Clinical severity caused by infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) can range from a mild self-limiting upper respiratory tract illness to severe and life-threatening acute lung injury with requirement for intensive care (1). Hyperinflammation is a major driver of severity in coronavirus disease (COVID-19), with multiple studies to date highlighting the key role played by proinflammatory cytokines such as IL-6 and IL-1β (2). This disease feature is amenable to therapeutic inhibition using antiinflammatory approaches such as dexamethasone or tocilizumab, which have been shown to confer therapeutic benefit in clinical trials (3, 4).
The nucleocapsid protein (N-protein) of SARS-CoV-2 is a putative inducer of these inflammatory pathways through activation of NF-κB, but the mechanisms leading to this activation are poorly characterized (5, 6). One potentially important upstream mediator is RAGE (receptor for advanced glycation products), a transmembrane glycoprotein highly expressed in multiple pulmonary cell types (7, 8). Although the involvement of RAGE in COVID-19 pathogenesis has been postulated (9), its specific role in N-protein–induced acute lung injury has been unknown. A better understanding of the precise cellular mechanisms underlying the activation and propagation of the proinflammatory response in severe COVID-19 could have profound implications for developing more effective therapies.
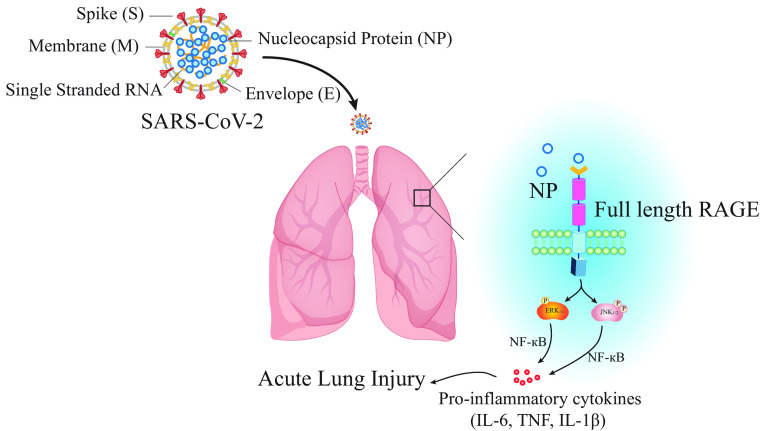
In this issue of the Journal, Xia and colleagues (pp. 508–520) report on a study that elucidates a key role for RAGE in N-protein–mediated acute lung injury (10). Initially, the authors determined that RAGE is a receptor for N-protein through a combination of flow cytometric assays, surface plasmon resonance analysis, and demonstration that N-protein binding is attenuated in bone marrow–derived macrophages isolated from RAGE-deficient (RAGE−/−) mice. A pull-down assay in mouse lung lysate was used to show that N-protein contains two different binding sites for RAGE (N-terminal domain and C-terminal domain), both of which bind to the receptor with high affinity.
Having established the ligand–receptor interaction between N-protein and RAGE, the authors next interrogated the signaling cascade that begins with the binding of N-protein to RAGE and culminates in the activation of the NF-κB pathway to stimulate inflammation. Treatment of macrophages with recombinant N-protein led to phosphorylation of ERK1/2, p38 MAPK (mitogen-activated kinase), and JNK1/2, critical activators of the proinflammatory response. Importantly, inhibition of RAGE using an antagonist or through the stimulation of macrophages from RAGE−/− mice led to abrogated phosphorylation of ERK1/2 and NF-κB p65.
Previously, the authors had demonstrated that intratracheal N-protein administration induces acute lung injury in mice (6). Here, they used this model to study the in vivo effects of RAGE in N-protein–induced acute lung injury, by showing that RAGE−/− mice have attenuated pulmonary proinflammatory responses and immunopathology. Inhibition of the NLRP3 pathway, which has also been previously implicated in N-protein–mediated acute lung injury, had similar effects, indicating that more than one pathway may be responsible (11). Finally, the authors show that the administration of a RAGE antagonist one hour before treatment with recombinant N-protein in mice similarly alleviates pulmonary immunopathology, hinting at the potential value of RAGE inhibitors as therapeutic options for patients with severe COVID-19.
The authors should be commended for presenting a focused and robust study that combines detailed mechanistic experiments in vitro with confirmation that the pathways identified are also important in vivo within a preclinical animal model. The authors used N-protein administration as a surrogate to model COVID-19 acute lung injury, and further studies are required to confirm that similar effects occur in small animal models of live SARS-CoV-2 infection (12). From a translational point of view, the next step would be to provide evidence that the key role played by the RAGE–ERK1/2–NF-κB pathway is also relevant in the context of human in vivo disease. Whether this pathway is differentially activated in individuals with risk factors that predispose to severe COVID-19 (e.g., older age, comorbidities) also requires further focused assessment. Furthermore, given that similar proinflammatory pathways may also drive long-term post–COVID-19 pulmonary complications (13), it would be interesting to study RAGE in this context. This may ultimately pave the way for human experimental studies testing the utility of targeting this pathway for acute and chronic disease.
The authors also correctly point out that although RAGE inhibition in vitro and in vivo leads to a significant decrease in the intensity of the proinflammatory response after exposure to N-protein, inflammation is not completely abrogated, suggesting that other redundant pathways may also contribute to this response. Although NLRP3 was briefly studied here, future studies may seek to identify the other receptors and pathways that also contribute to the immunopathology of COVID-19. Moreover, other RAGE ligands, such as S100A8 and S100A9, have been shown to be upregulated in COVID-19 (14) and also to exacerbate neutrophilic lung infiltration in other contexts (e.g., tuberculosis) (15); the relative roles played by these ligands also warrant future exploration.
