, Neill F H, Romalde J L, Le Guyader F, Woodley C M, Metcalf T G, Estes M K. Detection of Norwalk virus and hepatitis A virus in shellfish tissues with the PCR. Appl Environ Microbiol. 1995;61:3014–3018. doi: 10.1128/aem.61.8.3014-3018.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cubitt W D, Bradley D W, Carter M J, Chiba S, Estes M K, Saif L J, et al. Virus taxonomy. Sixth report of the International Committee on Taxonomy of Viruses. Arch Virol. 1995;S10:359–363. [Google Scholar]
- 4.Dingle K E, Lambden P R, Caul E O, Clarke I N. Human enteric Caliciviridae: the complete genome sequence and expression of virus-like particles from a genetic group II small round-structured virus. J Gen Virol. 1995;76:2349–2355. doi: 10.1099/0022-1317-76-9-2349. [DOI] [PubMed] [Google Scholar]
- 5.Green J, Clegg J C S, Lewis D, Brown D W G. Fourth International Symposium on Positive Strand RNA Viruses, Utrecht, The Netherlands. 1995. Analysis of diversity in the capsid and ORF3 proteins of some small round structured viruses circulating in the UK, abstr. W03-4; p. 12. [Google Scholar]
- 6.Green J, Gallimore C I, Norcott J P, Lewis D, Brown D W G. Broadly reactive reverse-transcription polymerase chain reaction (RT-PCR) for the diagnosis of SRSV-associated gastroenteritis. J Med Virol. 1995;47:392–398. doi: 10.1002/jmv.1890470416. [DOI] [PubMed] [Google Scholar]
- 7.Green S M, Lambden P R, Caul E O, Ashley C R, Clarke I N. Capsid diversity in small round-structured viruses: molecular characterization of an antigenically distinct human enteric calicivirus. Virus Res. 1995;37:271–283. doi: 10.1016/0168-1702(95)00041-n. [DOI] [PubMed] [Google Scholar]
- 8.Green S M, Dingle K E, Lambden P R, Caul E O, Ashley C R, Clarke I N. Human enteric Caliciviridae: a new prevalent small round-structured group defined by RNA-dependent RNA polymerase and capsid diversity. J Gen Virol. 1994;75:1883–1888. doi: 10.1099/0022-1317-75-8-1883. [DOI] [PubMed] [Google Scholar]
- 9.Jiang X, Wang M, Wang K, Estes M. Sequence and genomic organization of Norwalk virus. Virology. 1993;195:51–61. doi: 10.1006/viro.1993.1345. [DOI] [PubMed] [Google Scholar]
- 10.Jiang X, Matson D O, Velazquez R F, Calva J J, Zhong W, Palacios G M R, Pickering L K. A study of Norwalk-related viruses in Mexican children. J Med Virol. 1995;47:309–316. doi: 10.1002/jmv.1890470404. [DOI] [PubMed] [Google Scholar]
- 11.Lambden P R, Caul O, Ashley C, Clarke I N. Sequence and genome organization of a human small round structured (Norwalk-like) virus. Science. 1993;259:516–519. doi: 10.1126/science.8380940. [DOI] [PubMed] [Google Scholar]
- 12.Lees D N, Henshilwood K, Dore W. Development of a method for detection of enteroviruses in shellfish by PCR with poliovirus as a model. Appl Environ Microbiol. 1994;60:2999–3005. doi: 10.1128/aem.60.8.2999-3005.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lees D N, Henshilwood K, Butcher S. Development of a PCR-based method for the detection of enteroviruses and hepatitis A virus in molluscan shellfish and its application to polluted field samples. Water Sci Technol. 1995;31:457–464. [Google Scholar]
- 14.Lees D N, Henshilwood K, Green J, Gallimore C I, Brown D W G. Detection of small round structured viruses in shellfish by reverse transcription-PCR. Appl Environ Microbiol. 1995;61:4418–4424. doi: 10.1128/aem.61.12.4418-4424.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lew J F, Kapikian A Z, Valdesuso J, Green K Y. Molecular characterization of Hawaii virus and other Norwalk-like viruses: evidence for genetic polymorphism among human caliciviruses. J Infect Dis. 1994;170:535–542. doi: 10.1093/infdis/170.3.535. [DOI] [PubMed] [Google Scholar]
- 16.Lew J F, Kapikian A Z, Jiang X, Estes M K, Green K Y. Molecular characterization and expression of the capsid protein of a Norwalk-like virus recovered from a Desert Shield troop with gastroenteritis. Virology. 1994;200:319–325. doi: 10.1006/viro.1994.1194. [DOI] [PubMed] [Google Scholar]
- 17.Lew J F, Petric M, Kapikian A Z, Jiang X, Estes M K, Green K Y. Identification of minireovirus as a Norwalk-like virus in pediatric patients with gastroenteritis. J Virol. 1994;68:3391–3396. doi: 10.1128/jvi.68.5.3391-3396.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsui S M, Kim J P, Greenberg H B, Su W, Sun Q, Johnson P C, DuPont H L, Oshiro L S, Reyes G R. The isolation and characterization of a Norwalk virus-specific cDNA. J Clin Invest. 1991;87:1456–1461. doi: 10.1172/JCI115152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moe C L, Gentsch J, Ando T, Grohmann G S, Monroe S, Jiang X, Wang J, Estes M K, Seto Y, Humphrey C, Stine S, Glass R I. Application of PCR to detect Norwalk virus in fecal specimens from outbreaks of gastroenteritis. J Clin Microbiol. 1994;32:642–648. doi: 10.1128/jcm.32.3.642-648.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Norcott J P, Green J, Lewis D, Estes M K, Barlow K L, Brown D W G. Genomic diversity of small round structured viruses in the UK. J Med Virol. 1993;44:280–286. doi: 10.1002/jmv.1890440312. [DOI] [PubMed] [Google Scholar]
- 21.Utagawa E T, Takeda N, Inouye S, Kasuga K, Yamazaki S. 3′-terminal sequence of a small round structured virus (SRSV) in Japan. Arch Virol. 1994;135:185–192. doi: 10.1007/BF01309777. [DOI] [PubMed] [Google Scholar]
- 22.Viral Gastroenteritis Sub-Committee of the PHLS Virology Committee. Outbreaks of gastroenteritis associated with SRSV’s. PHLS Microbiol Dig. 1993;10(1):2–8. [Google Scholar]
- 23.Wang J, Jiang X, Madore H P, Desselberger U, Gray J, Ando T, Seto Y, Yamazaki K, Oishi I, Estes M K. Sequence diversity of small, round-structured viruses in the Norwalk virus group. J Virol. 1994;68:5982–5990. doi: 10.1128/jvi.68.9.5982-5990.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]