Abstract
The main bacteria in peaty, acid grassland soils in the Netherlands were investigated by ribosome isolation, temperature gradient gel electrophoresis, hybridization, cloning, and sequencing. Instead of using only 16S rDNA to determine the sequences present, we focused on rRNA to classify and quantify the most active bacteria. After direct ribosome isolation from soil, a partial amplicon of bacterial 16S rRNA was generated by reverse transcription-PCR. The sequence-specific separation by temperature gradient gel electrophoresis yielded soil-specific fingerprints, which were compared to signals from a clone library of genes coding for 16S rRNA. Cloned 16S rDNA sequences matching with intense bands in the fingerprint were sequenced. The relationships of the sequences to those of cultured organisms of known phylogeny were determined. Most of the amplicons originated from organisms closely related to Bacillus species. Such sequences were also detected by direct dot blot hybridization on soil rRNA: a probe specific for Firmicutes with low G+C content counted for about 50% of all bacterial rRNA. The bacterial activity in Drentse A grassland soil could be estimated by direct dot blot hybridization and sequencing of clones; it was found that about 65% of all the bacterial ribosomes originated from Firmicutes. The most active bacteria apparently were Bacillus species, from which about half of the sequences derived. Other sequences similar to those of gram-positive bacteria were only remotely related to known Firmicutes with a high G+C content. Other sequences were related to Proteobacteria, mainly the alpha subclass.
During the last few years, microbial ecologists have switched more and more to molecular strategies to study the distribution and activity of microorganisms in the environment. The earlier culture-dependent surveys used to describe bacterial communities were suspected of suffering from the “great plate count anomaly” (48). Most natural bacterial cells apparently were not accessible for the cultivation methods used today (2, 42). Explanations for these observations have fluctuated between the presence of cells which were not viable (nonculturability) and the hitherto unknown specific medium requirements of most bacteria (not yet cultured). Supporting both of these explanations, recent molecular studies of terrestrial and aquatic environments indicated on the one hand the presence of extremely small, possibly nonviable cells (3, 43) but on the other hand the presence of rRNA sequences of unknown species which have never been described as a cultured strain.
Thousands of different bacterial genomes per gram of soil were estimated to occur in terrestrial environments (52). Even comprehensive culture collections could hardly compete with such an extensive bacterial diversity in soil. Around the world, several culture-independent surveys of the microbial diversity in soil had been performed (5, 6, 23–25, 29, 39, 40, 44, 46, 53). They all were based principally on the PCR amplification of the small-subunit (SSU) rDNA from directly extracted soil DNA with universal primers. These amplicons were used for the subsequent generation of more or less comprehensive SSU rDNA clone libraries, allowing subsequent sequencing analysis. Unfortunately, all the studies used different cell lysis methods and primer sets. Although the comparability is thus limited, all these sequences provide the first indication of microbial diversity based on “real environmental” 16S rDNA data. Analysis of such 16S rDNA clone libraries demonstrated the presence of hitherto unidentified bacteria that were only remotely related to known strains (5, 6, 24, 25, 29, 40, 57). In fact, only a minority of sequences retrieved from directly isolated soil DNA could be closely related to cultured organisms. The major conclusion was that bacterial communities in the environment were composed mainly of uncultured species. Hence, the structure and function of bacterial communities in terrestrial and aquatic environments must have been mainly unknown. To date, this fact has prevented deeper insights into most basic nutrient fluxes in the ecosystems, where bacteria are suspected to contribute major functions.
Beyond the present collection of 16S rDNA sequences, our investigations are intended to reveal the metabolically most active members of the bacterial community in soil. A promising strategy for this had to be based on the direct isolation of suitable marker molecules. Genomic DNA could not be considered suitable, because detection of the DNA neither indicated activity nor proved the viability or even the presence of the corresponding cells (20, 28). The ribosome appeared to be a more useful marker, since the amount of ribosomes (and their rRNA) per cell was found to be roughly proportional to the growth activity of bacteria in pure culture (55). The 16S rRNA sequences were used as a marker for bacterial activity (59), a providing universal presence (in all cellular organisms) and species-specific sequence information (36, 37, 56). Starting point of our strategy consequently was the direct isolation of ribosomes and the subsequent purification of their rRNA from environmental soil samples (8). Then the major taxa represented by this ribosome fraction were identified by the application of different group-specific probes to the membrane-bound rRNA samples. Subsequent quantification of the probe signals and their comparison to a universal Bacteria probe yielded relative quantities of taxa. Another, more specific approach was used to compare the rRNA fraction with a 16S rDNA clone library generated from directly extracted soil DNA. Universal Bacteria primers were used to amplify the 16S rRNA target by reverse transcription-PCR (RT-PCR) and cloned 16S rDNA amplicons by PCR. The resulting amplicons were separated into a banding pattern of single sequences by temperature gradient gel electrophoresis (TGGE) (41), a technique that detects single changes in sequences. This technique, like the comparable denaturing gradient gel electrophoresis (DGGE) (15), was useful to reveal sequence diversity by generating fingerprints specific for the bacterial community (14, 33, 51). Comparison of single clones to the ribosomal soil band pattern indicated possible matches to particular bands within the soil rRNA fingerprints. Subsequent sequencing allowed initial interpretation of the organisms from which the single bands in the soil band pattern were derived. Then a detailed phylogenetic analysis could be added, because the clones were obtained by amplification of the almost complete 16S rDNA sequence (58).
This paper comprises the results of a comprehensive survey of most of the active bacteria in soil. The site we used is located in the Drentse A research area in The Netherlands and consisted of fields of peaty, acid grassland. Based on the approach focusing on metabolic activity, we identify most prominent bacteria in the upper soil layer and their distribution among major taxa, as indicated by their 16S rRNA sequences.
MATERIALS AND METHODS
Soil sampling.
Peaty, acid grasslands of the Drentse A agricultural research area in The Netherlands (06°41′E, 53°03′N), were the sites of sample collection. They covered a geologically homogeneous stretch of approximately 1.5 km along the Anlooër Diepje River. The different cultivation history of the Drentse A plots was taken into account by sampling six plots representing different final years of fertilization for agricultural hay production. One plot was within the still fertilized area (type F), while another plot was part of an area that had not been fertilized since 1967 (type K). On the other plots, the fertilization stopped between 1985 and 1991 (type A). Details of the soil properties have been published (49). In total, 360 surface samples (<10 cm deep) were obtained in March and October 1996. Soil cores of approximately 50 g were obtained with a drill (0 to 10 cm deep) and transferred into sterile sample bags. Two types of samples were prepared: single soil samples were used for ribosome isolation to check the variability of the 16S rRNA community fingerprints per plot (11), and homogenized, pooled samples were used to compare the different areas. The pooled samples were obtained by pooling the single samples from each plot by sieving and mixing 10 single samples (5 g each) to end up with four samples.