Overall, the study by Xia and colleagues (10) provides unique insight into the biology of SARS-CoV-2–related acute lung injury by shedding light on the cellular mechanisms underlying the activation of the inflammatory response (a summary of mechanistic pathways is shown in Figure 1). The authors provide compelling evidence to implicate RAGE as a key driver of this pathway. Despite this, the complex pathways governing hyperinflammation in COVID-19 remain incompletely understood, and further studies are now needed to gain more detailed mechanistic insight to ultimately facilitate the development of effective new therapies.
Figure 1.
Schematic figure depicting the role played by RAGE in driving inflammation during COVID-19. RAGE = receptor for advanced glycation products; SARS-CoV-2 = severe acute respiratory syndrome coronavirus 2.
Footnotes
Originally Published in Press as DOI: 10.1165/rcmb.2023-0227ED on July 25, 2023
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1. Lamers MM, Haagmans BL. SARS-CoV-2 pathogenesis. Nat Rev Microbiol . 2022;20:270–284. doi: 10.1038/s41579-022-00713-0. [DOI] [PubMed] [Google Scholar]
- 2. Gustine JN, Jones D. Immunopathology of hyperinflammation in COVID-19. Am J Pathol . 2021;191:4–17. doi: 10.1016/j.ajpath.2020.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Horby P, Lim WS, Emberson JR, Mafham M, Bell JL, Linsell L, et al. RECOVERY Collaborative Group Dexamethasone in hospitalized patients with COVID-19. N Engl J Med . 2021;384:693–704. doi: 10.1056/NEJMoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Salama C, Han J, Yau L, Reiss WG, Kramer B, Neidhart JD, et al. Tocilizumab in patients hospitalized with COVID-19 pneumonia. N Engl J Med . 2021;384:20–30. doi: 10.1056/NEJMoa2030340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gao T, Zhu L, Liu H, Zhang X, Wang T, Fu Y, et al. Highly pathogenic coronavirus N protein aggravates inflammation by MASP-2-mediated lectin complement pathway overactivation. Signal Transduct Target Ther . 2022;7:318. doi: 10.1038/s41392-022-01133-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Xia J, Tang W, Wang J, Lai D, Xu Q, Huang R, et al. SARS-CoV-2 N protein induces acute lung injury in mice via NF-κB activation. Front Immunol . 2021;12:791753. doi: 10.3389/fimmu.2021.791753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pickering RJ, Tikellis C, Rosado CJ, Tsorotes D, Dimitropoulos A, Smith M, et al. Transactivation of RAGE mediates angiotensin-induced inflammation and atherogenesis. J Clin Invest . 2019;129:406–421. doi: 10.1172/JCI99987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Katsuoka F, Kawakami Y, Arai T, Imuta H, Fujiwara M, Kanma H, et al. Type II alveolar epithelial cells in lung express receptor for advanced glycation end products (RAGE) gene. Biochem Biophys Res Commun . 1997;238:512–516. doi: 10.1006/bbrc.1997.7263. [DOI] [PubMed] [Google Scholar]
- 9. Chiappalupi S, Salvadori L, Vukasinovic A, Donato R, Sorci G, Riuzzi F. Targeting RAGE to prevent SARS-CoV-2-mediated multiple organ failure: hypotheses and perspectives. Life Sci . 2021;272:119251. doi: 10.1016/j.lfs.2021.119251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xia J, Wang J, Ying L, Huang R, Zhang K, Zhang R, et al. RAGE is a receptor for SARS-CoV-2 N protein and mediates N protein–induced acute lung injury. Am J Respir Cell Mol Biol . 2023;69:508–520. doi: 10.1165/rcmb.2022-0351OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pan P, Shen M, Yu Z, Ge W, Chen K, Tian M, et al. SARS-CoV-2 N protein promotes NLRP3 inflammasome activation to induce hyperinflammation. Nat Commun . 2021;12:4664. doi: 10.1038/s41467-021-25015-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dinnon KH, III, Leist SR, Schäfer A, Edwards CE, Martinez DR, Montgomery SA, et al. A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. Nature . 2020;586:560–566. doi: 10.1038/s41586-020-2708-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. George PM, Reed A, Desai SR, Devaraj A, Faiez TS, Laverty S, et al. A persistent neutrophil-associated immune signature characterizes post-COVID-19 pulmonary sequelae. Sci Transl Med . 2022;14:eabo5795. doi: 10.1126/scitranslmed.abo5795. [DOI] [PubMed] [Google Scholar]
- 14. Chen L, Long X, Xu Q, Tan J, Wang G, Cao Y, et al. Elevated serum levels of S100A8/A9 and HMGB1 at hospital admission are correlated with inferior clinical outcomes in COVID-19 patients. Cell Mol Immunol . 2020;17:992–994. doi: 10.1038/s41423-020-0492-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gopal R, Monin L, Torres D, Slight S, Mehra S, McKenna KC, et al. S100A8/A9 proteins mediate neutrophilic inflammation and lung pathology during tuberculosis. Am J Respir Crit Care Med . 2013;188:1137–1146. doi: 10.1164/rccm.201304-0803OC. [DOI] [PMC free article] [PubMed] [Google Scholar]