Isolation of ribosomes and rRNA purification.
Ribosomes were isolated from Drentse A soil samples by a previously described method (8). Briefly, the ribosomes were released from 1 g of soil by bead beater treatment in the presence of ribosome buffer. Subsequent centrifugations cleared the suspension of cell debris and soil particles. Then the ribosomes were precipitated by an ultra-high-speed centrifugation (2 h at 100,000 × g). The rRNA was extracted and purified by phenol extractions, ethanol precipitations, and DNase digestion to obtain suitable templates for RT-PCR. From 1 g of soil, we eventually obtained 100 μl of solution containing approximately 15 ng of rRNA per μl.
Specific quantitation of rRNA by dot blot hybridization.
The Bacteria-specific probe EUB338 (1) was used to estimate the amount of bacterial rRNA per gram of soil in 24 rRNA samples (4 per plot). The average value was taken as 100% for subsequent comparison. Probe EUK1379 was used to detect eukaryotic SSU rRNA (18), and probe ARC915 was used to detect SSU rRNA of Archaea (47). The probes ALF1b, BET42a, and GAM42a were applied to quantify rRNA of the alpha, beta, and gamma classes of the Proteobacteria, respectively (31). Probe HGC was specific for high-G+C Gram-positive organisms (54). The LGC probe set has been applied to quantify Gram-positive organisms with a low content of G and C nucleotides (32). Another probe set called PLA has been applied to quantify Planctomycetes (34). The procedures have been published previously (31, 32, 34).
Partial amplification of 16S rRNA.
RT-PCR was performed with the rTth DNA polymerase kit from Perkin-Elmer Cetus. RT reaction mixtures (10 μl) contained 10 mM Tris-HCl (pH 8.3), 90 mM KCl, 1 mM MnCl2, 200 μM each dATP, dCTP, dGTP, and dTTP, 750 nM primer L1401 (35), 2.5 U of rTth DNA polymerase, and 1 μl of 10-fold-diluted template RNA (approximately 1.5 ng). After incubation for 15 min at 68°C, 40 μl of the PCR additive containing 10 mM Tris-HCl (pH 8.3), 100 mM KCl, 0.75 mM EGTA, 0.05% (vol/vol) Tween 20, 3.75 mM MgCl2, 50 μM each dATP, dCTP, dGTP, and dTTP, 190 nM primer U968-GC (35) was added. Amplification was performed in a GeneAmp PCR System 2400 thermocycler (Perkin-Elmer Cetus), with 35 cycles of 94°C for 10 s, 56°C for 20 s, and 68°C for 40 s.
Screening of a 16S rDNA clone library for matching sequences.
Total DNA was isolated from Drentse A soil samples as previously described (8). 16S rDNA sequences were amplified with a GeneAmp PCR System 2400 thermocycler, using 35 cycles of 94°C for 10 s, 54°C for 20 s, and 68°C for 2 min. The PCR mixtures (50 μl) contained 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 3 mM MgCl2, 0.05% detergent W-1 (Life Technologies), 150 μM each dATP, dCTP, dGTP, and dTTP, 30 pmol of primers 8f and 1512r (10), 2.5 U of Taq DNA polymerase (Life Technologies), and 1 μl of template DNA (approximately 10 pg). The amplification products were confirmed by agarose gel electrophoresis (1.4% agarose) and then separated from primers and deoxynucleoside triphosphates on a low-melting-point agarose gel. Then they were cloned in pGEM-T linear plasmid vector and Escherichia coli JM109 competent cells as specified by the manufacturer (Promega, Madison, Wis.). After the transformants were grown overnight, single-clone colonies were taken up with sterile toothpicks and transferred into 1.5-ml microcentrifuge tubes containing 50 μl of TE buffer. The tubes were heated for 15 min at 95°C to lyse the cells and then chilled on ice. Insert sequences were amplified with a thermocycler (as above), using 25 cycles of 94°C for 10 s, 46°C for 20 s, and 68°C for 100 s. The PCR mixtures (10 μl) contained 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 3 mM MgCl2, 150 μM each dATP, dCTP, dGTP, and dTTP, 3 pmol of primers T7 and SP6 (21), 0.25 U of Taq DNA polymerase (Life Technologies), and 1 μl of cell lysate. The vector-specific primers T7 and SP6 amplified the region between the multiple cloning sites where the amplicons should be inserted. Clones providing an amplicon of the correct size (approximately 1.6 kb) were identified by agarose gel electrophoresis. Cell lysates of positively identified clones were again amplified with a thermocycler (as above), using 25 cycles of 94°C for 10 s, 56°C for 20 s, and 68°C for 40 s. The PCR mixtures (10 μl) contained 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 3 mM MgCl2, 50 μM each dATP, dCTP, dGTP, and dTTP, 1 pmol of primer U968-GC and L1401 (35), 0.25 U of Taq DNA polymerase (Life Technologies) and 1 μl of template DNA (approximately 10 pg). A 1-μl sample of each amplification product was separated by TGGE next to RT-PCR amplicons from soil rRNA (see above).
The Diagen TGGE system (Diagen, Düsseldorf, Germany) was used for sequence-specific separation of PCR products. Electrophoresis took place in a 0.8-mm polyacrylamide gel (6% [wt/vol] acrylamide, 0.1% [wt/vol] bisacrylamide, 8 M urea, 20% [vol/vol] formamide, 2% [vol/vol] glycerol) with 1x TA buffer (40 mM Tris acetate [pH 8.0]) at a fixed current of 9 mA (approximately 120 V) for 16 h. A temperature gradient from 37 to 46°C was built up in the direction of electrophoresis. After the run, the gels were silver stained (7). Then the gels could be screened for matches between clone signals and the bands of the RT-PCR fingerprints from soil. Apparent visual matches were confirmed with clone-specific V6 probe Southern blot hybridizations (16, 17). RT-PCR fingerprints from soil and clone signals were transferred to a nylon membrane. A clone-specific probe was used to detect the cloned sequence within the RT-PCR fingerprints from soil. The detailed procedure has been published previously (10).
Sequencing of PCR products from cloned inserts.
Insert sequences were amplified with a thermocycler (as above), using 30 cycles of 94°C for 10 s, 46°C for 20 s, and 68°C for 100 s. The PCR mixtures (two 100-μl samples) contained 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 3 mM MgCl2, 150 μM each dATP, dCTP, dGTP, and dTTP, 100 pmol of primer T7 and SP6, 2.5 U of Taq DNA polymerase (Life Technologies), and 1 μl of cell lysate (see above). The PCR products were purified and concentrated (from 200 to 50 μl) on fiberglass spin columns as specified by the manufacturer (High Pure PCR Product purification kit; Boehringer, Mannheim, Germany). Purified DNA was eluted from the columns with 50 μl of deionized water. The sequencing was done with a Sequenase (T7) sequencing kit (Amersham, Slough, England). Each 4-μl reaction mixture (A, C, G, and T) contained 2.5 μl of template, 0.5 μl of labelled primer (Infra-Red Dye 41; MWG-Biotech, Ebersberg, Germany), and 1 μl of reaction mix (A, C, G, or T; Amersham). The inserts were read in two directions: primer seqT7 and seqSP6 (sequence-like primers T7 and SP6) read from the plasmid into the insert, and primers seq515 (5′-ATCGTATTACCGCGGCTGCTGGCA-3′), seq338 (inverted sequence of probe EUB338), and seq968 (primer U968-GC without the GC clamp) read from inside the insert to its borders. The reaction was performed in a thermocycler (as above) with 35 cycles at 94°C for 5 s, 56°C for 10 s, and 68°C for 10 s. After the addition of 3 μl of loading dye (Amersham), the reactions were run on a no. 4000L sequencer (Li-Cor, Lincoln, Neb.).
Phylogenetic analyses.
The environmental sequences were analyzed with ARB software (50). The ARB package is a combination of alignment and dendrogram tools, allowing alignments to a comprehensive SSU rDNA database (of 8,000 sequences) and detailed phylogenetic analysis. Distance matrices were calculated by the neighbor-joining method (45), and phylogenetic trees were constructed by using maximum parsimony criteria with nearest-neighbor optimization. Sequences with less than 90% similarity to any other known sequence were checked for chimera formation with the CHECK_CHIMERA software of the Ribosomal Database Project (30).
Nucleotide sequence accession numbers.
The sequences of the soil rDNA clones were deposited in the EMBL database. Clones of other clone libraries usually were cited as a short combination of site-specific letters and clone numbers. Clones from forested soil from the Mount Coot-tha region in Australia were assigned MC sequence numbers (24, 25); those from a peat bog sample from Germany were assigned TM sequence numbers (40); those from a soybean field in Japan were assigned FIE or PAD sequence numbers (53); those from the Amazonia rainforest soil were assigned P or M sequence numbers (6). The new DA sequences and their accession numbers are as follows: DA001 (X99967), DA004 (Y07647), DA007 (Y07583), DA008 (Y12597), DA011 (Y07580), DA014 (Y07585), DA015 (Y07605), DA016 (Y07606), DA018 (Y07581), DA022 (Y07579), DA023 (Y07586), DA032 (Y07574), DA036 (AJ000981), DA038 (AJ000986), DA040 (AJ000985), DA052 (Y07646), DA054 (Y07575), DA056 (X99966), DA057 (AJ000988), DA066 (AJ000982), DA067 (Y07582), DA079 (Y11555), DA101 (Y07576), DA111 (Y12596), DA114 (AJ000980), DA115 (Y07578), DA116 (AJ000984), DA134 (AJ000983), DA136 (Y07577), and DA154 (AJ001222).
RESULTS
Ribosome isolation.
The rRNA yield from Drentse A grassland soil samples was estimated by direct dot blot hybridization with the Bacteria-specific EUB338 probe to be approximately 1.5 ± 0.6 μg of bacterial rRNA per g (dry weight) of soil. The original ribosome isolation protocol (8) was modified so that the amount of soil material used was reduced from 1.5 to 1.0 g to reduce the size of the ultracentrifugation pellets and their resistance to resuspension. It has been found that soil input reductions can overcome such overloading problems with precipitates (9). The final rRNA solutions (100 μl per g of soil) were of suitable purity for dot blot hybridization (10 μl of input per dot). A 10-fold-diluted solution for RT-PCR (1 μl of input) was used to generate amplicons of reproducibly high yield and quality (data not shown). The rRNA solutions for RT-PCR were successfully checked for the absence of genomic DNA as previously described (8).
Group-specific quantification of soil rRNA.
The hybridization experiments gave the first indications of the most active bacterial groups in Drentse A grassland soils. The Bacteria-specific EUB338 probe gave, for all plots, an average value of 1.55 μg of bacterial rRNA per g (dry weight) of soil, which was taken to be 100% for subsequent comparisons. The Archaea probe ARC915 gave only 0.5% ± 0.2%, and the Eucarya probe EUK1379 gave between 0 and 2%. As a theoretical part of the EUB338 signal, the ALF1b probe for the alpha Proteobacteria detected 22% ± 5% of all bacterial rRNA. The probe BET42a for the beta Proteobacteria found 1.5%, and the GAM42a probe for the gamma Proteobacteria gave no rRNA. The probe HGC for Firmicutes with a high G+C content counted approximately 19% ± 6%. The Planctomycetes probe set PLA gave between 0 and 4%. For the PLA probes and also for the Eucarya probe, the separation between background and signal was not clearly significant. The strongest signals appeared with the probe set LGC for Firmicutes with a low G+C content. With 49% ± 11%, about half of all bacterial ribosomes in the Drentse A grassland soils appeared to be from gram-positive bacteria with a low G+C content. The results from the different plots showed slight but not significant differences within the ratio of the ALF1b, HGC, and LGC signals (data not shown).
Identification of cloned 16S rRNA sequences in RT-PCR fingerprints from soil.
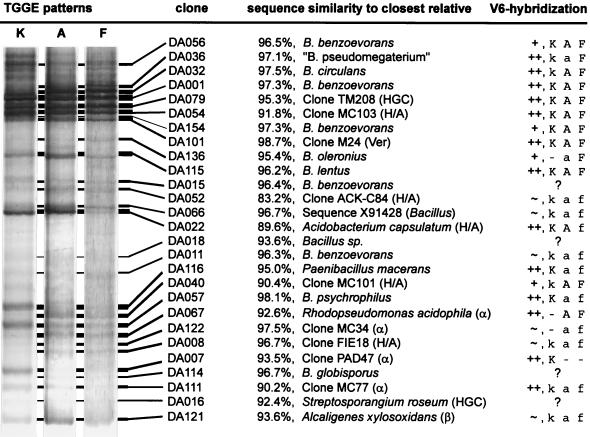
TGGE analysis of RT-PCR products gave specific fingerprints for the rRNA population in soil. In a previous study (11), it was demonstrated that selected plots of the Drentse A area gave highly reproducible fingerprints. During our studies, three types of fingerprints could be distinguished for the surface soil layer (<10 cm deep) of Drentse A grasslands (Fig. 1). Fingerprints of type F originated from the still cultivated section of the Drentse A area. Type K was found in a plot taken out of production in 1967. The most abundant type, type A, represented the areas where fertilization stopped between 1985 and 1991.
FIG. 1.
Matching of clones with TGGE fingerprints of types A, F, and K generated from soil rRNA. The sequenced clones and their closest relatives are indicated. The 16S rRNA clusters are indicated in parentheses: HGC, high-G+C gram-positive bacteria; H/A, Holophaga/Acidobacterium cluster; Ver, Verrucomicrobium cluster; α and β, alpha and beta Proteobacteria. The column V6-hybridization summarizes the results of the V6 probe hybridization approach. Symbols: ++, positive identification by highly specific hybridization signal; +, positive identification by specific hybridization signal with minor cross-reactions; ∼, tentative identification by specific hybridization signal with major cross-reactions; ?, hybridization signals within the TGGE pattern too faint; K, A, or F, prominent sequence in type K, A or F; k, a or f, less abundant sequence in type K, A or F; −, not detected by the V6 probe.
Many of the predominat bands can be found in types K, A, and F. The distribution of the main bacteria appeared to be relatively homogeneous. Only a minority of the strong bands were area specific; most variable bands showed reproducible variations in intensity but were present everywhere.
Clone signals matching soil fingerprint bands indicated the identity of the sequences within the clone library and the soil fingerprints. Also, clone redundancy was indicated by TGGE analysis, where several clones showed the same migration distance. Redundant clones were most commonly found for clones matching the most intense fingerprint bands. Redundancy of the presented sequences is indicated in the phylogenetic trees (Fig. 2 to 5). For example, clone DA001 matched the most intense band of the RT-PCR fingerprint from soil (Fig. 1) and also represented another eight identical sequences of the 16S rDNA clone library (Fig. 3B). Of 165 clones, 37 could be identified as redundant by TGGE and subsequent partial sequencing (approximately 500 bp with primer seq968). The complete sequencing analysis could be limited to only different clones, which were found in the RT-PCR fingerprint from soil.
FIG. 2.
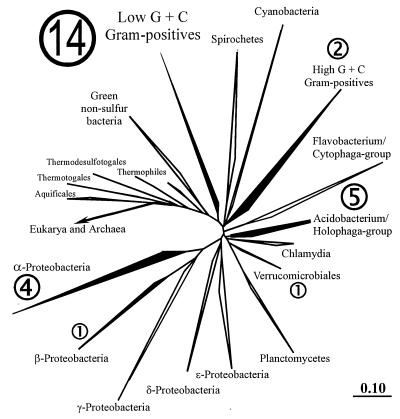
Phylogenetic tree of almost 8,000 SSU rRNA sequences within the ARB database. The clusters containing the DA sequences are highlighted. The numbers indicate the distribution of the DA sequences as compiled in Fig. 1. The alpha Proteobacteria and Cyanobacteria clusters also represent mitochondrial and chloroplast sequences, respectively. The Archaea and Eucarya branches are hidden. The bar in the lower right corner indicates the branch length and represents 0.1 base substitution per nucleotide.
FIG. 5.
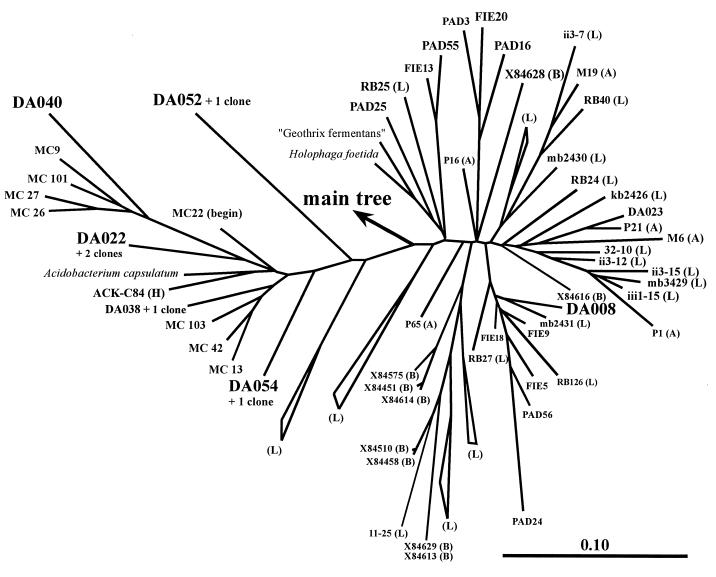
Zoom into the cluster of Holophaga and Acidobacterium within the main tree in Fig. 2. The DA sequences found back in the TGGE fingerprints are highlightened, and two more DA sequences (DA023, DA038) are also presented. Beyond the MC sequences from Australian forested soil (24) and the Japanese FIE and PAD sequences (53), other environmental sequences are marked with (A) from Amazonian forested soil (6), (H) from American mountain lakes (19), (B) from Australian sludge (4), and (L) from German agricultural soil (29). The 31 (L) sequences were assigned to (L) clusters where possible. The bar in the lower right corner indicates the branch-length.
FIG. 3.
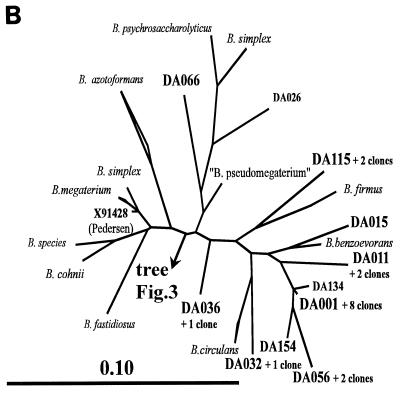
(A) Zoom into the cluster of the low-G+C gram-positive bacteria within the main tree in Fig. 2. The major clusters are represented by their best-known genera or species. The clusters containing the DA sequences are resolved, and the DA sequences are highlighted. One cluster is hidden but is presented in panel B. The bar in the lower right corner indicates the branch length. Abbreviations: B., Bacillus; P., Paenibacillus; B. sporoth., Bacillus sporothermodurans. (B) The hidden Bacillus cluster in panel A. The DA sequences found back in the TGGE fingerprints are highlighted, and two more DA sequences (DA026 and DA134) are also presented. One unnamed environmental sequence from Swedish groundwater is given by its accession number X91428 (38). The bar in the lower left corner indicates the branch length.
Figure 1 shows all the matches of clones with intense and also some faint fingerprint bands. Although TGGE has high resolution and the identities of clones with the same migration distance on TGGE are known, it could not be excluded that quite different sequences accidentally migrated to the same position. Hence sequence identity had to be verified by V6 probe hybridization. This approach could be used for most of the intense fingerprint bands (Fig. 1). Perfect probe specificity was demonstrated for clones DA079 and DA101 (10, 12) but could not always be achieved for the others. Due to cross-reactions, some results remained ambiguous. Weak fingerprint bands often could not clearly be identified as the matching clone sequence because the hybridization signals were too faint (data not shown).
Sequence analysis and phylogenetic assignment of clones.
The partial 16S rRNA sequences covered a stretch of approximately 1500 nucleotides. About half of the sequences found in the clone library showed only slight relationships to other known sequences, while the other half were highly similar (approximately 95% sequence identity) to other database entries (mainly Bacillus species). The average sequencing error could be estimated by screening the latter. The cloned sequences were checked for “impossible nucleotides” by alignment to the next relatives. Less than 0.5% of all nucleotides were found to be unique within conserved regions of the cloned sequence and could almost always be related to reading errors in ambiguous regions of the sequencing gel. Sequences with less than 90% similarity to any other complete sequence of cultured organisms were checked for chimera formation. In all sequences, the beginning and the end of the sequences showed highly similar alignment results; therefore, chimera formation was not indicated. Only sequence DA052 remained questionable, because it (or any part of it) was not closely related to any other known sequence.
The sequences found in the clone library were not randomly distributed over the main 16S rRNA phylogeny clusters of bacteria. Most of the sequences fell into the cluster of low G+C gram-positive bacteria (mainly Bacillus relatives). Other groups were the alpha and beta Proteobacteria, the Verrucomicrobiales, the Holophaga/Acidobacterium cluster, and the high-G+C gram-positive bacteria (Fig. 2 to 5).
DISCUSSION
Experimental strategy.
Bacterial communities in soil were found to be extremely complex (52). Hence, one could not expect to gain a serious understanding of the general bacterial diversity on the basis of only sequence analysis of a few hundred 16S rDNA clones (5, 46). This could not indicate all the bacteria present, since this would demand comprehensive clone libraries, or allow any quantitative conclusions. Surveys on such complex bacterial communities should be limited to more specific goals such as a revealing uncultured bacteria (24, 25) or investigating the diversity of particular phylogenetic taxons (29, 40) or physiological groups like the most active species (this study).
When Muyzer et al. (33) introduced the DGGE approach (which is comparable to TGGE) to molecular microbial ecology, they proposed this as an easier and much faster alternative to the sometimes tedious and expensive cloning procedure. Amplified environmental sequence populations could be specifically separated by DGGE, at once indicating the relative abundance of each sequence. Single bands could be excised, reamplified, and sequenced. However, two drawbacks had to be considered. First, the excised band would represent only a few hundred nucleotides of the target sequence. A detailed phylogenetic analysis might be hampered by this limitation. Second, it could never be excluded that one particular band might contain more than one sequence and consequently confuse the sequencing analysis. This had to be considered, especially when complex environmental bacterial communities were analyzed. Cloning of the excised and reamplified material must then be used to demonstrate its singularity. This cloning of single bands of interest and subsequent screening of the clones might become tedious and expensive. The only remaining advantage of TGGE or DGGE was the greatly enhanced semiquantitative assessment of sequence abundance by comparing band intensities.
We found that both approaches, cloning and TGGE, could complement each other and give a rather powerful combination if they were applied in parallel from the beginning. The possible drawbacks of TGGE and DGGE were erased because the cloned sequences were unique and represented the almost complete 16S rDNA sequences. TGGE fingerprint bands did not have to be excised, which might have been difficult when bands were very close to each other (see, e.g., the fingerprints in Fig. 1). Comparing amplicons of the cloned inserts next to the environmental fingerprint on TGGE gels indicated possible matches. Southern blot hybridization with clone-specific probes could prove the presence of the cloned sequence within the fingerprint (10). The whole approach could possibly be biased by irregular cell lysis and primer or probe specificity. These general drawbacks of molecular microbial ecology might be determined for cultured organisms, but they cannot be estimated for unknown organisms. Hence, it could not be excluded that the cell lysis techniques and Bacteria primers missed some important, hitherto unknown prokaryotes in the soil.
Diversity of the most active bacteria in soil.
The 16S rDNA clone library from Drentse A grassland soils comprised 165 clones, representing 128 different types and 37 redundant sequences. Other studies of environmental clone libraries found a smaller number of or even no redundant clones. This was interpreted as an indication of the high bacterial diversity (5, 59). Compared to these studies, the Drentse A clone library contained a relatively high sequence redundancy. This might be the result of the combination of high-resolution TGGE clone-screening and accurate sequencing. However, it could also indicate a limited bacterial diversity caused by selective influences of the environment. More arguments for the latter possibility were the defined small number of intense bands in the TGGE fingerprint and the unequal presence of the major bacterial taxa. The clear dominance of Bacillus species and the limited number of other taxa (Fig. 2) were remarkable. Borneman et al. (5), for example, found much more diversity of cloned sequences from an agricultural soil from Wisconsin. Maybe the peaty, acid (pH ∼ 4) Drentse A grasslands are a highly selective environment for bacteria. A comparable redundancy of 16S rDNA clones has been found in German peat bog samples (40). The question remained whether these acid environments caused a comparable selective pressure. The phylogeny of their 16S rDNA sequences could hardly be related to each other: only one cloned sequence from the Drentse A soil, DA079, could be related to the ones from German peat bog (10).
Dominance of Bacillus sequences.
Of the 72 sequenced clones, 37 could be related to cultured species of the genus Bacillus. Most of the Bacillus sequences fell into one particular Bacillus branch of the low-G+C gram-positive organism tree (Fig. 3). Most of them were members of novel, hitherto uncultured phylogenetic lines within the B. benzoevorans line of descent (Fig. 3B). These B. benzoevorans relatives apparently were the most important group of soil bacteria. They were represented by approximately 20% of all sequenced clones, including clone DA001. This clone was the most abundant one in the 16S rDNA library (9 of 165 clones) and corresponded to the strongest band in the TGGE fingerprints (Fig. 1). The multiple appearance of closely related B. benzoevorans-like sequences in the TGGE fingerprint raised the question whether this could be due to 16S rDNA sequence heterogeneity, in which one bacterial strain produces more than one signal on TGGE. Such a finding was first described for Paenibacillus polymyxa (35). This cannot be excluded for our case, but it appears to be relatively rare among bacteria (13).
An interesting link to the B. benzoevorans relatives could be found in an SSU rDNA clone library of isolates from agricultural soil in Wisconsin (5). Clone DA001 had up to 99% sequence identity (270-nucleotide overlap) to four clones from Wisconsin (clones 102, 112, 122, and 132). This indicated a worldwide presence and maybe also importance of this Bacillus group.
Unidentified groups of environmental bacteria.
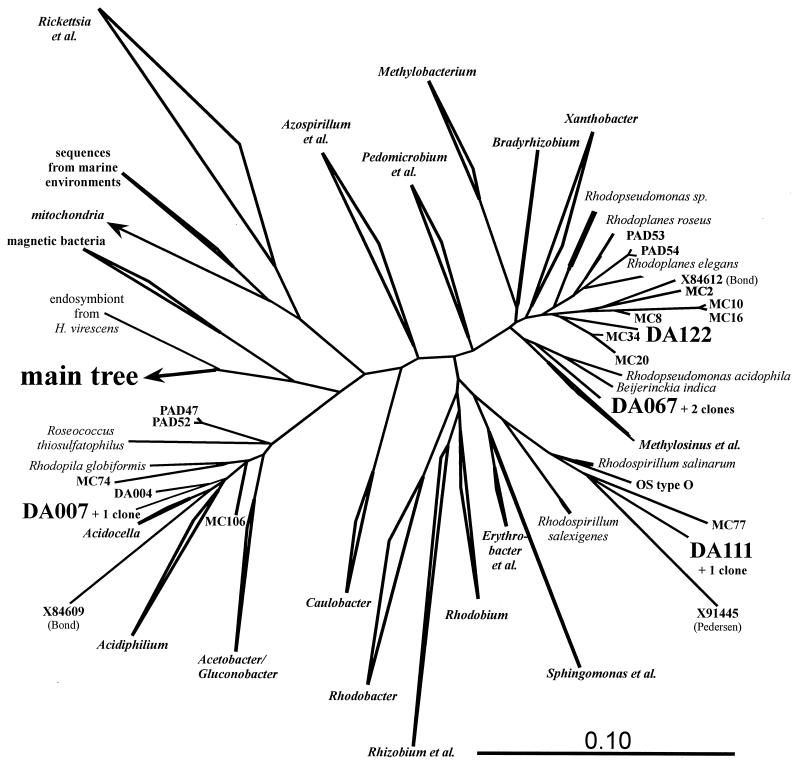
Several distinct groups of unidentified environmental bacteria could be found in Drentse A grassland soils. Most of the alpha Proteobacteria-like sequences showed similarities to sequences from other environmental clone libraries. Clone DA007 fell into the neighborhood of Acidiphilium, together with two clones from Australian forested soil and another two from Japanese soybean field soil (Fig. 4). Clones from the same sources and Swedish groundwater (4) were grouped with clone DA122 within the Rhodoplanes line of descent. The sequence DA111 also had relatives in Australian forest soil and, surprisingly, also in the Octopus Spring microbial mat (59), all of which fell into the Rhodospirillum branch.
FIG. 4.
Zoom into the cluster of alpha Proteobacteria within the main tree in Fig. 2. The major clusters are represented by their best-known genera. The clusters containing the DA sequences are resolved, and the DA sequences found back in the TGGE fingerprints are highlighted. One more DA sequence (DA004) is also presented. The mitochondrial branch is hidden. Three unnamed environmental sequences are given by their accession numbers: those from Swedish groundwater (X91445) and Australian sludge (X84609 and X84612). Sequence “OS type O” originated from an Octopus Spring cyanobacterial mat (56). The bar in the lower right corner indicates the branch length.
Other clones, which were not closely related to cultured organisms, fell into a group of German peat bog clones (DA079) (10), the Verrucomicrobiales cluster (DA101) (12), and, especially, the Holophaga/Acidobacterium cluster (Fig. 5), which probably represents a major taxon on its own (29). Other clones falling into this line of descent again were derived from Australian forested soil (24), Japanese soybean field soil (53), American mountain lakes (19), and, especially, the Roggenstein agricultural test fields in Germany (29). Apart from these several dozen environmental sequences, only three cultured strains were described in this cluster: Acidobacterium capsulatum (22), Holophaga foetida (26), and “Geothrix fermentans” (27). All these species and sequences were isolated from terrestrial environments and might represent one of the most important groups of soil bacteria. This study demonstrated their prominent presence not only in 16S rDNA clone libraries but also within the ribosome fraction from soil. Hence, they also might constitute a major part of the microbial activity in soil.
Conclusions.
TGGE-supported clone screening was a convenient and efficient way to detect and retrieve the most abundant 16S rRNA sequences. This study yielded environmental sequence data of high quality, allowing detailed phylogenetic analysis of the main soil bacteria. A lot of work is done in the field of molecular microbial ecology to retrieve environmental 16S rDNA sequences as the first data about the real bacterial world. The most exciting aspect of gathering 16S rDNA sequences from environments around the world springs from the possibility of revealing phylogenetic relationships to each other and to cultured bacteria. The use of cultured relatives of an unidentified sequence could point to selective approaches to cultivating the organism, and relationships between cloned sequences from different habitats could give the first indications of their potential importance and spatial distribution in the environment. This study once more detected unidentified bacterial lines of descent as already found on other sites of the world. Beyond that, they were also indicated as being metabolically active by their ribosomes. Now it seems likely that these uncultured bacteria are some of the most important metabolizers in soil. Their stage of activity also promises the possibility to grow them in culture. Revealing these rulers of our environment, isolating them, and finally studying them in vitro and in situ will certainly give important insights into the major nutrient fluxes of our planet.
ACKNOWLEDGMENTS
This work was supported by a grant from the European Communities EC project High Resolution Automated Microbial Identification (EC-HRAMI project BIO2-CT94-3098).
Alexander Neef and Harald Meier are especially acknowledged for making the LGC and PLA probes available before publication. We also thank the State Forestry Commission for allowing us access to the nature reserve.
REFERENCES
- 1.Amann R I, Krumholz L, Stahl D A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann R I, Ludwig W, Schleifer K H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Appl Environ Microbiol. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bakken L R, Olsen R A. The relationship between cell size and viability of soil bacteria. Microb Ecol. 1987;13:103–114. doi: 10.1007/BF02011247. [DOI] [PubMed] [Google Scholar]
- 4.Bond P L, Hugenholtz P, Keller J, Blackall L. Bacterial community structures of phosphate-removing and non-phosphate-removing activated sludges from sequencing batch reactors. Appl Environ Microbiol. 1995;61:1910–1916. doi: 10.1128/aem.61.5.1910-1916.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borneman J, Skroch P W, O’Sullivan K M, Palus J A, Rumjanek N G, Jansen J L, Nienhuis J, Triplett E W. Molecular microbial diversity of an agricultural soil in Wisconsin. Appl Environ Microbiol. 1996;62:1935–1943. doi: 10.1128/aem.62.6.1935-1943.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borneman J, Triplett E W. Molecular microbial diversity in soils from Eastern Amazonia: evidence for unusual microorganisms and microbial population shifts associated with deforestation. Appl Environ Microbiol. 1997;63:2647–2653. doi: 10.1128/aem.63.7.2647-2653.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cairns M J, Murray V. Rapid silver staining and recovery of PCR products separate on polyacrylamide gels. BioTechniques. 1994;17:915–919. [PubMed] [Google Scholar]
- 8.Felske A, Engelen B, Nübel U, Backhaus H. Direct ribosome isolation from soil to extract bacterial rRNA for community analysis. Appl Environ Microbiol. 1996;62:4162–4167. doi: 10.1128/aem.62.11.4162-4167.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Felske A, Backhaus H, Akkermans A D L. Direct ribosome isolation from soil. In: Akkermmans A D L, van Elsas J D, de Bruijn F J, editors. Molecular microbial ecology manual, suppl. 3. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1997. pp. 1.2.4/1–1.2.4/10. [Google Scholar]
- 10.Felske A, Rheims H, Wolterink A, Stackebrandt E, Akkermans A D L. Ribosome analysis reveals prominent activity of an uncultured member of the class Actinobacteria in grassland soils. Microbiology. 1997;143:2983–2989. doi: 10.1099/00221287-143-9-2983. [DOI] [PubMed] [Google Scholar]
- 11.Felske, A., and A. D. L. Akkermans. Spatial homogeneity of the most abundant bacterial 16S rRNA molecules in grassland soils. Microb. Ecol., in press. [DOI] [PubMed]
- 12.Felske A, Akkermans A D L. Prominent occurrence of ribosomes from an uncultured bacterium of the Verrucomicrobiales-cluster in grassland soils. 1997. Lett. Appl. Microbiol., in press. [DOI] [PubMed] [Google Scholar]
- 13.Felske, A., M. Vancanneyt, K. Kersters and A. D. L. Akkermans. Application of temperature gradient gel electrophoresis in taxonomy of coryneform bacteria. Submitted for publication. [DOI] [PubMed]
- 14.Ferris M J, Muyzer G, Ward D M. Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl Environ Microbiol. 1996;62:340–346. doi: 10.1128/aem.62.2.340-346.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fischer S G, Lerman L S. Length-independent separation of DNA restriction fragments in two-dimensional gel electrophoresis. Cell. 1979;16:191–200. doi: 10.1016/0092-8674(79)90200-9. [DOI] [PubMed] [Google Scholar]
- 16.Hartung K. Diploma thesis. Lippe, Germany: Fachhochschule Lippe; 1996. [Google Scholar]
- 17.Heuer H, Smalla K. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) for studying soil microbial communities. In: van Elsas J D, Wellington E M H, Trevors J T, editors. Modern soil microbiology. New York, N.Y: Marcel Dekker, Inc.; 1997. pp. 353–373. [Google Scholar]
- 18.Hicks R, Amann R I, Stahl D A. Dual staining of natural bacterioplankton with 4′,6-diamidino-2-phenylindole and fluorescent oligonucleotide probes targeting kingdom-level 16S rRNA sequences. Appl Environ Microbiol. 1992;58:2158–2163. doi: 10.1128/aem.58.7.2158-2163.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hiorns W D, Methé B A, Nierzwicki-Bauer S A, Zehr J P. Bacterial diversity in Adirondack mountain lakes as revealed by 16S rRNA sequences. Appl Environ Microbiol. 1997;63:2957–2960. doi: 10.1128/aem.63.7.2957-2960.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Josephson K L, Gerba C P, Pepper T L. Polymerase chain reaction of nonviable bacterial pathogens. Appl Environ Microbiol. 1993;59:3513–3515. doi: 10.1128/aem.59.10.3513-3515.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kimmerly W J, Kyle A L, Lustre V M, Martin C H, Palazzolo M J. Direct sequencing of terminal regions of genomic P1 clones: a general strategy for the design of sequence-tagged site markers. GATA. 1994;11(5–6):117–128. doi: 10.1016/1050-3862(94)90032-9. [DOI] [PubMed] [Google Scholar]
- 22.Kishimoto N, Kosako Y, Tano T. Acidobacterium capsulatum, new genus new species: An acidophilic chemoorganotrophic bacterium containing menaquinone from acidic mineral environment. Curr Microbiol. 1991;22:1–8. [Google Scholar]
- 23.Lee S-Y, Bollinger J, Bezdicek D, Ogram A. Estimation of the abundance of an uncultured soil bacterial strain by a competitive quantitative PCR method. Appl Environ Microbiol. 1996;62:3787–3793. doi: 10.1128/aem.62.10.3787-3793.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liesack W, Stackebrandt E. Occurrence of novel groups of the domain Bacteria as revealed by analysis of genetic material isolated from an Australian terrestrial environment. J Bacteriol. 1992;174:5072–5078. doi: 10.1128/jb.174.15.5072-5078.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liesack W, Stackebrandt E. Unculturable microbes detected by molecular sequences and probes. Biodivers Conserv. 1992;1:250–262. [Google Scholar]
- 26.Liesack W, Bak F, Kreft J U, Stackebrandt E. Holophaga foetida gen. nov., sp. nov., a new, homoacetogenic bacterium degrading methoxylated aromatic compounds. Arch Microbiol. 1994;162:85–90. doi: 10.1007/BF00264378. [DOI] [PubMed] [Google Scholar]
- 27.Lonergan D J, Jenter H L, Coates J D, Philips E J, Schmidt T M, Lovley D R. Phylogenetic analysis of dissimilatory Fe(III)-reducing bacteria. J Bacteriol. 1996;178:2402–2408. doi: 10.1128/jb.178.8.2402-2408.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lorenz M G, Wackernagel W. Adsorption of DNA to sand and variable degradation rates of adsorbed DNA. Appl Environ Microbiol. 1987;53:2948–2952. doi: 10.1128/aem.53.12.2948-2952.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ludwig W, Bauer S H, Bauer M, Held I, Kirchhof G, Schulze R, Huber I, Spring S, Hartmann A, Schleifer K H. Detection and in situ identification of representatives of a widely distributed new bacterial phylum. FEMS Microbiol Lett. 1997;153:181–190. doi: 10.1111/j.1574-6968.1997.tb10480.x. [DOI] [PubMed] [Google Scholar]
- 30.Maidak B L, Larsen N, McCaughey M J, Overbeck R, Olsen G J, Fogel K, Blandy J, Woese C R. The Ribosomal Database Project. Nucleic Acids Res. 1994;22:3483–3487. doi: 10.1093/nar/22.17.3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manz W, Amann R, Ludwig W, Wagner M, Schleifer K-H. Phylogenetic oligodeoxynucleotide probes for the major subclasses of proteobacteria: problems and solutions. Syst Appl Microbiol. 1992;15:593–600. [Google Scholar]
- 32.Meier H. Ph.D. thesis. Munich, Germany: Technical University of Munich; 1997. [Google Scholar]
- 33.Muyzer G, de Waal E C, Uitterlinden A G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neef A. Ph.D. thesis. Munich, Germany: Technical University of Munich; 1997. [Google Scholar]
- 35.Nübel U, Engelen B, Felske A, Snaidr J, Wieshuber A, Amann R I, Ludwig W, Backhaus H. Sequence heterogeneities of genes encoding 16S rRNAs in Paenibacillus polymyxa detected by temperature gradient gel electrophoresis. J Bacteriol. 1996;178:5636–5643. doi: 10.1128/jb.178.19.5636-5643.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olsen G J, Lane D J, Giovannoni S J, Pace N R. Microbial ecology and evolution: a ribosomal RNA approach. Annu Rev Microbiol. 1986;40:337–365. doi: 10.1146/annurev.mi.40.100186.002005. [DOI] [PubMed] [Google Scholar]
- 37.Pace N R, Stahl D A, Lane D J, Olsen G J. The analysis of natural microbial communities by ribosomal RNA sequences. Microb Ecol. 1986;9:1–55. [Google Scholar]
- 38.Pedersen K, Arlinger J, Ekendahl S, Hallbeck L. 16S rRNA gene diversity of attached and unattached bacteria in boreholes along the access tunnel to the Aspo hard rock laboratory, Sweden. FEMS Microbiol Ecol. 1996;194:249–262. [Google Scholar]
- 39.Rainey F A, Ward N L, Stackebrandt E. Thermophiles ’93. Hamilton, New Zealand: University of Waikato; 1993. Molecular ecology study of a New Zealand acidothermal soil, abstr. A7. [Google Scholar]
- 40.Rheims H, Spröer C, Rainey F A, Stackebrandt E. Molecular biological evidence for the occurrence of uncultured members of the Actinomycete line of descent in different environments and geographic locations. Microbiology. 1996;142:2863–2870. doi: 10.1099/13500872-142-10-2863. [DOI] [PubMed] [Google Scholar]
- 41.Rosenbaum V, Riesner D. Temperature-gradient gel electrophoresis—thermodynamic analysis of nucleic acids and proteins in purified form and in cellular extracts. Biophys Chem. 1987;26:235–246. doi: 10.1016/0301-4622(87)80026-1. [DOI] [PubMed] [Google Scholar]
- 42.Rosswall T, Kvillner E. Principal-components and factor analysis for the description of microbial populations. Adv Microb Ecol. 1978;2:1–48. [Google Scholar]
- 43.Roszak D B, Grimes D J, Colwell R R. Viable but nonrecoverable stage of Salmonella enteritidis in aquatic systems. Can J Microbiol. 1984;30:334–338. doi: 10.1139/m84-049. [DOI] [PubMed] [Google Scholar]
- 44.Saano A, Lindström K, van Elsas J D. 7th International Symposium on Microbial Ecology. Brazilian Society for Microbiology, Santos, Brazil. 1995. Eubacterial diversity in Finnish forest soil, abstr. P1–3.9. [Google Scholar]
- 45.Saitou N, Nei M. The neighbor-joining method: a new method for constructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 46.Stackebrandt E, Liesack W, Goebel B M. Bacterial diversity in a soil sample from a subtropical Australian environment as determined by 16S rDNA analysis. FASEB J. 1993;7:232–236. doi: 10.1096/fasebj.7.1.8422969. [DOI] [PubMed] [Google Scholar]
- 47.Stahl D A, Amann R. Development and application of nucleic acid probes in bacterial systematics. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. Chichester, England: John Wiley & Sons Ltd.; 1991. pp. 205–248. [Google Scholar]
- 48.Staley J T, Konopka A. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu Rev Microbiol. 1985;39:321–346. doi: 10.1146/annurev.mi.39.100185.001541. [DOI] [PubMed] [Google Scholar]
- 49.Stienstra A W, Klein Gunnewiek P, Laanbroek H J. Repression of nitrification in soils under climax grassland vegetation. FEMS Microbiol Ecol. 1994;14:45–52. [Google Scholar]
- 50.Strunk O, Ludwig W. ARB—a software environment for sequence data. Munich, Germany: Department of Microbiology, Technical University of Munich; 1995. [Google Scholar]
- 51.Teske A, Wawer C, Muyzer G, Ramsing N B. Distribution of sulfate-reducing bacteria in a stratified fjord (Mariager Fjord, Denmark) as evaluated by most-probable-number counts and DGGE of PCR-amplified ribosomal DNA fragments. Appl Environ Microbiol. 1996;62:1405–1415. doi: 10.1128/aem.62.4.1405-1415.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Torsvik V, Goksøyr J, Daae F L. High diversity in DNA of soil bacteria. Appl Environ Microbiol. 1990;56:782–787. doi: 10.1128/aem.56.3.782-787.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ueda T, Suga Y, Matsuguchi T. Molecular phylogenetic analysis of a soil microbial community in a soybean field. Eur J Soil Sci. 1995;46:415–421. [Google Scholar]
- 54.Wagner M, Erhart R, Manz W, Amann R, Lemmer H, Wede D, Schleifer K-H. Development of an rRNA-targeted oligonucleotide probe specific for the genus Acinetobacter and its application for in situ monitoring in activated sludge. Appl Environ Microbiol. 1994;60:792–800. doi: 10.1128/aem.60.3.792-800.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wagner R. The regulation of ribosomal RNA synthesis and bacterial cell growth. Arch Microbiol. 1994;161:100–106. doi: 10.1007/BF00276469. [DOI] [PubMed] [Google Scholar]
- 56.Ward D M, Bateson M M, Weller R, Ruff-Roberts A L. Ribosomal RNA analysis of microorganisms as they occur in nature. Adv Microb Ecol. 1992;12:219–286. [Google Scholar]
- 57.Ward-Rainey N, Rainey F A, Schlesner H, Stackebrandt E. Assignment of hitherto unidentified 16S rDNA species to a main line of descent within the domain Bacteria. Microbiology. 1995;141:3247–3250. [Google Scholar]
- 58.Weisburg W G, Barns S M, Pelletier D A, Lane D J. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weller R, Weller J W, Ward D M. 16S rRNA sequences of uncultivated hot spring cyanobacterial mat inhabitants retrieved as randomly primed complementary DNA. Appl Environ Microbiol. 1991;57:1146–1151. doi: 10.1128/aem.57.4.1146-1151.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]